Two pathways increase the capacity of the Chlamydomonas chloroplast to detoxify superoxide during Fe limitation stress. In one, a novel plastid-localized MnSOD is expressed in response to Fe limitation. In a second pathway, the plastid FeSOD is preferentially retained over other abundant Fe proteins, demonstrating prioritized allocation of Fe within the chloroplast.
Abstract
Fe deficiency is one of several abiotic stresses that impacts plant metabolism because of the loss of function of Fe-containing enzymes in chloroplasts and mitochondria, including cytochromes, FeS proteins, and Fe superoxide dismutase (FeSOD). Two pathways increase the capacity of the Chlamydomonas reinhardtii chloroplast to detoxify superoxide during Fe limitation stress. In one pathway, MSD3 is upregulated at the transcriptional level up to 103-fold in response to Fe limitation, leading to synthesis of a previously undiscovered plastid-specific MnSOD whose identity we validated immunochemically. In a second pathway, the plastid FeSOD is preferentially retained over other abundant Fe proteins, heme-containing cytochrome f, diiron magnesium protoporphyrin monomethyl ester cyclase, and Fe2S2-containing ferredoxin, demonstrating prioritized allocation of Fe within the chloroplast. Maintenance of FeSOD occurs, after an initial phase of degradation, by de novo resynthesis in the absence of extracellular Fe, suggesting the operation of salvage mechanisms for intracellular recycling and reallocation.
INTRODUCTION
Plants require water, CO2, light, and a few inorganic nutrients for growth. Changes in the availability of each/any of these essential ingredients of plant life generate system-wide changes in gene expression and metabolism. N, P, and K are major macronutrients and key components of fertilizer, whereas Cu, Fe, Mn, Zn, and other elements are classified as micronutrients, being required only in trace amounts in some cases. (The elemental symbols are used as abbreviations with no implication as to speciation.) Each of these chemical elements has a specific biochemical role (such as P as a structural component of nucleic acids and lipids or Fe as a catalyst in redox proteins), and an organism’s quota for each element is relatively constant (Merchant and Helmann, 2012). Nevertheless, availability is neither constant nor uniform across growth habitats. Availability is determined by the absolute amount in the environment, and this varies across the globe, by its chemical state (speciation), which impacts assimilation, and by the amount of other (nonuseful) species competing for assimilation pathways. Therefore, organisms have developed mechanisms for acclimating to changes in elemental nutrition (Merchant and Helmann, 2012). One approach involves more effective assimilation, but when this fails (because of the absence of the nutrient), mechanisms that operate to reduce the quota selectively are brought into play.
This can be accomplished by replacing one element with another (usually less effective or more “expensive”) element (Merchant and Helmann, 2012). Examples include replacement of metal cofactors in enzymes, Cd- or Co-containing carbonic anhydrases in place of the usual Zn enzymes, or use of different chemical species, sulfolipids in place of phospholipids in P-deficient bacteria and plants (Benning et al., 1995; Güler et al., 1996; Lane and Morel, 2000a, 2000b; Xu et al., 2007; Yu et al., 2002). Mechanisms of Cu sparing in the chloroplast, involving substitution of plastocyanin with cytochrome c6 and Cu/Zn superoxide dismutase (Cu/ZnSOD) with Fe superoxide dismutase (FeSOD), have been well documented in Cu-deficient algae and plants, respectively (Merchant et al., 2006; Burkhead et al., 2009; Pilon et al., 2011), but the sparing phenomenon has not been as well studied for chloroplast Fe.
Fe is an important micronutrient for photosynthetic organisms due to its extensive use as a cofactor in proteins of the photosynthetic apparatus (Raven et al., 1999), and it is commonly a limiting nutrient for photosynthetic organisms in both marine and terrestrial environments (Marschner, 1995; Morel and Price, 2003). As a result, photosynthetic organisms in particular have evolved sophisticated mechanisms for Fe assimilation and homeostasis, and these are well documented through studies in several reference organisms, such as Arabidopsis thaliana and Chlamydomonas reinhardtii in the laboratory or diatoms in the field (reviewed in Guerinot and Yi, 1994; Marschner and Römheld, 1994; Walker and Connolly, 2008, Jeong and Guerinot, 2009; Armbrust, 2009, Marchetti and Cassar, 2009; Blaby-Haas and Merchant, 2012). The mechanisms underlying metabolic acclimation in Fe-deficient cells are, by comparison, less investigated.
One of the advantages of the green alga C. reinhardtii as a reference organism is the ease of manipulation of its Fe status (Merchant et al., 2006). We previously defined four stages of Fe nutrition in photoheterotrophic C. reinhardtii, Fe replete (18 μM), Fe deficient (1 to 3 μM, depending on the strain), Fe limited (0.5 μM or less), and Fe excess (200 μM or more), based on growth rate, the appearance of chlorosis, the expression of genes encoding Fe assimilation components, and acquired sensitivity to high light (La Fontaine et al., 2002, Moseley et al., 2002a; Long and Merchant, 2008). An immediate response to Fe deficiency is upregulation of Fe assimilatory components (Merchant et al., 2006). C. reinhardtii appears to have two pathways for Fe acquisition. One consists of a plasma membrane ferroxidase, FOX1, coupled to a trivalent cation transporter, FTR1 (La Fontaine et al., 2002; Herbik et al., 2002; Kosman, 2003), and another consists of two ZIP family transporters, IRT1 and IRT2, which function in an alternate assimilation route (Allen et al., 2007b; Chen et al., 2008). Other components that facilitate Fe uptake are a ferrireductase, FRE1, and periplasmic proteins FEA1/2 (Allen et al., 2007b). The upregulation of the assimilation pathway is an indication that cells are experiencing Fe deficiency, and FOX1 has served as useful marker of intracellular Fe status.
In Fe-poor photoheterotrophic growth conditions, C. reinhardtii initiates a program to degrade various Fe-containing chloroplast proteins, including photosystem I (PSI), the cytochrome b6f complex, and ferredoxin. In addition, the PSI-associated light-harvesting complex is remodeled to ameliorate photooxidative stress resulting from compromised function of its Fe4S4 clusters (Moseley et al., 2002a; Naumann et al., 2005). The signal for the remodeling program may be the state of occupancy of individual chlorophyll proteins (like PsaK), which depend on the activity of the di-Fe–containing magnesium protoporphyrin monomethyl ester cyclase in the chlorophyll biosynthetic pathway whose activity is reduced in Fe deficiency (Spiller et al., 1982; Tottey et al., 2003). By contrast, mitochondrial respiratory complex proteins (also Fe-containing) are maintained, suggesting a hierarchy of Fe distribution to respiration in the mitochondria versus photosynthesis in the chloroplast in photoheterotrophically growing cells (Naumann et al., 2007; Terauchi et al., 2010). Plastid ferritins may buffer the Fe during the transition (Busch et al., 2008).
Fe-containing SOD, which is located in the chloroplast (Sakurai et al., 1993; Chen et al., 1996; Kitayama et al., 1999) is another potential target of Fe deficiency in C. reinhardtii. The SODs are a polyphyletic family of metalloproteins, the most common of which contain Fe (FeSODs), Mn (MnSODs), Cu plus Zn (Cu/ZnSODs), or Ni (NiSODs) as active site catalysts. The FeSODs and MnSODs exhibit very similar sequence and structure, reflecting a common ancestry, while the Cu/ZnSODs and NiSODs are distinct from the FeSODs and MnSODs and from each other (Fink and Scandalios, 2002; Priya et al., 2007). The FeSODs, MnSODs, and Cu/ZnSODs are widely distributed, and organisms typically possess enzymes of more than one class; many cyanobacteria have both FeSODs and MnSODs, metazoa and fungi produce both MnSODs and Cu/ZnSODs, and embryophytes synthesize all three forms (Fink and Scandalios, 2002; reviewed in Grace, 1990; Bowler et al., 1992; Pilon et al., 2011). Charophyte green algae also have all three forms, but chlorophyte green algae, including C. reinhardtii, lack Cu/ZnSODs, perhaps as a means of reducing their Cu quota (Asada et al., 1977; de Jesus et al., 1989; Egashira et al., 1989; Merchant and Helmann, 2012).
SODs represent the first line of antioxidant defense (reviewed in Imlay, 2008), and their importance can be seen in the severe oxygen-dependent phenotypic defects exhibited by mutants of Escherichia coli and Saccharomyces cerevisiae that lack one or more of these enzymes (Biliński et al., 1985; Carlioz and Touati, 1986; Farr et al., 1986; van Loon et al., 1986). Plant mutants lacking chloroplast FeSODs also show strong phenotypes: They are pale and grow poorly in the light because of damage to the plastome, emphasizing their importance in photoprotection (Myouga et al., 2008).
The degradation of Fe-containing proteins during the onset of Fe deficiency may release free Fe, which is known to contribute to oxidative stress by catalyzing the conversion of superoxide and hydrogen peroxide (H2O2) to the highly reactive hydroxyl radical via Fenton chemistry (reviewed in Halliwell and Gutteridge, 1984; Liochev and Fridovich, 1999). Indeed, symptoms of oxidative stress are evident in Fe-deficient cyanobacteria (Latifi et al., 2005). Therefore, any reduction in FeSOD activity during Fe deficiency in C. reinhardtii could be particularly deleterious, especially under illumination. Here, we show that two mechanisms serve to not only maintain but increase SOD activity in the chloroplast during Fe deficiency: First, a hierarchy of Fe allocation acting at the level of de novo synthesis favors FeSOD over other plastid proteins; second, a previously undiscovered chloroplast MnSOD is induced at the transcriptional level to augment the FeSOD activity.
RESULTS
Hierarchy of Fe Allocation to Various Chloroplast Proteins
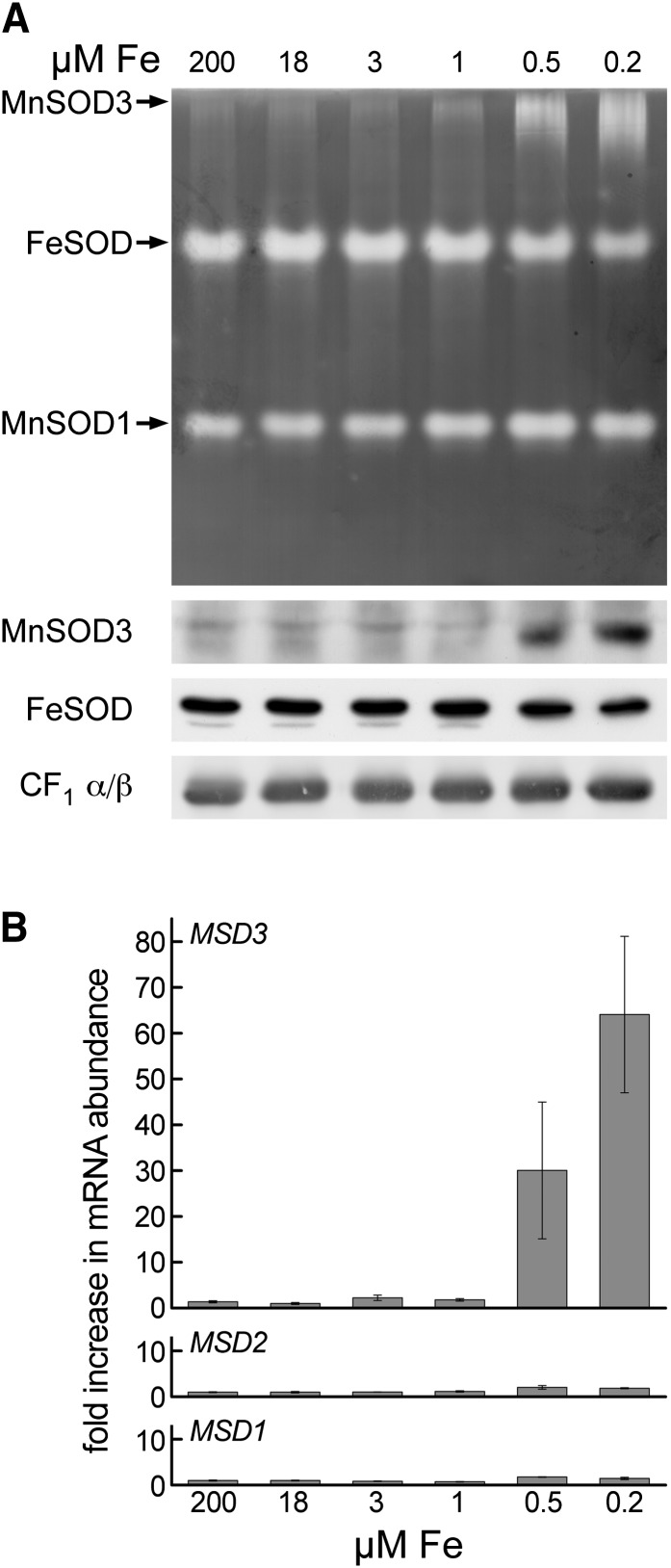
To distinguish the impact of Fe deficiency on FeSOD versus other chloroplast Fe proteins, we monitored the abundance of various Fe-containing chloroplast proteins by immunoblotting of extracts from cells nourished continuously in batch culture with different amounts of Fe. In keeping with the sacrifice of PSI and photosynthesis in photoheterotrophic C. reinhardtii, cytochrome f abundance was drastically reduced in Fe-limited cells (Figure 1). Similarly, the abundance of the magnesium protoporphyrin monomethyl ester cyclase (cyclase) was also markedly reduced. The cyclase has a diiron center and may be a key target of Fe deficiency (Spiller et al., 1982). The abundance of the major classic leaf-type ferredoxin (corresponding to the petF gene product) was also very sensitive to Fe nutrition; even mild Fe deficiency resulted in a substantial decrease in its abundance (cf. 1 μM to 3 μM Fe), but, perhaps because of its function in multiple essential metabolic pathways in the chloroplast, a low amount was maintained even in severely Fe-limited cells. The abundances of two other ferredoxins, Fdx3 and Fdx6, are also progressively reduced in Fe-deficient cells (Terauchi et al., 2009). By contrast, the abundance of FeSOD in Fe-limited cells (0.5 and 0.2 μM Fe) was not markedly reduced compared with that in Fe-replete cells as assessed by immunoblotting. This also contrasts with the substantial reduction in the major MnSOD that is observed in Mn-limited cells (Allen et al., 2007a). To assess whether the immunoreactive material is active, we analyzed the samples on nondenaturing gels for SOD activity and confirmed that the activity was retained as well (Figure 1). We conclude that FeSOD is preferentially retained against a background of loss of other chloroplast Fe proteins in photoheterotrophic cells acclimated to Fe limitation, demonstrating a selective impact of Fe deficiency on Fe proteins within a single compartment.
Figure 1.
Hierarchy of Fe Allocation to Chloroplast Fe Proteins.
Wild-type (CC125) C. reinhardtii cultures (shown in the top row) were grown in TAP medium containing the indicated concentration of supplemental Fe supplied as an Fe-EDTA chelate. Cultures were grown at 24°C for 4 d, with 50 μmol m−2 s−1 of constant light, and the abundance of FeSOD and various Fe-containing proteins in C. reinhardtii was determined at the different stages of Fe nutrition. Total proteins (20 μg) were separated by denaturing gel electrophoresis and transferred to PVDF or nitrocellulose for immunoblot analysis. Soluble proteins (60 μg) were separated by nondenaturing gel electrophoresis and FeSOD visualized by activity staining (FeSOD, bottom panel).
De Novo1 Synthesis of FeSOD during Fe Limitation
To address the question of how individual proteins might be targeted during Fe deficiency, we monitored the temporal transition from Fe replete to Fe starvation. Cells were grown Fe replete (i.e., 20 µM added Fe) to a density of ∼6 to 9 × 105 cells/mL and transferred to fresh Fe-free medium (Figure 2; labeled −Fe) or as a control to fresh replete medium (20 µM Fe, labeled +Fe). The cultures were sampled at time 0 or at 24-h intervals for 5 d to measure cell density, chlorophyll content, Fe content of cells and media, and the activity and abundance of various Fe-containing chloroplast proteins and SODs. Cell growth was highly reproducible between three independent experiments. Fe-starved cells reached a density of ∼5 to 6 × 106 cells/mL (indicative of growth limitation), while Fe-replete cultures reached 1.2 × 107 cells/mL (Figure 2A). The chlorophyll content in the –Fe cultures was reduced already after 1 d corresponding to the decrease in intracellular Fe content and eventually reached a new steady state, as noted previously (Figure 2B). In the first 24 h of Fe starvation, there was no impact on cell division; there was a nearly fourfold increase in cell density resulting in a fourfold decrease in Fe content per cell (Figure 2C), and the cells were entering Fe limitation by the end of this period as evidenced by the markedly reduced growth rate after this point (Figure 2A). The Fe content of spent −Fe medium was <1.5 nM (corresponding to the detection limit), 4.2 nM, and 42 nM for three individual experiments (Figure 2D; averages of technical triplicates). The concentration of Fe in +Fe medium fell by around 4 μM by the end of the experiment as the cultures reached stationary phase, in line with the amount of Fe taken up to meet the Fe quota of replete cells. Interestingly, the Fe-replete cultures experience reproducible transient Fe deficiency (accompanied by slightly reduced chlorophyll content) during the period of rapid exponential growth even in fully Fe replete medium because the assimilation pathway cannot keep up with intracellular demand (Figures 2B and 2C).
Figure 2.
Fe Reallocation to FeSOD during Transition to Fe Starvation.
Strain CC125 was grown in TAP medium with 18 μM Fe, washed, inoculated into TAP medium with 0 μM or 18 μM supplemental Fe (labeled −Fe and +Fe, respectively), and allowed to grow for 5 d.
(A) to (D) Comparison of cell density, measured by counting on a hemocytometer (A), chlorophyll content, measured spectrophotometrically (B), Fe content of cells (C), and spent media, measured by inductively coupled plasma–mass spectrometry (D). Data from three individual experiments are shown, represented by circles, triangles, and squares; chlorophyll content was calculated for a single experiment (the data series represented by square symbols in the other three panels).
(E) Abundance of MnSOD3 and hierarchy of Fe allocation. Soluble proteins (60 μg) were separated by nondenaturing gel electrophoresis and SODs visualized by activity staining (top panels for MnSOD3 and FeSOD). Total proteins (20 μg) were separated by denaturing gel electrophoresis and transferred to PVDF or nitrocellulose for immunoblot analysis.
When we monitored the abundance of various Fe-containing proteins, we noted that expression of the Fe deficiency biomarkers ferroxidase and ferritin were dramatically increased over days 0 to 5 of Fe starvation (Figure 2E). The continuous increase in levels of ferritin and ferroxidase throughout this time course indicates clearly that the cells were still fully capable of de novo protein synthesis and that they became progressively more Fe limited. By contrast, there was a progressive decrease in ferredoxin, cytochrome f, and the diiron-containing cyclase in the −Fe samples. Cytochrome f and ferredoxin showed a similar pattern on day 1 with a rapid decrease, but while the former continued to decrease to nearly undetectable levels, the latter was subsequently maintained at a new steady state level, consistent with its essential role in plastid metabolism. The abundance of the cyclase increased over the first 24 h, while the cells continued to divide (although not to the same extent as in the +Fe controls), but then decreased markedly relative to the day 0 level by day 5 of the experiment like ferredoxin and cytochrome f did.
Surprisingly, FeSOD abundance also decreased dramatically in response to Fe starvation in the first 24 h, but in contrast with cytochrome f and ferredoxin, the response was not sustained (Figure 2E); protein abundance starts to recover on day 2 until by day 5 it was restored to the day 0 level. We monitored the activity of FeSOD in nondenaturing gels and noted that the activity exactly paralleled polypeptide abundance. These results indicate that the mechanism of selective maintenance of FeSOD involves selective resynthesis after a period of nonselective degradation of multiple Fe-containing proteins. We hypothesize that the resynthesis relies on Fe that is recycled from expendable proteins, such as cytochrome f, ferredoxin, and the cyclase, since there is very little if any exogenous Fe available (Figure 2D). In the case of the cyclase, the impact of degradation was offset by increased synthesis in response to cell division (Figure 2E, +Fe, right panel). The virtually identical pattern of activity and immunoblot analyses indicate de novo synthesis of FeSOD in days 2 through 5 as opposed to remetallation of the apoprotein. It is possible that the Fe that populates the FeSOD active site may come from plastid ferritin, which serves as the buffer or depot for Fe released from degradation of PSI and other proteins (Busch et al., 2008).
A Previously Undiscovered SOD Expressed under Fe Limitation
A previously uncharacterized SOD activity was apparent in the activity staining gels. This activity was distinct, based on its migration in nondenaturing gels (Figure 3, the low mobility band at the top of the gels labeled MnSOD3) from the previously described two major SOD activities of Fe-replete cells: the plastid FeSOD and a possibly dual-localized MnSOD (Egashira et al., 1989; Sakurai et al., 1993; Kitayama et al., 1999). The activity was evident only in cells acclimated to Fe limitation (≤0.5 μM Fe) or after prolonged Fe starvation in the time course experiment (2 d in –Fe). FeSODs are sensitive to H2O2, whereas MnSODs are not (Asada et al., 1975); incubation of the gel with 3 mM H2O2 prior to staining almost abolished the FeSOD activity previously reported but did not significantly diminish the novel, Fe limitation–inducible activity, indicating that the enzyme responsible for this activity is not an FeSOD (see Supplemental Figure 1 online).
Figure 3.
A Novel SOD Induced under Fe Limitation Is Expressed Coordinately with the MSD3 Gene.
C. reinhardtii strain CC125 was grown in TAP medium containing the indicated Fe supplement.
(A) Soluble proteins (60 μg) were separated by nondenaturing gel electrophoresis and SODs visualized by activity staining (top panel). Total proteins (20 μg) were separated by denaturing gel electrophoresis and transferred to PVDF or nitrocellulose for immunoblot analysis (bottom three panels).
(B) RNA was analyzed by quantitative real-time PCR for the abundance of transcripts encoding MSD1, MSD2, and MSD3. Primers used are listed in Supplemental Table 5 online. The fold difference in abundance of each mRNA is shown after normalization to CBLP. Error bars represent the variation (sd) between experimental triplicates.
Six Candidate SOD Genes in C. reinhardtii
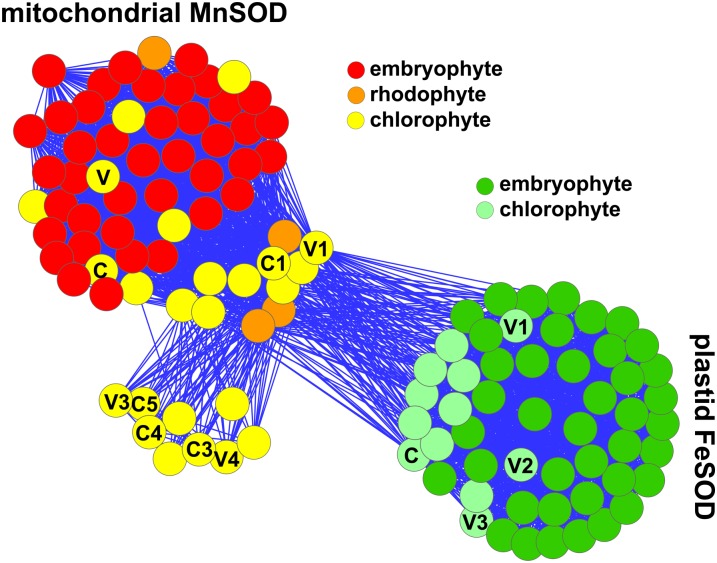
We surveyed the C. reinhardtii genome to identify the gene responsible for the newly discovered activity. Genes coding for SODs were identified by querying the version 3.0 draft genome using BLAST to distinguish gene models corresponding to homologs of SODs of other organisms, including fungi, plants, and animals. We identified six genes encoding candidate Fe/MnSODs, corresponding to protein IDs 182933, 53941, 193511, 196649, 114882, and 115390 in the version 3.0 genome. All six residues identified as perfectly conserved in Fe/MnSODs are present in the predicted proteins, and they were assigned as putative Fe- versus MnSODs on the basis of distinguishing sequence characteristics (Figure 4; Wintjens et al., 2004). The genes were named FSD1, MSD1, MSD2, MSD3, MSD4, and MSD5, respectively. The major MnSOD (H2O2 insensitive) detected by activity staining was identified by sequencing of tryptic peptides as the MSD1 gene product and named MnSOD1. The FeSOD (H2O2 sensitive) was confirmed as the FSD1 gene product by the same method (see Supplemental Figure 2 and Supplemental Tables 1 and 2 online). We did not identify any candidate genes encoding Cu/ZnSOD or any chaperone involved in its assembly (Culotta et al., 1997), consistent with previous reports that C. reinhardtii lacks this form of SOD, nor any gene potentially encoding a nickel SOD. The assignment of FeSOD1 as the FSD1 gene product was also validated in protein similarity network analysis (Figure 5), which grouped FeSOD1 with other FeSODs.
Figure 4.
Fe and Mn SODs of C. reinhardtii.
C. reinhardtii FeSOD and MnSOD1 through MnSOD5 were aligned with E. coli SodA (Mn; Swiss-Prot P00448) and SodB (Fe; Swiss-Prot P0AGD3) using MultAlin (http://prodes.toulouse.inra.fr/multalin/multalin.html; Corpet, 1988) and shaded using GeneDoc (http://www.nrbsc.org/gfx/genedoc/index.html; Nicholas et al., 1997; four levels of shading). Asterisks mark every 10th residue, and tilde symbols indicate that the N terminus of MnSOD5 remains to be characterized. Conserved residues participating in Mn/Fe binding are indicated by a yellow background and other residues perfectly conserved in Mn/FeSODs by a red background. Residues distinguishing MnSODs and FeSODs are indicated by blue and green backgrounds, respectively. Predicted cleavable N-terminal signal sequences are underlined. The N terminus of the mature form of FeSOD has been determined experimentally (Sakurai et al., 1993).
Figure 5.
Protein Similarity Network of Plant SODs.
A network was generated from an all-versus-all BLAST analysis (pairwise alignment between all pairs of proteins) of SOD sequences in a local sequence database, using an E-value of 1e-45. N termini were trimmed to the equivalent of Q3 of C. reinhardtii MnSOD1. Nodes representing embryophyte FeSODs are colored dark green, and green algal (Chlorophyceae and Trebouxiophyceae) FeSODs are colored light green, while nodes representing embryophyte MnSODs are colored red, green algal MnSODs are colored yellow, and red algal MnSODs orange. SODs from C. reinhardtii and Volvox are labeled C and V (for FeSODs and mitochondrial MnSODs), and C1, C3, C4, and C5 (for C. reinhardtii MnSOD1, 3, 4, and 5) and V1, V3, and V4 (for Volvox MnSOD1, 3, and 4).
Direct sequencing of cDNAs confirmed that all six genes are expressed (Table 1). In nutrient-replete Tris-acetate medium, FSD1, MSD1, and MSD2 were expressed at high levels, and MSD3, MSD4, and MSD5 at much lower levels (Figure 6). Transcript abundance has been shown to predict cognate protein abundance for 70 to 75% of transcripts in S. cerevisiae and mouse (Kislinger et al., 2006; Lu et al., 2007). Therefore, MSD3, MSD4, and MSD5 were thought to be candidates for encoding the new Fe deficiency–inducible SOD.
Table 1. Genes Encoding SODs in C. reinhardtii.
| Gene | Protein IDa | RPKMb |
|---|---|---|
| FSD1 | Cre10.g436050 | 481.0 |
| MSD1 | Cre02.g096150 | 281.0 |
| MSD2 | Cre13.g605150 | 82.8 |
| MSD3 | Cre16.g676150 | 4.6 |
| MSD4 | Cre12.g490300 | 0.2 |
| MSD5 | Cre17.g703150 | 0.2 |
SOD transcript abundance determined by RNA-sequencing in C. reinhardtii strain 2137 (data from the National Center for Biotechnology Information Gene Expression Omnibus, accession number GSE25124; Castruita et al., 2011). Cells were grown photoheterotrophically in nutrient-replete TAP medium (18 μM Fe).
Corresponding to the Au10.2 models in the version 4.0 genome assembly.
Reads per kilobase of exon per million mapped sequence reads (Mortazavi et al., 2008); paired-end reads mapped to Augustus 10.2 gene models.
Figure 6.
MSD3 Is Induced by Fe Limitation, Mn Deficiency, or H2O2 Stress.
RNA from Fe-deficient (closed circles) and Mn-deficient (open circles) cells and from cells treated with 1 mM H2O2 (triangles) was analyzed by quantitative real-time PCR for the abundance of transcripts encoding FSD1 and MSD1 through MSD5. Primers used are listed in Supplemental Table 5 online. C. reinhardtii strains 2137, CC125, and CC425 were grown in TAP medium with either 0.5 or 18 μM added Fe and either 0 or 25 μM added Mn to ∼3 × 106 cells/mL. For H2O2 treatment, cells were grown to ∼2 × 106 cells/mL, H2O2 added to 1 mM final concentration, and cells harvested 2 h later. This concentration of H2O2 was chosen based on previous studies (Leisinger et al., 2001).The fold difference in abundance of each mRNA is shown after normalization to CBLP. Each data point is the average of technical triplicates and represents an individual experiment.
Improved gene models, shown schematically in Supplemental Figure 3 online (see also Supplemental Methods 1 online), have been incorporated into the version 4.0 assembly of the C. reinhardtii genome at the Joint Genome Institute and the sequence data submitted to the GenBank/EMBL/DDBJ databases under accession numbers GQ413964 (FSD1), GQ413965, GQ413966, GQ415402, GU134344, and GU134345 (MSD1-5, respectively). Sequence and protein similarity network analyses (Figure 5) supported the assignment of MnSOD2 as a typical mitochondrial MnSOD, consistent with the detection of this protein in the C. reinhardtii mitochondrial proteome (Atteia et al., 2009), but the separation of MnSOD1 and MnSOD3 through MnSOD5 in the network is suggestive of specialized function for these algal isoforms.
MSD3 Is Induced under Fe Limitation, Mn Deficiency, and H2O2 Stress
When we tested the expression of the SOD-encoding genes in response to Fe limitation by real-time PCR, we noted that MSD3 was dramatically upregulated under conditions of Fe limitation, with its mRNA increasing up to 103-fold (Figures 3B and 6). By contrast, expression of the other SOD-encoding genes was either little changed (FSD1, MSD1, and MSD2) or increased only slightly (≤10-fold) (MSD4 and MSD5). When the mRNAs were quantified in RNA-sequencing experiments, we noted that MSD3 mRNA became the most abundant of the SOD-encoding transcripts in Fe-deficient cells (Figure 7). We observed a similar pattern of expression in cells grown under conditions of Mn deficiency, with levels of MSD3 mRNA increasing up to 200-fold over those measured in Mn-replete cells (Figure 6). This counterintuitive response results from secondary Fe deficiency in Mn-deficient C. reinhardtii (Allen et al., 2007a, 2007b). When RNAs were sampled from cells acclimated to Fe deficiency and Fe-limited conditions, the MSD3 transcript abundance paralleled the activity of the novel MnSOD (Figure 3). Expression of the other two highly expressed MnSOD-encoding genes, MSD1 and MSD2, changed little (less than or equal to twofold) across the range of Fe concentrations tested, and mRNAs for MSD4 and MSD5 were not detected in this experiment. The MSD3 gene was thus considered a likely candidate for encoding the newly discovered inducible MnSOD activity.
Figure 7.
Impact of Fe Nutrition on Abundance of SOD-Encoding mRNAs.
SOD transcript abundance determined by RNA-sequencing in C. reinhardtii Strain 2137 (data from the National Center for Biotechnology Information Gene Expression Omnibus, accession number GSE35305; methods described by Castruita et al., 2011). Cells were grown photoheterotrophically in TAP medium (acetate) or phototrophically in TP medium (CO2) containing 20, 1, or 0.25 μM Fe. Reads were mapped to Augustus v10.2 gene models.
To determine this unambiguously, we generated mono-specific antibodies against the recombinant protein (see Supplemental Figure 4 online). The antibodies recognized a protein with an apparent molecular mass of ∼35 kD only in Fe-limited cells (0.5 or 0.2 μM added Fe) (i.e., samples exhibiting the novel MnSOD activity) but not in cells grown with higher concentrations of added Fe (Figure 3). To relate the novel activity to the immunoreactive band, we subjected the native gel to immunoblotting and demonstrated that the low mobility activity and the immune-staining band comigrate (Figure 8).
Figure 8.
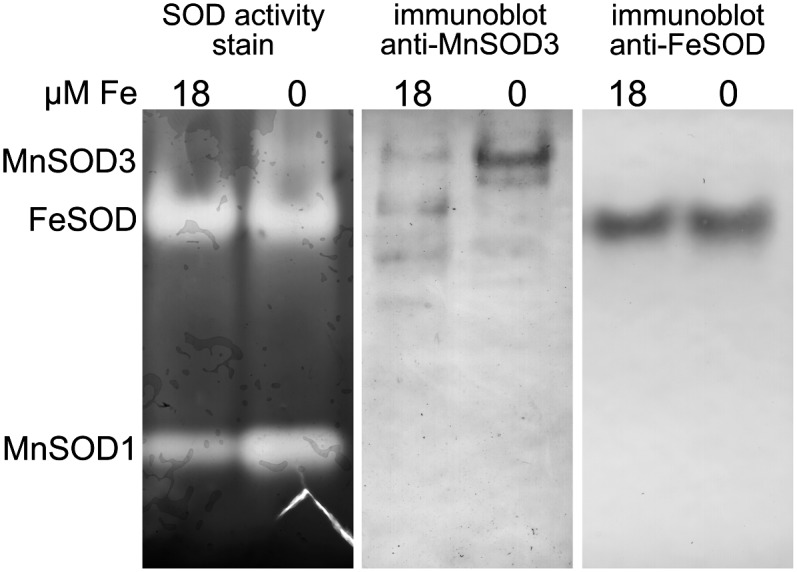
Identification of MnSOD3 as the MSD3 Gene Product.
Soluble proteins (60 and 20 μg) isolated from cells grown under conditions of Fe repletion (18 μM Fe) or starvation (0 μM added Fe, 5 d) were separated by nondenaturing gel electrophoresis and the gel divided in three. For one part of the gel (60 μg protein), SODs were visualized by activity staining; for the other parts (20 μg protein), proteins were transferred to nitrocellulose for immunoblotting with anti-MnSOD3 (1:1000) or anti-FeSOD (1:2000).
The MSD3 gene responds also to H2O2 stress (Figure 6). MSD3 RNA was increased in abundance up to 20- to 60-fold in response to H2O2, while expression of the other five SOD-encoding genes was either little changed (MSD1, MSD2, and MSD5) or increased only two- to sixfold (FSD1 and MSD4).
MSD3 Is Transcriptionally Regulated by Fe and H2O2
To distinguish the mechanism of regulation of the MSD3 gene by Fe and H2O2, reporter constructs (Figure 9A) were introduced into the Arg auxotrophic C. reinhardtii strain CC425 (cw15 arg7) by cotransformation with pARG7.8 (carrying the complementing gene). Arg prototrophs were tested for arylsulfatase expression on plates containing either high (18 µM), medium (3 µM), or low (1 µM) concentrations of added Fe using the chromogenic substrate X-sulfate (de Hostos et al., 1988). Cotransformants were screened by PCR to check cotransformation frequencies and for the presence of full-length reporter construct DNAs. Approximately 50% of pJD100GW-5′MSD3 cotransformants carried full-length reporter DNA, and ∼35% of these were arylsulfatase expressers (see Supplemental Table 3 online). A single representative reporter-expressing colony from the transformation is shown in Figure 9B (blue halo indicates arylsulfatase activity). All cotransformants exhibiting Fe-responsive arylsulfatase expression carried full-length reporter construct DNA. By contrast, no colony arising from the pJD100 control transformation showed significant arylsulfatase activity. Quantitation of arylsulfatase expression by enzyme assay showed clearly that the 5′ flanking DNA from MSD3 confers Fe-responsive expression on the reporter gene (Table 2). The average increases in arylsulfatase expression in Fe-deficient and Fe-limited over Fe-replete cells were ∼15-, ∼26-, ∼79-, and ∼297-fold for cells grown with 3, 1, 0.5, and 0.2 µM added Fe, respectively. Arylsulfatase expression from the pJD100GW-5′MSD3 reporter construct was also increased when transformants were grown under conditions of Mn deficiency (which generates secondary Fe deficiency). The increase in arylsulfatase expression in deficient over replete cells was ∼14-fold (Table 2), similar to that observed for the endogenous MSD3 gene, and consistent with the observation that the amount of Fe in Mn-deficient cells is similar to the Fe content of cells grown in medium containing 3 μM added Fe (Allen et al., 2007a). When transformants carrying the pJD100GW-5′MSD3 construct were challenged with H2O2, expression of the ARS2 mRNA was strongly upregulated (Figure 9C) with kinetics similar to those observed for the endogenous MSD3 gene.
Figure 9.
Transcriptional Regulation of MSD3 by Poor Fe Nutrition or by H2O2 Stress.
(A) Schematic of reporter gene construct. White segments represent the ARS reporter gene, black the minimal TUB2 promoter, red the MSD3 5′ flanking DNA. The numbers beneath the MSD3 5′ flanking segment indicate positions relative to the transcript start site determined by 5′-rapid amplification of cDNA ends.
(B) Arylsulfatase activity on plates. Single representative transformants growing on agar-solidified TAP medium with the indicated Fe supplement were stained for arylsulfatase activity (visualized as a blue halo around colonies) by spraying with a solution of the chromogenic substrate 5-bromo-4-chloro-3-indoxyl sulfate.
(C) Coordinate accumulation of mRNAs for MSD3 (closed symbols) and the ARS2 reporter gene driven by the MSD3 promoter (open symbols) in cells challenged with H2O2. Transformants carrying either the MSD3 5′ flanking DNA fused to the ARS2 reporter gene (circles), or the ARS2 reporter gene alone (squares), were grown to early log phase (2 × 106 cells/mL) in TAP medium, and H2O2 was added to 1 mM. Cells were collected at the time points shown and RNA analyzed by real-time PCR for the abundance of transcripts encoding MSD3 and ARS2. Data shown are for representative individual transformants.
Table 2. Fe Nutrition Status Responsive Arylsulfatase Expression from an MSD3-ARS2 Reporter Gene Construct.
| Construct | Arylsulfatase Activity |
||||||
|---|---|---|---|---|---|---|---|
| μM Added Fe |
µM Added Mn |
||||||
| 18 | 3 | 1 | 0.5 | 0.2 | 25 | 0 | |
| pJD100GW | 6 ± 2 | 12 ± 1 | 5 ± 1 | 7 ± 1 | 5 ± 1 | 1 ± 0 | 2 ± 1 |
| pJD100GW−5′MSD3 | 11 ± 2 | 169 ± 39 | 285 ± 79 | 869 ± 303 | 3272 ± 854 | 11 ± 2 | 151 ± 53 |
Transformants carrying the MSD3 5′ flanking DNA fused to the ARS2 reporter gene (n = 3) or a control construct (n = 3) were grown to mid to late log phase (2 × 106 to 2 × 107 cells/mL) in TAP media with various amounts of added Fe (as indicated), or with (25 µM) or without (0 µM) added Mn, and culture supernatants assayed for arylsulfatase activity using p-nitrophenyl sulfate. Activities are expressed as picomoles of p-nitrophenol liberated per 106 cells per minute and are given as mean ± se.
We conclude that Fe deficiency and H2O2 act at the transcriptional level on the MSD3 gene. To investigate whether posttranscriptional mechanisms contribute to changes in MSD3 mRNA abundance, we monitored the decay of MSD3 transcript levels after blocking transcription with Actinomycin D. We observed no difference in persistence of the MSD3 transcript between cells resupplied with Fe and those left without (see Supplemental Figure 5 online) and conclude that regulation of MSD3 transcript abundance by Fe nutrition occurs primarily at the transcriptional level.
MnSOD3 Is Localized to the Chloroplast
The specific transcriptional upregulation of a previously undiscovered MnSOD in Fe deficiency raised a question concerning its localization because MnSOD is found in the mitochondrial matrix in most eukaryotes. If MnSOD3 functions as a backup for FeSOD, as suggested by its increased expression in Fe-limited cells, it must be localized in the same compartment as FeSOD because the superoxide anion cannot freely cross biological membranes (Takahashi and Asada, 1983; Missirlis et al., 2003; Lynch and Fridovich, 1978). Therefore, we localized MnSOD3 by biochemical fractionation. Chloroplasts and mitochondria were isolated from strain CC425 cells grown under conditions of Fe limitation and purified by density gradient centrifugation. The efficiency of this fractionation was assessed by immunoblotting with antibodies to alternative oxidase 1, a protein of the inner mitochondrial membrane, and the chloroplast-localized ketol-acid reductoisomerase and FeSOD (Vanlerberghe and McIntosh, 1997; Dumas et al., 1989; Kitaya ma et al., 1999). This analysis showed that the mitochondrial fraction was free of chloroplast material and the chloroplast fraction only slightly contaminated with mitochondria. When we probed the fractions by immunoblotting with antibodies against MnSOD3, we found that it was present in the chloroplast fraction (Figure 10).
Figure 10.

MnSOD3 Is Localized to the Chloroplast.
Strain CC425 was grown in TAP medium containing 0.5 μM Fe. Soluble proteins (60 μg) from whole cells, and from isolated chloroplasts (CP) and mitochondria, were separated by nondenaturing gel electrophoresis and SODs visualized by activity staining (top panels). Total proteins (20 μg) were separated by denaturing gel electrophoresis and transferred to PVDF or nitrocellulose for immunoblot analysis with anti-MnSOD3, anti-FeSOD (chloroplast), and anti-KARI (chloroplast stroma), and antialternative oxidase (mitochondrial inner membrane).
Based on the prevailing view of Fe sparing, the simplest interpretation would be that a new chloroplast-localized MnSOD compensates for the potential loss of a FeSOD in Fe-deficient cells. However, as shown above, the FeSOD activity appears to be maintained. And in fact, MnSOD3 abundance and activity does not mirror the decrease in FeSOD. Instead, the activity and abundance increase as the cells begin to experience Fe limitation and continue to increase up to day 5 even though FeSOD activity is restored (Figure 2E). This suggests that the function of MnSOD3 may be to increase the capacity of the chloroplast to detoxify superoxide rather than to compensate for reduced FeSOD activity.
We measured total SOD activity during the transition to Fe starvation in an independent set of experiments involving several different wild-type strains. Total SOD activity per cell increased 1.5- to twofold after 4 d of Fe starvation in all four strains (Figure 11A). More strikingly, the specific activity of total SOD in soluble extracts increased threefold to 3.5-fold over the same period (Figure 11B), due to a reduction in the amount of soluble protein per cell, which declined to about half its initial level within the first 24 h of Fe starvation and then stayed stable over the remaining 4 d of the experiment (Figure 11C). FeSOD abundance exhibited a similar pattern of loss and recovery as observed previously (Figure 11D).
Figure 11.

Increased SOD Activity in Fe-Starved Cells.
Strain CC425 was grown in TAP medium with 18 μM Fe, washed, inoculated into TAP medium with 0 μM added Fe, and allowed to grow for 5 d. Data from three individual experiments are shown, represented in (A) to (C) by circles, triangles, and squares.
(A) Total soluble SOD activity per cell, normalized to day 0. SOD activities at day 0 were 1.95, 1.59, and 1.46 units per 106 cells for the three replicates.
(B) SOD specific activities in soluble protein extracts, normalized to day 0. SOD specific activities at day 0 were 39.2, 31.6, and 28.5 units per mg protein for the three replicates.
(C) Protein concentration in soluble extracts, normalized to day 0.
(D) Abundance of FeSOD. Soluble proteins (1 μg) were separated by denaturing gel electrophoresis and transferred to nitrocellulose for immunoblot analysis.
DISCUSSION
Maintenance of FeSOD
When C. reinhardtii cells get less Fe than they need to maintain the cellular Fe quota, the precious nutrient is selectively distributed. The mitochondria are prioritized over the chloroplast, especially when a reduced carbon source is available (Naumann et al., 2007; Terauchi et al., 2010). In acetate-grown cells, a program of proteolysis, particularly of the photosynthetic apparatus, is initiated in the chloroplasts (Moseley et al., 2002a). Fe released from the degradation of these proteins is most likely used for maintenance of the mitochondrial respiratory chain (Naumann et al., 2007). Here, we show that prioritization of Fe use occurs even within an organellar compartment, with FeSOD being maintained at its usual abundance in Fe-deficient cells while Fd abundance decreases to a new steady state (Figure 2E). How is this achieved? The steady state level of any macromolecule is determined by a balance between rates of synthesis and degradation. FeSOD could be maintained simply by being protected from degradation, but the time course experiment (Figure 2) argues against this model. At day 1, FeSOD activity and abundance are greatly reduced, comparable to the reduction in ferredoxin and cytochrome f abundances. Nevertheless, while cytochrome f and ferredoxin either continue to decrease or are simply maintained at a new lower steady state level, respectively, FeSOD abundance actually increases. The virtually identical pattern of the activity staining and the immunoblotting indicates that the increase is attributed to specific resynthesis of the polypeptide and de novo metallation. Apo-FeSOD does not incorporate Fe unless the protein is unfolded and then refolded in the presence of Fe2+ (Yamakura and Suzuki, 1976), suggesting that in vivo, Fe incorporation occurs during folding of the nascent polypeptide, as demonstrated for Mn incorporation into yeast mitochondrial MnSOD (Luk et al., 2005). Because the activity of FeSOD is H2O2 sensitive, we know that the increase can be attributed to metallation by Fe rather than mismetallation by Mn ion.
This raises the question of how FeSOD is selectively targeted for resynthesis. One mechanism might be through increased synthesis of the apoprotein. This could occur at the level of gene expression generating more template for synthesis of the apoprotein; however, FSD1 mRNA levels are not impacted by Fe deficiency. Translational regulation of the FSD1 mRNA is another model that could account for increased synthesis. A second mechanism, which we favor, is by selective Fe allocation to the FeSOD assembly pathway. Increased expression of a specific assembly chaperone would increase FeSOD synthesis and balance the ongoing degradation. In the absence of a nutritional supply of Fe, increased accumulation would result only when there is an adequate supply of Fe salvaged from the degradation of PSI, ferredoxin, cytochrome f, and other Fe-containing proteins. Indeed, accumulation of FeSOD is evident only at day 2, after there is an opportunity for Fe recycling from the degradation of other proteins. Thus, maintenance of a particular protein (FeSOD) might occur by relying on continuous resynthesis, dependent on a pool of cofactor that is salvaged from degradation of other abundant Fe proteins and selectively allocated to FeSOD. The upregulation of plastid ferritins that serve as an Fe depot in Fe-deficient C. reinhardtii is consistent with this idea (Busch et al., 2008; Long et al., 2008).
The balancing of synthesis and degradation is nicely evident from comparing the pattern of accumulation of the diiron-containing cyclase (Figure 2E). CTH1 mRNA (encoding the cyclase) increases transiently upon transfer of cells to fresh medium, most likely to accommodate the rapid growth rate. The amplitude of the increase in Fe-starved cells is damped relative to the +Fe control, presumably because of ongoing degradation in the former situation, and eventually the cyclase abundance is also decreased substantially like ferredoxin and cytochrome f.
Why is FeSOD prioritized? We expect an increased propensity for O2∙ − generation by the Mehler reaction in Fe-deficient chloroplasts because its high Fe content renders PSI particularly susceptible to Fe deficiency stress (Sandmann and Malkin, 1983). Reduced energy input via remodeling of the PSI antenna is one mechanism to circumvent the problem (i.e., by reducing the production of O2∙ −) (Moseley et al., 2002a; Naumann et al., 2005). Increased dissipation of the superoxide is another mechanism. Many studies have established the importance of chloroplast SOD activity for plant survival during oxidative stress (Tsang et al., 1991; Bowler et al., 1991; Herbert et al., 1992; Van Camp et al., 1996; Phee et al., 2004; Sunkar et al., 2006; Myouga et al., 2008). For example, when FeSOD was overexpressed in Arabidopsis chloroplasts, the transgenic plants were more tolerant of methyl viologen–induced stress, whereas loss of the chloroplast FeSODs resulted in pale or albino plants, depending on the severity of the loss (Myouga et al., 2008). Since the phenotype of the FeSOD-deficient plants is dependent on light intensity, it suggests that plastid SODs are important for handling O2∙ − generated by the light reactions.
How are Fe proteins recognized for degradation? In the case of those with labile FeS clusters, their steady state abundance relies on the operation of cluster repair mechanisms, and these are sensitive to Fe availability (Garland et al., 1999; Jensen and Culotta, 2000; Djaman et al., 2004). In Fe deficiency, the apoproteins would not be remetallated, making them susceptible to proteases (Reyda et al., 2008). For other Fe proteins, a recognition mechanism is not obvious, but their degradation might be a consequence of oxidative stress damage from reaction of the active site Fe with H2O2 (Halliwell and Gutteridge, 1984; reviewed in Imlay, 2008). That there is some level of selectivity for Fe-containing proteins (although not the type of Fe center) is evident from the observation that many other proteins like CF1 and plastocyanin are retained.
Increased Capacity for Detoxifying Superoxide
Even though FeSOD is retained in Fe-limited cells, a new SOD activity is induced in chloroplasts (Figures 2, 3, and 10). In previous work, the product of the MSD3 gene could not be unambiguously related to a particular isoform on the activity gel. In this work, we used isoform-specific antibodies to relate the inducible chloroplast-localized activity with low electrophoretic mobility in nondenaturing gels to the MSD3 gene. Upregulation of MnSOD by Fe limitation has been observed in many microorganisms and is viewed as a Fe-sparing mechanism (i.e., replacement of an abundant Fe-containing enzyme with an Fe-independent one) (Campbell and Laudenbach, 1995; Kim et al., 1999). In this work, we show that the new activity is an addition to the existing activity rather than a replacement, resulting in a net increase in the capacity to detoxify superoxide (Figure 11). In fact, its activity continues to increase at day 3 even when FeSOD activity is already recovering. When we compare the total count of SOD-encoding mRNAs, the increased capacity for synthesizing SODs is also evident (Figure 7). The use of a MnSOD to provide this extra capacity is sensible in this situation because Fe is in limiting supply. Another possibility is that the MnSOD has a different function. In cyanobacteria, which have both Fe- and MnSODs, the two isoforms have different localizations suggestive of distinct, and probably complementary, functions (Hopkin et al., 1992; Thomas et al., 1998; Li et al., 2002; Regelsberger et al., 2004). In this context, we note that the predicted isoelectric points of C. reinhardtii FeSOD and MnSOD3 are very different (6.3 and 9.5, respectively), which might well lead to differential localization within the chloroplast, although clearly both are soluble enzymes. Therefore, we suggest that upregulation of MnSOD3 in Fe-deficient C. reinhardtii may occur to provide additional antioxidant defense rather than only as a replacement for FeSOD. This is suggested by the pattern of MnSOD3 expression, which is high even when FeSOD abundance is unaffected or only marginally affected (Figures 2 and 3; see Supplemental Figure 6 online). Could the inducible Mn-containing form be a safer option for protection of nucleic acids in the Fe-limited cell? The fact that the MSD3 gene is regulated by peroxide as well is consistent with this idea.
Chloroplast MnSOD
A diatom, Thalassiosira pseudonana, with two genes for MnSODs and two for FeSODs (but none for Cu/ZnSOD; Armbrust et al., 2004), was also found to house a chloroplast-located MnSOD, and Fe deficiency promoted the assembly of more MnSOD (Peers and Price, 2004; Wolfe-Simon et al., 2006). Given the typical low level of Fe in 30% of the world’s oceans and their habitation by a variety of diatom species, the use of a MnSOD during Fe limitation stress might be widespread in nature.
The occurrence of structurally related Fe- and MnSODs in chloroplasts does raise the question of the mechanism of selective metal incorporation. In E. coli and S. cerevisiae mitochondria, the MnSOD can be isolated as a mixture of correctly and incorrectly metallated enzyme; the degree of mismetallation depends on the status of Fe and Mn homeostasis in the assembly compartment (Beyer and Fridovich, 1991; Culotta et al., 2006; Yang et al., 2006; Naranuntarat et al., 2009). Fe incorporation into the MnSOD appears to be more of a problem than Mn incorporation into FeSOD. Whereas this might appear to be a non-issue in Fe-depleted chloroplasts, we note that both proteins are actually being synthesized simultaneously (day 3 in Figure 2), and correct allocation of a limited resource (i.e., Fe) might be quite important. The mechanism by which this occurs awaits further investigation.
The SODs are among the most important antioxidant defenses in plants (Alscher et al., 2002). Genome sequences show that there are multiple SOD isoforms in most plants and algae. The chlorophyte algae lack Cu/ZnSOD, but instead have several forms of the Fe and Mn type. This may be an adaptation to low-oxygen environments; species of the genus C. reinhardtii are found in naturally hypoxic or even anoxic habitats (Harris, 2009), in which the availability of copper is limited by the insolubility of Cu(I) salts (Österberg, 1974), whereas Mn becomes more soluble and bioavailable [as Mn(II)] as oxygen tension decreases (Wollast et al., 1979). In C. reinhardtii, sequence analyses indicate that MnSOD2 is a typical mitochondria-localized protein, but the divergence of MnSOD1 and MnSOD3 through MnSOD5 is suggestive of specialized functions for these isoforms, as demonstrated here for MnSOD3. The regulation of C. reinhardtii MSD3 by peroxide stress as well as Fe deficiency could indicate the existence of a peroxide-responsive element in C. reinhardtii, but the response might also be mediated via peroxide destruction of an Fe/S-containing Fe sensor (Kumánovics et al., 2008; Mühlenhoff et al., 2010) (Figures 6 and 9).
METHODS
Strains and Culture Conditions
Chlamydomonas reinhardtii strains CC125, CC425 (cw15 arg2), and 2137 were obtained from the C. reinhardtii culture collection (Duke University) and grown in Tris-acetate-phosphate (TAP) medium (Harris, 2009) at 24°C and a photon flux density of 50 to 100 μmol m−2 s−1 provided by a mixture of cool white fluorescent bulbs at 4100K and warm white fluorescent bulbs at 3000K used in the ratio of 2:1. For experiments involving Fe nutrition, strains were maintained in standard TAP medium (18 μM Fe) and transferred to −Fe TAP supplemented with Fe-EDTA at the concentrations indicated (Moseley et al., 2002a). Cells were made Mn deficient as described by Allen et al. (2007a); deficiency was confirmed by examination of chlorophyll fluorescence decay kinetics. For H2O2 treatment, cells were grown to 106 or 2 × 106 cells/mL and treated with 1 mM H2O2 for 2 h prior to protein or RNA isolation. In the case of CC425, media were supplemented with Arg (1.15 mM, added prior to autoclaving). Transformants of strain CC425 were grown as above or on TAP agar plates supplemented with Fe-EDTA at the concentrations indicated (22 to 25°C, ∼100 μmol m−2 s−1 light intensity).
Metal and Chlorophyll Measurements
Total metal content of cells was measured for 2 × 108 cells as described by Allen et al. (2007a), with the exception that nitric acid was used at a final concentration of 2.4%. Spent medium was transferred to acid-washed tubes, and any remaining cells removed by further centrifugation before nitric acid was added (to 2.4%). Total metal content was measured by the standard addition method. Chlorophyll was measured as described by Moseley et al. (2000).
Nucleic Acid Isolation and Analysis
Total C. reinhardtii RNA was prepared as described previously (Quinn and Merchant, 1998). C. reinhardtii DNA for colony PCR was isolated as described by Berthold et al. (1993). DNA sequencing was performed by Agencourt Biosciences. Plasmids intended for transformation into C. reinhardtii were isolated using the Marligen High-Purity Plasmid Purification System. Improved gene models were generated as described in Supplemental Methods 1 online, Supplemental Figures 7 and 8 online, and Supplemental Table 4 online.
Real-Time PCR
cDNA preparation and quantitative RT-PCR were performed as described by Allen et al. (2007a). Primers used are listed in Supplemental Table 5 online. Data are presented as the fold change in mRNA abundance, normalized to the endogenous reference gene CBLP. Abundance of the CBLP mRNA does not change under the conditions tested.
RNA Abundance
Sequence data from Gene Expression Omnibus accessions were analyzed as described previously (Castruita et al., 2011).
RNA Half-Life Determination
Strain CC125 was grown to a cell density of ∼2 × 106 cells/mL in TAP medium with either 20 or 0 μM supplemental Fe for 2 d. The 0 μM Fe culture was then divided between two flasks and to one of these Fe was added to 20 μM. Actinomycin D (40 μg/mL) was added to all cultures, and RNA isolated from cells harvested at various time points was analyzed by real-time PCR. A second experiment was performed identically except that Actinomycin D was not added to the cultures.
Organelle Isolation
Chloroplasts and mitochondria were isolated as described by Terauchi et al. (2009). Isolated chloroplasts and mitochondria were resuspended in 10 mM sodium phosphate, pH 7.0, containing 20% (w/v) glycerol. When required, organelles were lysed as described by Allen et al. (2008).
SOD Activity Gels and Activity Assays
Soluble proteins were quantitatively released from cells by two freeze-thaw cycles (Merchant et al., 1991) and separated by nondenaturing PAGE (7.5 to 9% acrylamide monomer) using the system of Davis (1964). Gels were photopolymerized as described by Lyubimova et al. (1993). SOD activity was visualized as described by Beauchamp and Fridovich (1971). Activity due to FeSOD was identified by sensitivity of the enzyme to pretreatment of the gel with 3 mM H2O2 prior to activity staining. Total SOD activity was measured by the xanthine oxidase/xanthine/cytochrome c method as described by McCord (1999).
Antibody Production and Immunoblotting
A DNA segment encoding the N-terminal 60% (A1 to Q122) of the mature FeSOD was amplified from the cDNA clone LC046e10 using primers FeSOD-F1 plus FeSOD-R2, digested with BamHI plus KpnI, and cloned in pTrxFus (Invitrogen) to create in-frame fusions to Escherichia coli TrxA. The DNA region coding for the fusion protein, plus upstream DNA incorporating the TrxA ribosome binding site, was amplified using primers TrxA-F1 plus FeSOD-R3, digested with BamHI plus XhoI, and cloned in pET23(+) (EMD Biosciences/Novagen) to create an in-frame fusion to the His6-coding region of the vector. Similarly, a DNA segment encoding the region E93 to A183 of the predicted MnSOD3 precursor was amplified from the cDNA clone MX248H08 using primers MSD3F-ab plus MSD3R-ab, digested with BamHI plus PstI, and cloned in pTrxFus. The DNA region coding for the fusion protein, plus upstream DNA, was amplified using primers TrxA-F1 plus MSD3-R1, digested with NdeI plus XhoI, and cloned in pET22b(+) to create an in-frame fusion to the His6-coding region of the vector. All primers used are listed in Supplemental Table 6 online.
The resulting plasmids were transformed into E. coli BL21(DE3) for isopropyl-β-d-1 thiogalactopyranoside–inducible expression. The majority (>90%) of the TrxA-FeSOD-His6 fusion protein was soluble, while the TrxA-MnSOD3-His6 fusion protein formed inclusion bodies that could be solubilized with 5 M urea. Both fusion proteins were purified by immobilized metal affinity chromatography on a Ni2+ charged His-Trap column (GE Healthcare), the TrxA-MnSOD3-His6 fusion protein under denaturing conditions (6 M urea). Details of the purifications are given in Supplemental Methods 1 online. The fusion proteins were further purified by preparative SDS-PAGE; the regions of the gels containing the fusion proteins (visualized by Zn-imidazole staining) were excised and used directly for antiserum production in rabbits (service provided by Agrisera and Pacific Immunology). The antisera were shown to be specific for FeSOD and MnSOD3, respectively (see Supplemental Figure 4 online).
For immunoblot analysis, proteins were separated by denaturing PAGE (10 to 15% monomer) and transferred to polyvinylidene fluoride and detected as described by Allen et al. (2007a). Primary antibodies were used at the following dilutions: antialternative oxidase, 1:5000 (Agrisera AB AS06 152); anticytochrome f, 1:2000 (Agrisera AB AS06 119); antiferredoxin, 1:1000 (Terauchi et al., 2009); anti-FeSOD, 1:5000. Alternatively, proteins were transferred to nitrocellulose and detected as described by Page et al. (2009). In this case, primary antibodies were used at the following dilutions: anti-CF1 α/β, 1;10,000 (Merchant and Selman, 1983); anticytochrome f, 1:2000; antiferritin1, 1:1000 (Long et al., 2008); antiferroxidase, 1:300 (La Fontaine et al., 2002); anti-FeSOD, 1:1500; anti-MnSOD3, 1:1000; anti-KARI, 1:1000; anti- magnesium protoporphyrin monomethyl ester cyclase, 1:1000 (Moseley et al., 2002b).
Reporter Constructs and Their Analysis
All primers are listed in Supplemental Table 7 online. The DNA region upstream of the MSD3 gene 5′ end was amplified from C. reinhardtii CC125 genomic DNA using primers M3PF1 and M3PR1, cloned via recombination in the Gateway entry vector pDONR221 (Invitrogen), and verified by sequencing. The cloned segment was transferred via recombination into the reporter vector pJD100GW (constructed by introduction of the Gateway Cmr-ccdB cassette into the Klenow-blunted KpnI site of pJD100; ptubB2Δ3,2,1/ars; Davies and Grossman, 1994) to generate the construct pJD100GW-5′MSD3. Plasmids pJD100 and pJD100GW-5′MSD3 were linearized with PsiI and BsaI, respectively, and cotransformed with pArg7.8 into strain CC425 as described previously (Quinn and Merchant, 1995). Arg prototrophs were picked, streaked onto agar-solidified TAP containing 1, 3, and 18 μM added Fe, and screened for arylsulfatase expression using 5-bromo-4-chloro-3-indolyl sulfate (X-sulfate; Gold BioTechnology) as described by de Hostos et al. (1988). Cotransformation frequency and intactness of integrated reporter constructs were assessed by colony PCR using primers pJD100-tub-f plus ARS1 (for the TUB2-ARS2 region) and ARS-3′-F plus ARS-3′-R (for the ARS2 3′ end).
Selected transformants were grown in liquid TAP medium containing various amounts of added Fe, or TAP containing no added Mn, and arylsulfatase in the culture medium determined after removal of the cells by centrifugation (de Hostos et al., 1988). In the case of pJD100GW-5′MSD3, the three strongest arylsulfatase expressers were analyzed. In the case of pJD100, three transformants carrying full-length reporter constructs were analyzed.
Sequence Analysis
Protein similarity networks were generated from an all-versus-all BLAST analysis (pairwise alignment between all pairs of proteins) of SOD sequences in a local sequence database. Protein sequences were trimmed to the equivalent of Q3 (the Q residue at position 3) of C. reinhardtii MnSOD1 to remove targeting sequences. The network was created in Cytoscape version 2.8 (Smoot et al., 2011) with the BLAST2SimilarityGraph plug-in (Wittkop et al., 2011) and the yFiles Organic layout engine provided with Cytoscape. Nodes (representing a protein) are connected with an edge (line) if the E-value between two sequences was at least as good as the given value.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers GQ413964 (FSD1), GQ413965 (MSD1), GQ413966 (MSD2), GQ415402 (MSD3), GU134344 (MSD4), and GU134345 (MSD5) and in the National Center for Biotechnology Information Gene Expression Omnibus under accession numbers GES25124 and GSE35305.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. The Newly Discovered Fe Limitation-Inducible SOD Is Not Sensitive to Hydrogen Peroxide.
Supplemental Figure 2. Chemical Identification of the Two Major SODs in C. reinhardtii.
Supplemental Figure 3. Gene Models for FSD1, MSD1, MSD2, MSD3, MSD4, and MSD5.
Supplemental Figure 4. Specificity of Antibodies Raised to FeSOD and MnSOD3.
Supplemental Figure 5. Time Course of mRNA Decay for MSD3 and Known Fe Homeostasis Genes.
Supplemental Figure 6. Accumulation of SOD mRNAs in Response to Fe Starvation.
Supplemental Figure 7. A Single MSD3 Transcript Is Detected by Hybridization and PCR Amplification.
Supplemental Figure 8. Determination of MSD5 Intron 2 Size.
Supplemental Table 1. Tryptic Peptides Recovered from the Major MnSOD Activity Band.
Supplemental Table 2. Tryptic Peptides Recovered from the FeSOD Activity Band.
Supplemental Table 3. Summary of Transformant Analyses.
Supplemental Table 4. Primers Used to Generate Improved Gene Models.
Supplemental Table 5. Primers Used for Real-Time PCR.
Supplemental Table 6. Primers Used to Generate Protein Expression Constructs.
Supplemental Table 7. Primers Used for Reporter Construct Assembly and Analysis.
Supplemental Methods 1. Assessment of Anti-FeSOD and Anti-MnSOD3 Antibody Specificity and Generation of Improved Gene Models for MSD3, MSD4, and MSD5.
Acknowledgments
We thank Maryse Block for the gift of antibodies to the spinach chloroplast ketol-acid reductoisomerase (KARI), Carla Koehler for antibodies to E. coli glutathione S-transferase, and Joanne Long for RNA samples. We are grateful to Arthur Grossman’s laboratory, the Kazusa DNA Research Institute for providing cDNA clones, and Crysten Blaby-Haas for help with the protein similarity network analysis. This work was supported by the Department of Energy (DE-FG02-04ER15529). M.D.A. was supported in part by Institutional and Individual Ruth L. Kirchstein National Research Service Awards (GM07185 and GM077066), and S.K. was supported in part by an Institutional Ruth L. Kirchstein National Research Service Award (GM07185).
AUTHOR CONTRIBUTIONS
S.S.M. conceived the project. S.S.M., M.D.P., and M.D.A. designed the experiments. M.D.P., M.D.A, J.K., E.I.U., S.J.K., and S.I.H. performed the experiments. J.A.L. contributed new analytic tools. S.S.M., M.D.P, and M.D.A. analyzed the data. S.S.M. and M.D.P. prepared the article.
Glossary
- PSI
photosystem I
- SOD
superoxide dismutase
- H2O2
hydrogen peroxide
- TAP
Tris-acetate-phosphate
- PVDF
to be defined
Footnotes
Online version contains Web-only data.
References
- Allen M.D., del Campo J.A., Kropat J., Merchant S.S. (2007b). FEA1, FEA2, and FRE1, encoding two homologous secreted proteins and a candidate ferrireductase, are expressed coordinately with FOX1 and FTR1 in iron-deficient Chlamydomonas reinhardtii. Eukaryot. Cell 6: 1841–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen M.D., Kropat J., Merchant S.S. (2008). Regulation and localization of isoforms of the aerobic oxidative cyclase in Chlamydomonas reinhardtii. Photochem. Photobiol. 84: 1336–1342 [DOI] [PubMed] [Google Scholar]
- Allen M.D., Kropat J., Tottey S., Del Campo J.A., Merchant S.S. (2007a). Manganese deficiency in Chlamydomonas results in loss of photosystem II and MnSOD function, sensitivity to peroxides, and secondary phosphorus and iron deficiency. Plant Physiol. 143: 263–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alscher R.G., Erturk N., Heath L.S. (2002). Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. J. Exp. Bot. 53: 1331–1341 [PubMed] [Google Scholar]
- Armbrust E.V. (2009). The life of diatoms in the world’s oceans. Nature 459: 185–192 [DOI] [PubMed] [Google Scholar]
- Armbrust E.V., et al. (2004). The genome of the diatom Thalassiosira pseudonana: Ecology, evolution, and metabolism. Science 306: 79–86 [DOI] [PubMed] [Google Scholar]
- Asada K., Kanematsu S., Uchida K. (1977). Superoxide dismutases in photosynthetic organisms: Absence of the cuprozinc enzyme in eukaryotic algae. Arch. Biochem. Biophys. 179: 243–256 [DOI] [PubMed] [Google Scholar]
- Asada K., Yoshikawa K., Takahashi M., Maeda Y., Enmanji K. (1975). Superoxide dismutases from a blue-green alga, Plectonema boryanum. J. Biol. Chem. 250: 2801–2807 [PubMed] [Google Scholar]
- Atteia A., et al. (2009). A proteomic survey of Chlamydomonas reinhardtii mitochondria sheds new light on the metabolic plasticity of the organelle and on the nature of the α-proteobacterial mitochondrial ancestor. Mol. Biol. Evol. 26: 1533–1548 [DOI] [PubMed] [Google Scholar]
- Beauchamp C., Fridovich I. (1971). Superoxide dismutase: Improved assays and an assay applicable to acrylamide gels. Anal. Biochem. 44: 276–287 [DOI] [PubMed] [Google Scholar]
- Benning C., Huang Z.H., Gage D.A. (1995). Accumulation of a novel glycolipid and a betaine lipid in cells of Rhodobacter sphaeroides grown under phosphate limitation. Arch. Biochem. Biophys. 317: 103–111 [DOI] [PubMed] [Google Scholar]
- Berthold D.A., Best B.A., Malkin R. (1993). A rapid DNA preparation for PCR from Chlamydomonas reinhardtii and Arabidopsis thaliana. Plant Mol. Biol. Rep. 11: 338–344 [Google Scholar]
- Beyer W.F., Jr., andFridovich I. (1991). In vivo competition between iron and manganese for occupancy of the active site region of the manganese-superoxide dismutase of Escherichia coli. J. Biol. Chem. 266: 303–308 [PubMed] [Google Scholar]
- Biliński T., Krawiec Z., Liczmański A., Litwińska J. (1985). Is hydroxyl radical generated by the Fenton reaction in vivo? Biochem. Biophys. Res. Commun. 130: 533–539 [DOI] [PubMed] [Google Scholar]
- Blaby-Haas C.E., Merchant S.S. (May 1, 2012). The ins and outs of algal metal transporters. Biochim. Biophys. Acta http://dx.doi.org/10.1016/j.bbamcr.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler C., Slooten L., Vandenbranden S., De Rycke R., Botterman J., Sybesma C., Van Montagu M., Inzé D. (1991). Manganese superoxide dismutase can reduce cellular damage mediated by oxygen radicals in transgenic plants. EMBO J. 10: 1723–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler C., Van Montagu M., Inzé D. (1992). Superoxide dismutase and stress tolerance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 43: 83–116 [Google Scholar]
- Burkhead J.L., Reynolds K.A., Abdel-Ghany S.E., Cohu C.M., Pilon M. (2009). Copper homeostasis. New Phytol. 182: 799–816 [DOI] [PubMed] [Google Scholar]
- Busch A., Rimbauld B., Naumann B., Rensch S., Hippler M. (2008). Ferritin is required for rapid remodeling of the photosynthetic apparatus and minimizes photo-oxidative stress in response to iron availability in Chlamydomonas reinhardtii. Plant J. 55: 201–211 [DOI] [PubMed] [Google Scholar]
- Campbell W.S., Laudenbach D.E. (1995). Characterization of four superoxide dismutase genes from a filamentous cyanobacterium. J. Bacteriol. 177: 964–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlioz A., Touati D. (1986). Isolation of superoxide dismutase mutants in Escherichia coli: Is superoxide dismutase necessary for aerobic life? EMBO J. 5: 623–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castruita M., Casero D., Karpowicz S.J., Kropat J., Vieler A., Hsieh S.I., Yan W., Cokus S., Loo J.A., Benning C., Pellegrini M., Merchant S.S. (2011). Systems biology approach in Chlamydomonas reveals connections between copper nutrition and multiple metabolic steps. Plant Cell 23: 1273–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Romo-Leroux P.A., Salin M.L. (1996). The iron-containing superoxide dismutase-encoding gene from Chlamydomonas reinhardtii obtained by direct and inverse PCR. Gene 168: 113–116 [DOI] [PubMed] [Google Scholar]
- Chen J.C., Hsieh S.I., Kropat J., Merchant S.S. (2008). A ferroxidase encoded by FOX1 contributes to iron assimilation under conditions of poor iron nutrition in Chlamydomonas. Eukaryot. Cell 7: 541–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet F. (1988). Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16: 10881–10890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culotta V.C., Klomp L.W., Strain J., Casareno R.L., Krems B., Gitlin J.D. (1997). The copper chaperone for superoxide dismutase. J. Biol. Chem. 272: 23469–23472 [DOI] [PubMed] [Google Scholar]
- Culotta V.C., Yang M., O’Halloran T.V. (2006). Activation of superoxide dismutases: Putting the metal to the pedal. Biochim. Biophys. Acta 1763: 747–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J.P., Grossman A.R. (1994). Sequences controlling transcription of the Chlamydomonas reinhardtii beta 2-tubulin gene after deflagellation and during the cell cycle. Mol. Cell. Biol. 14: 5165–5174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis B.J. (1964). Disc electrophoresis. II. Method and application to human serum proteins. Ann. N. Y. Acad. Sci. 121: 404–427 [DOI] [PubMed] [Google Scholar]
- de Hostos E.L., Togasaki R.K., Grossman A. (1988). Purification and biosynthesis of a derepressible periplasmic arylsulfatase from Chlamydomonas reinhardtii. J. Cell Biol. 106: 29–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jesus M.D., Tabatabai F., Chapman D.J. (1989). Taxonomic distribution of copper-zinc superoxide dismutase in green algae and its phylogenetic importance. J. Phycol. 25: 767–772 [Google Scholar]
- Djaman O., Outten F.W., Imlay J.A. (2004). Repair of oxidized iron-sulfur clusters in Escherichia coli. J. Biol. Chem. 279: 44590–44599 [DOI] [PubMed] [Google Scholar]
- Dumas R., Joyard J., Douce R. (1989). Purification and characterization of acetohydroxyacid reductoisomerase from spinach chloroplasts. Biochem. J. 262: 971–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egashira T., Takahama U., Nakamura K. (1989). A reduced activity of catalase as a basis for light dependent methionine sensitivity of a Chlamydomonas reinhardtii mutant. Plant Cell Physiol. 30: 1171–1175 [Google Scholar]
- Farr S.B., D’Ari R., Touati D. (1986). Oxygen-dependent mutagenesis in Escherichia coli lacking superoxide dismutase. Proc. Natl. Acad. Sci. USA 83: 8268–8272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink R.C., Scandalios J.G. (2002). Molecular evolution and structure-function relationships of the superoxide dismutase gene families in angiosperms and their relationship to other eukaryotic and prokaryotic superoxide dismutases. Arch. Biochem. Biophys. 399: 19–36 [DOI] [PubMed] [Google Scholar]
- Garland S.A., Hoff K., Vickery L.E., Culotta V.C. (1999). Saccharomyces cerevisiae ISU1 and ISU2: Members of a well-conserved gene family for iron-sulfur cluster assembly. J. Mol. Biol. 294: 897–907 [DOI] [PubMed] [Google Scholar]
- Grace S.C. (1990). Phylogenetic distribution of superoxide dismutase supports an endosymbiotic origin for chloroplasts and mitochondria. Life Sci. 47: 1875–1886 [DOI] [PubMed] [Google Scholar]
- Guerinot M.L., Yi Y. (1994). Iron: Nutritious, noxious, and not readily available. Plant Physiol. 104: 815–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güler S., Seeliger A., Härtel H., Renger G., Benning C. (1996). A null mutant of Synechococcus sp. PCC7942 deficient in the sulfolipid sulfoquinovosyl diacylglycerol. J. Biol. Chem. 271: 7501–7507 [DOI] [PubMed] [Google Scholar]
- Harris, E.H. (2009). The Genus Chlamydomonas. In The Chlamydomonas Sourcebook, 2nd ed, Vol. 1, E.H. Harris, D.B. Stern, and G.B. Whitman, eds (San Diego, CA: Academic Press), pp. 1–24.
- Halliwell B., Gutteridge J.M.C. (1984). Oxygen toxicity, oxygen radicals, transition metals and disease. Biochem. J. 219: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert S.K., Samson G., Fork D.C., Laudenbach D.E. (1992). Characterization of damage to photosystems I and II in a cyanobacterium lacking detectable iron superoxide dismutase activity. Proc. Natl. Acad. Sci. USA 89: 8716–8720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbik A., Bölling C., Buckhout T.J. (2002). The involvement of a multicopper oxidase in iron uptake by the green algae Chlamydomonas reinhardtii. Plant Physiol. 130: 2039–2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkin K.A., Papazian M.A., Steinman H.M. (1992). Functional differences between manganese and iron superoxide dismutases in Escherichia coli K-12. J. Biol. Chem. 267: 24253–24258 [PubMed] [Google Scholar]
- Imlay J.A. (2008). Cellular defenses against superoxide and hydrogen peroxide. Annu. Rev. Biochem. 77: 755–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen L.T., Culotta V.C. (2000). Role of Saccharomyces cerevisiae ISA1 and ISA2 in iron homeostasis. Mol. Cell. Biol. 20: 3918–3927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J., Guerinot M.L. (2009). Homing in on iron homeostasis in plants. Trends Plant Sci. 14: 280–285 [DOI] [PubMed] [Google Scholar]
- Kim Y.C., Miller C.D., Anderson A.J. (1999). Transcriptional regulation by iron of genes encoding Fe- and manganese-superoxide dismutases from Pseudomonas putida. Gene 239: 129–135 [DOI] [PubMed] [Google Scholar]
- Kislinger T., et al. (2006). Global survey of organ and organelle protein expression in mouse: Combined proteomic and transcriptomic profiling. Cell 125: 173–186 [DOI] [PubMed] [Google Scholar]
- Kitayama K., Kitayama M., Osafune T., Togasaki R.K. (1999). Subcellular localization of iron and manganese superoxide dismutase in Chlamydomonas reinhardtii (Chlorophyceae). J. Phycol. 35: 136–142 [Google Scholar]
- Kosman D.J. (2003). Molecular mechanisms of iron uptake in fungi. Mol. Microbiol. 47: 1185–1197 [DOI] [PubMed] [Google Scholar]
- Kumánovics A., Chen O.S., Li L., Bagley D., Adkins E.M., Lin H., Dingra N.N., Outten C.E., Keller G., Winge D., Ward D.M., Kaplan J. (2008). Identification of FRA1 and FRA2 as genes involved in regulating the yeast iron regulon in response to decreased mitochondrial iron-sulfur cluster synthesis. J. Biol. Chem. 283: 10276–10286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Fontaine S., Quinn J.M., Nakamoto S.S., Page M.D., Göhre V., Moseley J.L., Kropat J., Merchant S. (2002). Copper-dependent iron assimilation pathway in the model photosynthetic eukaryote Chlamydomonas reinhardtii. Eukaryot. Cell 1: 736–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane T.W., Morel F.M. (2000a). A biological function for cadmium in marine diatoms. Proc. Natl. Acad. Sci. USA 97: 4627–4631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane T.W., Morel F.M. (2000b). Regulation of carbonic anhydrase expression by zinc, cobalt, and carbon dioxide in the marine diatom Thalassiosira weissflogii. Plant Physiol. 123: 345–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latifi A., Jeanjean R., Lemeille S., Havaux M., Zhang C.C. (2005). Iron starvation leads to oxidative stress in Anabaena sp. strain PCC 7120. J. Bacteriol. 187: 6596–6598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leisinger U., Rüfenacht K., Fischer B., Pesaro M., Spengler A., Zehnder A.J., Eggen R.I. (2001). The glutathione peroxidase homologous gene from Chlamydomonas reinhardtii is transcriptionally up-regulated by singlet oxygen. Plant Mol. Biol. 46: 395–408 [DOI] [PubMed] [Google Scholar]
- Li T., Huang X., Zhou R., Liu Y., Li B., Nomura C., Zhao J. (2002). Differential expression and localization of Mn and Fe superoxide dismutases in the heterocystous cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 184: 5096–5103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liochev S.I., Fridovich I. (1999). Superoxide and iron: Partners in crime. IUBMB Life 48: 157–161 [DOI] [PubMed] [Google Scholar]
- Long J.C., Merchant S.S. (2008). Photo-oxidative stress impacts the expression of genes encoding iron metabolism components in Chlamydomonas. Photochem. Photobiol. 84: 1395–1403 [DOI] [PubMed] [Google Scholar]
- Long J.C., Sommer F., Allen M.D., Lu S.F., Merchant S.S. (2008). FER1 and FER2 encoding two ferritin complexes in Chlamydomonas reinhardtii chloroplasts are regulated by iron. Genetics 179: 137–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P., Vogel C., Wang R., Yao X., Marcotte E.M. (2007). Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat. Biotechnol. 25: 117–124 [DOI] [PubMed] [Google Scholar]
- Luk E., Yang M., Jensen L.T., Bourbonnais Y., Culotta V.C. (2005). Manganese activation of superoxide dismutase 2 in the mitochondria of Saccharomyces cerevisiae. J. Biol. Chem. 280: 22715–22720 [DOI] [PubMed] [Google Scholar]
- Lynch R.E., Fridovich I. (1978). Permeation of the erythrocyte stroma by superoxide radical. J. Biol. Chem. 253: 4697–4699 [PubMed] [Google Scholar]
- Lyubimova T., Caglio S., Gelfi C., Righetti P.G., Rabilloud T. (1993). Photopolymerization of polyacrylamide gels with methylene blue. Electrophoresis 14: 40–50 [DOI] [PubMed] [Google Scholar]
- Marchetti A., Cassar N. (2009). Diatom elemental and morphological changes in response to iron limitation: A brief review with potential paleoceanographic applications. Geobiology 7: 419–431 [DOI] [PubMed] [Google Scholar]
- Marschner, H. (1995). Mineral Nutrition of Higher Plants, 2nd ed. (Orlando, FL: Academic Press)
- Marschner H., Römheld V. (1994). Strategies of plants for acquisition of iron. Plant Soil 165: 261–274 [Google Scholar]
- McCord, J.M. (1999). Measurement of superoxide dismutase activity. Curr. Protoc. Toxicol. 8 (suppl.): 7.3.1–7.3.9. [DOI] [PubMed]
- Merchant S., Hill K., Howe G. (1991). Dynamic interplay between two copper-titrating components in the transcriptional regulation of cyt c6. EMBO J. 10: 1383–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant S., Selman B.R. (1983). Identification of the alpha and beta subunits of the chloroplast coupling factor one in Chlamydomonas reinhardi. Eur. J. Biochem. 137: 373–376 [DOI] [PubMed] [Google Scholar]
- Merchant S.S., Allen M.D., Kropat J., Moseley J.L., Long J.C., Tottey S., Terauchi A.M. (2006). Between a rock and a hard place: Trace element nutrition in Chlamydomonas. Biochim. Biophys. Acta 1763: 578–594 [DOI] [PubMed] [Google Scholar]
- Merchant S.S., Helmann J.D. (2012). Elemental economy: Microbial strategies for optimizing growth in the face of nutrient limitation. Adv. Microb. Physiol. 60: 91–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missirlis F., Hu J., Kirby K., Hilliker A.J., Rouault T.A., Phillips J.P. (2003). Compartment-specific protection of iron-sulfur proteins by superoxide dismutase. J. Biol. Chem. 278: 47365–47369 [DOI] [PubMed] [Google Scholar]
- Morel F.M.M., Price N.M. (2003). The biogeochemical cycles of trace metals in the oceans. Science 300: 944–947 [DOI] [PubMed] [Google Scholar]
- Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Moseley J., Quinn J., Eriksson M., Merchant S. (2000). The Crd1 gene encodes a putative di-iron enzyme required for photosystem I accumulation in copper deficiency and hypoxia in Chlamydomonas reinhardtii. EMBO J. 19: 2139–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley J.L., Allinger T., Herzog S., Hoerth P., Wehinger E., Merchant S., Hippler M. (2002a). Adaptation to Fe-deficiency requires remodeling of the photosynthetic apparatus. EMBO J. 21: 6709–6720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley J.L., Page M.D., Alder N.P., Eriksson M., Quinn J., Soto F., Theg S.M., Hippler M., Merchant S. (2002b). Reciprocal expression of two candidate di-iron enzymes affecting photosystem I and light-harvesting complex accumulation. Plant Cell 14: 673–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlenhoff U., Molik S., Godoy J.R., Uzarska M.A., Richter N., Seubert A., Zhang Y., Stubbe J., Pierrel F., Herrero E., Lillig C.H., Lill R. (2010). Cytosolic monothiol glutaredoxins function in intracellular Fe sensing and trafficking via their bound iron-sulfur cluster. Cell Metab. 12: 373–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myouga F., Hosoda C., Umezawa T., Iizumi H., Kuromori T., Motohashi R., Shono Y., Nagata N., Ikeuchi M., Shinozaki K. (2008). A heterocomplex of iron superoxide dismutases defends chloroplast nucleoids against oxidative stress and is essential for chloroplast development in Arabidopsis. Plant Cell 20: 3148–3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naranuntarat A., Jensen L.T., Pazicni S., Penner-Hahn J.E., Culotta V.C. (2009). The interaction of mitochondrial iron with manganese superoxide dismutase. J. Biol. Chem. 284: 22633–22640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumann B., Busch A., Allmer J., Ostendorf E., Zeller M., Kirchhoff H., Hippler M. (2007). Comparative quantitative proteomics to investigate the remodeling of bioenergetic pathways under iron deficiency in Chlamydomonas reinhardtii. Proteomics 7: 3964–3979 [DOI] [PubMed] [Google Scholar]
- Naumann B., Stauber E.J., Busch A., Sommer F., Hippler M. (2005). N-terminal processing of Lhca3 Is a key step in remodeling of the photosystem I-light-harvesting complex under iron deficiency in Chlamydomonas reinhardtii. J. Biol. Chem. 280: 20431–20441 [DOI] [PubMed] [Google Scholar]
- Nicholas K.B., Nicholas H.B., Jr., andDeerfield D.W., II (1997). GeneDoc: Analysis and visualization of genetic variation. EMBnet.news 4: 1–4 [Google Scholar]
- Österberg R. (1974). Origins of metal ions in biology. Nature 249: 382–383 [DOI] [PubMed] [Google Scholar]
- Page M.D., Kropat J., Hamel P.P., Merchant S.S. (2009). Two Chlamydomonas copper transporters with a novel Cys-Met motif are localized to the plasma membrane and function in copper assimilation. Plant Cell 21: 928–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peers G., Price N.M. (2004). A role for manganese in superoxide dismutases and growth of iron-deficient diatoms. Limnol. Oceanogr. 49: 1774–1783 [Google Scholar]
- Phee B.K., Cho J.H., Park S., Jung J.H., Lee Y.H., Jeon J.S., Bhoo S.H., Hahn T.R. (2004). Proteomic analysis of the response of Arabidopsis chloroplast proteins to high light stress. Proteomics 4: 3560–3568 [DOI] [PubMed] [Google Scholar]
- Pilon M., Ravet K., Tapken W. (2011). The biogenesis and physiological function of chloroplast superoxide dismutases. Biochim. Biophys. Acta 1807: 989–998 [DOI] [PubMed] [Google Scholar]
- Priya B., Premanandh J., Dhanalakshmi R.T., Seethalakshmi T., Uma L., Prabaharan D., Subramanian G. (2007). Comparative analysis of cyanobacterial superoxide dismutases to discriminate canonical forms. BMC Genomics 8: 435–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn J.M., Merchant S. (1995). Two copper-responsive elements associated with the Chlamydomonas Cyc6 gene function as targets for transcriptional activators. Plant Cell 7: 623–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn J.M., Merchant S. (1998). Copper-responsive gene expression during adaptation to copper deficiency. Methods Enzymol. 297: 263–279 [DOI] [PubMed] [Google Scholar]
- Raven J.A., Evans M.C.W., Korb R.E. (1999). The role of trace metals in photosynthetic electron transport in O2-evolving organisms. Photosynth. Res. 60: 111–149 [Google Scholar]
- Regelsberger G., Laaha U., Dietmann D., Rüker F., Canini A., Grilli-Caiola M., Furtmüller P.G., Jakopitsch C., Peschek G.A., Obinger C. (2004). The iron superoxide dismutase from the filamentous cyanobacterium Nostoc PCC 7120. Localization, overexpression, and biochemical characterization. J. Biol. Chem. 279: 44384–44393 [DOI] [PubMed] [Google Scholar]
- Reyda M.R., Dippold R., Dotson M.E., Jarrett J.T. (2008). Loss of iron-sulfur clusters from biotin synthase as a result of catalysis promotes unfolding and degradation. Arch. Biochem. Biophys. 471: 32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai H., Kusumoto N., Kitayama K., Togasaki R.K. (1993). Isoenzymes of superoxide dismutase in Chlamydomonas and purification of one of the major isozymes containing Fe. Plant Cell Physiol. 34: 1133–1137 [Google Scholar]
- Sandmann G., Malkin R. (1983). Iron-sulfur centers and activities of the photosynthetic electron transport chain in iron-deficient cultures of the blue-green alga aphanocapsa. Plant Physiol. 73: 724–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoot M.E., Ono K., Ruscheinski J., Wang P.L., Ideker T. (2011). Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics 27: 431–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiller S.C., Castelfranco A.M., Castelfranco P.A. (1982). Effects of iron and oxygen on chlorophyll biosynthesis: I. In vivo observations on iron and oxygen-deficient plants. Plant Physiol. 69: 107–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R., Kapoor A., Zhu J.K. (2006). Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 18: 2051–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M.-A., Asada K. (1983). Superoxide anion permeability of phospholipid membranes and chloroplast thylakoids. Arch. Biochem. Biophys. 226: 558–566 [DOI] [PubMed] [Google Scholar]
- Terauchi A.M., Lu S.-F., Zaffagnini M., Tappa S., Hirasawa M., Tripathy J.N., Knaff D.B., Farmer P.J., Lemaire S.D., Hase T., Merchant S.S. (2009). Pattern of expression and substrate specificity of chloroplast ferredoxins from Chlamydomonas reinhardtii. J. Biol. Chem. 284: 25867–25878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terauchi A.M., Peers G., Kobayashi M.C., Niyogi K.K., Merchant S.S. (2010). Trophic status of Chlamydomonas reinhardtii influences the impact of iron deficiency on photosynthesis. Photosynth. Res. 105: 39–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D.J., Avenson T.J., Thomas J.B., Herbert S.K. (1998). A cyanobacterium lacking iron superoxide dismutase is sensitized to oxidative stress induced with methyl viologen but Is not sensitized to oxidative stress induced with norflurazon. Plant Physiol. 116: 1593–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tottey S., Block M.A., Allen M., Westergren T., Albrieux C., Scheller H.V., Merchant S., Jensen P.E. (2003). Arabidopsis CHL27, located in both envelope and thylakoid membranes, is required for the synthesis of protochlorophyllide. Proc. Natl. Acad. Sci. USA 100: 16119–16124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang E.W., Bowler C., Hérouart D., Van Camp W., Villarroel R., Genetello C., Van Montagu M., Inzé D. (1991). Differential regulation of superoxide dismutases in plants exposed to environmental stress. Plant Cell 3: 783–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Camp W., Capiau K., Van Montagu M., Inzé D., Slooten L. (1996). Enhancement of oxidative stress tolerance in transgenic tobacco plants overproducing Fe-superoxide dismutase in chloroplasts. Plant Physiol. 112: 1703–1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlerberghe G.C., McIntosh L. (1997). Alternative oxidase: From gene to function. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48: 703–734 [DOI] [PubMed] [Google Scholar]
- van Loon A.P.G.M., Pesold-Hurt B., Schatz G. (1986). A yeast mutant lacking mitochondrial manganese-superoxide dismutase is hypersensitive to oxygen. Proc. Natl. Acad. Sci. USA 83: 3820–3824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker E.L., Connolly E.L. (2008). Time to pump iron: Iron-deficiency-signaling mechanisms of higher plants. Curr. Opin. Plant Biol. 11: 530–535 [DOI] [PubMed] [Google Scholar]
- Wintjens R., Noël C., May A.C., Gerbod D., Dufernez F., Capron M., Viscogliosi E., Rooman M. (2004). Specificity and phenetic relationships of iron- and manganese-containing superoxide dismutases on the basis of structure and sequence comparisons. J. Biol. Chem. 279: 9248–9254 [DOI] [PubMed] [Google Scholar]
- Wittkop T., Emig D., Truss A., Albrecht M., Böcker S., Baumbach J. (2011). Comprehensive cluster analysis with Transitivity Clustering. Nat. Protoc. 6: 285–295 [DOI] [PubMed] [Google Scholar]
- Wolfe-Simon F., Starovoytov V., Reinfelder J.R., Schofield O., Falkowski P.G. (2006). Localization and role of manganese superoxide dismutase in a marine diatom. Plant Physiol. 142: 1701–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollast R., Billon G., Duinker J.C. (1979). Behaviour of manganese in the Rhine and Scheldt estuaries. I. Physico-chemical aspects. Estuar. Coast. Mar. Sci. 9: 161–169 [Google Scholar]
- Xu Y., Degui T., Shaked Y., Morel F.M.M. (2007). Zinc, cadmium, and cobalt interreplacement and relative use efficiencies in the coccolithophore Emiliania huxleyi. Limnol. Oceanogr. 52: 2294–2305 [Google Scholar]
- Yamakura F., Suzuki K. (1976). Reconstitution of iron-superoxide dismutase. Biochem. Biophys. Res. Commun. 72: 1108–1115 [DOI] [PubMed] [Google Scholar]
- Yang M., Cobine P.A., Molik S., Naranuntarat A., Lill R., Winge D.R., Culotta V.C. (2006). The effects of mitochondrial iron homeostasis on cofactor specificity of superoxide dismutase 2. EMBO J. 25: 1775–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B., Xu C., Benning C. (2002). Arabidopsis disrupted in SQD2 encoding sulfolipid synthase is impaired in phosphate-limited growth. Proc. Natl. Acad. Sci. USA 99: 5732–5737 [DOI] [PMC free article] [PubMed] [Google Scholar]