Vegetative propagation of plants from shoots requires the development of adventitious roots that regenerate from stem cuttings. Multiple endogenous and environmental factors control this complex process; this study in the model plant Arabidopsis thaliana shows that adventitious rooting is an adaptive developmental response involving crosstalk between auxin and jasmonate signaling pathways.
Abstract
Vegetative shoot-based propagation of plants, including mass propagation of elite genotypes, is dependent on the development of shoot-borne roots, which are also called adventitious roots. Multiple endogenous and environmental factors control the complex process of adventitious rooting. In the past few years, we have shown that the auxin response factors ARF6 and ARF8, targets of the microRNA miR167, are positive regulators of adventitious rooting, whereas ARF17, a target of miR160, is a negative regulator. We showed that these genes have overlapping expression profiles during adventitious rooting and that they regulate each other’s expression at the transcriptional and posttranscriptional levels by modulating the homeostasis of miR160 and miR167. We demonstrate here that this complex network of transcription factors regulates the expression of three auxin-inducible Gretchen Hagen3 (GH3) genes, GH3.3, GH3.5, and GH3.6, encoding acyl-acid-amido synthetases. We show that these three GH3 genes are required for fine-tuning adventitious root initiation in the Arabidopsis thaliana hypocotyl, and we demonstrate that they act by modulating jasmonic acid homeostasis. We propose a model in which adventitious rooting is an adaptive developmental response involving crosstalk between the auxin and jasmonate regulatory pathways.
INTRODUCTION
Adventitious roots arise from aerial organs of the plant either naturally or in response to altered environmental conditions (Geiss et al., 2009; Li et al., 2009). They are required for vegetative propagation of plants and the successful clonal multiplication of elite genotypes of forest, horticultural, and agricultural plant species. The ability to form adventitious roots is a heritable quantitative trait controlled by multiple endogenous and environmental factors (Geiss et al., 2009; Li et al., 2009), among which the plant hormone auxin plays a central role. Auxin is often applied exogenously to promote the development of adventitious roots on stem cuttings of difficult-to-root genotypes. However, exogenous auxin is not always effective, and a better understanding of the molecular mechanisms through which it regulates adventitious rooting is needed in order to improve the rooting of recalcitrant genotypes. So far, only a few genes involved in the regulation of adventitious rooting have been identified. In rice (Oryza sativa), disruption of the auxin-inducible CROWN ROOTLESS1/ADVENTITIOUS ROOTLESS1 gene (CRL1/ARL1), which encodes a member of the plant-specific LATERAL ORGAN BOUNDARY DOMAIN (LBD) protein family, has been shown to prevent the initiation of adventitious crown root primordia (Inukai et al., 2005; Liu et al., 2005). The promoter of the CRL1/ARL1 gene contains specific cis-regulatory elements that interact with the rice transcription factor ARF16, a member of the AUXIN RESPONSE FACTOR (ARF) family (Inukai et al., 2005). ARF16 is the rice ortholog of both ARF7 and ARF19 in Arabidopsis thaliana (Wang et al., 2007). ARF7 and ARF19 regulate lateral root formation in Arabidopsis by activating the expression of LBD16 and LBD29, both of which are phylogenetically closely related to ARL1/CRL1 (Wilmoth et al., 2005; Okushima et al., 2007). ARF transcription factors mediate auxin signaling at the transcriptional level by regulating the expression of auxin-responsive genes (Guilfoyle and Hagen, 2007). The characterization of Arabidopsis argonaute1 (Bohmert et al., 1998) and superroot2 (Delarue et al., 1998; Barlier et al., 2000) single and double mutants allowed us to identify several genes potentially involved in the regulation of adventitious rooting, including one ARF transcription factor, ARF17 (At1g77850), that encodes the target of the microRNA miR160 and three auxin-inducible GH3-like genes, GH3.3 (At2g23170), GH3.5/GH3a/WES1 (At4g27260), and GH3.6/DFL1 (At5g54510; Nakazawa et al., 2001; Tanaka et al., 2002; Sorin et al., 2005, 2006; Park et al., 2007b, 2007a). The first GH3 gene was identified as an early auxin-responsive gene in soybean (Glycine max) (Hagen et al., 1984), and since then, GH3-like genes have been found in many plant species. However, it is only recently that they have been shown to encode enzymes able to conjugate various amino acids to jasmonates, auxins, and benzoates, leading to the activation, inactivation, or degradation of these molecules (Staswick et al., 2005; Chen et al., 2010; Wang et al., 2010; Westfall et al., 2010). We first showed that ARF17 was a negative regulator of adventitious rooting that could potentially integrate auxin and light signaling pathways affecting this process (Sorin et al., 2005). We also showed that ARF17 was a negative regulator of GH3.3, GH3.5, and GH3.6 expression (Sorin et al., 2005), and, in addition, we found that the GH3.3, GH3.5, and GH3.6 protein content was positively correlated with the number of adventitious roots but not with the endogenous auxin content (Sorin et al., 2006). More recently, we demonstrated that adventitious root initiation is controlled in Arabidopsis hypocotyls by a subtle balance between the negative regulator ARF17, which is the target of miR160, and the positive regulators ARF6 (At1g30330) and ARF8 (At5g37020), which are targets of miR167. These three ARFs display overlapping expression domains, interact genetically, and regulate each other’s expression at both the transcriptional and posttranscriptional levels by modulating the availability of the regulatory microRNAs miR160 and miR167 (Gutierrez et al., 2009). In this article, we show that, in contrast to ARF17, ARF6 and ARF8 are positive regulators of the auxin-inducible genes GH3.3, GH3.5, and GH3.6. We also demonstrate that these three GH3 genes have redundant functions and are required for fine-tuning adventitious root initiation in Arabidopsis hypocotyls by modulating jasmonic acid (JA) homeostasis.
RESULTS
The Number of Adventitious Roots in arf Mutants and ARF-Overexpressing Lines Does Not Correlate with Endogenous Auxin Content But Does Correlate with GH3.3, GH3.5, and GH3.6 Transcript Levels
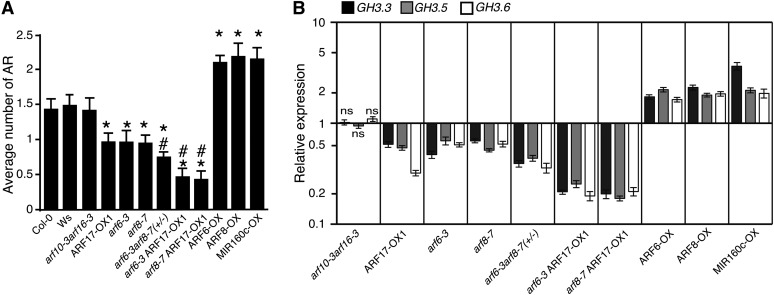
In an attempt to identify the pathway through which the microRNA-targeted genes ARF6, ARF8, and ARF17 regulate adventitious rooting, we selected for evaluation representative mutants and overexpressing lines that we previously characterized as having contrasting adventitious rooting phenotypes (Gutierrez et al., 2009) (Figure 1A). The double mutant arf10-3 arf16-3, which has no phenotype affecting adventitious root growth, was used as a control for the MIR160c-overexpressing line (MIR160c-OX), in which the accumulation of miR160 specifically targets ARF10, ARF16, and ARF17 transcripts for degradation (Wang et al., 2005). In the absence of an arf17 knockout mutant, we used MIR160-OX as an ARF17 downregulated line, as described previously (Gutierrez et al., 2009).
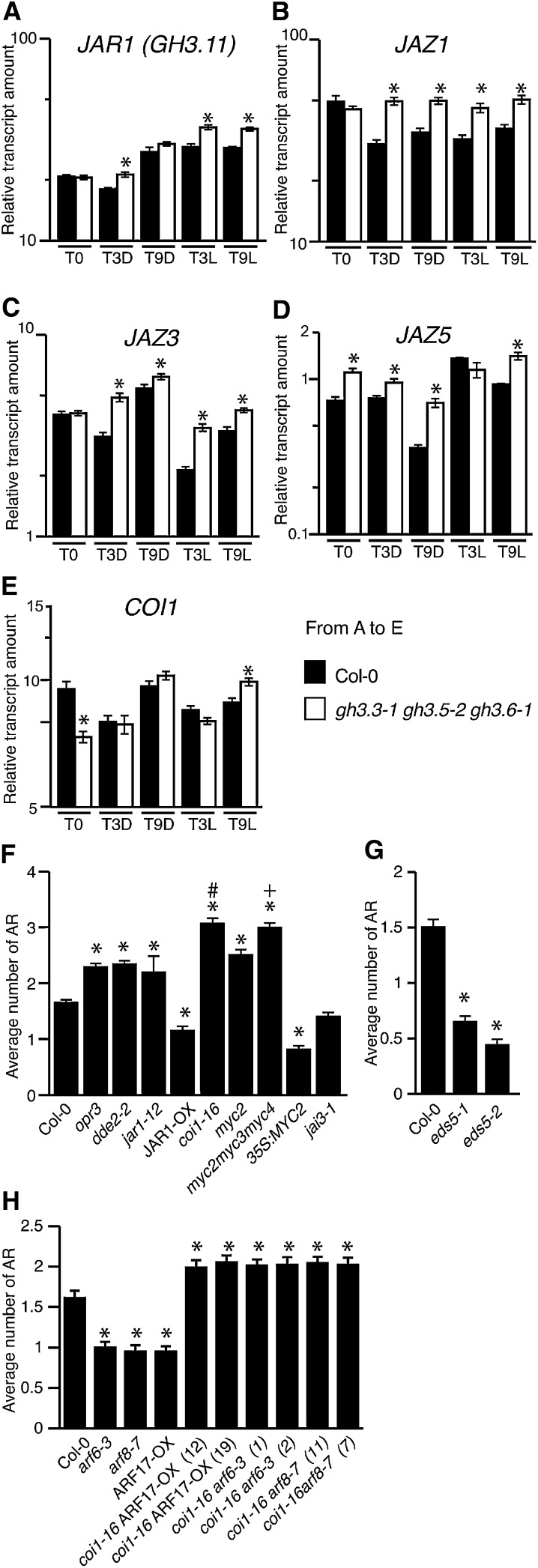
Figure 1.
Variation in the Number of Adventitious Roots in the Hypocotyl of arf Mutants and ARF-Overexpressing Lines Correlates with the Expression Levels of GH3.3, GH3.5, and GH3.6.
(A) Adventitious roots (AR) were counted on seedlings of arf mutant lines and ARF-overexpressing lines. Seedlings were first etiolated in the dark, until their hypocotyls were 6 mm long, and then transferred to the light for 7 d. Data from three independent biological replicates, each of at least 30 seedlings, were pooled and averaged. Error bars indicate se. A one-way analysis of variance combined with the Tukey’s multiple comparison posttest indicated that the values marked with asterisks were significantly different from wild-type values (P < 0.01; n > 90). In addition, those marked with hash signs were significantly different from values obtained from single mutants or the ARF17-OX1 line (P < 0.01; n > 90). Col-0, ecotype Columbia; Ws, ecotype Wassilewskija.
(B) The GH3.3, GH3.5, and GH3.6 transcript abundance was quantified in the hypocotyls of the different arf mutants or overexpressing lines, which had been etiolated and then transferred to the light for 72 h. Gene expression values are relative to expression in the wild type, for which the value is set to 1. Error bars indicate se obtained from three independent biological replicates. A one-way analysis of variance combined with the Dunnett’s comparison posttest indicated that the values marked with ns were not significantly different from wild-type values, whereas the others were significantly different (P < 0.05; n = 3).
Since auxin is a well-known inducer of adventitious rooting in Arabidopsis (Boerjan et al., 1995; Delarue et al., 1998), it was important to verify whether the increased or reduced number of adventitious roots was due to a higher or lower endogenous content of free indole-3-acetic acid (IAA) in the hypocotyl. Therefore, the level of endogenous free IAA and its major conjugates, IAAsp [N-(indole-3-ylacetyl)-aspartate] and IAGlu [N-(indole-3-ylacetyl)-glutamate], was quantified in the hypocotyls of seedlings grown as described previously (Gutierrez et al., 2009). Neither mutations in, nor overexpression of, ARF6, ARF8, or ARF17 significantly altered the endogenous content of free IAA, suggesting that the observed adventitious root phenotype is unlikely to be related to an increase or a reduction in IAA content (see Supplemental Figure 1 online).
To investigate the interactions between GH3.3, GH3.5, and GH3.6, whose expression levels were correlated with the number of adventitious roots (Sorin et al., 2006) and ARF genes, we first analyzed the expression of the three GH3 genes during the early steps of adventitious root formation using transcriptional fusion constructs containing β-glucuronidase (GUS) fused to the respective GH3 promoters. The promGH3.3:GUS, promGH3.5:GUS, and promGH3.6:GUS constructs have overlapping expression patterns with those of promARF6:GUS, promARF8:GUS, and promARF17:GUS and are likewise regulated by light (see Supplemental Figure 2 online) (Gutierrez et al., 2009). Second, we performed real-time quantitative RT-PCR analysis of the relative transcript amounts of the three GH3 genes in the hypocotyls of the various ARF mutant and overexpressing lines (Figure 1B). In the double mutant arf10-3arf16-3, which displayed no difference in adventitious rooting phenotype compared with the wild type, the GH3.3, GH3.5, and GH3.6 transcript levels were not affected. In contrast, the transcript levels of all three genes were reduced in all of the lines showing fewer adventitious roots than the wild type, whereas it was increased in those with a higher number of adventitious roots (Figures 1A and 1B). In agreement with previous data (Sorin et al., 2006), the smaller the number of adventitious roots, the lower the amount of GH3.3, GH3.5, and GH3.6 transcripts and vice versa. Since the arf6-3arf8-7 double mutant is sterile, we analyzed a population of progeny from the sesquimutant arf6-3/arf6-3 arf8-7/ARF8-7 segregating for 25% homozygous double mutants. The average number of adventitious roots was significantly lower than that of the wild type and of the single mutants arf6-3 and arf8-7, indicating an additive effect of the two positive regulators ARF6 and ARF8 (Figure 1A). This additive effect was confirmed by the reduced level of the total GH3.3+GH3.5+GH3.6 relative transcript amount in the sesquimutant compared with that in the single mutants (see Supplemental Figure 3A online). In contrast, the three GH3 genes were expressed at a higher level in ARF6- and ARF8-overexpressing lines. The three GH3 genes were also downregulated in the ARF17-OX1 line (Figure 1B) (Sorin et al., 2005) and, as expected, upregulated in the MIR160c-OX line, in which ARF17 is downregulated. When ARF17 was overexpressed in the knockout arf6-3 and arf8-7 mutant backgrounds, a significant decrease in the relative amount of transcript of each of the three GH3 genes was observed compared with the ARF17-overexpressing lines or the arf6-3 and arf8-7 single mutants (Figure 1B). This confirmed the existence of an additive effect due to the overexpression of the negative regulator ARF17 in mutants lacking one of the positive regulators (ARF6 or ARF8) of GH3.3, GH3.5, and GH3.6 expression.
GH3.3, GH3.5, and GH3.6 belong to a family of 19 genes. Therefore, we checked, in the hypocotyl of the ARF mutant and overexpressing lines, the expression of the other members of the GH3 gene family as well as that of selected auxin-inducible genes shown to be differentially expressed in the ago1-3 versus wild-type hypocotyls (http://urgv.evry.inra.fr/cgi-bin/projects/CATdb/catdb_index.pl). None of these genes were found to be coregulated with GH3.3, GH3.5, and GH3.6 in the different ARF mutant or overexpressing lines (see Supplemental Figures 3B and 3C online).
GH3.3, GH3.5, and GH3.6 Positively and Redundantly Regulate Adventitious Rooting
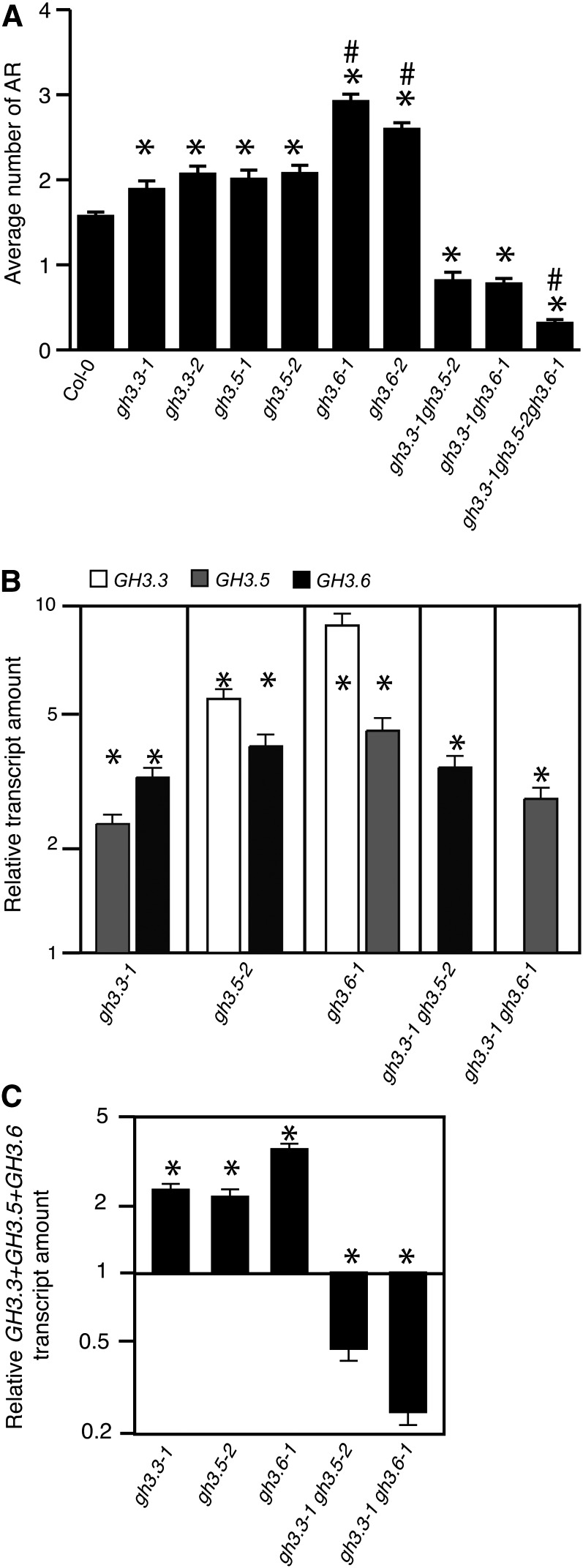
In order to assess the potential contributions of the three GH3 genes to the regulation of adventitious rooting, we analyzed adventitious root formation in gh3.3-1, gh3.3-2, gh3.5-1, gh3.5-2, gh3.6-1, and gh3.6-2 single knockout mutant alleles, in gh3.3-1gh3.5-2 and gh3.3-1gh3.6-1 double mutants, and in a gh3.3-1gh3.5-2gh3.6-1 triple mutant grown under conditions described previously (Sorin et al., 2005; Gutierrez et al., 2009) (Figure 2A). We confirmed that none of the single, double, or triple gh3 mutants showed a defect in primary root elongation or secondary root development when grown in our conditions (see Supplemental Figure 4 online).
Figure 2.
GH3.3, GH3.5, and GH3.6 Genes Have Redundant Functions in the Regulation of Adventitious Rooting.
(A) Average number of adventitious roots (AR) in two independent knockout alleles for gh3.3, gh3.5, and gh3.6 single mutants, in the double mutants gh3.3-1gh3.5-2 and gh3.3-1gh3.6-1, and the triple mutant gh3.3-1gh3.5-2gh3.6-1. Seedlings were first etiolated in the dark, until their hypocotyls were 6 mm long, and then transferred to the light for 7 d. Data from three independent biological replicates, each of at least 30 seedlings, were pooled and averaged. Error bars indicate se. A one-way analysis of variance combined with the Tukey’s multiple comparison posttest indicated that the differences observed in the mutants versus the wild type (*), in gh3.6 versus gh3.3 or gh3.5 (#), and in the triple mutant versus the double mutants (#) are significant (P < 0.01; n > 90). Col-0, ecotype Columbia.
(B) The steady state expression levels of GH3.3, GH3.5, and GH3.6 were quantified by quantitative RT-PCR in the hypocotyls of the different gh3 single and double mutants first etiolated in the dark, until their hypocotyls were 6 mm long, and then transferred to the light for 72 h.
(C) The relative amount of the total GH3.3+GH3.5+GH3.6 mRNAs in the hypocotyl of single and double gh3 mutants grown as in (B) was quantified by quantitative RT-PCR.
For (B) and (C) Gene expression values are relative to the expression in the wild type, for which the value is set to 1. Error bars indicate se obtained from three independent biological replicates. A one-way analysis of variance combined with the Dunnett’s comparison posttest confirmed that the differences between the wild type and the mutants (*) are significant (P < 0.001, n = 3).
Interestingly, all single mutants produced significantly more adventitious roots than did the wild type. This was unexpected, given that the results presented above, together with our previous data (Sorin et al., 2006), indicated a positive correlation between the number of adventitious roots and the GH3 transcript or GH3 protein content. This phenotype was not explained by an increased endogenous auxin content since, despite the fact that these three GH3 proteins were shown to conjugate IAA (Staswick et al., 2005), no modification of the endogenous free IAA content was observed in the single mutants (see Supplemental Figure 5 online). In contrast to the single mutants, the gh3.3-1gh3.5-2 and gh3.3-1gh3.6-1 double mutants developed only half the number of adventitious roots compared with the wild type. Also, the gh3.3-1gh3.5-2gh3.6-1 triple mutant produced significantly fewer adventitious roots than the double mutants (Figure 2A), showing the additive effect of the mutations and suggesting a redundant function for GH3.3, GH3.5, and GH3.6 in the regulation of adventitious rooting in Arabidopsis. In this particular context, this result was also contradictory with a role in IAA conjugation and a potential increase of free IAA, since one would expect to see the number of adventitious roots increasing and not decreasing in the double and triple mutants. This did not explain why the single mutants produced more adventitious roots than the wild type. Because the three GH3 genes were strictly coregulated in the different arf mutants and ARF-overexpressing lines, we performed quantitative RT-PCR analysis of the relative transcript amounts of these GH3 genes in the hypocotyl of each single and double gh3 mutant (Figure 2A). Interestingly, the expression of GH3.5 and GH3.6 was upregulated in gh3.3-1 mutant seedlings, whereas that of GH3.3 and GH3.6 was upregulated in gh3.5-2 mutant seedlings, and that of GH3.3 and GH3.5 was upregulated in gh3.6-1 mutant seedlings (Figure 2B). This compensatory effect led to a higher level of total GH3.5+GH3.6, GH3.3+GH3.6, and GH3.3+GH3.5 transcripts in the single mutants than the total GH3.3+GH3.5+GH3.6 transcript amount in the wild type (Figure 2C), which was likely to explain the increased average number of adventitious roots in the single mutants. The GH3.6 and GH3.5 transcripts also accumulated to higher than wild-type levels in the gh3.3-1gh3.5-2 and gh3.3-1gh3.6-1 double mutants, respectively, but this accumulation was not sufficient to be equivalent to the total GH3.3+GH3.5+GH3.6 transcript amount in the wild type. This likely explains the reduced number of adventitious roots produced by the gh3.3-1gh3.5-2 and gh3.3-1gh3.6-1 double mutants. In conclusion, these results suggest that GH3.3, GH3.5, and GH3.6 redundantly and positively regulate adventitious rooting and that this role is independent from their activity as IAA-amido synthetases.
GH3.3, GH3.5, and GH3.6 Are Likely to Stimulate Adventitious Rooting by Inactivating JA
The results described above suggest that GH3.3, GH3.5, and GH3.6, which are auxin-inducible genes, do not act by regulating auxin levels but might inactivate an inhibitor of adventitious rooting. Among the three proteins, GH3.5 was shown to adenylate salicylic acid (SA) as well as IAA in vitro (Staswick et al., 2005). Therefore, it is possible that, in planta, GH3.3 and GH3.6 proteins could also conjugate SA. Salicylate, therefore, was an obvious candidate to test as a potential target for GH3 proteins. Like SA, JA is a stress-related hormone that can be conjugated by GH3 proteins. The protein product of the gene GH3.11, which is also called JASMONIC ACID RESISTANT1 (JAR1), conjugates JA to Ile, producing jasmonoyl-l-isoleucine (JA-Ile), the active signaling molecule recognized by the F-box receptor CORONATINE INSENSITIVE1 (COI1) (Fonseca et al., 2009). In addition, methyl jasmonate was shown to inhibit root growth in Arabidopsis (Staswick et al., 1992), and JA transiently accumulated at the base of Petunia hybrida stem cuttings after mechanical wounding but very rapidly returned to its basal level before adventitious root formation took place (Ahkami et al., 2009). Fattorini et al. (2009) showed that submicromolar amounts of methyl jasmonate promoted adventitious root development from thin cell layers of tobacco (Nicotiana tabacum). This information prompted us to also investigate the role of JA in adventitious root formation on Arabidopsis hypocotyls.
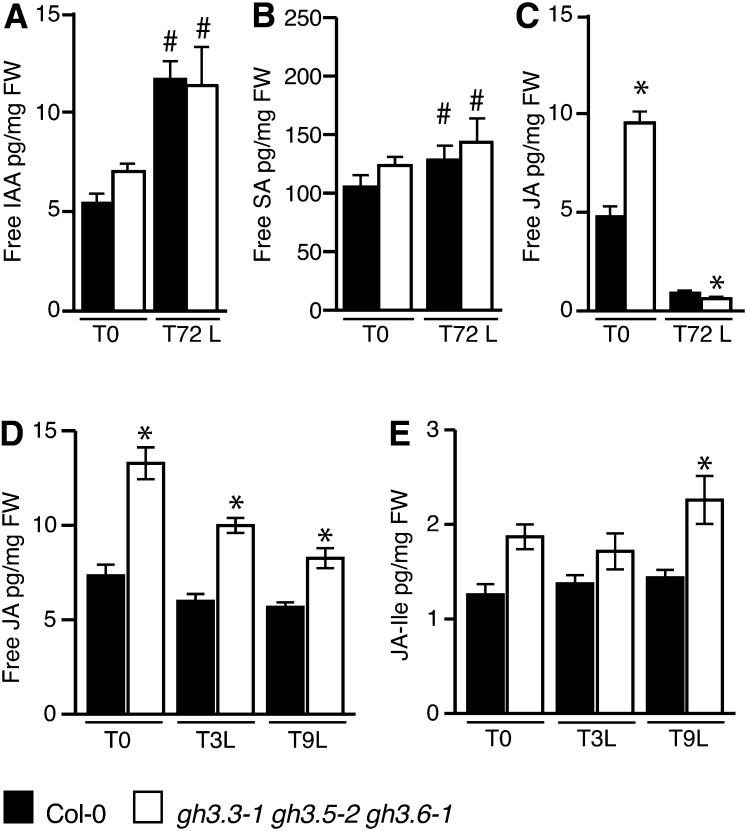
To verify whether or not the reduced number of adventitious roots observed in the gh3.3-1gh3.5-2gh3.6-1 triple mutant could be due to a modification of endogenous SA or JA content in the early stages of adventitious root formation, we quantified free IAA, free JA, and free SA in hypocotyls of wild-type seedlings and the gh3.3-1gh3.5-2gh3.6-1 triple mutant seedlings at time T0 (i.e., etiolated seedlings prior to transfer to the light) and after transfer to the light for 72 h (T72L). No significant differences in the free IAA or free SA content could be observed in the gh3.3-1gh3.5-2gh3.6-1 triple mutant compared with the wild type at T0 or T72L (Figures 3A and 3B). Both free IAA and free SA contents increased after transfer to the light in both the wild type and the gh3.3-1gh3.5-2gh3.6-1 triple mutant. In contrast, dark-grown seedlings of the gh3.3-1gh3.5-2gh3.6-1 triple mutant accumulated twice as much JA compared with the wild type (Figure 3C). Three days after transfer to the light, the JA level was significantly reduced in both wild-type seedlings and the triple mutant. This result indicates that light regulates JA homeostasis. Whether this is due to a downregulation of JA biosynthesis or an activation of JA degradation remains to be determined.
Figure 3.
GH3.3, GH3.5, and GH3.6 Regulate Adventitious Root Initiation by Modulating JA Homeostasis.
(A) to (C) Endogenous free IAA (A), free SA (B), and free JA (C) contents were measured in the hypocotyl of wild type ecotype Columbia (Col-0) and the gh3.3-1gh3.5-2gh3.6-1 triple mutant. Seedlings were first etiolated in the dark, until their hypocotyls were 6 mm long (T0), and then transferred to the light for 72 h (T72L). Error bars indicate sd of eight biological replicates. A one-way analysis of variance combined with the Bonferroni posttest indicated that the values indicated by asterisks are significantly different from wild-type Col-0 values and the values indicated by hash signs are significantly different from values observed at T0 (P < 0.05; n = 8). FW, fresh weight.
(D) and (E) Endogenous free JA (D) and JA-Ile (E) were measured in the hypocotyl of wild type Col-0 and the gh3.3-1gh3.5-2gh3.6-1 triple mutant, etiolated as in (A) (T0) or transferred to the light for 3 h (T3L) or 9 h (T9L). Error bars indicate sd of nine biological replicates. A one-way analysis of variance combined with the Bonferroni posttest indicated that the values indicated by asterisks are significantly different from wild-type Col-0 values (P < 0.05; n = 9).
We had previously shown that the early events of adventitious root initiation, such as the expression of the cell cycle gene CYCLIN B1, take place sometime between T0 and 48 h after transfer to the light (Sorin et al., 2005). Therefore, we tested whether GH3.3, GH3.5, and GH3.6 could play a role in the regulation of JA homeostasis by quantifying free JA and the active conjugate JA-Ile in the hypocotyl of wild type and gh3.3-1gh3.5-2gh3.6-1 triple mutant seedlings at T0 and soon after transfer to the light (Figures 3D and 3E). JA content was significantly increased in the triple mutant compared with the wild type, whereas that of JA-Ile was also higher in the triple mutant but was only at a statistically significant level 9 h after transfer to the light. Although low, this increase in endogenous JA-Ile might explain the decreased number of adventitious roots observed in the triple gh3 mutant, since concentration as low as 1 nM exogenously applied JA reduced by 50% the average number of adventitious roots in wild-type seedlings (see Supplemental Figure 6 online).
In conclusion, the accumulation of JA in the triple gh3.3gh3.5gh3.6 mutant suggests that GH3.3, GH3.5, and GH3.6 proteins might conjugate JA to amino acids whose identities have yet to be determined, thereby modulating its level or inactivating it.
GH3.3, GH3.5, and GH3.6 Proteins May Control JA Homeostasis by Conjugating It to Amino Acids
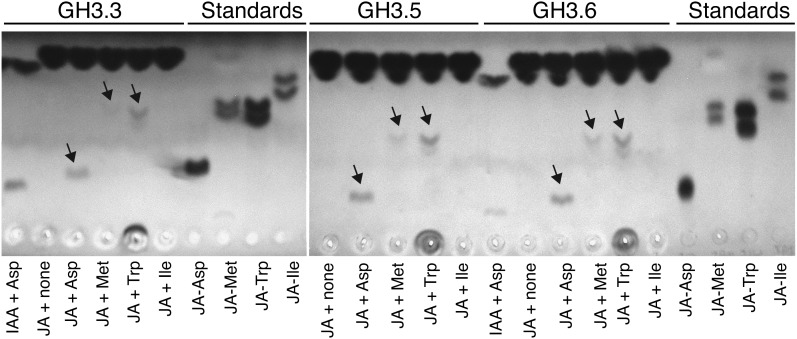
To establish whether GH3.3, GH3.5, and GH3.6 can conjugate JA, end-point assays with the respective glutathione S-transferase (GST) fusion proteins were done, and the products were analyzed by thin layer chromatography. These enzymes conjugate IAA to several amino acids in vitro, but Asp, Met, and Trp are among the better substrates and therefore were tested with JA (Staswick et al., 2005). All three enzymes produce products consistent with JA-Asp, JA-Met, and JA-Trp (Figure 4). In contrast, no detectable product was seen with Ile, in agreement with the fact that relatively little JA-Ile is produced with these enzymes. Our data show that GH3.3, GH3.5, and GH3.6 are able to conjugate JA to amino acids; therefore, the accumulation of JA in the gh3.3-1gh3.5-2gh3.6-1 triple mutant might be explained by a reduction of the conjugation process.
Figure 4.
GH3.3, GH3.5, and GH3.6 Can Conjugate JA to Amino Acids in Vitro.
Thin layer chromatography analysis of JA-amino acid conjugates synthesized by recombinant GST:GH3.3, GST:GH3.5, and GST:GH3.6 proteins. Substrates added to each enzyme reaction mixture are indicated below the image. Concentrations for JA and IAA were 10 and 1 mM, respectively. JA conjugate standards, shown to the right, migrated with somewhat greater mobility because they were loaded on the plate in organic solvent rather than the enzyme reaction mixture. Free IAA and JA migrated to near the top of the plate.
Nevertheless, because an accumulation of JA could also be due to an upregulation of the biosynthesis pathway, we checked the expression of genes encoding the main enzymes involved in JA biosynthesis (see Supplemental Figure 7 online). All genes tested were upregulated in the triple mutant compared with the wild type. JA accumulation in the triple mutant could be due to a combination of increased biosynthesis and reduced conjugation.
Jasmonate Regulates Adventitious Rooting through the COI1 Signaling Pathway
The higher level of JA-Ile that accompanied the increase of JA might activate the COI1 signaling pathway and thereby inhibit the early stages of adventitious root initiation. To confirm that the JA signaling pathway was upregulated, we analyzed the expression of JAR1, JAZ1, JAZ3, JAZ5, and COI1 genes in the hypocotyl of the wild type and the triple gh3.3-1gh3.5-2gh3.6-1 mutant. JAR1 was upregulated in the hypocotyl of the gh3.3-1gh3.5-2gh3.6-1 mutant compared with wild-type seedlings after they had been transferred to the light for 3 and 9 h (Figure 5A). The relative transcript amounts of the three JA-inducible genes JAZ1, JAZ3, and JAZ5 were upregulated in the hypocotyl of the triple mutant whether it was transferred to the light or not (Figures 5B to 5D), and last, the relative transcript amount of COI1 was upregulated in the triple mutant hypocotyl 9 h after transfer to the light (Figure 5E). We also looked at the expression pattern of promJAZ1:GUS in wild-type seedlings grown either in the dark or transferred to the light for 24, 48, or 72 h (see Supplemental Figure 8 online). promJAZ1:GUS was expressed in the hypocotyl and root of seedlings either kept in the dark or transferred to the light. In seedlings transferred to the light for 72 h, the expression was restricted to the stele. No staining was observed in the adventitious root primordia (see Supplemental Figure 8C online). This expression pattern overlapped with that of promARF6:GUS, promARF8:GUS, and promARF17:GUS, which we have described previously (Gutierrez et al., 2009).
Figure 5.
The COI1 Signaling Pathway Is Required for the Inhibition of Adventitious Rooting by Jasmonate.
(A) to (E) Quantification by quantitative RT-PCR of JAR1 (GH3.11) JAZ1, JAZ3, JAZ5, and COI1 transcripts in hypocotyls of wild-type etiolated seedlings (T0), after an additional 3 or 9 h in the dark (T3D or T9D), and after transfer to the light for 3 or 9 h (T3L or T9L). Values are relative to the expression level of APT1, which was used as a reference gene as described in Methods. Error bars indicate se obtained from three independent biological replicates. A one-way analysis of variance combined with the Tukey’s multiple comparison posttest confirmed that the differences between the wild type and the mutants (*) are significant (P < 0.05, n = 3). Col-0, ecotype Columbia.
(F) and (G) Average number of adventitious roots (AR) in several mutants altered in jasmonate biosynthesis or signaling (F) and in the SA-deficient mutant lines eds5-1 and eds5-2 (G). Data from three independent biological replicates, each of at least 30 seedlings, were pooled and averaged. Error bars indicate se. A one-way analysis of variance combined with the Tukey’s multiple comparison test showed that the values indicated by asterisks are significantly different from wild-type values (P < 0.01; n > 90); the Bonferroni posttest indicates that the value indicated by the plus sign is significantly different from that of the myc2 mutant and that indicated by the hash mark is significantly different from that of jar1-12 (P < 0.01; n > 90).
(H) Average number of adventitious roots in single arf6-3 and arf8-7 mutants and the ARF17-OX line and in the corresponding double mutants with coi1-16. Data from three independent biological replicates, each of at least 30 seedlings, were pooled and averaged. Error bars indicate se. A one-way analysis of variance combined with the Dunnett’s multiple comparison test showed that the values indicated by asterisks are significantly different from wild-type values (P < 0.05; n > 90).
To further confirm that JA and the COI1 signaling pathways were negatively regulating adventitious root development, we analyzed the phenotypes of several mutants affected in JA biosynthesis and signaling (Figure 5F). We analyzed two mutants altered in JA biosynthesis, opr3/dde1 (Sanders et al., 2000; Stintzi and Browse, 2000) and dde2-2 (von Malek et al., 2002). Among the T-DNA insertion lines publicly available, we identified a new knockout mutant allele in the GH3.11/JAR1 gene that we called jar1-12 and a GH3.11/JAR1-overexpressing line, JAR1-OX. We also analyzed the coi1-16 allele (Ellis and Turner, 2002) altered in the expression of COI1, the jai1/myc2 mutant (Lorenzo et al., 2004) altered in the expression of MYC2, a bHLH transcription factor that is a direct target of JAZ repressors and part of the core signaling module in the jasmonate signaling pathway (Chini et al., 2007), a 35S:MYC2 line, the triple mutant myc2myc3myc4 that is as impaired as coi1-1 in the activation of several, but not all, JA-mediated responses (Fernández-Calvo et al., 2011), and last, the JA-insensitive mutant jai3-1 altered in the expression of JAZ3 (Chini et al., 2007). These mutants were characterized under the same conditions used previously (Figure 2A). Both opr3/dde1 and dde2-2, which are affected in JA biosynthesis, developed more adventitious roots than the wild type, confirming a negative effect of JA on adventitious root development (Figure 5F). Interestingly, two SA-deficient mutants, eds5-1 and eds5-2 (Rogers and Ausubel, 1997), that we also analyzed produced fewer adventitious roots than did the wild type (Figure 5G), suggesting that SA is possibly a positive regulator of adventitious rooting. The jasmonate-insensitive jai3-1 mutant did not show any difference in the average number of adventitious roots compared with the wild type, whereas lines in which the JA signaling pathway was upregulated (JAR1-OX and 35S:MYC2) had significantly fewer adventitious roots than the wild type, and those lines with a downregulated JA signaling pathway (jar1-12, coi1-16, myc2, and myc2myc3myc4) all had more adventitious roots than the wild type (Figure 5F). Noticeably, coi1-16 and the myc2myc3myc4 triple mutant developed the highest average number of adventitious roots, confirming that the COI1 signaling pathway plays a role in the control of adventitious rooting.
The upregulated JA signaling pathway in the hypocotyl of the gh3.3-1gh3.5-2gh3.6-1 mutant explains the reduced number of adventitious roots, since the double mutants arf6-3coi1-16, arf8-7coi1-16, and coi1-16ARF17-OX developed more adventitious roots than the wild type, whereas the single arf mutants and the ARF17-OX line had fewer (Figure 5H). This last result confirms that JA negatively regulates adventitious root initiation through COI1, which acts downstream of the ARF/GH3 module.
DISCUSSION
In a previous study, we showed that adventitious rooting on the Arabidopsis hypocotyl is controlled by a complex interaction between three transcription factors from the ARF family, ARF6, ARF8, and ARF17, and the regulatory microRNAs miR167 and miR160. ARF6 and ARF8 are positive regulators of adventitious rooting, whereas ARF17 is a negative regulator (Gutierrez et al., 2009). In addition, the levels of the proteins encoded by three auxin-responsive genes from the GH3 family (GH3.3, GH3.5, and GH3.6) were positively correlated with the number of adventitious roots, suggesting a role for these genes in the regulation of adventitious rooting (Sorin et al., 2006).
The GH3 family is part of the broader acyl-adenylate/thioester-forming enzyme family, also called the firefly luciferase family (Staswick et al., 2002). In Arabidopsis, there are 19 members that fall into three distinct phylogenetic groups (Staswick et al., 2002, 2005). In group I, GH3.10/DFL2 was described as being involved in red light-specific hypocotyl elongation (Takase et al., 2004), and GH3.11/JAR1 was first shown to be required for the root growth inhibition response to methyl jasmonate (Staswick et al., 1992) and later to catalyze the formation of JA-Ile, the bioactive form of JA (Staswick and Tiryaki, 2004). In addition to GH3.3, GH3.5, and GH3.6/DFL1, group II of the GH3 family comprises five additional members that catalyze the formation of auxin-amino acid conjugates (Staswick et al., 2005). And last, group III is so far the least well characterized. It is composed of nine proteins, one of which (GH3.12/PBS3) has recently been shown to conjugate amino acids to 4-substituted benzoates (Okrent et al., 2009). We analyzed the relative transcript amount of all GH3 genes and a few additional auxin-responsive genes, and with the exception of GH3.3, GH3.5, and GH3.6/DFL1, it was not correlated with the adventitious root phenotype of the arf mutants and ARF-overexpressing lines. Transcripts were either not detected in Arabidopsis hypocotyls or their level was either not affected or significantly increased in the different arf mutants or ARF-overexpressing lines. We could then conclude that overall auxin signaling was not affected in the arf mutants and ARF-overexpressing lines and that GH3.3, GH3.5, and GH3.6 are the only GH3 genes out of 19 that are strictly coregulated in the Arabidopsis hypocotyl during adventitious rooting. Furthermore, a decrease or increase in the GH3 transcript level leads to a lower or higher average number of adventitious roots, respectively, confirming the positive correlation we described previously (Sorin et al., 2006).
Exogenous auxin is often used to induce adventitious rooting on stem cuttings of different species (de Klerk et al., 1999; Geiss et al., 2009; Li et al., 2009). In Arabidopsis, mutants overproducing auxin, such as superroot1, superroot2, or yucca1, spontaneously develop numerous adventitious roots on the hypocotyl and, in the case of superroot1, on cuttings of different organs (Boerjan et al., 1995; Delarue et al., 1998; Zhao et al., 2001). Nevertheless, in this work, we show that the variation in the average number of adventitious roots developing on the hypocotyl of representative arf mutants, ARF-overexpressing lines, and the gh3.3, gh3.5, and gh3.6 single and triple mutants does not correlate with a change in the free IAA content. Rather, the data are consistent with our previous results showing that the number of adventitious roots is correlated to the level of GH3.3, GH3.5, and GH3.6 proteins (Sorin et al., 2005, 2006). This was unexpected, given the evidence that these GH3s inactivate IAA by conjugating it to amino acids (Staswick et al., 2005). Consistent with this function, the levels of IAGlu and IAAsp were reduced in the single gh3.3-1, gh3.5-2, and gh3.6-1 mutant hypocotyls compared with the wild type. However, this did not increase the endogenous level of free IAA. Surprisingly, in each single mutant, the other two GH3 genes were upregulated, so one might actually have predicted a reduction of free IAA and a reduced number of adventitious roots, which did not occur. Together, these results suggest that these three GH3 proteins are not directly regulating IAA homeostasis in hypocotyls in a way that controls adventitious rooting.
The single gh3 mutants developed more adventitious roots than the wild type likely because of the upregulation of the other two remaining GH3 genes, which resulted in a total amount of the three GH3 transcripts that was higher in the single mutants than the total amount in the wild type. In contrast, the double knockout mutants (gh3.3-1gh3.5-2 and gh3.3-1gh3.6-1) developed fewer adventitious roots compared with the wild type, and the gh3.3-1gh3.5-2gh3.6-1 triple mutant developed even fewer than the double mutants. This additive effect indicates that the GH3.3, GH3.5, and GH3.6 genes have redundant functions, but none of them alone can fully compensate for the lack of expression of the other two.
These results indicate that ARF6, ARF8, and ARF17 transcription regulators and their downstream GH3 targets do not control adventitious rooting by modulating free IAA content. Rather, GH3.3, GH3.5, and GH3.6 appear to be promiscuous enzymes that affect the homeostasis of one or more other compounds that negatively regulate adventitious rooting.
Because GH3.5 was described as acting at the crosstalk of auxin homeostasis and SA signaling (Zhang et al., 2007, 2008), we investigated whether SA was an inhibitor of adventitious rooting. The two mutant alleles eds5-1 and eds5-2, which are deficient in SA biosynthesis (Rogers and Ausubel, 1997), produced fewer adventitious roots than the wild type, suggesting that SA was not a negative, but likely a positive, regulator of adventitious rooting. Furthermore, free IAA and SA contents were not altered in the triple gh3 mutant compared with the wild type, either in the dark or 72 h after transfer to the light, although in both genotypes the IAA and SA contents significantly increased after transfer to the light. Thus, the phenotype of the triple gh3 mutant was not due to a modification in the level of free IAA or SA.
In contrast with SA mutants, JA-deficient mutants opr3/dde1 (Sanders et al., 2000; Stintzi and Browse, 2000) and dde2-2 (von Malek et al., 2002) produced more adventitious roots than the wild type, suggesting that jasmonate is an inhibitor of adventitious rooting. Free JA in the hypocotyl of the gh3.3-1gh3.5-2gh3.6-1 triple mutant was twice that in the wild type at T0, and after 3 and 9 h in the light, the percentage of decline in endogenous JA was greater for the triple mutant than for the wild type, but it was still significantly higher than in the wild type at 9 h. Three days after transfer to the light, the amount was markedly decreased in both the wild type and the triple mutant, in agreement with published results showing that JA biosynthesis is regulated by light (Zhai et al., 2007). One way to explain the higher JA levels in the triple gh3 mutant is if the three GH3 enzymes regulate JA homeostasis by conjugating it to amino acids, leading either to JA catabolism or its unavailability for synthesis of the JA-Ile signal by GH3.11/JAR1. All three enzymes produced JA conjugates in vitro with three of the amino acids tested in this study, but importantly, none synthesized appreciable amounts of JA-Ile. The JA concentration used in the enzyme assays here was 10-fold higher for JA than for IAA, because previous studies indicated that GH3.3 and GH3.5 were considerably less active on JA (Staswick et al., 2002). Nevertheless, the results suggest that JA could be an alternative substrate for GH3.3, GH3.5, and GH3.6, pointing to a role in controlling the level of free JA at least in hypocotyl tissues. In this study, JA-Trp was not detected in the hypocotyl of the wild type or gh3.3-1gh3.5-2gh3.6-1. JA-Trp is an inhibitor of auxin responses, and we previously found low levels in Arabidopsis (Staswick, 2009). In any case, the auxin-inhibiting activity of JA-Trp is not likely responsible for regulating adventitious rooting, because the latter is COI1 dependent whereas JA-Trp activity is COI1 independent. JA-Met and JA-Asp have not been reported to occur in Arabidopsis, but even if synthesized, they may not accumulate appreciably if they are further metabolized, as is the case for IAAsp.
We also observed that the expression of genes encoding enzymes involved in JA biosynthesis was upregulated in the triple mutant, suggesting that the increase of JA could be due to an upregulation of its biosynthesis. We cannot at present discriminate between these two hypotheses, and indeed, the increase in JA might result from a combination of decreased conjugation and increased biosynthesis in the triple gh3 mutant. The elevated JA content was accompanied by a small increase in JA-Ile at 9 h after transfer to the light, and this coincided with the upregulation of the GH3.11/JAR1 transcript. Several JA biosynthesis genes are well established to be transcriptionally upregulated by JA-Ile/COI1 signaling, so reduced conjugation of JA to other amino acids in the triple mutant might free up more JA for JA-Ile synthesis, which in turn could enhance JA production. This is consistent with a model that includes JA-Ile signaling as a negative regulator of adventitious rooting.
JA-Ile promotes the interaction of the F-box receptor protein COI1 with the JAZ coreceptors, which also function as transcriptional repressors of jasmonate response genes (Thines et al., 2007). JAZ proteins are then ubiquitinated, targeting them for degradation by the 26S proteasome, followed by the activation of associated JA-dependent responses (Chini et al., 2007; Yan et al., 2007; Chung et al., 2008). Further evidence that JA-Ile/COI1 signaling negatively regulates adventitious rooting is the finding that, with the exception of the jasmonate-insensitive jai3-1 mutant, all of the tested downregulation mutants in this pathway (jar1-12, coi1-16, myc2, and myc2myc3myc4) developed significantly more adventitious roots than the wild type, while the upregulation lines (JAR1-OX and 35S:MYC2) were deficient in root production. The lack of a phenotype in jai3-1 could be explained if this particular JAZ protein (JAZ3) is not involved in adventitious rooting or if other JAZ proteins are redundant for its activity. The JAZ1, JAZ3, and JAZ5 transcripts were elevated in the gh3.3-1gh3.5-2gh3.6-1 triple mutant kept in the dark or transferred to the light, indicating that jasmonate signaling was indeed upregulated in the hypocotyl at this time. The COI1 transcript was also higher in the hypocotyl of the triple mutant 9 h after transfer to the light. The increase in adventitious roots in the double mutants coi1-16arf6-7, coi1-16ar8-7, and coi1-16ARF17-OX, in contrast to the reduced rooting in the single arf6-3, arf8-7, and ARF17-OX lines, supports the idea that the COI1-dependent jasmonate signaling pathway acts downstream of the regulatory module composed of three ARFs (ARF6, ARF8, and ARF17) and the three GH3s they regulate (GH3.3, GH3.5, and GH3.6).
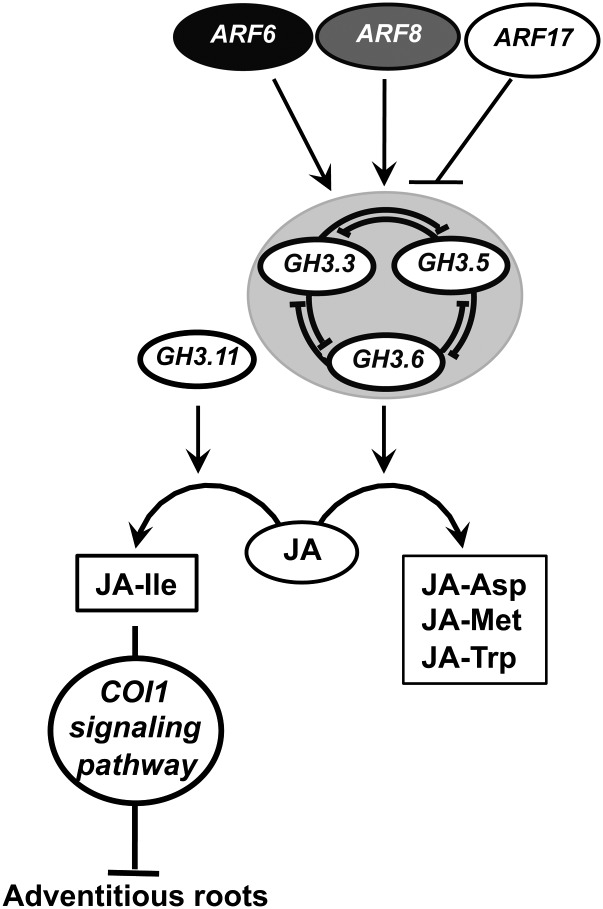
In conclusion, our data strongly support the hypothesis that the JA-Ile/COI1 signaling pathway negatively regulates adventitious rooting in Arabidopsis hypocotyls. We propose that auxin stimulates adventitious rooting by inducing GH3.3, GH3.5, and GH3.6 gene expression, via the positive regulators ARF6 and ARF8, leading to an increase in JA conjugation and, as a consequence, a reduction in free JA level. These interactions can be added to our previous model that integrated the interactions between the three ARFs and their regulatory microRNAs (Gutierrez et al., 2009) and are summarized in the model presented in Figure 6. ARF6 and ARF8 have previously been described as positive regulators of JA biosynthesis during flower development (Nagpal et al., 2005; Tabata et al., 2010); here, we show that they are likely to regulate the conjugation of jasmonate to amino acids. Our results highlight a regulatory pathway at the crosstalk of IAA and JA, in which ARF6, ARF8, and ARF17 and their downstream targets GH3.3, GH3.5, and GH3.6 are involved, and demonstrate that JA homeostasis is under auxin control.
Figure 6.
Adventitious Root Initiation: A Proposed Model for a Regulatory Pathway.
ARF6, ARF8, and ARF17 and their regulatory microRNAs interact in a complex network (Gutierrez et al., 2009) and act upstream of GH3.3, GH3.5, and GH3.6. ARF6 and ARF8 are positive regulators of the three GH3s, whereas ARF17 is a negative regulator. GH3.3, GH3.5, and GH3.6 control each other’s expression through a pathway yet to be identified. The three GH3 genes control JA homeostasis. The JA-Ile level in the hypocotyl is determined by the JA level. The level of JA-Ile negatively regulates adventitious rooting through the activation of the COI1 signaling pathway. In conclusion, auxin controls adventitious root initiation through the activation of ARF6 and ARF8 proteins, leading to a downregulation of the inhibiting COI1 signaling pathway.
METHODS
Plant Material and Growth Conditions
The ARF6-OX, ARF8-OX, and ARF17-OX1 lines have been described previously (Nagpal et al., 2005). Line MIR160c-OX, which overexpresses the gene MIR160c, was described by Wang et al. (2005). The knockout mutants arf8-7 and arf6-3 were described by Gutierrez et al. (2009). The SA-deficient mutant lines eds5-1 and eds5-2 were obtained from Frederick Ausubel. Seeds from the two JA-deficient mutants dde2-2 and opr3 were gifts from Beate Keller and John Browse, respectively. Seeds from coi1-16, jai3-1, myc2, 35S:MYC2, and the myc2myc3myc4 triple mutant were gifts from Laurens Pauwels.
Insertion lines for the GH3.3, GH3.5, GH3.6, and GH3.11/JAR1 genes were obtained from the Nottingham Arabidopsis Stock Centre. The presence of each insertion was verified using the gene-specific primers listed in Supplemental Table 1 online. The absence or the overexpression of a full-length mRNA was verified for each line by RT-PCR using primers for the coding region (see Supplemental Table 2 online). We identified two gh3.3 mutant alleles, gh3.3-1 (SM_3-39271) and gh3.3-2 (SM_3-17798); two gh3.5 mutant alleles, gh3.5-1 (SALK_014376) and gh3.5-2 (SALK_151766), which was also described by Zhang et al. (2007); two gh3.6 mutant alleles, gh3.6-1 (SALK_133707) and gh3.6-2 (SALK_060813); one gh3.11 mutant allele (SALK_011510) that we called jar1-12 according to Suza and Staswick (2008); and a GH3.11-overexpressing line, JAR1-OX (SALK_013425).
For generating double mutants with arf6-3, arf8-7, and ARF17-OX1 lines, the coi1-16 allele described by Ellis and Turner (2002) was used. In the F2 population, homozygous arf6-3, arf8-7, and ARF17-OX1 were identified by PCR using the genotyping primers listed in Supplemental Table 1 online. To genotype the coi1-16 point mutation, new derived cleaved-amplified polymorphic sequence primers were designed using the dCAPS Finder 2.0 software (Neff et al., 2002; http://helix.wustl.edu/dcaps/dcaps.html). A mismatch (underlined) was introduced in the F primer to incorporate a restriction site in the PCR product of one allele (see Supplemental Table 1 online). After amplification, the PCR products were digested with XbaI (Fermentas Fast Digest) following the manufacturer’s recommendations and separated on a 4% agarose gel. The wild-type allele yielded two fragments of 146 and 19 bp, while coi1-16 gave one band of 165 bp.
Adventitious rooting experiments were performed as described previously (Gutierrez et al., 2009).
Expression Profiling of GH3 Genes
A 1.5-kb-long fragment upstream from the start codon of GH3.3, GH3.5, and GH3.6 was amplified from genomic DNA by PCR, cloned using a pENTR/D-TOPO Cloning Kit (Invitrogen), and transferred into the pKGWFS7 binary vector (Karimi et al., 2002) using a Gateway LR Clonase Enzyme Mix (Invitrogen) according to the manufacturer’s instructions. Transgenic Arabidopsis thaliana plants expressing the different promGH3:GUS fusions were produced, and the expression pattern was checked in the T2 progeny of 10 to 15 independent transgenic lines. One representative homozygous line for each gene was used for further characterization. Histochemical assays for GUS expression were performed as described previously (Sorin et al., 2005).
RNA Isolation and cDNA Synthesis
RNAs from arf and gh3 mutants and ARF-overexpressing lines were prepared as described by Gutierrez et al. (2009). Portions (5 μg) of the resulting RNA preparations were treated with DNaseI using a DNAfree Kit (Ambion), and cDNA was synthesized by reverse transcribing 5 μg of total RNA using SuperScript III reverse transcriptase (Invitrogen) with 2.5 μg of random hexamers and 500 ng of oligo(dT)12 primer according to the manufacturer’s instructions. The reaction was stopped by incubation at 70°C for 10 min; the reaction mixture was then treated with RNaseH (Invitrogen) according to the manufacturer’s instructions and diluted by adding 1 mL of distilled water. All cDNA samples were tested by PCR using specific primers flanking an intron sequence to confirm the absence of genomic DNA contamination.
Real-Time RT-PCR Experiments
Transcript levels for Figures 1 and 2 were assessed by quantitative RT-PCR as described by Gutierrez et al. (2009). Transcript levels for Supplemental Figures 2 and 3 online were assessed by quantitative RT-PCR, in assays with triplicate reaction mixtures (final volume, 20 μL) containing 5 μL of cDNA, 0.3 μM of both forward and reverse primers, and 1× FastStart DNA MasterPLUS SYBR Green I (Roche), using a Roche LightCycler, and crossing threshold (CT) values for each sample were acquired with LightCycler software 3.5 (Roche) using the second derivative maximum method. Transcript levels for Figure 5 and Supplemental Figure 7 online were assessed by quantitative RT-PCR, in assays with triplicate reaction mixtures (final volume, 10 μL) containing 2 μL of cDNA, 0.5 μM of both forward and reverse primers, and 1× LightCycler 480 SYBR Green I Master (Roche) on 384-well plates. The plates were filled by an EVO150 robotic liquid handling platform (Tecan), and quantitative PCR was performed with a LightCycler 480 (Roche). The CT values for each sample were acquired with the LightCycler 480 software (Roche) using the second derivative maximum method. All quantifications were repeated with three independent biological replicates.
Steady state levels of uncleaved ARF transcripts were quantified using primers spanning the microRNA target site. The following standard protocol was applied for the amplification of each mRNA: 10 min at 95°C, followed by 40 cycles of 10 s at 95°C, 15 s at 60°C (except for GH3.5, for which the annealing temperature was 65°C), and 15 s at 72°C. The sequences of primers used for all target genes are presented in Supplemental Table 2 online. Due to the high sequence conservation, note that the primers for GH3.13 might also produce an amplicon from GH3.15 transcripts, which was not a problem in our analysis since GH3.15 transcripts are not detectable.
Each amplicon was first sequenced to ensure the specificity of the amplified sequence, and in order to check that the fluorescence signal was derived from the single intended amplicon in the following runs, a melting curve analysis was added to each PCR program.
Real-Time RT-PCR Data Analysis
APT1 and TIP41 had previously been validated as the most stably expressed genes among 11 tested in our experimental procedures (Gutierrez et al., 2009) and were used to normalize the real-time RT-PCR data. The normalized expression patterns obtained using both reference genes were similar, so only the data normalized with APT1 are shown. For Figures 1 and 2 and Supplemental Figure 3 online, CT and PCR efficiency (E) values were used to calculate expression using the formula ET(CTWT-CTM)/ER(CTWT-CTM), where T is the target gene and R is the reference gene, CT is the crossing threshold value, M refers to cDNA from the mutant line, and WT refers to cDNA from the wild type. In these figures, the data for mutants are presented as relative to the wild type, the calibrator. For Figure 5 and Supplemental Figures 2 and 7 online, the expression in the wild type and/or mutants was calculated using the formula ERCTWT/ETCTWT [i.e., (1/ETCTWT)/(1/ERCTWT): the normalized relative quantity of template in the original sample], the expression levels of target genes being relative to those of the reference gene. All real-time RT-PCR results presented are means from three independent biological replicates. For each independent biological replicate, the relative transcript amount was calculated as the mean of three technical replicates, using the method for calculation of se values in relative quantification recommended by Rieu and Powers (2009).
IAA and IAA Conjugate Analysis
Seedlings from each genotype were grown in vitro as described above. Seventy-two hours after transfer to the light, cotyledons and roots were removed, and an average of 150 hypocotyls (50 to 70 mg) per replicate were harvested, dried on tissue paper, and frozen in liquid nitrogen. Up to nine replicates from three independent experiments were harvested. Samples were extracted, purified, and analyzed by liquid chromatography–multiple reaction monitoring–mass spectrometry as described previously (Kowalczyk and Sandberg, 2001).
Free IAA, JA, JA-Ile, and SA Quantification
Hypocotyls from the wild type and triple gh3 mutants were collected as described above. JA, JA-Ile, SA, and IAA extraction, purification, and quantification were performed according to Bergougnoux et al. (2009), with modifications. To each extract, 20 pmol of [2H6]JA and [2H2]JA-Ile (Olchemim) and 100 pmol of [2H4]SA and [13C6]IAA (Cambridge Isotope Laboratories) were added as internal standards to check recovery during purification and to validate the quantification. Purified samples were analyzed by the liquid chromatography–tandem mass spectrometry system consisting of an ACQUITY UPLC System (Waters) and a Xevo TQ (Waters) triple quadrupole mass spectrometer. Samples were dissolved in 15% acetonitrile, injected onto the ACQUITY UPLC BEH C18 column (100 × 2.1 mm, 1.7 µm; Waters), and eluted with a linear gradient (0 to 3 min, 15% B; 3 to 10 min, 20% B; 10 to 20 min, 30% B; flow rate of 0.25 mL/min) of 7.5 mM formic acid (A) and acetonitrile (B). Quantification was obtained using a multiple reaction monitoring mode of [M-H]+/[M-H]− and the appropriate product ion. The multiple reaction monitoring transitions 215.1>59, 326.2>151.1, 141.1>97, and 182.1>136.1 were used for labeling, and 209.1>59, 324.2>151.1, 137.1>93, and 176.1>130.1 were used for authentic JA, JA-Ile, SA, and IAA, respectively. For selective multiple reaction monitoring experiments, optimal conditions were as follows: capillary/cone voltage, 1.0 kV/25 V; source/desolvation gas temperature, 120/550°C; cone/desolvation gas, 70/1000 L/h; collision gas, 0.21 mL/min; low mass/high mass resolution, 2.8/12.5; collision energy, 14 eV; ion energy 1/2, 1.0/0.7 V; entrance/exit, 2.0 V. The limits of detection (signal-to-noise ratio of 1:3) for all analytes were close to 50.0 fmol. The linear range was at least over the 5 orders of magnitude with a correlation coefficient of 0.9989 to 0.9998. For each mutant, three independent experiments were performed, each composed of three biological replicates.
Enzymatic Assays
Enzyme assays were done using GH3-GST fusion proteins produced in Escherichia coli as described previously (Staswick et al., 2005). Reactions were for 16 h at 23°C in 50 mM Tris-HCl, pH 8.6, 1 mM MgCl2, 1 mM ATP, 1 mM DTT, and 2 mM amino acid. Either IAA (1 mM) or JA (10 mM) was included in each reaction. Reactions were analyzed on silica gel 60 F260 plates developed in chloroform:ethyl acetate:formic acid (35:55:10, v/v) and then stained with vanillin reagent (6% vanillin [w/v], 1% sulfuric acid [v/v] in ethanol).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: ACX1 (At4g16760) AIR12 (At3g07390), AOC2 (At3g25770), AOS (At5g42650) APT1 (At1g27450), ARF6 (At1g30330), ARF8 (At5g37020), ARF17 (At1g77850), COI1 (At2g39940), GH3.1 (At2g14960), GH3.2 (At4g37390), GH3.3 (At2g23170), GH3.4 (At1g59500), GH3.5 (At4g27260), GH3.6 (At5g54510), GH3.7 (At1g23160), GH3.8 (At5g51470), GH3.9 (At2g47750), GH3.10 (At4g03400), GH3.11 (At2g46370), GH3.12 (At5g13320), GH3.13 (At5g13350), GH3.14 (At5g13360), GH3.15 (At5g13370), GH3.16 (At5g13380), GH3.17 (At1g28130), GH3.18 (At1g48670), GH3.19 (At1g48660), GH3.20 (At1g48690), IAA7 (At3g23050), IAA17 (At1g04250), IAA19 (At3g15540), IAA28 (At5g25890), JAZ1 (At1g19180), JAZ3 (At3g17860), JAZ5 (At1g17380), KAT2 (At2g33150), LOX2 (At3g45140), OPCL1 (At1g20510), OPR3 (At2g06050), and TIP41 (At4g34270).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Content of IAA and Its Major Conjugates in Hypocotyls of the Wild Type, arf Mutants, and ARF-Overexpressing Lines.
Supplemental Figure 2. Expression Pattern of GH3.3, GH3.5, and GH3.6.
Supplemental Figure 3. Relative Amount of the Sum of GH3.3, GH3.5, and GH3.6 mRNAs in the Single arf6-3 and arf8-7 Mutants and the arf6-3arf8-7(+/−) Sesquimutant.
Supplemental Figure 4. Root Length and Lateral Root Number in gh3 Single, Double, and Triple Mutants.
Supplemental Figure 5. Content of IAA and Its Major Conjugates in Hypocotyls of the Wild Type and gh3.3-1, gh3.5-1, and gh3.6-1 Mutants.
Supplemental Figure 6. Effect of Exogenously Applied JA on Adventitious Rooting.
Supplemental Figure 7. Jasmonate Biosynthesis is Upregulated in the Triple gh3.3gh3.5gh3.6 Mutants.
Supplemental Figure 8. Expression Pattern of promJAZ1:GUS.
Supplemental Table 1. Sequences of Primers Used for Genotyping Newly Identified gh3 Mutants and Overexpressing Lines.
Supplemental Table 2. Sequences of Primers Used for Quantifying Target Genes by Real-Time RT-PCR.
Acknowledgments
We thank the Nottingham Arabidopsis Stock Centre for providing seeds. We are very grateful to Kotaro T. Yamamoto (Hokkaido University), Jason W. Reed (University of North Carolina), Xiao-Ya Chen (Shanghai Institutes for Biological Sciences), Frederick Ausubel (Harvard Medical School), Beate Keller (Institute of Plant Biology, University of Zurich), John Browse (Institute of Biological Chemistry, Washington State University), and Laurens Pauwels (Flemish Institute for Biotechnology, Department of Plant Systems Biology, Ghent University Technologiepark) for kindly providing mutant or overexpressing lines. This work was supported by the Swedish Natural Sciences Research Council, the Swedish Foundation for Strategic Research, the Swedish Research Council for Research and Innovation for Sustainable Growth, the K.&A. Wallenberg Foundation, the Carl Trygger Foundation, the University of Picardie Jules Verne, the Regional Council of Picardie, the European Regional Development Fund, and the Grant Agency of the Academy of Sciences of the Czech Republic (KAN 200380801).
AUTHOR CONTRIBUTIONS
K.F. and D.I.P. contributed equally to this work. L.G. and G.M. characterized the mutant phenotypes and performed the quantitative PCR analysis. L.G., G.M., and D.I.P. performed the GUS assays. D.I.P. produced the multiple mutants. D.I.P. and M.P. genotyped the mutants. K.F., O.N., and M.K. performed the hormone measurements. P.S. performed the enzymatic assays. H.D. produced the biological material for quantitative PCR analysis and hormone measurements. G.G. produced the prom:GUS lines. L.G. and C.B. designed the research and analyzed the data. C.B. conceptualized and supervised the overall project. C.B. wrote the article with inputs from L.G., O.N., M.K., and P.S. P.S. edited the article.
Glossary
- IAA
indole-3-acetic acid
- GUS
β-glucuronidase
- SA
salicylic acid
- JA
jasmonic acid
- CT
crossing threshold
- ANOVA
analysis of variance
Footnotes
Online version contains Web-only data.
References
- Ahkami A.H., Lischewski S., Haensch K.T., Porfirova S., Hofmann J., Rolletschek H., Melzer M., Franken P., Hause B., Druege U., Hajirezaei M.R. (2009). Molecular physiology of adventitious root formation in Petunia hybrida cuttings: involvement of wound response and primary metabolism. New Phytol. 181: 613–625 [DOI] [PubMed] [Google Scholar]
- Barlier I., Kowalczyk M., Marchant A., Ljung K., Bhalerao R., Bennett M., Sandberg G., Bellini C. (2000). The SUR2 gene of Arabidopsis thaliana encodes the cytochrome P450 CYP83B1, a modulator of auxin homeostasis. Proc. Natl. Acad. Sci. USA 97: 14819–14824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerjan W., Cervera M.T., Delarue M., Beeckman T., Dewitte W., Bellini C., Caboche M., Van Onckelen H., Van Montagu M., Inzé D. (1995). Superroot, a recessive mutation in Arabidopsis, confers auxin overproduction. Plant Cell 7: 1405–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohmert K., Camus I., Bellini C., Bouchez D., Caboche M., Benning C. (1998). AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J. 17: 170–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Westfall C.S., Hicks L.M., Wang S., Jez J.M. (2010). Kinetic basis for the conjugation of auxin by a GH3 family indole-acetic acid-amido synthetase. J. Biol. Chem. 285: 29780–29786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Fernández G., Adie B., Chico J.M., Lorenzo O., García-Casado G., López-Vidriero I., Lozano F.M., Ponce M.R., Micol J.L., Solano R. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 [DOI] [PubMed] [Google Scholar]
- Chung H.S., Koo A.J.K., Gao X.L., Jayanty S., Thines B., Jones A.D., Howe G.A. (2008). Regulation and function of Arabidopsis JASMONATE ZIM-domain genes in response to wounding and herbivory. Plant Physiol. 146: 952–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Klerk G.J., van der Krieken W., de Jong J.C. (1999). The formation of adventitious roots: New concepts, new possibilities. In Vitro Cell. Dev. Biol. Plant 35: 189–199 [Google Scholar]
- Delarue M., Prinsen E., Onckelen H.V., Caboche M., Bellini C. (1998). Sur2 mutations of Arabidopsis thaliana define a new locus involved in the control of auxin homeostasis. Plant J. 14: 603–611 [DOI] [PubMed] [Google Scholar]
- Ellis C., Turner J.G. (2002). A conditionally fertile coi1 allele indicates cross-talk between plant hormone signalling pathways in Arabidopsis thaliana seeds and young seedlings. Planta 215: 549–556 [DOI] [PubMed] [Google Scholar]
- Fattorini L., Falasca G., Kevers C., Rocca L.M., Zadra C., Altamura M.M. (2009). Adventitious rooting is enhanced by methyl jasmonate in tobacco thin cell layers. Planta 231: 155–168 [DOI] [PubMed] [Google Scholar]
- Bergougnoux V., Hlavácková V., Plotzová R., Novák O., Fellner M. (2009). The 7B-1 mutation in tomato (Solanum lycopersicum L.) confers a blue light-specific lower sensitivity to coronatine, a toxin produced by Pseudomonas syringae pv. tomato. J. Exp. Bot. 60: 1219–1230 [DOI] [PubMed] [Google Scholar]
- Fernández-Calvo P., et al. (2011). The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23: 701–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca S., Chico J.M., Solano R. (2009). The jasmonate pathway: The ligand, the receptor and the core signalling module. Curr. Opin. Plant Biol. 12: 539–547 [DOI] [PubMed] [Google Scholar]
- Geiss, G., Gutierrez, L., and Bellini, C. (2009). Adventitious root formation: New insights and perspectives. In Root Development, T. Beekman, ed. (Oxford: Wiley-Blackwell), p. 376. [Google Scholar]
- Guilfoyle T.J., Hagen G. (2007). Auxin response factors. Curr. Opin. Plant Biol. 10: 453–460 [DOI] [PubMed] [Google Scholar]
- Gutierrez L., Bussell J.D., Pacurar D.I., Schwambach J., Pacurar M., Bellini C. (2009). Phenotypic plasticity of adventitious rooting in Arabidopsis is controlled by complex regulation of AUXIN RESPONSE FACTOR transcripts and microRNA abundance. Plant Cell 21: 3119–3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen G., Kleinschmidt A., Guilfoyle T. (1984). Auxin-regulated gene-expression in intact soybean hypocotyl and excised hypocotyl sections. Planta 162: 147–153 [DOI] [PubMed] [Google Scholar]
- Inukai Y., Sakamoto T., Ueguchi-Tanaka M., Shibata Y., Gomi K., Umemura I., Hasegawa Y., Ashikari M., Kitano H., Matsuoka M. (2005). Crown rootless1, which is essential for crown root formation in rice, is a target of an AUXIN RESPONSE FACTOR in auxin signaling. Plant Cell 17: 1387–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kowalczyk M., Sandberg G. (2001). Quantitative analysis of indole-3-acetic acid metabolites in Arabidopsis. Plant Physiol. 127: 1845–1853 [PMC free article] [PubMed] [Google Scholar]
- Li S.W., Xue L., Xu S., Feng H., An L. (2009). Mediators, genes and signaling in adventitious rooting. Bot. Rev. 75: 230–247 [Google Scholar]
- Liu H.J., Wang S.F., Yu X.B., Yu J., He X.W., Zhang S.L., Shou H.X., Wu P. (2005). ARL1, a LOB-domain protein required for adventitious root formation in rice. Plant J. 43: 47–56 [DOI] [PubMed] [Google Scholar]
- Lorenzo O., Chico J.M., Sánchez-Serrano J.J., Solano R. (2004). JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16: 1938–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal P., Ellis C.M., Weber H., Ploense S.E., Barkawi L.S., Guilfoyle T.J., Hagen G., Alonso J.M., Cohen J.D., Farmer E.E., Ecker J.R., Reed J.W. (2005). Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 132: 4107–4118 [DOI] [PubMed] [Google Scholar]
- Nakazawa M., Yabe N., Ichikawa T., Yamamoto Y.Y., Yoshizumi T., Hasunuma K., Matsui M. (2001). DFL1, an auxin-responsive GH3 gene homologue, negatively regulates shoot cell elongation and lateral root formation, and positively regulates the light response of hypocotyl length. Plant J. 25: 213–221 [DOI] [PubMed] [Google Scholar]
- Neff M.M., Turk E., Kalishman M. (2002). Web-based primer design for single nucleotide polymorphism analysis. Trends Genet. 18: 613–615 [DOI] [PubMed] [Google Scholar]
- Okrent R.A., Brooks M.D., Wildermuth M.C. (2009). Arabidopsis GH3.12 (PBS3) conjugates amino acids to 4-substituted benzoates and is inhibited by salicylate. J. Biol. Chem. 284: 9742–9754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y., Fukaki H., Onoda M., Theologis A., Tasaka M. (2007). ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 19: 118–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.E., Park J.Y., Kim Y.S., Staswick P.E., Jeon J., Yun J., Kim S.Y., Kim J., Lee Y.H., Park C.M. (2007a). GH3-mediated auxin homeostasis links growth regulation with stress adaptation response in Arabidopsis. J. Biol. Chem. 282: 10036–10046 [DOI] [PubMed] [Google Scholar]
- Park J.E., Seo P.J., Lee A.K., Jung J.H., Kim Y.S., Park C.M. (2007b). An Arabidopsis GH3 gene, encoding an auxin-conjugating enzyme, mediates phytochrome B-regulated light signals in hypocotyl growth. Plant Cell Physiol. 48: 1236–1241 [DOI] [PubMed] [Google Scholar]
- Rieu I., Powers S.J. (2009). Real-time quantitative RT-PCR: design, calculations, and statistics. Plant Cell 21: 1031–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers E.E., Ausubel F.M. (1997). Arabidopsis enhanced disease susceptibility mutants exhibit enhanced susceptibility to several bacterial pathogens and alterations in PR-1 gene expression. Plant Cell 9: 305–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders P.M., Lee P.Y., Biesgen C., Boone J.D., Beals T.P., Weiler E.W., Goldberg R.B. (2000). The Arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell 12: 1041–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorin C., Bussell J.D., Camus I., Ljung K., Kowalczyk M., Geiss G., McKhann H., Garcion C., Vaucheret H., Sandberg G., Bellini C. (2005). Auxin and light control of adventitious rooting in Arabidopsis require ARGONAUTE1. Plant Cell 17: 1343–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorin C., Negroni L., Balliau T., Corti H., Jacquemot M.P., Davanture M., Sandberg G., Zivy M., Bellini C. (2006). Proteomic analysis of different mutant genotypes of Arabidopsis led to the identification of 11 proteins correlating with adventitious root development. Plant Physiol. 140: 349–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick P.E. (2009). The tryptophan conjugates of jasmonic and indole-3-acetic acids are endogenous auxin inhibitors. Plant Physiol. 150: 1310–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick P.E., Serban B., Rowe M., Tiryaki I., Maldonado M.T., Maldonado M.C., Suza W. (2005). Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17: 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick P.E., Su W., Howell S.H. (1992). Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc. Natl. Acad. Sci. USA 89: 6837–6840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick P.E., Tiryaki I. (2004). The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell 16: 2117–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick P.E., Tiryaki I., Rowe M.L. (2002). Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell 14: 1405–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stintzi A., Browse J. (2000). The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc. Natl. Acad. Sci. USA 97: 10625–10630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suza W.P., Staswick P.E. (2008). The role of JAR1 in jasmonoyl-L-isoleucine production during Arabidopsis wound response. Planta 227: 1221–1232 [DOI] [PubMed] [Google Scholar]
- Tabata R., Ikezaki M., Fujibe T., Aida M., Tian C.E., Ueno Y., Yamamoto K.T., Machida Y., Nakamura K., Ishiguro S. (2010). Arabidopsis auxin response factor6 and 8 regulate jasmonic acid biosynthesis and floral organ development via repression of class 1 KNOX genes. Plant Cell Physiol. 51: 164–175 [DOI] [PubMed] [Google Scholar]
- Takase T., Nakazawa M., Ishikawa A., Kawashima M., Ichikawa T., Takahashi N., Shimada H., Manabe K., Matsui M. (2004). ydk1-D, an auxin-responsive GH3 mutant that is involved in hypocotyl and root elongation. Plant J. 37: 471–483 [DOI] [PubMed] [Google Scholar]
- Tanaka S., Mochizuki N., Nagatani A. (2002). Expression of the AtGH3a gene, an Arabidopsis homologue of the soybean GH3 gene, is regulated by phytochrome B. Plant Cell Physiol. 43: 281–289 [DOI] [PubMed] [Google Scholar]
- Thines B., Katsir L., Melotto M., Niu Y., Mandaokar A., Liu G.H., Nomura K., He S.Y., Howe G.A., Browse J. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665 [DOI] [PubMed] [Google Scholar]
- von Malek B., van der Graaff E., Schneitz K., Keller B. (2002). The Arabidopsis male-sterile mutant dde2-2 is defective in the ALLENE OXIDE SYNTHASE gene encoding one of the key enzymes of the jasmonic acid biosynthesis pathway. Planta 216: 187–192 [DOI] [PubMed] [Google Scholar]
- Wang D., Pei K., Fu Y., Sun Z., Li S., Liu H., Tang K., Han B., Tao Y. (2007). Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa). Gene 394: 13–24 [DOI] [PubMed] [Google Scholar]
- Wang J.W., Wang L.J., Mao Y.B., Cai W.J., Xue H.W., Chen X.Y. (2005). Control of root cap formation by microRNA-targeted auxin response factors in Arabidopsis. Plant Cell 17: 2204–2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Bai Y., Shen C., Wu Y., Zhang S., Jiang D., Guilfoyle T.J., Chen M., Qi Y. (2010). Auxin-related gene families in abiotic stress response in Sorghum bicolor. Funct. Integr. Genomics 10: 533–546 [DOI] [PubMed] [Google Scholar]
- Westfall C.S., Herrmann J., Chen Q., Wang S., Jez J.M. (2010). Modulating plant hormones by enzyme action: The GH3 family of acyl acid amido synthetases. Plant Signal. Behav. 5: 1607–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmoth J.C., Wang S., Tiwari S.B., Joshi A.D., Hagen G., Guilfoyle T.J., Alonso J.M., Ecker J.R., Reed J.W. (2005). NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. Plant J. 43: 118–130 [DOI] [PubMed] [Google Scholar]
- Yan Y.X., Stolz S., Chételat A., Reymond P., Pagni M., Dubugnon L., Farmer E.E. (2007). A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 19: 2470–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Q.Z., Li C.B., Zheng W.G., Wu X.Y., Zhao J.H., Zhou G.X., Jiang H.L., Sun J.Q., Lou Y.G., Li C.Y. (2007). Phytochrome chromophore deficiency leads to overproduction of jasmonic acid and elevated expression of jasmonate-responsive genes in Arabidopsis. Plant Cell Physiol. 48: 1061–1071 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Li Q., Li Z., Staswick P.E., Wang M., Zhu Y., He Z. (2007). Dual regulation role of GH3.5 in salicylic acid and auxin signaling during Arabidopsis-Pseudomonas syringae interaction. Plant Physiol. 145: 450–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Wang M., Li Z., Li Q., He Z. (2008). Arabidopsis GH3.5 regulates salicylic acid-dependent and both NPR1-dependent and independent defense responses. Plant Signal. Behav. 3: 537–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Christensen S.K., Fankhauser C., Cashman J.R., Cohen J.D., Weigel D., Chory J. (2001). A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291: 306–309 [DOI] [PubMed] [Google Scholar]