The CCA1 MYB transcription factor is a component of the circadian clock, which plays an important role in plant responses to cold temperatures. This work shows that cold temperatures regulate CCA1 activity by modulating CCA1 alternative splicing and thus altering the molecular ratios of alternatively spliced isoforms to induce cold tolerance responses.
Abstract
The circadian clock synchronizes biological processes to daily cycles of light and temperature. Clock components, including CIRCADIAN CLOCK-ASSOCIATED1 (CCA1), are also associated with cold acclimation. However, it is unknown how CCA1 activity is modulated in coordinating circadian rhythms and cold acclimation. Here, we report that self-regulation of Arabidopsis thaliana CCA1 activity by a splice variant, CCA1β, links the clock to cold acclimation. CCA1β interferes with the formation of CCA1α-CCA1α and LATE ELONGATED HYPOCOTYL (LHY)-LHY homodimers, as well as CCA1α-LHY heterodimers, by forming nonfunctional heterodimers with reduced DNA binding affinity. Accordingly, the periods of circadian rhythms were shortened in CCA1β-overexpressing transgenic plants (35S:CCA1β), as observed in the cca1 lhy double mutant. In addition, the elongated hypocotyl and leaf petiole phenotypes of CCA1α-overexpressing transgenic plants (35S:CCA1α) were repressed by CCA1β coexpression. Notably, low temperatures suppressed CCA1 alternative splicing and thus reduced CCA1β production. Consequently, whereas the 35S:CCA1α transgenic plants exhibited enhanced freezing tolerance, the 35S:CCA1β transgenic plants were sensitive to freezing, indicating that cold regulation of CCA1 alternative splicing contributes to freezing tolerance. On the basis of these findings, we propose that dynamic self-regulation of CCA1 underlies the clock regulation of temperature responses in Arabidopsis.
INTRODUCTION
Low temperatures profoundly influence the overall growth and development of plants, including reproductive success and crop yields. Therefore, plants have evolved versatile strategies to rapidly sense temperature fluctuations and activate adaptive responses under temperature extremes. The best-understood cold signaling mediators include a small group of C-repeat/dehydration-responsive element binding factors (CBFs/DREBs). These factors bind to the cis-acting elements in the promoters of many COLD-REGULATED (COR) genes to enhance freezing tolerance (Gilmour et al., 1998, 2000; Shinozaki and Yamaguchi-Shinozaki, 2000).
INDUCER OF CBF EXPRESSION1 (ICE1) is a basic helix-loop-helix transcription factor that plays a central role in the CBF cold response pathway (Chinnusamy et al., 2003; Dong et al., 2006; Miura et al., 2007). It binds directly to the promoter of the CBF3 gene (Chinnusamy et al., 2003). Posttranslational regulation of the ICE1 protein facilitates rapid induction of CBF3 upon exposure to cold (Dong et al., 2006; Miura et al., 2007), and the ICE1-CBF-COR module is generally considered a major regulator in eliciting freezing tolerance (Chinnusamy et al., 2004).
The circadian clock is the endogenous molecular machinery that synchronizes biochemical, metabolic, physiological, and behavioral cycles to daily environmental changes, such as light, temperature, and nutrient availability, in all living organisms (Alabadí et al., 2001; Dodd et al., 2005; Niwa et al., 2007; Más and Yanovsky, 2009; Song et al., 2010; Thines and Harmon, 2010). The circadian clock is regulated by multiple negative regulatory feedback loops in Arabidopsis thaliana. Coordinated regulation of circadian oscillators and input and output genes at various steps is essential in establishing and maintaining circadian rhythms (Mizoguchi et al., 2002; Farré et al., 2005; Pruneda-Paz et al., 2009; Yakir et al., 2009).
Recently, evidence that the circadian clock also regulates CBF gene expression and thus is related to freezing tolerance has accumulated. The expression of CBF and its target genes exhibits circadian rhythmic patterns (Harmer et al., 2000; Franklin and Whitelam, 2007; Dong et al., 2011). In addition, cold induction of CBF genes is dependent on the time of day (Fowler et al., 2005). The gating effects on CBF gene expression are abolished in CCA1-overexpressing plants, consistent with the clock control of cold acclimation. More direct evidence that circadian clock components contribute to cold acclimation has recently been reported. The CCA1 transcription factor regulates the expression of CBF genes by binding directly to the gene promoters and thereby induces cold tolerance (Dong et al., 2011).
Consistent with the close relationship between temperature responses and the circadian clock (Harmer et al., 2000; Hotta et al., 2007; Espinoza et al., 2008), Arabidopsis plants with mutations in clock genes exhibit altered responses to freezing temperatures (Nakamichi et al., 2009; Espinoza et al., 2010; Dong et al., 2011). The GIGANTEA (GI)-deficient gi-3 mutant is susceptible to freezing (Cao et al., 2005), which is probably caused by impaired sugar metabolism (Cao et al., 2007). Arabidopsis cca1-11 and lhy-21 mutants are also sensitive to freezing (Espinoza et al., 2010; Dong et al., 2011). However, it is currently unclear how low temperatures regulate CCA1 activity in inducing freezing tolerance.
A number of known clock components are transcription factors, underscoring that gene expression regulation is a critical part of clock control. Transcription factors are regulated at various steps, including transcriptional, posttranscriptional, and posttranslational controls. Dynamic dimer formation also plays a role in regulating transcription factor activities by modulating their functional specificities and diversities (Baxevanis and Vinson, 1993; Izawa et al., 1993; Vinson et al., 1993).
A conceptually similar but biochemically distinct mechanism regulating transcription factors has emerged in recent years. A group of small proteins possesses dimerization domains, which have limited sequence similarity to those of transcription factors, but lacks DNA binding domains and/or transcriptional regulation domains. Therefore, they are able to form heterodimers with target transcription factors and attenuate their activities (Wenkel et al., 2007; Kim et al., 2008; Hong et al., 2011; Seo et al., 2011a). LITTLE ZIPPER proteins consisting of 67 to 105 residues contain Leu zipper motifs and interact with class III homeodomain-Leu zipper transcription factors, inhibiting their transcriptional regulation activities (Wenkel et al., 2007; Kim et al., 2008). Similarly, MINI FINGER proteins interfere with Zn finger-homeodomain transcription factors functioning in multiple hormone signaling pathways and in floral development (Hu and Ma, 2006; Hong et al., 2011).
An additional intriguing example of competitive inhibitors is found in the alternative splicing of transcription factor genes. Alternative RNA splicing is thought to be a means of enhancing the diversity of the transcriptome and proteome in eukaryotes. However, in many cases, alternatively spliced isoforms of transcription factors apparently lack the functional domains required for transcriptional regulation, indicating that they are transcriptionally nonfunctional. Notably, it has been demonstrated that a splice variant (IDD14β) of Arabidopsis INDETERMINATE DOMAIN14 (IDD14) transcription factor inhibits the function of IDD14α in starch metabolism by forming heterodimers (Seo et al., 2011b), demonstrating a distinct role for alternative splicing in regulating transcription factor activity.
In this study, we found that alternative RNA splicing modulates CCA1’s functions in clock regulation and freezing tolerance. A splice variant of the CCA1 transcription factor (CCA1β) has a structural organization similar to the small competitive inhibitors in that it has a protein domain required for dimerization but lacks the MYB DNA binding motif. The CCA1β isoform inhibits CCA1α activity by forming nonfunctional heterodimers. Interestingly, CCA1 alternative splicing is suppressed by cold, derepressing the CCA1α transcription factor and allowing it to be fully functional in promoting freezing tolerance in Arabidopsis. This regulatory scheme would explain the disruption of circadian rhythms accompanied by enhanced freezing tolerance under cold conditions.
RESULTS
Alternative Pre-mRNA Splicing Produces Two CCA1 Isoforms
A recent genome-wide comparative analysis of transcription factors and alternatively spliced genes in Arabidopsis estimated that ∼340 transcription factor genes are alternatively spliced (Seo et al., 2011b). After comparing the predicted protein domain organizations of the transcription factors and their alternatively spliced isoforms, we chose the CCA1 gene for further analysis.
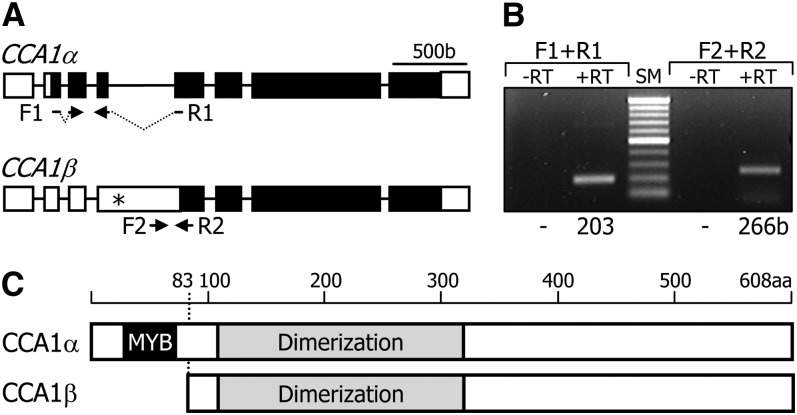
RT-PCR analysis detected two CCA1-specific transcripts (Figures 1A and 1B; see Supplemental Figure 1 online). Sequence comparison of the two CCA1 gene transcripts, designated CCA1α and CCA1β, revealed that the CCA1β transcript is produced by retention of the fourth intron (see Supplemental Figure 2 online). A recent in silico analysis of plant genomes has suggested that CCA1 alternative splicing is a conserved molecular event in different plant species and the shorter splice variant encodes a truncated CCA1 form containing the N-terminal MYB motif (Filichkin et al., 2010). The above prediction is based on the appearance of stop codon following the MYB-coding sequence region in the CCA1β transcript (Figure 1A; see Supplemental Figure 2 online). However, our sequence analysis of the two CCA1 splice variants and protein domain prediction of the CCA1 isoforms revealed that the smaller isoform, CCA1β, has a dimerization domain like the CCA1α form but lacks the N-terminal MYB motif (Figure 1C; see Supplemental Figure 3 online), which is involved in DNA binding (Wang et al., 1997; Daniel et al., 2004). The CCA1α and CCA1β sequences and the type of alternative splicing are identical to those predicted in The Arabidopsis Information Resource database.
Figure 1.
Alternative Splicing of the CCA1 Gene.
(A) Genomic structure of CCA1 splice variants. White boxes indicate untranslated regions, and black boxes indicate exons. Asterisk indicates an in-frame stop codon. F1 and F2 are forward primers. R1 and R2 are reverse primers (see Supplemental Table 1 online). b, base pairs.
(B) Detection of alternatively spliced transcripts. Wild-type cDNA was subjected to RT-PCR. Sizes of the PCR products are indicated at the bottom. RT, reverse transcription; SM, size marker.
(C) Protein structures of two CCA1 isoforms. The CCA1β isoform lacks the MYB DNA binding domain. aa, amino acid.
CCA1β Interacts with CCA1α and LATE ELONGATED HYPOCOTYL
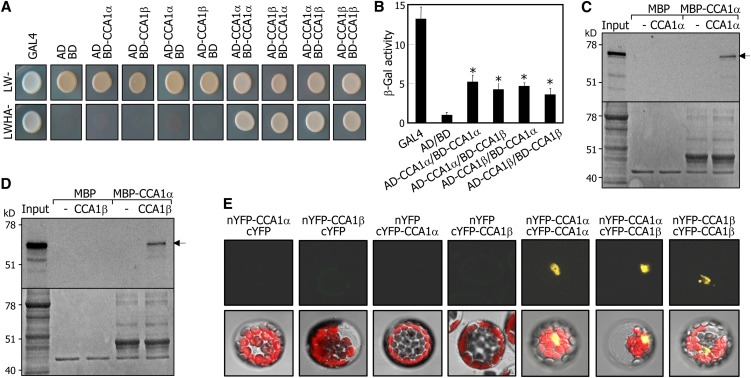
On the basis of the domain organizations of the CCA1α and CCA1β proteins, it was hypothesized that they would interact with each other to form homodimers and heterodimers. To examine this hypothesis, yeast two-hybrid assays were employed, and cell growth on selective media lacking Leu, Trp, His, and adenine (−LWHA) and β-galactosidase (β-Gal) activities were measured. The CCA1α and CCA1β proteins formed homodimers as well as CCA1α-CCA1β heterodimers (Figures 2A and 2B). We also examined the CCA1α-CCA1β interactions by in vitro pull-down assays using a recombinant maltose binding protein (MBP)-CCA1α fusion protein and in vitro–translated CCA1α and CCA1β polypeptides. The recombinant MBP-CCA1α protein interacted efficiently with both CCA1α (Figure 2C) and CCA1β polypeptides (Figure 2D).
Figure 2.
Formation of Homodimers and Heterodimers of CCA1α and CCA1β.
(A) Yeast coexpression assays. Cell growth of yeast transformants on selective media without Leu, Trp, His, and Ade (LWHA-) indicates positive interactions.
(B) β-Gal activity assays in yeast cells. β-Gal activities were normalized by dividing total activity by optical cell density. Three measurements of β-Gal activities were averaged and statistically treated using a Student’s t test (*P < 0.01). Bars indicate the se.
(C) and (D) In vitro pull-down assays. A recombinant MBP-CCA1α fusion protein prepared in E. coli cells and in vitro–translated radiolabeled CCA1α (∼67 kD) (C) and CCA1β (∼58 kD) (D) were used. Arrows indicate the positions of expected bands of CCA1α and CCA1β. MBP protein was also included as a control in the assays. Bottom panels are parts of Coomassie blue–stained gels.
(E) BiFC assays. Partial YFP fusion constructs containing either CCA1α or CCA1β were transiently coexpressed in Arabidopsis protoplasts. Vectors without CCA1 genes (cYFP and nYFP) were also included in the assays. Chloroplasts appear red.
We next examined the CCA1α–CCA1β interactions in vivo by bimolecular fluorescence complementation (BiFC) assays in Arabidopsis protoplasts. Split yellow fluorescent protein (YFP)-CCA1 fusions were coexpressed transiently in Arabidopsis protoplasts. The fluorescence was detected exclusively in the nucleus in all combinations of coexpression (Figure 2E), indicating that CCA1α-CCA1β heterodimers as well as CCA1α-CCA1α and CCA1β-CCA1β homodimers are formed in the nucleus.
Since CCA1α and LATE ELONGATED HYPOCOTYL (LHY) have partially redundant functions and form heterodimers (Mizoguchi et al., 2002; Lu et al., 2009; Yakir et al., 2009), we wanted to know whether CCA1β also interacted with LHY. Yeast two-hybrid assays revealed that CCA1β interacted with LHY (see Supplemental Figure 4A online). In addition, BiFC assays showed that the formation of CCA1β-LHY heterodimers occurred in the nucleus (see Supplemental Figure 4B online), just like the interaction of CCA1β with CCA1α.
CCA1β Acts as a Dominant-Negative Self-Regulator
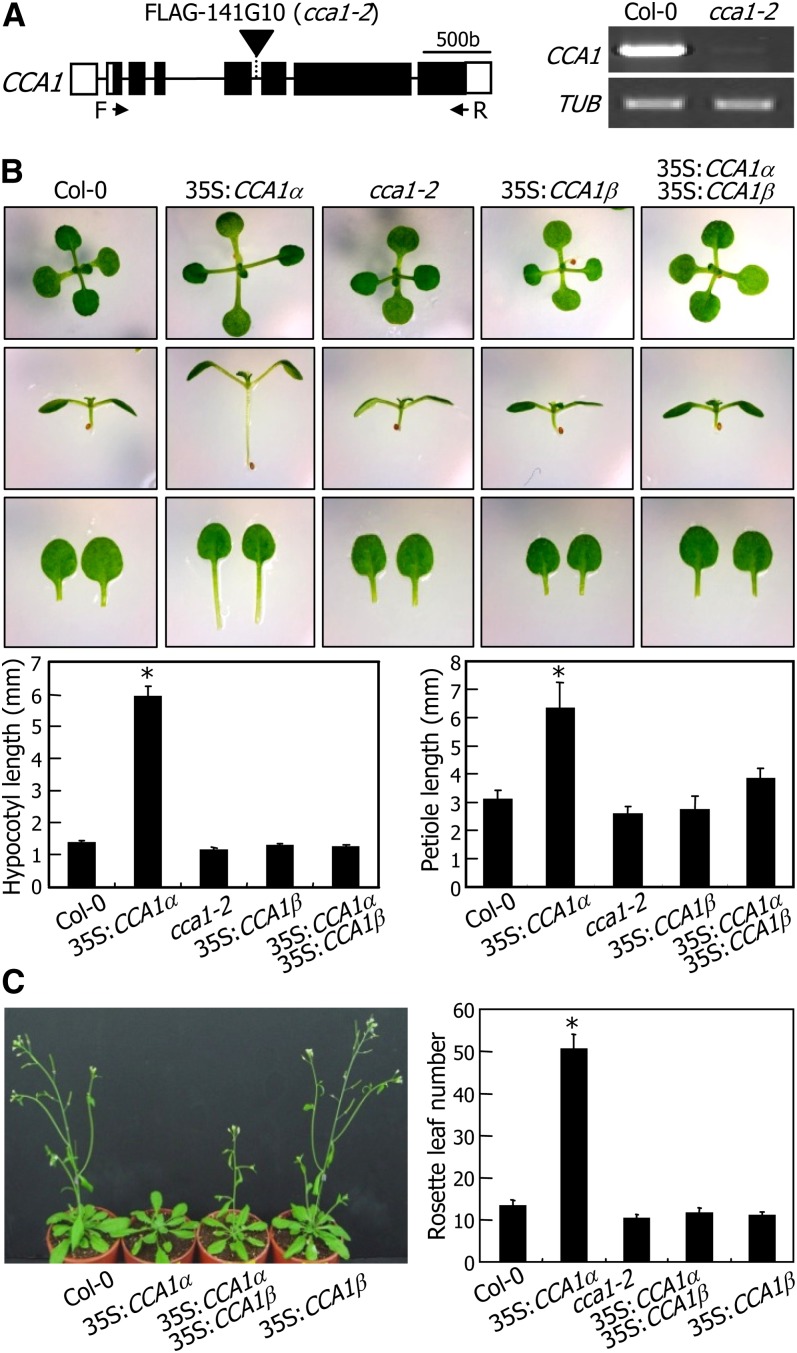
The CCA1β isoform is able to interact with the CCA1α transcription factor. However, it does not have the MYB motif that mediates DNA–protein interactions. Therefore, one plausible hypothesis would be that CCA1β attenuates CCA1α activity by forming CCA1α-CCA1β heterodimers. To examine this hypothesis, we produced transgenic plants that overexpress either CCA1α or CCA1β cDNA under control of the cauliflower mosaic virus (CaMV) 35S promoter. We also obtained a CCA1-deficient mutant in the Columbia-0 (Col-0) background (Figure 3A), which was designated cca1-2 to distinguish it from the previously characterized cca1-1 mutant in the Wassilewskija-2 background (Green and Tobin, 1999).
Figure 3.
Attenuation of CCA1α Activity by CCA1β.
(A) Mapping of T-DNA insertion in cca1-2 mutant. A single copy of the T-DNA element is inserted into the fifth intron of the CCA1 gene (left panel). Absence of CCA1 gene expression in the mutant was verified by RT-PCR (right panel) using the indicated forward (F/CCA1α-B1) and reverse (R/CCA1α-B2) primers. b, base pairs.
(B) Suppression of 35S:CCA1α phenotypes by CCA1β coexpression. Plants were grown on MS-agar plates for 3 weeks before photographs were taken (top panel). The lengths of hypocotyls (bottom left panel) and petioles (bottom right panel) of ∼30 plants were measured and averaged. Statistical significance was determined using a Student’s t test (*P < 0.01). Bars indicate the se.
(C) Flowering times. Five-week-old plants grown in soil were photographed (left panel). Rosette leaf numbers at bolting were counted using 30 plants and averaged for each plant genotype (right panel). Bars indicate the se (Student’s t test, *P < 0.01).
The cca1-2 mutant did not exhibit any visible phenotypes (Figure 3B), as has been observed in the cca1-1 mutant. The 35S:CCA1β transgenic plants were also phenotypically indistinguishable from Col-0 plants. By contrast, the CCA1α-overexpressing transgenic plants (35S:CCA1α) had elongated hypocotyls and leaf petioles at the seedling stage, as described previously (Wang and Tobin, 1998; Lu et al., 2012). Next, we crossed the 35S:CCA1α transgenic plants with the 35S:CCA1β transgenic plants, resulting in 35S:CCA1α × 35S:CCA1β plants. Interestingly, the phenotypes of the 35S:CCA1α transgenic plants, such as long hypocotyls and leaf petioles, were repressed by CCA1β coexpression (Figure 3B). Moreover, the late flowering phenotype of the 35S:CCA1α transgenic plants was also suppressed in the 35S:CCA1α × 35S:CCA1β plants (Figure 3C). In addition, we detected no cosuppression in the CCA1α- and CCA1β-overexpressing plants (see Supplemental Figure 5 online). These observations indicate that the CCA1β isoform negatively regulates CCA1α activity possibly by competitively forming CCA1α-CCA1β heterodimers, which may have impaired transcription factor activity.
CCA1β Competes with CCA1α and LHY for Dimer Formation
Our data suggested that CCA1β attenuates CCA1α transcription factor activity. We next asked how CCA1β regulates CCA1α activity. Dimer formation is known to enhance the DNA binding affinity and specificity of CCA1α and LHY (Daniel et al., 2004; Lu et al., 2009; Yakir et al., 2009). Therefore, it was hypothesized that CCA1β would inhibit the activity of CCA1α and LHY by forming nonfunctional heterodimers.
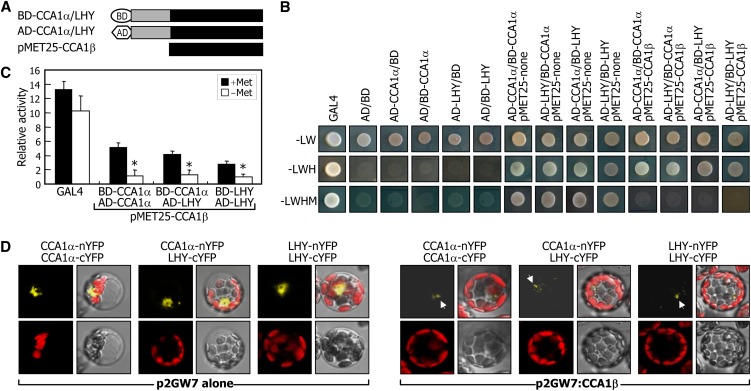
We first employed yeast three-hybrid assays, in which CCA1β cDNA was expressed under control of a Met-suppressible promoter (pMET25) in yeast cells expressing the DNA binding domain (BD)-CCA1α/LHY fusions and the activation domain (AD)-CCA1α/LHY fusions (Figure 4A). Cell growth assays on selective media and β-Gal activities showed that induction of the CCA1β expression substantially repressed the formation of CCA1α-LHY heterodimers as well as CCA1α-CCA1α and LHY-LHY homodimers (Figures 4B and 4C). CCA1β-mediated competitive inhibition was also examined by BiFC assays in Arabidopsis protoplasts. The formation of CCA1α-CCA1α, CCA1α-LHY, and LHY-LHY dimers was strongly suppressed by CCA1β coexpression (Figure 4D).
Figure 4.
Inhibition of Dimer Formation of CCA1α and LHY by CCA1β.
(A) Expression constructs used in yeast three-hybrid assays. The CCA1β cDNA was expressed under control of the Met-suppressible promoter (pMET25-CCA1β). Gray boxes represent sequence regions containing MYB domains.
(B) and (C) Inhibition of CCA1α dimer formation by CCA1β in yeast cells. Yeast three-hybrid assays were performed, and cell growth on selective media (-LWHM) (B) and β-Gal activities (C) were examined. Note that the CCA1β gene is not expressed on selective media without Leu, Trp, and His (-LWH) but is expressed on selective media without Leu, Trp, His, and Met (-LWHM). In (C), β-Gal activities were measured in the presence or absence of Met. Five measurements were averaged, and statistical significance of the measurements was determined using a Student’s t test by comparing the β-Gal activity in the presence of Met (*P < 0.01). Bars indicate the se.
(D) Inhibition of CCA1α-CCA1α, LHY-LHY, and CCA1α-LHY dimer formations by CCA1β in Arabidopsis protoplasts. The BiFC assays were performed as described in Figure 2E. Each photograph is a representative of protoplasts (n > 30) that exhibit similar patterns of fluorescence signals. White arrows indicate YFP fluorescence.
CCA1β Prevents CCA1α from Binding DNA
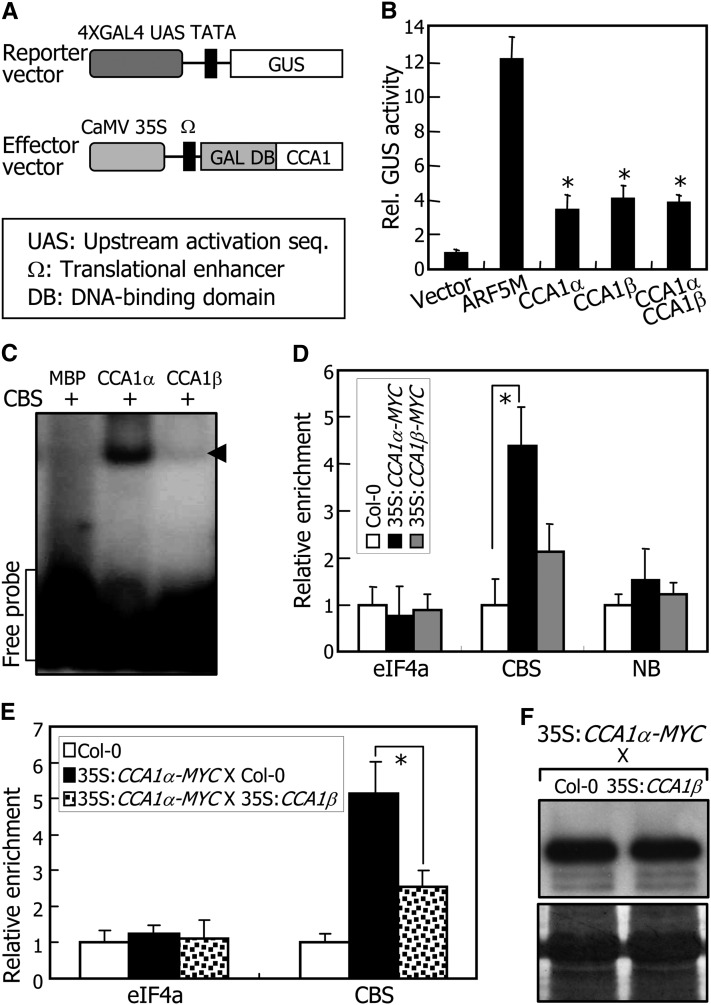
Except for the lack of the N-terminal MYB motif, the CCA1β protein is identical to the CCA1α protein in that both have the dimerization domain and the C-terminal region. Transient β-glucuronidase (GUS) expression assays in Arabidopsis protoplasts revealed that the CCA1α and CCA1β proteins exhibit similar transcriptional regulation activities (Figures 5A and 5B). In addition, coexpression of CCA1β did not influence the transcriptional regulation activity of CCA1α (Figure 5B), showing that the CCA1β inhibition of CCA1α activity does not occur at the level of transcriptional regulation.
Figure 5.
Inhibition of DNA Binding of CCA1α by CCA1β.
(A) Reporter and effector vectors used in transient expression assays using Arabidopsis protoplasts. GAL4 transient expression assays were performed as previously described (Miura et al., 2007). The Renilla luciferase gene was used as an internal control to normalize values in individual assays.
(B) Transcriptional regulation activities of CCA1 isoforms. ARF5M is a transcriptional activator (Miura et al., 2007) and used as positive control. Five independent measurements of GUS activities were averaged. Statistical significance of the measurements was determined using a Student’s t test by comparing the vector control (*P < 0.01). Bars indicate the se.
(C) Electrophoretic mobility shift assays. Recombinant MBP-CCA1α (CCA1α) and MBP-CCA1β (CCA1β) fusion proteins and radiolabeled DNA were used. The CBS has been described previously (Pruneda-Paz et al., 2009). MBP alone was also included as a control in the assays. Arrows indicate protein-DNA complexes.
(D) ChIP assays. qRT-PCR primers were designed on the basis of the sequences flanking the CBS element of the CHE gene (Pruneda-Paz et al., 2009). The nonbinding site (NB) covering the region of 926 to 1035 bp downstream of the CBS element was amplified as a negative control. Three-week-old plants were used for the assays. Biological triplicates were averaged. The statistical significance of the measurements was determined using a Student’s t test by comparing with the values for CBS in Col-0 plants (*P < 0.01). Bars indicate the se.
(E) ChIP assays in 35S:CCA1α-MYC X 35S:CCA1β plants. Biological triplicates were averaged and statistically treated (t test, *P < 0.01). Bars indicate the se.
(F) Relative levels of CCA1α proteins. CCA1α proteins were detected immunologically using an anti-MYC antibody (top panel). Part of a Coomassie blue–stained gel is shown (bottom panel).
The CCA1 transcription factor binds directly to a conserved CCA1 binding site (CBS), which consists of AAAAATCT and exists in the promoters of several genes, including TIMING OF CAB EXPRESSION1 (TOC1), CCA1 HIKING EXPEDITION (CHE), and CBFs (Alabadí et al., 2001; Pruneda-Paz et al., 2009; Dong et al., 2011). Electrophoretic mobility shift assays using recombinant MBP-CCA1 fusion proteins produced in Escherichia coli cells revealed that the CCA1α protein bound efficiently to the CBS motif, but the CCA1β protein did not (Figure 5C).
Chromatin immunoprecipitation (ChIP) assays were also employed to confirm the binding of CCA1α to the CHE promoter (Pruneda-Paz et al., 2009) using transgenic plants overexpressing CCA1-MYC gene fusions, in which a MYC-coding sequence was fused in frame to the 3′ end of either CCA1α or CCA1β cDNA. Quantitative real-time ChIP-PCR assays demonstrated that the CCA1α-MYC protein bound to the gene promoters in planta, whereas the CCA1β-MYC protein did not have any discernible DNA binding affinity (Figure 5D). In addition, no cosuppression was observed in the 35S:CCA1a-MYC and 35S:CCA1β-MYC transgenic plants (see Supplemental Figure 6 online), supporting the dominant-negative effects of CCA1β on the CCA1α binding to DNA.
CCA1β interacted with CCA1α but did not bind to target DNA, suggesting that CCA1β prevents CCA1α from binding DNA. We performed ChIP assays using 35S:CCA1α-MYC transgenic plants that were crossed with 35S:CCA1β transgenic plants. Binding of CCA1α to the CHE gene promoter was significantly reduced in the 35S:CCA1α-MYC × 35S:CCA1β plants (Figure 5E). We also produced 35S:LHY-HA transgenic plants, in which LHY cDNA was fused in frame to a nucleotide sequence encoding four hemagglutinin (HA) tags and expressed under control of the CaMV 35S promoter. The transgenic plants were crossed with 35S:CCA1β transgenic plants. ChIP assays revealed that LHY binding to the TOC1 gene promoter was also reduced in the 35S:LHY-HA × 35S:CCA1β plants (see Supplemental Figure 7 online). Levels of CCA1α and LHY proteins and CCA1β transcripts were unaltered in the plants used for ChIP assays (Figure 5F; see Supplemental Figure 8 online), demonstrating that CCA1β inhibits CCA1α and LHY activities by reducing its DNA binding affinity via the formation of nonfunctional heterodimers.
Circadian Rhythms Are Altered in 35S:CCA1β Transgenic Plants
CCA1-deficient mutants are phenotypically indistinguishable from wild-type plants but display disturbed rhythmic expression patterns in the clock-regulated genes (Green and Tobin, 1999; Dong et al., 2011). Similar to the cca1-1 and cca1-11 mutants in the Wassilewskija-2 background, the cca1-2 mutant in the Col-0 background also exhibited shortened oscillation periods (see Supplemental Figure 9 online), suggesting that the CCA1 function is conserved in the two ecotypes.
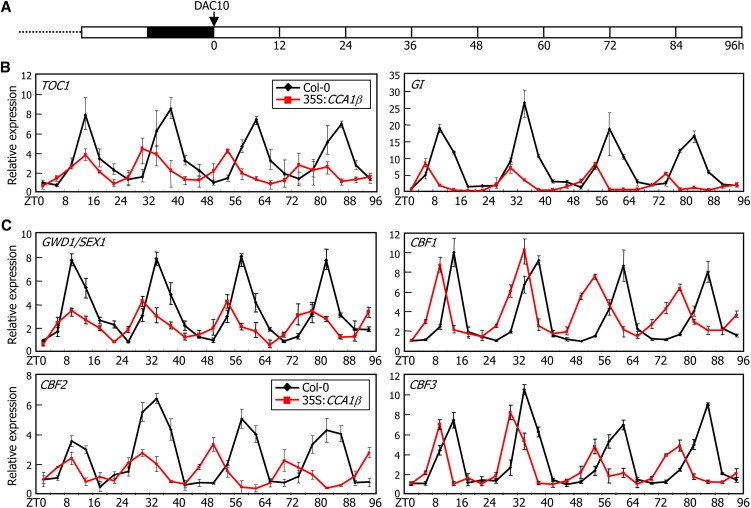
Since CCA1β negatively regulates the CCA1α and LHY transcription factors, the 35S:CCA1β transgenic plants were expected to be physiologically identical to cca1 lhy mutants. To examine this, plants grown on Murashige and Skoog (MS)-agar plates under neutral day cycles (12 h light and 12 h dark) for 10 d were transferred to continuous light conditions at zeitgeber time (ZT) 0 (Figure 6A), and the rhythmic expression patterns of clock genes, such as TOC1, LHY, and GI, were investigated by quantitative real-time RT-PCR (qRT-PCR). The results showed that the periods of circadian oscillations were shortened, and the rhythmic amplitudes were also considerably reduced in the 35S:CCA1β transgenic plants (Figure 6B; see Supplemental Figure 10 online), similar to the patterns observed in the cca1-11 lhy-21 double mutant (Lu et al., 2009). Furthermore, the rhythmic expression pattern of circadian clock–controlled, cold-responsive genes, such as CBFs and α-GLUCAN WATER DIKINASE1 (GWD1)/STARCH EXCESS1 (SEX1) (Yano et al., 2005), were also considerably altered in the 35S:CCA1β transgenic plants (Figure 6C). The rhythmic amplitude was reduced, and its rhythmic period was also shortened. These observations indicate that the CCA1β isoform antagonizes CCA1α and LHY activities in the circadian clock.
Figure 6.
Circadian Traces of TOC1, GI, GWD1/SEX1, and CBF Gene Expression in 35S:CCA1β Transgenic Plants.
Plants grown on MS-agar plates under neutral day cycles (12 h light and 12 h dark) for 10 d were transferred to continuous light conditions (A). Whole plants were harvested at ZT points up to 96 h, and gene transcript levels of clock genes (B) and cold-responsive genes (C) were determined by qRT-PCR. Biological triplicates were averaged. Bars indicate the se. DAC, days after cold imbibition.
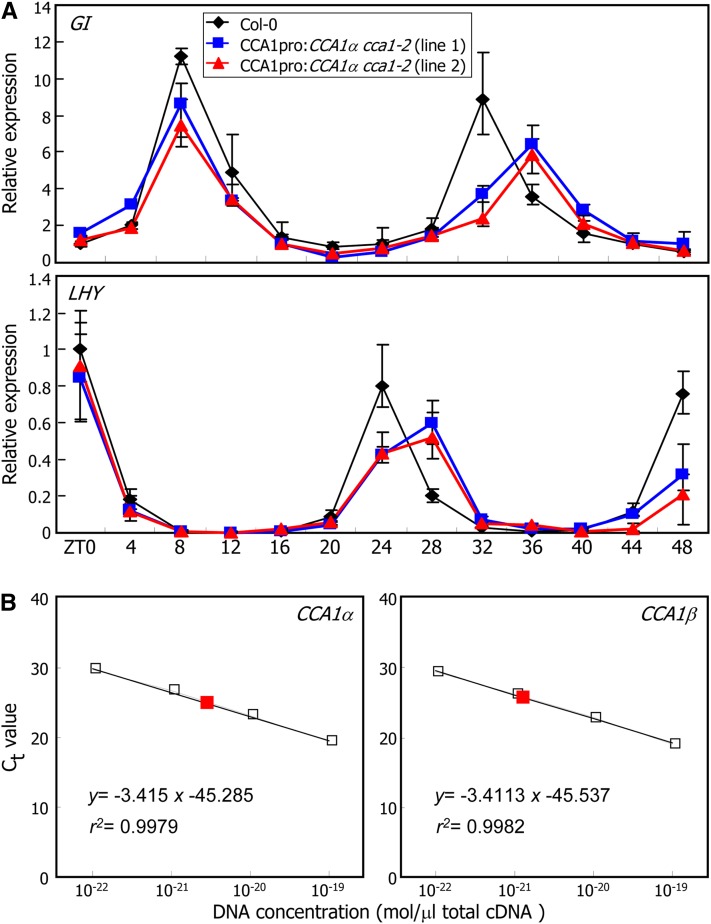
A critical question was whether naturally produced CCA1β proteins are sufficient to have a significant effect on CCA1α activity. To answer this question, we first analyzed the transcript accumulation of clock-regulated genes in CCA1β-deficient plants (CCA1pro:CCA1α cca1-2), in which the CCA1α cDNA was driven by the endogenous CCA1 gene promoter in the cca1-2 mutant background. qRT-PCR assays revealed that the periods of rhythmic oscillations of GI and LHY transcripts were elongated in the CCA1pro:CCA1α cca1-2 plants (Figure 7A). Levels of CCA1α transcripts in these plants were comparable to those in Col-0 plants (see Supplemental Figure 11 online), indicating that the elongated periods of rhythmic oscillations of the clock gene expression are not caused by CCA1α overproduction but due to the lack of CCA1β under normal growth conditions.
Figure 7.
Functional Relevance of CCA1β in Circadian Rhythms.
(A) Circadian rhythms of GI and LHY gene expression in CCA1pro:CCA1α cca1-2 plants that lack functional CCA1β. In the CCA1pro:CCA1α cca1-2 plants, CCA1α expression was driven by the endogenous CCA1 promoter in the cca1-2 mutant background. Plants grown on MS-agar plates under neutral day cycles for 10 d were transferred to continuous light conditions. Whole plants were harvested at ZT points up to 48 h, and gene transcript levels were determined by qRT-PCR. Biological triplicates were averaged. Bars indicate the se.
(B) Quantification of CCA1α and CCA1β transcripts. Two-week-old Col-0 plants grown on MS-agar plates were harvested at ZT4 and used for extraction of total RNA. A series of 10-fold dilutions of the CCA1 plasmid DNA was used to draw the absolute standard curve. The regression line from the dilution curve was used to determine the concentration of CCA1α and CCA1β. Red squares represent the absolute amount of CCA1α and CCA1β transcripts. Ct, threshold cycle.
We next performed absolute quantification of CCA1α and CCA1β transcripts by quantitative real-time PCR analysis (Bustin, 2000; Whelan et al., 2003). Comparison of the Ct (cycle threshold) values of CCA1α and CCA1β transcripts with the standard curve revealed that the molar level of CCA1β transcripts was ∼43% of that of CCA1α transcripts at ZT4: whereas the CCA1α transcripts were 2.597 × 10−21 mol/μL total cDNA, the CCA1β transcripts were 1.140 × 10−21 mol/μL total cDNA (Figure 7B). Time-course measurements of the relative amounts of CCA1α and CCA1β transcripts showed that the molar ratios fluctuate within a range of 35 to 60% (see Supplemental Figure 12 online).
Measurements of the translational efficiencies of CCA1α and CCA1β transcripts in Arabidopsis protoplasts showed that the two transcripts are translated at a similar level (see Supplemental Figure 13 online). In addition, assays on the turnover rates of CCA1α and CCA1β proteins showed that the stabilities of the two proteins are similar to each other (see Supplemental Figure 14 online). These observations support that the relative levels of CCA1α and CCA1β transcripts reflect those of CCA1α and CCA1β proteins. We therefore concluded that the endogenous level of CCA1β is relevant to its role in circadian clock oscillations.
Production of CCA1β Inhibitor Is Suppressed under Cold Conditions
We found that CCA1β inhibits transcriptionally active CCA1α and LHY transcription factors by competitively forming nonfunctional heterodimers in modulating clock-regulated genes. A critical question was how CCA1 alternative splicing is regulated.
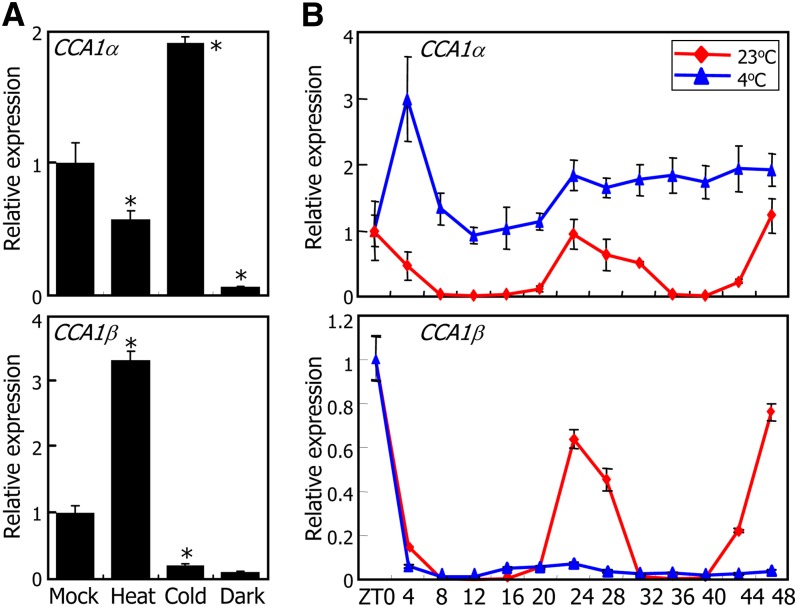
We measured the relative levels of CCA1α and CCA1β transcripts under various growth conditions, including heat (37°C), cold (4°C), and light-dark cycle. The levels of both CCA1α and CCA1β transcripts were reduced to a similar degree in the dark (Figure 8A), indicating that whereas the light-dark cycle regulates CCA1 transcription, it does not influence alternative pre-mRNA splicing. By contrast, the relative ratios of CCA1α and CCA1β transcripts were dramatically changed under both heat and cold conditions (Figure 8A). In heat-treated plants, the level of CCA1α transcripts was reduced by ∼40%, while that of CCA1β transcripts was elevated threefold. Notably, the changes in the relative transcript levels were reversed in cold-treated plants: The level of CCA1α transcripts was elevated approximately twofold, while that of CCA1β transcripts was reduced by more than 80% (Figure 8A).
Figure 8.
Effects of Cold (4°C) on CCA1 Alternative Splicing.
(A) Effects of cold, heat, and dark on accumulation of CCA1α and CCA1β transcripts. Two-week-old plants grown on MS-agar plates under long days were subjected to heat (37°C) or cold (4°C) under continuous light conditions at ZT0, and whole plant materials were harvested at ZT24. For dark treatments, plants were transferred to complete darkness at ZT0, and whole plant materials were harvested at ZT24. Transcript levels were determined by qRT-PCR. Biological triplicates were averaged and statistically treated using a Student’s t test (*P < 0.01). Bars indicate the se.
(B) Circadian traces of CCA1α and CCA1β transcript accumulation. Two-week-old plants grown on MS-agar plates under long days were transferred to cold conditions under continuous light conditions, and whole plants were harvested at ZT points for 2 d. Transcript levels were determined as described in (A). Bars indicate the se.
Considering the role of CCA1 in freezing tolerance (Espinoza et al., 2010; Dong et al., 2011), we further investigated the pattern of CCA1 alternative splicing under cold conditions. Two-week-old plants grown on MS-agar plates at 23°C were either maintained at 23°C or transferred to 4°C under continuous light conditions, and whole plants were harvested at ZT points for 2 d. qRT-PCR assays showed that both the CCA1α and CCA1β transcripts exhibited rhythmic patterns with peaks at ZT24 in plants grown at 23°C (Figure 8B). By contrast, the rhythmic patterns of CCA1 transcript oscillations were disturbed when plants were grown at 4°C. The CCA1α transcript levels were maintained at a high level at all ZT points, whereas the rhythmic peak of CCA1β transcripts at ZT24 disappeared completely, indicating that alternative splicing of CCA1 is suppressed by cold.
Suppression of CCA1 Alternative Splicing Is Required for Freezing Tolerance
We found that CCA1 activity is regulated at the posttranscriptional level by alternative pre-mRNA splicing under cold conditions. We therefore asked whether CCA1 alternative splicing is correlated with the role of the CCA1 gene in freezing tolerance.
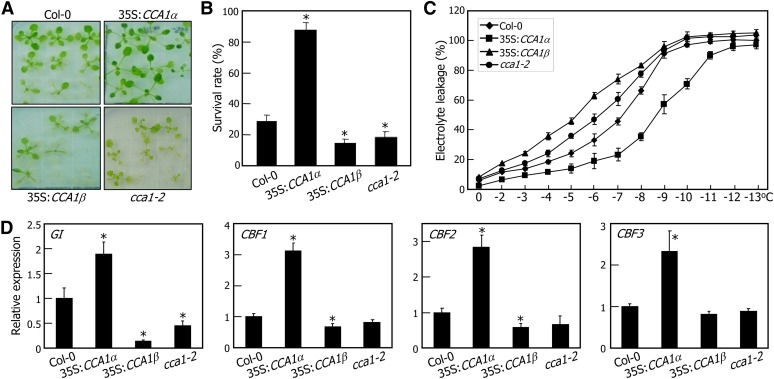
CCA1-overexpressing transgenic plants and the cca1-2 mutant were grown for 2 weeks on MS-agar plates and subsequently incubated at −7°C for 5 h. The plants were allowed to recover at 23°C for 1 week before surviving seedlings were counted. Whereas the 35S:CCA1α (Col-0) transgenic plants exhibited enhanced tolerance to freezing (Figures 9A and 9B), consistent with previous reports (Espinoza et al., 2010; Dong et al., 2011), the 35S:CCA1β (Col-0) transgenic and cca1-2 mutant plants were more sensitive to freezing than the Col-0 plants (Figures 9A and 9B). Freezing tolerance was further examined by electrolyte leakage assays. The temperature at 50% electrolyte leakage (T50) was about −7°C in the Col-0 plants (Figure 9C). By contrast, whereas the T50 was −9°C in the 35S:CCA1α (Col-0) transgenic plants, it was −5°C in the 35S:CCA1β (Col-0) transgenic plants. In particular, the survival rate of the 35S:CCA1α cca1-2 plants, in which the CCA1α gene is driven by the CaMV 35S promoter in the cca1-2 background, was higher than that of the 35S:CCA1α (Col-0) transgenic plants (see Supplemental Figure 15 online), which is explained by the negative regulation of the CCA1α activity by CCA1β in inducing freezing tolerance.
Figure 9.
Induction of Freezing Tolerance by CCA1α.
(A) and (B) Freezing tolerance assays. Two-week-old plants grown on MS-agar plates were incubated at −7°C for 5 h and allowed to recover at 23°C for 1 week before photographs were taken (A). Three measurements of survival rates, each consisting of ∼50 plants, were averaged (B). The statistical significance of the measurements was determined using a Student’s t test by comparing with the value of Col-0 plants (*P < 0.01). Bars indicate the se.
(C) Electrolyte leakage assay. Two-week-old plants grown on MS-agar plates were transferred to 4°C for 7 d under neutral day cycles and tested for freezing tolerance. Biological triplicates were averaged. Bars indicate the se.
(D) Expression of GI and CBF genes in CCA1-overexpressing plants. Plants grown on MS-agar plates under neutral day cycles for 10 d were transferred to continuous light conditions at 4°C. Whole plants were harvested at ZT48, and transcript levels were determined by qRT-PCR. Biological triplicates were averaged and statistically treated using a Student’s t test (*P < 0.01). Bars indicate the se.
We next examined the transcript accumulation of clock genes and cold-responsive genes, including GI and CBFs. Plants entrained to neutral day cycles (12 h light and 12 h dark) for 10 d were transferred to continuous light conditions at either 23 or 4°C. Whole plants were harvested at ZT points up to 48 h (see Supplemental Figure 16A online), and the transcript levels of clock-associated genes were determined by qRT-PCR. At 4°C, rhythmic oscillations of clock genes diminished, which correlated with cold acclimation (see Supplemental Figures 16A and 16B online; Ramos et al., 2005; Bieniawska et al., 2008). Notably, the expression of CBF and GI genes was maintained at higher levels in the 35S:CCA1α (Col-0) transgenic plants but was considerably lower in the 35S:CCA1β (Col-0) transgenic plants in cold-acclimated seedlings (Figure 9D; see Supplemental Figure 16B online), supporting that CCA1α regulates the amplitude of CBF and GI transcripts under cold conditions.
Together, our findings show that CCA1α activity is modulated in a temperature-dependent manner through a self-regulatory module provided by CCA1β. The CCA1β modulator integrates temperature signals into the clock, where CCA1α regulates an output pathway containing GI and CBF genes to induce freezing tolerance.
DISCUSSION
Environmental Stress and Alternative Splicing
It is estimated that more than 95% of multiexonic genes in humans undergo alternative splicing (Pan et al., 2008). In Arabidopsis, ∼42% of intron-containing genes are alternatively spliced (Filichkin et al., 2010). Alternative pre-mRNA splicing has evolved to overcome the limited coding capacities of eukaryotic genomes by producing multiple proteins from a single gene. This process enhances transcriptome plasticity and proteome diversity and is therefore manifested at different developmental stages and under various environmental conditions.
In plants, alternative splicing is often associated with environmental stress responses (Palusa et al., 2007). It affects a certain class of genes that is primarily involved in signal transduction (Ner-Gaon et al., 2004; Barbazuk et al., 2008). Transcription factor genes constitute a major group of these genes. The wheat (Triticum aestivum) WDREB2 gene, an Arabidopsis DREB2 homolog, produces three different transcripts through exon skipping under stress conditions (Egawa et al., 2006). The three isoforms have different accumulation patterns, and the relative ratio of the transcript isoforms is modulated via an abscisic acid–dependent pathway under drought and salt stresses as well as an abscisic acid–independent pathway at low temperatures. In addition, a subgroup of MYB transcription factor genes in Arabidopsis and rice (Oryza sativa) produces alternatively spliced transcripts, the accumulation of which is influenced by various stress signals. Alternative splicing of each MYB gene results in multiple spliced isoforms and produces putative proteins differing by the number of MYB repeats and thus their DNA binding affinities (Li et al., 2006). The Arabidopsis IDD14 transcription factor gene also undergoes alternative splicing, producing IDD14α and IDD14β isoforms (Seo et al., 2011b). Cold-induced intron retention produces the IDD14β isoform, which contains a dimerization domain but lacks a DNA binding domain. It attenuates the IDD14α activity through physical interactions, regulating starch metabolism under cold conditions.
Genes encoding other groups of signaling regulators are also influenced by alternative splicing. A small group of genes encoding putative ribokinases and C3H2C3 RING finger proteins in durum wheat (Triticum durum) undergoes alternative splicing in which a subset of introns is retained at low temperatures (Mastrangelo et al., 2005). Alternative splicing of genes encoding nuclear splicing factors is also influenced by abiotic stresses. Most of the 19 genes encoding Ser/Arg-rich proteins, which are classified as RNA binding proteins with roles as splicing regulators in Arabidopsis, undergo alternative splicing in response to environmental stimuli (Isshiki et al., 2006; Palusa et al., 2007). STABILIZED1 is a pre-mRNA splicing factor, and the sta1-1 mutant has alterations in the alternative splicing patterns of the COR15A gene that is influenced by cold, resulting in hypersensitivity to freezing (Lee et al., 2006). It is apparent that alternative splicing of specific transcription factors and signaling regulators contributes to a broad spectrum of abiotic stress responses in plants.
CCA1 Alternative Splicing in the Clock and Temperature Responses
Several clock genes are known to be regulated at the posttranscriptional level in both plants and animals (Petrillo et al., 2011; Staiger and Green, 2011). In plants, mutations in the PROTEIN ARGININE METHYLTRANSFERASE5 (PRMT5) gene disrupt clock oscillations (Hong et al., 2010; Sanchez et al., 2010). PRMT5 catalyzes symmetric dimethylation of Arg residues in splicing factors that mediate alternative splicing of PRR9 (Hong et al., 2010; Sanchez et al., 2010; Petrillo et al., 2011), which constitutes a negative feedback loop in the central oscillator. Alternative splicing of clock genes is also observed in animals. Roles of a PRMT5 homolog in the regulation of alternative splicing of clock genes have been demonstrated in Drosophila melanogaster (Sanchez et al., 2010). In addition, alternative splicing of FREQUENCY (FRQ) links the clock to ambient temperature responses in Neurospora crassa (Liu et al., 1997), suggesting that alternative splicing is a critical molecular scheme of the clock function.
Here, we present experimental evidence that alternative splicing serves as a self-regulatory scheme, in which regulation of CCA1α activity by a splice variant, CCA1β, plays a role in coordination of the circadian clock to cold acclimation. Dimerization of CCA1α and LHY is particularly important for their ability to regulate circadian rhythms (Daniel et al., 2004; Lu et al., 2009; Yakir et al., 2009). Notably, CCA1β negatively regulates the activities of CCA1α and LHY by competitively forming nonfunctional heterodimers. As a result, 35S:CCA1β transgenic plants exhibit impaired circadian rhythms, as observed in the cca1 lhy double mutant (Lu et al., 2009). By contrast, CCA1pro:CCA1α cca1-2 plants, which lack functional CCA1β but possess CCA1α to a level comparable to that in Col-0 plants, showed a long period. In addition, the molar ratio of CCA1β to CCA1α isoforms ranged from 35 to 60%, indicating that the level of naturally produced CCA1β protein is physiologically relevant in regulating CCA1α activity under normal growth conditions.
Alternative splicing of CCA1 is suppressed by low temperatures. Therefore, CCA1β production is reduced significantly at low temperatures, derepressing CCA1α activity. Consistent with this finding, 35S:CCA1α (Col-0) transgenic plants were tolerant to freezing, whereas 35S:CCA1β (Col-0) transgenic plants showed reduced freezing tolerance. In addition, freezing tolerance was further enhanced in 35S:CCA1α cca1-2 plants, in which CCA1β activity is absent.
It has been reported that PRR9 and LHY undergo alternative splicing in Arabidopsis (Sanchez et al., 2010; Seo et al., 2011b). We found that CCA1 also undergoes alternative splicing. It is remarkable that CCA1 alternative splicing is influenced by ambient temperatures, similar to that of FRQ, which links the clock with ambient temperature responses in N. crassa (Liu et al., 1997). More work is required to determine whether CCA1 plays a major role in the integration of temperature signals into the clock in plants.
Circadian Clock and Temperature Response
Light and temperature are two major determinants of circadian rhythms in both plants and animals (Rensing and Ruoff, 2002; Salomé and McClung, 2005; Yamashino et al., 2008). In plants, central circadian oscillators are also involved in freezing tolerance responses, in addition to temperature entrainment to circadian oscillations (Nakamichi et al., 2005; Salomé and McClung, 2005; Espinoza et al., 2008, 2010; Yamashino et al., 2008; Dong et al., 2011). The circadian clock regulates the expression of a significant portion of plant genomes (Harmer et al., 2000; Michael and McClung, 2003) and a number of physiological events, such as stress responses, hormone responses, and secondary metabolite biosynthesis (Covington and Harmer, 2007; Bieniawska et al., 2008; Covington et al., 2008; Espinoza et al., 2010), suggesting that the regulation of circadian clock components would provide an adaptive strategy by which endogenous physiology is adjusted under changing growth conditions, such as cold. Consistent with this view, Arabidopsis plants having mutations in circadian clock genes exhibit impaired cold acclimation (Cao et al., 2005; Nakamichi et al., 2009; Espinoza et al., 2010).
It is notable that the rhythmic expression of circadian oscillator genes is disrupted during cold acclimation (Ramos et al., 2005; Bieniawska et al., 2008), indicating that the arrhythmicity of circadian oscillation may be intimately related to freezing tolerance. In nature, continuous cold stress is occasionally followed by unexpected freezing shock. Therefore, plants should establish constitutive resistance to freezing during cold acclimation and minimize the dependence of gene expression on the circadian clock (Fowler et al., 2005), explaining the arrhythmicity that occur under cold conditions.
Constitutive expression of CCA1α not only minimizes the gating effects of the clock (Fowler et al., 2005) but also induces CBF and GI genes to enhance freezing tolerance during cold acclimation (Figure 9D). Because the rhythmic expression patterns of clock-regulated genes are attenuated during cold acclimation (Ramos et al., 2005; Bieniawska et al., 2008), it is likely that CCA1α regulation of the amplitude of gene expression is critical to plant adaptation to cold conditions. It has been known that CCA1α acts as a transcriptional repressor of several clock genes, such as GI and TOC1, under normal growth temperatures (Alabadí et al., 2001; Más and Yanovsky, 2009). Meanwhile, it positively regulates CBF and GI genes in response to cold stress (Dong et al., 2011; our data). These observations suggest that the transcriptional regulation activity of CCA1α and/or interacting proteins that affect expression of CBF and GI transcripts would be modulated under cold conditions. Under cold conditions, suppression of CCA1 alternative splicing enhances CCA1α activity, which would contribute to the arrhythmicity and induction of freezing tolerance by amplitude regulation of the expression of CBF and GI genes. We therefore conclude that regulation of CCA1 activity by alternative splicing is important to plant adaptation to cold conditions.
METHODS
Plant Materials and Growth Conditions
All Arabidopsis thaliana lines used were in the Col-0 background, unless otherwise specified. Plants were grown in a controlled culture room at 23°C with a relative humidity of 55% under long days (16 h light/8 h dark) with white light illumination (120 μmol photons/m2s) provided by fluorescent FLR40D/A tubes (Osram).
The cca1-2 mutant (FLAG-141G10) in the Col-0 background was isolated from an Arabidopsis mutant pool of T-DNA insertion lines deposited in the Institut National de la Recherche Agronomique (Versailles, France). Homozygotic lines were obtained by herbicide selection for three or more generations and by analysis of segregation ratios. Lack of CCA1 gene expression in the mutant was verified by RT-PCR.
To produce transgenic plants overexpressing CCA1α and CCA1β genes, full-length cDNAs were subcloned into the binary pB2GW7 vector under control of the CaMV 35S promoter (Invitrogen) through homologous recombination of attB1 and attB2 sites (see Supplemental Table 1 online). Agrobacterium tumefaciens–mediated Arabidopsis transformation was performed according to a modified floral dip method (Clough and Bent, 1998). T3 transgenic plants having single T-DNA insertional events were used in the assays.
Analysis of Gene Transcript Levels
qRT-PCR was employed to measure the transcript levels. RNA sample preparation, reverse transcription, and quantitative PCR were performed according to the rules recently proposed by Udvardi et al. (2008) to provide reproducible and accurate measurements. The extraction of total RNA samples from appropriate plant materials and RT-PCR conditions have been described previously (Kim et al., 2008). The RNA samples were pretreated extensively with an RNase-free DNase to remove any contaminating genomic DNA prior to use.
qRT-PCR reactions were performed in 96-well blocks with an Applied Biosystems 7500 real-time PCR system using the SYBR Green I master mix in a volume of 25 μL. The PCR primers were designed using the Primer Express Software installed in the system and listed in Supplemental Table 1 online. The two-step thermal cycling profile used was 15 s at 94°C and 1 min at 68°C. An eIF4A gene (At3g13920) was included in the reactions as an internal control to normalize the variations in the amounts of cDNA used (Gutierrez et al., 2008). All qRT-PCR reactions were performed in biological triplicates using RNA samples extracted from three independent plant materials grown under identical growth conditions. The comparative ΔΔCT method was employed to evaluate relative quantities of each product amplified from the samples. The threshold cycle (CT) was automatically determined for each reaction using the default parameters of the system. The specificity of the PCR reactions was determined by melt curve analysis of the amplified products using the standard methods installed in the system.
For absolute quantification of endogenous CCA1α and CCA1β transcripts, cDNAs of CCA1α and CCA1β were subcloned into the pDONR vector (Invitrogen) through homologous recombination of attB1 and attB2 sites, and an absolute standard curve of each transcript was generated by 10-fold serial dilutions covering 10−17 to 10−23 mol, as previously described (Bustin, 2000; Whelan et al., 2003). Quantitative RT-PCR was performed using the SYBR Green I master mix (Applied Biosystems), with CCA1α-specific primers (forward, 5′-GATCTGGTTATTAAGACTCGGAAGCCATATAC-3′; and reverse, 5′-GCCTCTTTCTCTACCTTGGAGA-3′) and CCA1β-specific primers (forward, 5′-GAATGTTCCTTGTGATAAGCCATAGAGG-3′, and reverse, 5′-AGGATCGTTCCACTTCCCGTCTT-3′).
Yeast Two-Hybrid Assays
Yeast two-hybrid assays were performed using the BD Matchmaker system (Clontech). The pGADT7 vector was used for GAL4 AD, and the pGBKT7 vector was used for GAL4 BD. Yeast strain AH109 (Leu-, Trp-, Ade-, His-), which has the chromosomally integrated reporter genes lacZ and HIS under control of the GAL1 promoter, was used for transformation. The PCR products were digested with EcoRI and BamHI (for CCA1α) and NdeI and EcoRI (for CCA1β) and subcloned into the pGBKT7 and pGADT7 vectors. Transformation of AH109 cells was performed according to the manufacturer’s instructions. Colonies obtained were streaked on medium without His, Ade, Leu, and Trp. To confirm the results, β-Gal assays were performed according to the system procedure.
The pBridge vector (Clontech) was used for yeast three-hybrid screening. The CCA1α cDNA was amplified by RT-PCR (see Supplemental Table 1 online), and the PCR product was digested with EcoRI and BamHI and then subcloned into the pBridge vector, resulting in the BD-CCA1α construct. The CCA1β cDNA was subcloned into the NotI and BglII-digested pBridge vector so that its expression was controlled by the Met-repressible pMET25 promoter. The expression constructs (BD-CCA1α and pMET25-CCA1β in the pBridge vector and AD-CCA1α in the pGADT7 vector) were cotransformed into AH109 cells. The colonies were streaked on media without Leu, Trp, and His and supplemented with or without Met.
In Vitro Pull-Down Assays
Recombinant MBP and MBP-CCA1α fusion proteins were produced in Escherichia coli BL21-CodonPlus (DE3)-RIL strains (Stratagene) and purified as follows. One-tenth volume of precultured cells in 5 mL of Luria-Bertani medium was added to 500 mL of fresh Luria-Bertani medium and cultured at 37°C until OD600 reached 0.3 to 0.6. Protein production was induced by adding isopropyl-β-d-thiogalactopyranoside at a final concentration of 0.5 mM and shaking at 37°C for 5 h. The cells were harvested and resuspended in buffer A (25 mM HEPES, pH 7.5, 20% glycerol, 1 mM DTT, 100 mM NaCl, and 0.2 mM EDTA) containing protease inhibitor cocktail (Sigma-Aldrich) and 1 mM PMSF. The cells were lysed using a French press (8500 p.s.i.; one time). The lysates were sonicated twice for 30 s each and then centrifuged at 20,000g for 20 min. The supernatants were stored at −80°C until use. The CCA1α and CCA1β cDNAs were amplified by RT-PCR and subcloned into the pGADT7 vector. The CCA1α and CCA1β polypeptides were labeled with 35S-Met using the TNT-coupled reticulocyte lysate system (Promega).
The MBP or MBP-CCA1α proteins were mixed with amylose resin (Sigma-Aldrich) and agitated for 15 min at room temperature. The beads were then washed three times with 1× PBS buffer and one time with buffer A. Five microliters of the 35S-labeled proteins was added, and the samples were incubated for 2 h at 4°C. Next, the beads were washed five times with buffer A. The bound proteins were eluted with 1× SDS-PAGE loading buffer by boiling for 5 min at 100°C and subjected to SDS-PAGE and autoradiography.
BiFC Assays
BiFC assays were performed by cotransfection of the CCA1α-nYFP and CCA1β-cYFP vectors or vice versa into Arabidopsis mesophyll protoplasts. The expression constructs were transformed into Arabidopsis protoplasts by polyethylene glycol-calcium transfection (Yoo et al., 2007). The subcellular distribution of CCA1 proteins was visualized by differential interference contrast microscopy and fluorescence microscopy. Reconstitution of YFP fluorescence was observed using a confocal microscope with the following YFP filter setup: excitation 515 nm, 458/514 dichroic, and emission 560- to 615-nm band-pass filter.
Transcriptional Regulation Activity Assays
For transient expression assays in Arabidopsis protoplasts, several reporter and effector plasmids were constructed. The reporter plasmids contain four copies of the GAL4 upstream activation sequence and the GUS gene. To construct the p35S:CCA1 effector plasmids, the CCA1α and CCA1β cDNAs were fused to the GAL4 BD-coding sequence and inserted into an expression vector containing the CaMV 35S promoter. The reporter and effector plasmids were cotransformed into Arabidopsis protoplasts by a polyethylene glycol–mediated transformation method. GUS activities were measured by the fluorometric method as previously described (Jefferson et al., 1987). A CaMV 35S promoter-luciferase construct was also cotransformed as an internal control. The luciferase assay was performed using the Luciferase Assay System (Promega).
Electrophoretic Mobility Shift Assays
The CCA1α and CCA1β cDNAs were subcloned into the pMAL-c2X E. coli expression vector (NEB) with an MBP-coding sequence. The MBP-CCA1 fusion proteins were purified according to the manufacturer’s instructions using the pMAL Protein Fusion and Purification System (#E8000S; New England BioLabs Inc.). DNA fragments were end labeled with [γ-32P]dATP using T4 polynucleotide kinase. Labeled probes were incubated with ∼0.5 μg of purified MBP-CCA1 proteins for 30 min at 25°C in binding buffer (10 mM Tris-HCl, pH 7.6, 50 mM NaCl, 1 mM EDTA, 5 mM DTT, and 5% glycerol). The reaction mixtures were electrophoresed on 6% native PAGE gels. The gels were dried on Whatman 3MM paper and exposed to x-ray films.
ChIP Assays
An MYC-coding sequence was fused in frame to the 3′ ends of the CCA1α and CCA1β cDNAs, and the fusions were subcloned under control of the CaMV 35S promoter (Kim et al., 2008). The expression construct was transformed into Arabidopsis plants. Three-week-old 35S:CCA1α-MYC and 35S:CCA1β-MYC transgenic plants grown on half-strength MS-agar plates were used for extraction of the total cellular extracts. Processing of plant materials and qRT-PCR were performed as described previously (Seo et al., 2011b). The qRT-PCR primers used have been described previously (Pruneda-Paz et al., 2009). The primers used to amplify the CBS-containing sequence region in the CHE promoter were, forward, 5′-AAAAATCTCGACGCAACGAC-3′, and reverse, 5′-CATTTGGAGCGTGGCATAAT-3′. A sequence region consisting of nucleotides 926 to 1035 downstream of the CBS element was amplified by the primer pair, forward, 5′-TGTCTCCACCAGCCTAGCTTC-3′, and reverse 5′-CATGGAATTAGGATTTCGTTATCA-3′, and used as a negative control.
Freezing Tolerance Assays
Approximately 30 plants grown for 2 weeks on MS-agar plates were incubated for 5 h at −7°C. After incubating at 4°C for 24 h in the dark, the plants were allowed to recover at 23°C for 1 week before surviving plants were counted. Three independent measurements of survival rates were averaged and statistically analyzed using a Student’s t test. Electrolyte leakage assays were performed as previously described (Doherty et al., 2009).
Accession Numbers
Sequence data from this article can be obtained from the Arabidopsis Genome Initiative databases under the following accession numbers: CCA1 (At2g46830), LHY (At1g01060), TOC1 (At5g61380), GI (At1g22770), GWD1/SEX1 (At1g10760), CBF1 (At4g25490), CBF2 (At4g25470), and CBF3 (At4g25480).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Alternatively Spliced Variants of CCA1 Gene.
Supplemental Figure 2. Nucleotide Sequences of CCA1α and CCA1β cDNAs.
Supplemental Figure 3. Amino Acid Sequence Comparison of CCA1α and CCA1β Proteins.
Supplemental Figure 4. Interactions between CCA1β and LHY.
Supplemental Figure 5. Levels of CCA1α and CCA1β Transcripts in CCA1α- and CCA1β-Overexpressing Transgenic Plants.
Supplemental Figure 6. Levels of CCA1α and CCA1β Transcripts in 35S:CCA1α-MYC and 35S:CCA1β-MYC Transgenic Plants.
Supplemental Figure 7. ChIP Assays on 35S:LHY-HA X 35S:CCA1β Plants.
Supplemental Figure 8. Relative Levels of CCA1β Transcripts.
Supplemental Figure 9. Altered Circadian Rhythm of GI Expression in the cca1-2 Mutant.
Supplemental Figure 10. Altered Circadian Rhythm of LHY Expression in 35S:CCA1β Transgenic Plants.
Supplemental Figure 11. Rhythmic Accumulation of CCA1α Transcripts in CCA1pro:CCA1α cca1-2 Plants.
Supplemental Figure 12. Ratios of CCA1α and CCA1β Transcripts.
Supplemental Figure 13. Downstream GUS Translational Efficiency of CCA1α and CCA1β Transcripts in Arabidopsis Protoplasts.
Supplemental Figure 14. Protein Turnover of CCA1α and CCA1β.
Supplemental Figure 15. Freezing Tolerance of 35S:CCA1α (Col-0) and 35S:CCA1α cca1-2 Plants.
Supplemental Figure 16. Altered Circadian Rhythms in 35S:CCA1α and 35S:CCA1β Transgenic Plants.
Supplemental Table 1. Primers Used in qRT-PCR and Subcloning.
Acknowledgments
We thank Jungmook Kim (Chonnam National University, Korea) for scientific discussions. This work was supported by the Leaping Research Program (20120005600) provided by the National Research Foundation of Korea, the Next-Generation BioGreen 21 program (Plant Molecular Breeding Center No. PJ008103) provided by the Rural Development Administration, and by grants from the National Research Foundation of Korea (20110027355) and the Agricultural R & D Promotion Center (309017-03), Korea Ministry for Food, Agriculture, Forestry, and Fisheries. P.J.S. is grateful for the Seoul Science Fellowship award.
AUTHOR CONTRIBUTIONS
C.-M.P. and P.J.S. designed the research. P.J.S., M.-J.P., M.-H.L., S.-G.K., and M.L. performed the experiments. P.J.S., S.-G.K., and I.T.B. analyzed the data. C.-M.P. and P.J.S. wrote the article.
Glossary
- β-Gal
β-galactosidase
- BiFC
bimolecular fluorescence complementation
- YFP
yellow fluorescent protein
- CaMV
cauliflower mosaic virus
- Col-0
Columbia-0
- BD
binding domain
- AD
activation domain
- GUS
β-glucuronidase
- CBS
CCA1 binding site
- ChIP
chromatin immunoprecipitation
- MS
Murashige and Skoog
- qRT-PCR
quantitative RT-PCR
- ZT
to be defined
References
- Alabadí D., Oyama T., Yanovsky M.J., Harmon F.G., Más P., Kay S.A. (2001). Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293: 880–883 [DOI] [PubMed] [Google Scholar]
- Barbazuk W.B., Fu Y., McGinnis K.M. (2008). Genome-wide analyses of alternative splicing in plants: Opportunities and challenges. Genome Res. 18: 1381–1392 [DOI] [PubMed] [Google Scholar]
- Baxevanis A.D., Vinson C.R. (1993). Interactions of coiled coils in transcription factors: Where is the specificity? Curr. Opin. Genet. Dev. 3: 278–285 [DOI] [PubMed] [Google Scholar]
- Bieniawska Z., Espinoza C., Schlereth A., Sulpice R., Hincha D.K., Hannah M.A. (2008). Disruption of the Arabidopsis circadian clock is responsible for extensive variation in the cold-responsive transcriptome. Plant Physiol. 147: 263–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin S.A. (2000). Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 25: 169–193 [DOI] [PubMed] [Google Scholar]
- Cao S., Song Y., Su L. (2007). Freezing sensitivity in the gigantea mutant of Arabidopsis is associated with sugar deficiency. Biol. Plant. 51: 359–362 [Google Scholar]
- Cao S., Ye M., Jiang S. (2005). Involvement of GIGANTEA gene in the regulation of the cold stress response in Arabidopsis. Plant Cell Rep. 24: 683–690 [DOI] [PubMed] [Google Scholar]
- Chinnusamy V., Ohta M., Kanrar S., Lee B.H., Hong X., Agarwal M., Zhu J.K. (2003). ICE1: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 17: 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V., Schumaker K., Zhu J.K. (2004). Molecular genetic perspectives on cross-talk and specificity in abiotic stress signalling in plants. J. Exp. Bot. 55: 225–236 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Covington M.F., Harmer S.L. (2007). The circadian clock regulates auxin signaling and responses in Arabidopsis. PLoS Biol. 5: e222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington M.F., Maloof J.N., Straume M., Kay S.A., Harmer S.L. (2008). Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 9: R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel X., Sugano S., Tobin E.M. (2004). CK2 phosphorylation of CCA1 is necessary for its circadian oscillator function in Arabidopsis. Proc. Natl. Acad. Sci. USA 101: 3292–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd A.N., Salathia N., Hall A., Kévei E., Tóth R., Nagy F., Hibberd J.M., Millar A.J., Webb A.A. (2005). Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309: 630–633 [DOI] [PubMed] [Google Scholar]
- Doherty C.J., Van Buskirk H.A., Myers S.J., Thomashow M.F. (2009). Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 21: 972–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C.H., Agarwal M., Zhang Y., Xie Q., Zhu J.K. (2006). The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc. Natl. Acad. Sci. USA 103: 8281–8286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong M.A., Farré E.M., Thomashow M.F. (2011). Circadian clock-associated 1 and late elongated hypocotyl regulate expression of the C-repeat binding factor (CBF) pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 7241–7246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egawa C., Kobayashi F., Ishibashi M., Nakamura T., Nakamura C., Takumi S. (2006). Differential regulation of transcript accumulation and alternative splicing of a DREB2 homolog under abiotic stress conditions in common wheat. Genes Genet. Syst. 81: 77–91 [DOI] [PubMed] [Google Scholar]
- Espinoza C., Bieniawska Z., Hincha D.K., Hannah M.A. (2008). Interactions between the circadian clock and cold-response in Arabidopsis. Plant Signal. Behav. 3: 593–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza C., Degenkolbe T., Caldana C., Zuther E., Leisse A., Willmitzer L., Hincha D.K., Hannah M.A. (2010). Interaction with diurnal and circadian regulation results in dynamic metabolic and transcriptional changes during cold acclimation in Arabidopsis. PLoS ONE 5: e14101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farré E.M., Harmer S.L., Harmon F.G., Yanovsky M.J., Kay S.A. (2005). Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr. Biol. 15: 47–54 [DOI] [PubMed] [Google Scholar]
- Filichkin S.A., Priest H.D., Givan S.A., Shen R., Bryant D.W., Fox S.E., Wong W.K., Mockler T.C. (2010). Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 20: 45–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler S.G., Cook D., Thomashow M.F. (2005). Low temperature induction of Arabidopsis CBF1, 2, and 3 is gated by the circadian clock. Plant Physiol. 137: 961–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin K.A., Whitelam G.C. (2007). Light-quality regulation of freezing tolerance in Arabidopsis thaliana. Nat. Genet. 39: 1410–1413 [DOI] [PubMed] [Google Scholar]
- Gilmour S.J., Sebolt A.M., Salazar M.P., Everard J.D., Thomashow M.F. (2000). Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol. 124: 1854–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour S.J., Zarka D.G., Stockinger E.J., Salazar M.P., Houghton J.M., Thomashow M.F. (1998). Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 16: 433–442 [DOI] [PubMed] [Google Scholar]
- Green R.M., Tobin E.M. (1999). Loss of the circadian clock-associated protein 1 in Arabidopsis results in altered clock-regulated gene expression. Proc. Natl. Acad. Sci. USA 96: 4176–4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez L., Mauriat M., Guénin S., Pelloux J., Lefebvre J.F., Louvet R., Rusterucci C., Moritz T., Guerineau F., Bellini C., Van Wuytswinkel O. (2008). The lack of a systematic validation of reference genes: A serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol. J. 6: 609–618 [DOI] [PubMed] [Google Scholar]
- Harmer S.L., Hogenesch J.B., Straume M., Chang H.S., Han B., Zhu T., Wang X., Kreps J.A., Kay S.A. (2000). Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290: 2110–2113 [DOI] [PubMed] [Google Scholar]
- Hong S., Song H.R., Lutz K., Kerstetter R.A., Michael T.P., McClung C.R. (2010). Type II protein arginine methyltransferase 5 (PRMT5) is required for circadian period determination in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 107: 21211–21216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S.Y., Kim O.K., Kim S.G., Yang M.S., Park C.M. (2011). Nuclear import and DNA binding of the ZHD5 transcription factor is modulated by a competitive peptide inhibitor in Arabidopsis. J. Biol. Chem. 286: 1659–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotta C.T., Gardner M.J., Hubbard K.E., Baek S.J., Dalchau N., Suhita D., Dodd A.N., Webb A.A. (2007). Modulation of environmental responses of plants by circadian clocks. Plant Cell Environ. 30: 333–349 [DOI] [PubMed] [Google Scholar]
- Hu W., Ma H. (2006). Characterization of a novel putative zinc finger gene MIF1: Involvement in multiple hormonal regulation of Arabidopsis development. Plant J. 45: 399–422 [DOI] [PubMed] [Google Scholar]
- Isshiki M., Tsumoto A., Shimamoto K. (2006). The serine/arginine-rich protein family in rice plays important roles in constitutive and alternative splicing of pre-mRNA. Plant Cell 18: 146–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa T., Foster R., Chua N.H. (1993). Plant bZIP protein DNA binding specificity. J. Mol. Biol. 230: 1131–1144 [DOI] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.S., Kim S.G., Lee M., Lee I., Park H.Y., Seo P.J., Jung J.H., Kwon E.J., Suh S.W., Paek K.H., Park C.M. (2008). HD-ZIP III activity is modulated by competitive inhibitors via a feedback loop in Arabidopsis shoot apical meristem development. Plant Cell 20: 920–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B.H., Kapoor A., Zhu J., Zhu J.K. (2006). STABILIZED1, a stress-upregulated nuclear protein, is required for pre-mRNA splicing, mRNA turnover, and stress tolerance in Arabidopsis. Plant Cell 18: 1736–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Li X., Guo L., Lu F., Feng X., He K., Wei L., Chen Z., Qu L.J., Gu H. (2006). A subgroup of MYB transcription factor genes undergoes highly conserved alternative splicing in Arabidopsis and rice. J. Exp. Bot. 57: 1263–1273 [DOI] [PubMed] [Google Scholar]
- Liu Y., Garceau N.Y., Loros J.J., Dunlap J.C. (1997). Thermally regulated translational control of FRQ mediates aspects of temperature responses in the neurospora circadian clock. Cell 89: 477–486 [DOI] [PubMed] [Google Scholar]
- Lu S.X., Knowles S.M., Andronis C., Ong M.S., Tobin E.M. (2009). CIRCADIAN CLOCK ASSOCIATED1 and LATE ELONGATED HYPOCOTYL function synergistically in the circadian clock of Arabidopsis. Plant Physiol. 150: 834–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S.X., Webb C.J., Knowles S.M., Kim S.H., Wang Z., Tobin E.M. (2012). CCA1 and ELF3 Interact in the control of hypocotyl length and flowering time in Arabidopsis. Plant Physiol. 158: 1079–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Más P., Yanovsky M.J. (2009). Time for circadian rhythms: Plants get synchronized. Curr. Opin. Plant Biol. 12: 574–579 [DOI] [PubMed] [Google Scholar]
- Mastrangelo A.M., Belloni S., Barilli S., Ruperti B., Di Fonzo N., Stanca A.M., Cattivelli L. (2005). Low temperature promotes intron retention in two e-cor genes of durum wheat. Planta 221: 705–715 [DOI] [PubMed] [Google Scholar]
- Michael T.P., McClung C.R. (2003). Enhancer trapping reveals widespread circadian clock transcriptional control in Arabidopsis. Plant Physiol. 132: 629–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K., Jin J.B., Lee J., Yoo C.Y., Stirm V., Miura T., Ashworth E.N., Bressan R.A., Yun D.J., Hasegawa P.M. (2007). SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 19: 1403–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi T., Wheatley K., Hanzawa Y., Wright L., Mizoguchi M., Song H.R., Carré I.A., Coupland G. (2002). LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev. Cell 2: 629–641 [DOI] [PubMed] [Google Scholar]
- Nakamichi N., Kita M., Ito S., Sato E., Yamashino T., Mizuno T. (2005). The Arabidopsis pseudo-response regulators, PRR5 and PRR7, coordinately play essential roles for circadian clock function. Plant Cell Physiol. 46: 609–619 [DOI] [PubMed] [Google Scholar]
- Nakamichi N., Kusano M., Fukushima A., Kita M., Ito S., Yamashino T., Saito K., Sakakibara H., Mizuno T. (2009). Transcript profiling of an Arabidopsis PSEUDO RESPONSE REGULATOR arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant Cell Physiol. 50: 447–462 [DOI] [PubMed] [Google Scholar]
- Ner-Gaon H., Halachmi R., Savaldi-Goldstein S., Rubin E., Ophir R., Fluhr R. (2004). Intron retention is a major phenomenon in alternative splicing in Arabidopsis. Plant J. 39: 877–885 [DOI] [PubMed] [Google Scholar]
- Niwa Y., Ito S., Nakamichi N., Mizoguchi T., Niinuma K., Yamashino T., Mizuno T. (2007). Genetic linkages of the circadian clock-associated genes, TOC1, CCA1 and LHY, in the photoperiodic control of flowering time in Arabidopsis thaliana. Plant Cell Physiol. 48: 925–937 [DOI] [PubMed] [Google Scholar]
- Palusa S.G., Ali G.S., Reddy A.S. (2007). Alternative splicing of pre-mRNAs of Arabidopsis serine/arginine-rich proteins: Regulation by hormones and stresses. Plant J. 49: 1091–1107 [DOI] [PubMed] [Google Scholar]
- Pan Q., Shai O., Lee L.J., Frey B.J., Blencowe B.J. (2008). Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 40: 1413–1415 [DOI] [PubMed] [Google Scholar]
- Petrillo E., Sanchez S.E., Kornblihtt A.R., Yanovsky M.J. (2011). Alternative splicing adds a new loop to the circadian clock. Commun. Integr. Biol. 4: 284–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruneda-Paz J.L., Breton G., Para A., Kay S.A. (2009). A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 323: 1481–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos A., Pérez-Solís E., Ibáñez C., Casado R., Collada C., Gómez L., Aragoncillo C., Allona I. (2005). Winter disruption of the circadian clock in chestnut. Proc. Natl. Acad. Sci. USA 102: 7037–7042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing L., Ruoff P. (2002). Temperature effect on entrainment, phase shifting, and amplitude of circadian clocks and its molecular bases. Chronobiol. Int. 19: 807–864 [DOI] [PubMed] [Google Scholar]
- Salomé P.A., McClung C.R. (2005). PSEUDO-RESPONSE REGULATOR 7 and 9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock. Plant Cell 17: 791–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez S.E., et al. (2010). A methyl transferase links the circadian clock to the regulation of alternative splicing. Nature 468: 112–116 [DOI] [PubMed] [Google Scholar]
- Seo P.J., Hong S.Y., Kim S.G., Park C.M. (2011a). Competitive inhibition of transcription factors by small interfering peptides. Trends Plant Sci. 16: 541–549 [DOI] [PubMed] [Google Scholar]
- Seo P.J., Kim M.J., Ryu J.Y., Jeong E.Y., Park C.M. (2011b). Two splice variants of the IDD14 transcription factor competitively form nonfunctional heterodimers which may regulate starch metabolism. Nat. Commun. 2: 303. [DOI] [PubMed] [Google Scholar]
- Shinozaki K., Yamaguchi-Shinozaki K. (2000). Molecular responses to dehydration and low temperature: Differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol. 3: 217–223 [PubMed] [Google Scholar]
- Song Y.H., Ito S., Imaizumi T. (2010). Similarities in the circadian clock and photoperiodism in plants. Curr. Opin. Plant Biol. 13: 594–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger D., Green R. (2011). RNA-based regulation in the plant circadian clock. Trends Plant Sci. 16: 517–523 [DOI] [PubMed] [Google Scholar]
- Thines B., Harmon F.G. (2010). Ambient temperature response establishes ELF3 as a required component of the core Arabidopsis circadian clock. Proc. Natl. Acad. Sci. USA 107: 3257–3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udvardi M.K., Czechowski T., Scheible W.R. (2008). Eleven golden rules of quantitative RT-PCR. Plant Cell 20: 1736–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinson C.R., Hai T., Boyd S.M. (1993). Dimerization specificity of the leucine zipper-containing bZIP motif on DNA binding: Prediction and rational design. Genes Dev. 7: 1047–1058 [DOI] [PubMed] [Google Scholar]
- Wang Z.Y., Kenigsbuch D., Sun L., Harel E., Ong M.S., Tobin E.M. (1997). A Myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene. Plant Cell 9: 491–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.Y., Tobin E.M. (1998). Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93: 1207–1217 [DOI] [PubMed] [Google Scholar]
- Wenkel S., Emery J., Hou B.H., Evans M.M., Barton M.K. (2007). A feedback regulatory module formed by LITTLE ZIPPER and HD-ZIPIII genes. Plant Cell 19: 3379–3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan J.A., Russell N.B., Whelan M.A. (2003). A method for the absolute quantification of cDNA using real-time PCR. J. Immunol. Methods 278: 261–269 [DOI] [PubMed] [Google Scholar]
- Yakir E., Hilman D., Kron I., Hassidim M., Melamed-Book N., Green R.M. (2009). Posttranslational regulation of CIRCADIAN CLOCK ASSOCIATED1 in the circadian oscillator of Arabidopsis. Plant Physiol. 150: 844–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashino T., Ito S., Niwa Y., Kunihiro A., Nakamichi N., Mizuno T. (2008). Involvement of Arabidopsis clock-associated pseudo-response regulators in diurnal oscillations of gene expression in the presence of environmental time cues. Plant Cell Physiol. 49: 1839–1850 [DOI] [PubMed] [Google Scholar]
- Yano R., Nakamura M., Yoneyama T., Nishida I. (2005). Starch-related alpha-glucan/water dikinase is involved in the cold-induced development of freezing tolerance in Arabidopsis. Plant Physiol. 138: 837–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572 [DOI] [PubMed] [Google Scholar]