β-Cyanoalanine synthase detoxifies cyanide, which is produced during ethylene biosynthesis. The three-dimensional structure of β-cyanoalanine synthase provides a molecular view of how this enzyme performs its protective function and reveals evolutionary changes in the active site that distinguish it from other related enzymes.
Abstract
Plants produce cyanide (CN−) during ethylene biosynthesis in the mitochondria and require β-cyanoalanine synthase (CAS) for CN− detoxification. Recent studies show that CAS is a member of the β-substituted alanine synthase (BSAS) family, which also includes the Cys biosynthesis enzyme O-acetylserine sulfhydrylase (OASS), but how the BSAS evolved distinct metabolic functions is not understood. Here we show that soybean (Glycine max) CAS and OASS form α-aminoacrylate reaction intermediates from Cys and O-acetylserine, respectively. To understand the molecular evolution of CAS and OASS in the BSAS enzyme family, the crystal structures of Gm-CAS and the Gm-CAS K95A mutant with a linked pyridoxal phosphate (PLP)-Cys molecule in the active site were determined. These structures establish a common fold for the plant BSAS family and reveal a substrate-induced conformational change that encloses the active site for catalysis. Comparison of CAS and OASS identified residues that covary in the PLP binding site. The Gm-OASS T81M, S181M, and T185S mutants altered the ratio of OASS:CAS activity but did not convert substrate preference to that of a CAS. Generation of a triple mutant Gm-OASS successfully switched reaction chemistry to that of a CAS. This study provides new molecular insight into the evolution of diverse enzyme functions across the BSAS family in plants.
INTRODUCTION
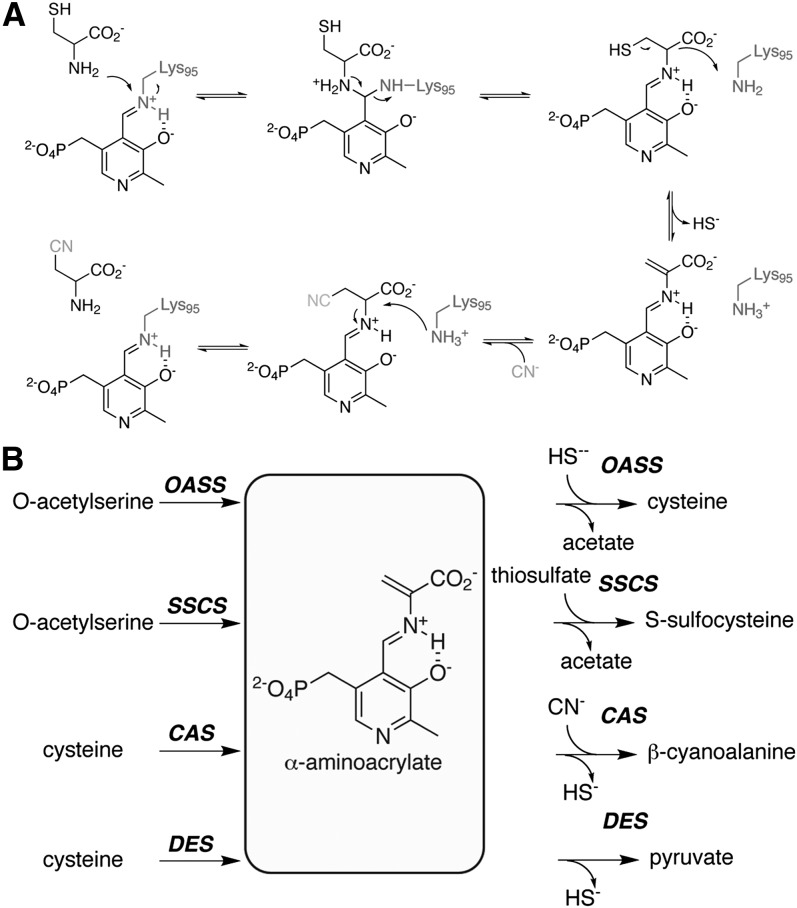
In plants, cyanide (CN−) is a naturally occurring molecule with important metabolic roles connected to plant protection. The toxicity of cyanide, through its inhibition of cytochrome c oxidase in the mitochondrial electron transport chain (Palmer, 1993), makes it a potent chemical deterrent. A variety of cyanogenic glucosides are present in many plant species, providing a chemical reservoir to release HCN in response to herbivores and pathogens that cause tissue damage. In addition, these same molecules are useful for nitrogen transport within the plant and nitrogen storage for primary metabolism (Møller, 2010). Biosynthetically, the production of cyanide in plants is directly coupled to ethylene synthesis (Bleecker and Kende, 2000). Conversion of 1-amino-cyclopropane-1-carboxylic acid to ethylene by 1-amino-cyclopropane-1-carboxylic acid oxidase results in the release of cyanoformic acid, which spontaneously decarboxylates to release CN− (Peiser et al., 1984). The generation of CN− in ethylene synthesis necessitates its detoxification and is linked to activation of β-cyanoalanine synthase (CAS; Blumenthal et al., 1968; Miller and Conn, 1980; Yip and Yang, 1988; Goudey et al., 1989).
CAS catalyzes the addition of CN− to Cys to yield β-cyanoalanine (Figure 1A), which can then be used for recycling of reduced nitrogen through metabolism to Asn (Castric et al., 1972). The activity of this enzyme is widespread in plant tissues (Wurtele et al., 1984, 1985) and localized to mitochondria (Wurtele et al., 1985; Hatzfeld et al., 2000; Maruyama et al., 2000; Warrilow and Hawkesford, 2000, 2002; Lai et al., 2009). Thus, CAS activity is induced and localized to effectively protect the electron transport chain at the site of ethylene production (Bleecker and Kende, 2000). Recent work by García et al. (2010) shows that CAS is essential for root hair development in Arabidopsis thaliana and suggests that its activity helps prevent the accumulation of HCN, which represses expression of genes encoding enzymes involved in cell wall maintenance and formation of the root hair tip.
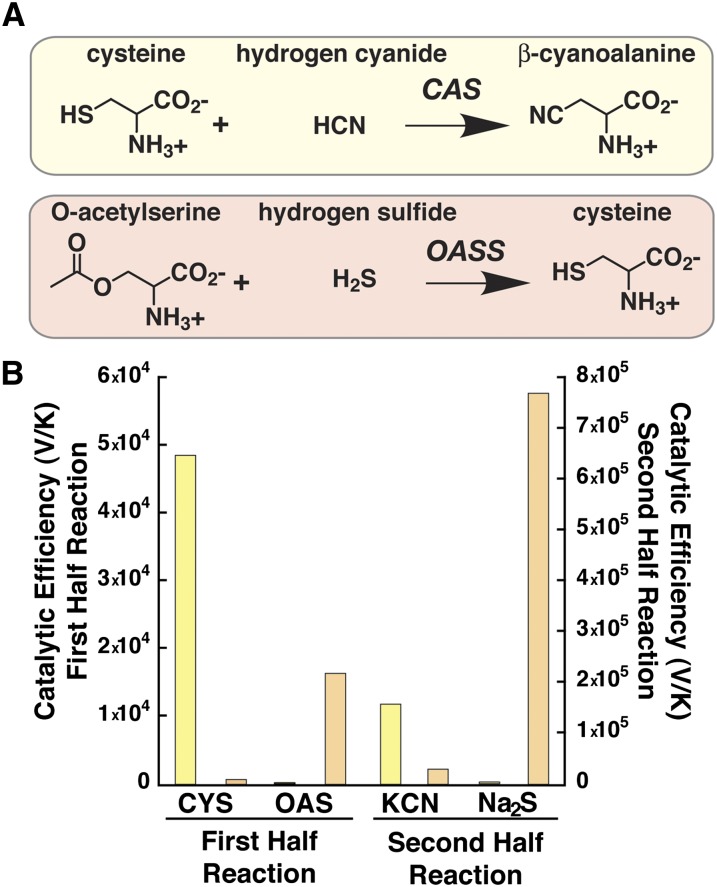
Figure 1.
Comparison of CAS and OASS Reactions and Substrate Specificity.
(A) Overall reactions catalyzed by CAS (Top) and OASS (Bottom). Note that the CAS and OASS reactions also result in the release of sulfide (HS−) and acetate, respectively, from the first half reaction.
(B) Comparison of the catalytic efficiencies (Vmax/Km) of Gm-CAS (yellow) and Gm-OASS (orange). Substrates for the first half reaction (left y axis) were Cys (CYS) and O-acetylserine (OAS). Substrates for the second half reaction (right y axis) were potassium cyanide (KCN) and sodium sulfide (Na2S). Bars plot Vmax/Km from Table 1.
Biochemically, CAS activity has been described in a variety of plant species, including blue lupine (Lupinus angustifolius) (Akopyan et al., 1975); spinach (Spinacia oleracea) (Ikegami et al., 1988b), peavine (Lathyrus latifolius) (Ikegami et al., 1988a), cassava (Manihot esculenta) (Elias et al., 1997), cocklebur (Xanthium pennsylvanicum) (Maruyama et al., 1998), potato (Solanum tuberosum) (Maruyama et al., 2000, 2001), apple (Malus domestica) (Yip and Yang, 1988), Arabidopsis (Warrilow and Hawkesford, 2000; Yamaguchi et al., 2000), and rice (Oryza sativa) (Lai et al., 2009). These same studies also identify CAS as a 50- to 70-kD dimeric pyridoxal phosphate (PLP)–dependent enzyme. Molecular cloning of CAS from Arabidopsis, spinach, and soybean (Glycine max) has established this enzyme as a member of the β-substituted alanine synthase (BSAS) family (Hatzfeld et al., 2000; Yamaguchi et al., 2000; Watanabe et al., 2008; Yi et al., 2010a, 2010b). The plant BSAS family also includes O-acetylserine sulfhydrylase (OASS), which catalyzes the synthesis of Cys from O-acetylserine and sulfide using a similar overall reaction as CAS (Figure 1A) (Bonner et al., 2005; Ravilious and Jez, 2012). Structural and functional studies of OASS indicate that the enzyme uses a two-step reaction mechanism in which O-acetylserine reacts with the PLP cofactor to yield a reactive α-aminoacrylate intermediate in the first half step. Binding of sulfide in the second step leads to reaction with the intermediate to produce Cys (Bonner et al., 2005). The kinetic mechanism reported for spinach CAS is also consistent with a similar two-step reaction for this enzyme (Warrilow and Hawkesford, 2002).
Although the metabolic role of CAS in plant CN− metabolism has long been established, little is known about how this protein functions at the molecular level. In other words, there are similarities in the overall reactions catalyzed by CAS and OASS, but it is unclear whether they use common α-aminoacrylate reaction intermediates. Moreover, as part of the BSAS enzyme family, the sequence and structural features that define physiological function and reaction specificity of various family members—OASS producing Cys and CAS using Cys as a substrate—remains to be understood. Using soybean CAS and OASS, we demonstrate that both enzymes rely on the same reaction intermediate derived from different substrates for cyanide detoxification and Cys biosynthesis. Furthermore, x-ray crystal structures of Gm-CAS provide the first views of this enzyme and reveal common structural features for the plant BSAS family. Crystallographic analysis also reveals a substrate-induced conformational change that encloses the active site for catalysis in CAS. In addition, amino acid residues that alter reaction specificity between OASS and CAS are identified. This article provides new molecular insight into the evolution of diverse enzyme functions across the BSAS family in plants.
RESULTS
Comparison of Gm-CAS and Gm-OASS Substrate Specificity and Reaction Intermediates
CAS activity in soybean has been previously reported (Goudey et al., 1989; Tittle et al., 1990). For functional comparison, Gm-CAS (Glyma09g39390; GmBSAS3;1) (Yi et al., 2010b) and Gm-OASS (Glyma11g00810; GmBSAS1;1) (Yi et al., 2010b) were expressed and purified as functional dimeric proteins, as previously described (Chronis and Krishnan, 2003; Bonner et al., 2005). Gm-CAS was expressed as a construct lacking the mitochondrial localization sequence, whereas the expressed Gm-OASS represents the full-length major cytosolic isoform of this enzyme in soybean (Chronis and Krishnan, 2003). Steady state kinetic analysis of Gm-CAS and Gm-OASS shows that each enzyme catalyzes the other's reaction, but with compromised catalytic efficiency (Vmax/Km) (Figure 1B, Table 1). Gm-CAS displays a 230-fold higher catalytic efficiency with Cys than O-acetylserine, but only a ninefold higher Vmax/Km for CN− versus sulfide (Figure 1B). By contrast, Gm-OASS prefers O-acetylserine versus Cys by 23-fold and sulfide by 270-fold over CN− (Figure 1B).
Table 1. Kinetic Comparison of Gm-CAS and Gm-OASS Substrate Specificities.
| Enzyme | CAS Reaction Substrates | OASS Reaction Substrates | ||||
|---|---|---|---|---|---|---|
| Cys |
O-acetylserine |
|||||
| kcat (s−1) | Km (mM) | kcat/Km (m−1 s−1) | kcat (s−1) | Km (mM) | kcat/Km (m−1 s−1) | |
| Gm-CAS | 38.9 ± 4.5 | 0.81 ± 0.25 | 48,024 | 1.82 ± 0.09 | 8.87 ± 1.31 | 205 |
| Gm-OASS | 0.21 ± 0.09 | 0.30 ± 0.01 | 700 | 57.5 ± 11.8 | 3.60 ± 0.40 | 15,972 |
| KCN | Na2S | |||||
| kcat (s−1) | Km (mM) | kcat/Km (m−1 s−1) | kcat (s−1) | Km (mM) | kcat/Km (m−1 s−1) | |
| Gm-CAS | 39.2 ± 0.03 | 0.26 ± 0.03 | 150,769 | 1.78 ± 0.13 | 0.11 ± 0.06 | 16,182 |
| Gm-OASS | 0.23 ± 0.03 | 0.08 ± 0.02 | 2,875 | 39.2 ± 5.7 | 0.05 ± 0.02 | 784,000 |
All values are expressed as a mean ± se (n = 3).
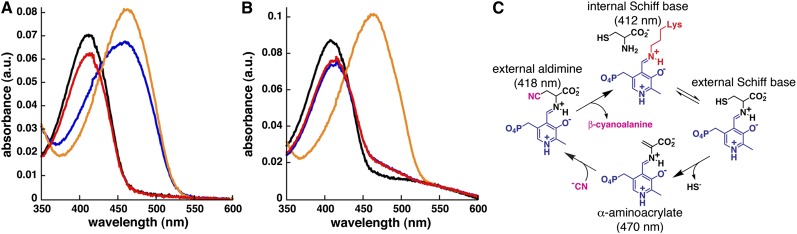
To determine whether Gm-CAS and Gm-OASS share common reaction mechanisms involving an α-aminoacrylate intermediate, each protein was incubated with Cys, O-acetylserine, and β-cyanoalanine and then analyzed by UV/visible absorption spectroscopy (Figure 2). Addition of Cys or O-acetylserine to Gm-CAS shifted the maximum absorbance (Amax) of PLP from 412 nm to 470 nm (Figure 2A), which is consistent with formation of an α-aminoacrylate intermediate from either molecule (Bonner et al., 2005). As reported with Arabidopsis OASS (Bonner et al., 2005), signal for the α-aminoacrylate intermediate (Amax = 470 nm) was observed after addition of O-acetylserine to Gm-OASS (Figure 2B). Incubation of Gm-CAS with β-cyanoalanine and Gm-OASS with either Cys or β-cyanoalanine yielded a shift to the external aldimine form of PLP (Amax = 418 nm), which is nonreactive. These experiments demonstrate that both CAS and OASS stably form a common α-aminoacrylate intermediate in their respective reactions (Figure 2C). To better understand the molecular function of CAS, we determined the x-ray crystal structure of the soybean enzyme.
Figure 2.
Spectroscopic Analysis of CAS and OASS.
(A) Absorption spectra of PLP in Gm-CAS alone (black) and after addition of 100 μM Cys (blue), 1 mM O-acetylserine (orange), or 1 mM β-cyanoalanine (red). a.u., arbitrary units.
(B) Absorption spectra of PLP in Gm-OASS alone (black) and after addition of 1 mM Cys (blue), 100 μM O-acetylserine (orange), or 1 mM β-cyanoalanine (red).
(C) Proposed catalytic cycle for CAS showing the initial Schiff base form of PLP (Amax = 412 nm), formation of the α-aminoacrylate intermediate (Amax = 470 nm) after the first half reaction, nucleophilic attack in the second half reaction leading to the external aldimine (Amax = 418 nm), and release of product.
Three-Dimensional Structure of Gm-CAS
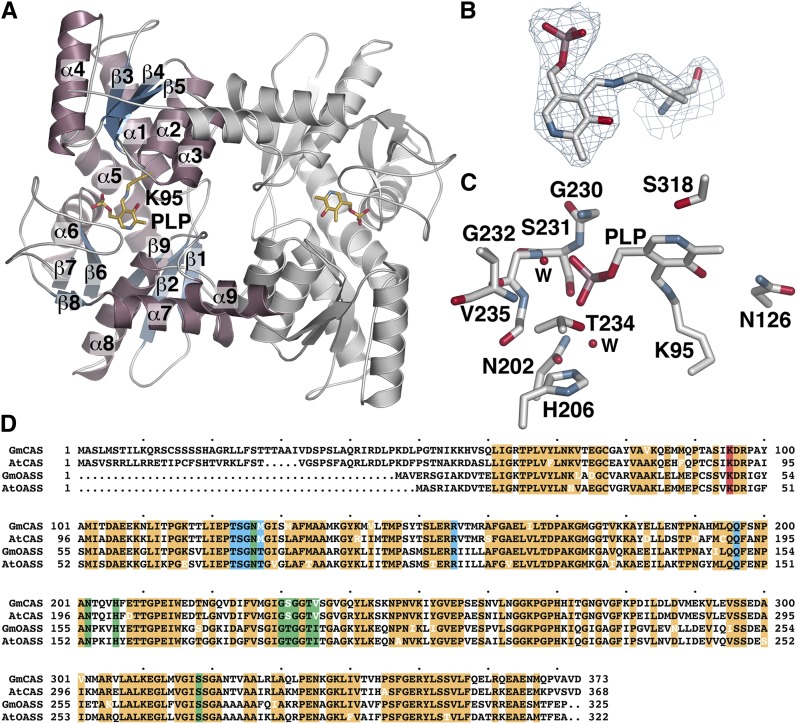
The x-ray crystal structure of Gm-CAS was determined by molecular replacement, with four molecules forming two dimers in the asymmetric unit (Table 2). Each monomer consists of two mixed α/β domains with a PLP cofactor at the active site separating the two halves (Figure 3A). The N-terminal domain (residues 94 to 197) is formed by a central β-sheet (β3 to β5) surrounded by four α-helices (α1 to α4). In between the two domains, α5 (residues 195 to 2019) spans both halves of the structure. The C-terminal domain consists of a short β-sheet (β1 to β2; residues 51 to 93) and a larger β-sheet (β6 to β8) surrounded by three α-helices (α6 to α8). An additional α-helix (α9) forms part of the dimer interface by packing against α2 and α3 of the adjoining monomer. The structure of Gm-CAS shares the same fold as Arabidopsis OASS (Bonner et al., 2005) with a 0.9 Å2 root mean square deviation for the aligned 316 Cα atoms. This common scaffold is also found in other PLP-dependent enzymes, including cystathionine β-synthase, Thr dehydratase, and Trp synthase (Meier et al., 2001; Simanshu et al., 2006; Ngo et al., 2007).
Table 2. Crystallographic Statistics.
| Crystal | Gm-CAS | Gm-CAS-K95A |
|---|---|---|
| Space group | C2 | P22121 |
| Cell dimensions | a = 190.32 Å, b = 154.58 Å, c =70.80 Å; β = 101.9° | a = 71.09 Å, b = 154.33 Å, c =198.33 Å |
| Data Collection | ||
| Wavelength (Å) | 0.979 | 0.979 |
| Resolution range (Å) (highest shell resolution) | 49.2 to 2.50 (2.54 to 2.50) | 47.2 to 1.77 (1.80 to 1.77) |
| Reflections (total/unique) | 327,779/68,674 | 805,528/201,382 |
| Completeness (highest shell) | 99.8% (100.0%) | 96.4% (98.2%) |
| <I/σ> (highest shell) | 29.8 (4.1) | 25.6 (1.9) |
| Rsyma (highest shell) | 8.5% (57.7%) | 10.8% (62.6%) |
| Model and refinement | ||
| Rcrystb / Rfreec | 16.7%/21.0% | 17.0%/20.2% |
| No. of protein atoms | 9,699 | 14,670 |
| No. of water molecules | 233 | 1,731 |
| No. of ligand atoms | 60 | 132 |
| Root mean square deviation, bond lengths (Å) | 0.008 | 0.006 |
| Root mean square deviation, bond angles (°) | 1.14 | 1.05 |
| Average B-factor (Å2) | 51.1, 46.7, 41.1 | 20.5, 33.9, 17.1 |
| Stereochemistry: most favored, allowed, generously allowed | 96.4, 3.6, 0.0% | 97.4, 2.6, 0.0% |
Rsym = Σ|Ih - <Ih>|/ΣIh, where <Ih> is the average intensity over symmetry.
Rcryst = Σ|Fo - <Fc>|/ΣFo, where summation is more than the data used for refinement.
Rfree is defined the same as Rcryst but was calculated using 5% of data excluded from refinement.
Figure 3.
Crystal Structure of Gm-CAS.
(A) Ribbon diagram showing the overall structure of the Gm-CAS dimer. The left-hand monomer shows secondary structure features with α-helices and β-strands colored pink and blue, respectively. The location of Lys-95 and PLP are shown as stick models (gold).
(B) Electron density of the PLP-Lys-95 Schiff base. A 2Fo-Fc omit map (1.5 σ) is shown.
(C) The Gm-CAS active site. Residues interacting with PLP are shown as stick models. Two water molecules are shown as red spheres marked with a “W”.
(D) Sequence comparison of soybean CAS (GmCAS), Arabidopsis CAS (AtCAS); soybean OASS (GmOASS), and Arabidopsis OASS (AtOASS). The mitochondrial localization sequence of Gm-CAS spans the first 50 amino acids. Conserved residues are highlighted in orange. Residues in white indicate a variation from the highly conserved sequence. The Schiff base Lys is highlighted in red. Residues in the PLP and amino acid binding sites are highlighted in green and blue, respectively.
The location of PLP in the structure identifies the active site. Clear electron density shows that the cofactor forms a Schiff base linkage with Lys-95, which is part of α1 (Figure 3B). Multiple interactions are formed to anchor PLP in the active site (Figure 3C; see Supplemental Figure 1A online). Asn-126 and Ser-318 form hydrogen bonds with the oxygen and nitrogen atoms, respectively, in the pyridine ring of PLP. Extensive hydrogen bond interactions with the phosphate moiety are made by the backbone nitrogens of Gly-230, Ser-231, and Gly-232 and the side chain hydroxyl groups of Ser-231 and Thr-234. Two additional contacts with the phosphate group are mediated by water molecules that hydrogen bond to the backbone nitrogen of Val-235 and the side chains of Asn-202 and His-206. Across the plant BSAS enzyme family, the PLP-attachment site corresponding to Lys-95 is invariant (Hatzfeld et al., 2000; Yamaguchi et al., 2000; Watanabe et al., 2008; Yi et al., 2010a, 2010b). In addition to this critical Lys, the residues forming the PLP binding site are highly conserved in the CAS and OASS from soybean and Arabidopsis (Figure 3D).
Ligand-Induced Conformational Change in Gm-CAS
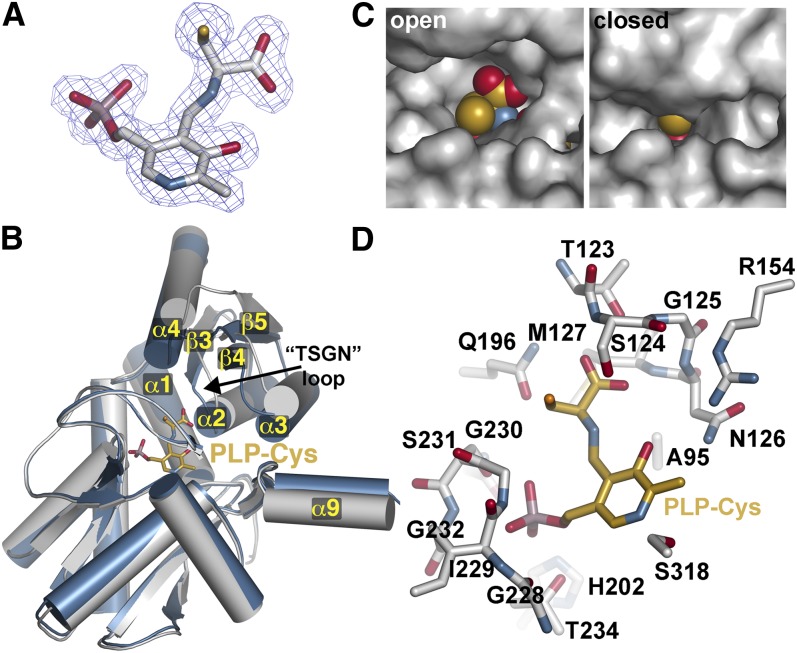
To understand recognition of amino acid substrates by Gm-CAS, the K95A mutant was generated, expressed, purified, and used for crystallization. In the structure of Arabidopsis OASS, mutation of the Schiff base Lys results in retention of PLP binding to the enzyme, but with a Met covalently linked as an external aldimine to the cofactor in the active site (Bonner et al., 2005). This same approach was used to generate a trapped substrate–intermediate complex of Gm-CAS. The 1.77-Å resolution structure of the Gm-CAS K95A mutant was solved by molecular replacement using the wild-type Gm-CAS as a search model. Within the active site of the K95A mutant, unambiguous electron density showed that PLP was trapped in an external aldimine linkage with a Cys molecule, which is the natural substrate of Gm-CAS (Figure 4A).
Figure 4.
Ligand-Induced Domain Movement in Gm-CAS.
(A) Electron density of the PLP-Cys external aldimine in the K95A mutant structure. A 2Fo-Fc omit map (1.5 σ) is shown.
(B) Domain movement in Gm-CAS. Comparison of the open active site wild-type Gm-CAS (gray) and the closed active site K95A mutant (blue) monomer structures. The position of the PLP-Cys external aldimine in the K95A mutant is shown as a stick model (gold). Secondary structure features of the N-terminal domain are labeled. The position of the substrate interaction or TSGN loop (residues 123 to 126) is indicated.
(C) Comparison of active site clefts in the open and closed structures. In the left panel, PLP-Cys (spheres) was modeled into the wild-type structure.
(D) Active site view of PLP-Cys in the Gm-CAS K95A mutant. Residues interacting with PLP-Cys (gold) are shown as stick models.
Comparison of the two Gm-CAS structures showed that the N-terminal domain undergoes a rigid body shift in position by ∼15° toward the PLP-Cys bound in the active site (Figure 4B). In addition, α9, which forms part of the dimerization interface by packing against α2 and α3 of the adjacent monomer, also coordinately changes position. The conformational change of the N-terminal domain alters the size of the active site entrance with the structural transition from an open conformation (wild-type structure) to a closed conformation (K95A mutant) (Figure 4C). In the open form, the entrance is wide enough to allow for binding of Cys in the first half reaction. In comparison, the active site cleft is solvent inaccessible in the closed form, although the reactive side chain of the amino acid attached to PLP is oriented toward a narrow opening at the surface (Figure 4C).
Within the K95A active site, the binding interactions between the protein and PLP are similar to those observed in the wild-type structure (see Supplemental Figures 1A and 1B online), although the pyridine ring rotates ∼15° toward the protein interior (see Supplemental Figure 1C online). Extensive hydrogen bonds are formed between the Cys carboxylate group and side chain and main chain atoms of Thr-123, Ser-124, Gly-125, and Asn-126 in the substrate interaction or TSGN loop (residues 123-126) to anchor the ligand in the active site (Figure 4D; see Supplemental Figure 1B online). Additional hydrogen bonds from the backbone nitrogen of Met-127 and the side chain amide of Gln-196 contribute to Cys binding. In addition, two water molecules mediate contacts between PLP and Arg-154 and Gln-196 (see Supplemental Figure 1B online). This intricate interaction network between the substrate and the TSGN loop drives the shift in position of the N-terminal domain to enclose the active site entrance during catalysis.
Specificity of CAS versus OASS Activity
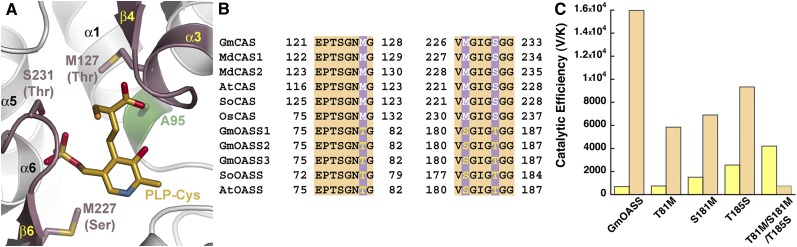
The three-dimensional structures and reaction chemistry of CAS and OASS are conserved; however, each protein is specialized for metabolic function (i.e., CN− detoxification and Cys biosynthesis, respectively). Structural and sequence comparison of the residues around the PLP-Cys external aldimine in the K95A mutant structure revealed only a few differences between CAS and OASS from a variety of species (Figures 5A and 5B). Of the residues interacting with the Cys moiety of PLP-Cys, Met-127 is conserved in CAS from different plant species but is a Thr residue in OASS. This residue is positioned at the end of the TSGN substrate interaction loop and may alter how the loop interacts with different amino acid ligands. Near the PLP phosphate group, Met-227 in β6 and Ser-231 in the β6/α6 loop of Gm-CAS also covary as a Ser and Thr, respectively, in the plant OASS proteins.
Figure 5.
Active Site Residues That Guide Cys versus O-Acetylserine Preference between CAS and OASS.
(A) Active site view showing the positions of residues around the PLP-Cys external aldimine in Gm-CAS K95A, which differ between CAS and OASS in the plant BSAS enzyme family. The corresponding changes from CAS to OASS are indicated in parentheses. The position of the K95A mutation is highlighted in green. The two regions of sequence compared in (B) are colored pink.
(B) Targeted sequence comparison of CAS from soybean (GmCAS), apple (MdCAS1 and MdCAS2), Arabidopsis (AtCAS), spinach (SoCAS), and rice (OsCAS) and OASS from soybean (GmOASS1, GmOASS2, GmOASS3), spinach (SoOASS), and Arabidopsis (AtOASS). The first block of the sequence (residues 121 to 128 in Gm-CAS) corresponds to the residues in α3 and β4, as shown in (A). The second block of the sequence (residues 226 to 233) corresponds to residues in the β6 to α6 loop shown in (A).
(C) Comparison of catalytic efficiencies (Vmax/Km) for Cys (yellow) and O-acetylserine (orange) of wild-type Gm-OASS and the Gm-OASS T81M, S181M, T185S, and T81M/S181M/T185S (triple) mutants. Bars plot Vmax/Km from Table 3.
To test whether any of these three amino acid differences alter CAS versus OASS activity, site-directed mutants of Gm-OASS were generated with the goal of switching its reaction sequence to prefer Cys and function as a CAS. Spectroscopic analysis of Gm-CAS (Figure 2A) showed that the α-aminoacrylate was chemically accessible for both Cys and O-acetylserine, whereas Gm-OASS at comparable concentrations of these substrates formed the reactive intermediate only with O-acetylserine (Figure 2B). Because stable formation of this intermediate from Cys is required for CAS activity, we targeted Gm-OASS for mutagenesis to introduce formation of the α-aminoacrylate intermediate from Cys.
Each position in the Gm-OASS active site was mutated to the corresponding residue from Gm-CAS, and the resulting Gm-OASS T81M, S181M, and T185S mutants were expressed and purified to homogeneity. Kinetic analysis of the three Gm-OASS point mutants showed that each change decreased catalytic efficiency (Vmax/Km) with O-acetylserine by less than threefold (Figure 5C, Table 3). With Cys as a substrate, the T81M mutation did not alter substrate preference but modestly improved the spectroscopic signal associated with formation of the α-aminoacrylate intermediate compared with Gm-OASS (Figure 2B; see Supplemental Figure 2A online). The S181M and T185S mutants showed twofold to fourfold improvements in Vmax/Km. Moreover, comparison of the catalytic efficiencies of each mutant for the two substrates indicated that each retained a fourfold to eightfold preference for OASS activity (Figure 5C). Spectroscopic analysis of each mutant showed that the absorption signal of PLP shifted to the α-aminoacrylate after addition of 100 μM O-acetylserine (see Supplemental Figure 2 online). However, the T185S mutant showed the highest efficiency for the CAS reaction among the mutants tested and displayed a similar shift after incubation with 100 μM Cys (see Supplemental Figure 2 online), which was distinct from that observed with wild-type Gm-OASS (Figure 2B).
Table 3. Kinetic Comparison of Wild-Type and Mutant Gm-OASS with Amino Acid Substrates.
| Type of Gm-OASS | Cys |
O-acetylserine |
||||
|---|---|---|---|---|---|---|
| kcat (s−1) | Km (mM) | kcat/Km (m−1 s−1) | kcat (s−1) | Km (mM) | kcat/Km (m−1 s−1) | |
| Gm-OASS | 0.21 ± 0.09 | 0.30 ± 0.01 | 700 | 57.5 ± 11.8 | 3.60 ± 0.40 | 15,972 |
| T81M | 0.45 ± 0.11 | 0.60 ± 0.10 | 750 | 15.8 ± 3.7 | 2.71 ± 0.14 | 5,830 |
| S181M | 1.06 ± 0.34 | 0.73 ± 0.19 | 1,452 | 15.2 ± 2.5 | 2.19 ± 0.26 | 6,940 |
| T185S | 0.77 ± 0.09 | 0.31 ± 0.04 | 2,484 | 28.1 ± 2.2 | 2.94 ± 0.32 | 9,558 |
| Triple | 0.59 ± 0.13 | 0.13 ± 0.01 | 4,538 | 1.57 ± 0.28 | 2.16 ± 0.15 | 726 |
All values are expressed as a mean ± se (n = 3).
Because each of the individual mutations altered the ratio of CAS:OASS activity, the Gm-OASS T81M/S181M/T185S triple mutant was generated. Kinetic analysis showed that this protein converts Gm-OASS into a CAS with a 6.3-fold preference for Cys over O-acetylserine (Figure 5C, Table 3). Consistent with its improved catalytic efficiency for CAS activity, the triple mutant protein also displayed stable α-aminoacrylate formation when Cys was added as substrate for the first half reaction (see Supplemental Figure 2 online). This result demonstrates that multiple mutations are required for evolution of reaction specificity in the BSAS family of plant enzymes.
DISCUSSION
CAS plays a central role in the detoxification of CN− produced as a byproduct of ethylene biosynthesis (Blumenthal et al., 1968; Miller and Conn, 1980; Yip and Yang, 1988; Goudey et al., 1989), but how this enzyme performs its critical function was unclear. Although parallels between the reactions catalyzed by CAS in CN− detoxification and OASS in Cys synthesis have been suggested (Figure 1A), the structural and functional studies presented here provide the first molecular insight into the mechanism of CN− detoxification in plants and evidence that both enzymes use the same chemistry to generate a common reaction intermediate from different substrates. Comparison of CAS and OASS also reveal how subtle active site variations lead to specialization of function between these two BSAS family members and suggest how the biochemical functions of other BSAS proteins in plants evolved.
The structure of Gm-CAS (Figures 3 and 4) shares a common three-dimensional fold with other PLP-dependent enzymes, including OASS from Arabidopsis and bacteria (Burkhard et al., 1999; Meier et al., 2001; Bonner et al., 2005; Claus et al., 2005; Francois et al., 2006; Simanshu et al., 2006; Ngo et al., 2007). The conserved structure is important for maintaining the PLP binding site across the BSAS family in plants and for providing core reaction chemistry for different metabolic functions. Although Gm-CAS adopts a similar structure as the OASS from Arabidopsis (Bonner et al., 2005), kinetic analysis of Gm-CAS and Gm-OASS confirms that each enzyme prefers distinct substrates (Table 1), as observed in the BSAS from other plant species (Hatzfeld et al., 2000; Yamaguchi et al., 2000; Watanabe et al., 2008; Yi et al., 2010a, 2010b). The ability of CAS and OASS to perform the other’s reaction, albeit with a lower catalytic efficiency, implies a common chemical mechanism. Spectroscopic analysis of Gm-CAS demonstrates formation of the α-aminoacrylate intermediate from either Cys or O-acetylserine (Figure 2A). By contrast, Gm-OASS forms the reaction intermediate with O-acetylserine but not with Cys (Figure 2B), ensuring efficient production of Cys without significant diversion to the unproductive external aldimine form of PLP. These results show that even though each enzyme can bind both ligands, there are differences in their active sites that guide the reaction chemistry along two possible paths that result in formation of either useful reactive α-aminoacrylate intermediate that leads to product formation or a nonreactive external aldimine (Figure 2C).
The reaction catalyzed by CAS begins with binding of Cys to the active site, which is in the open conformation (Figure 3). This positions the carboxylate group of Cys near the substrate interaction or TSGN loop to trigger the N-terminal domain movement that leads to the closed form of the enzyme (Figure 4). In the first half reaction catalyzed by CAS (Figure 6A), the α-amine of the Cys reacts with the C4′ of the PLP-Lys-95 Schiff base to release the Lys residue. Formation of PLP-Cys allows Lys-95 to act as a general base in the α,β-elimination of sulfide that results in the α-aminoacrylate intermediate (Figure 6A). This is similar to the role that the corresponding Lys plays in the elimination of acetate from O-acetylserine in the OASS reaction (Tai et al., 1995; Burkhard et al., 1999; Bonner et al., 2005). Mutation of Lys-95 to an Ala in Gm-CAS prevents this residue from providing a general base in the reaction, which allows for trapping of the PLP-Cys external aldimine (Figure 4). This result presented in our study confirms the critical catalytic role for this residue in the CAS reaction mechanism. At the same time, trapping of the transient PLP intermediate bound to the natural substrate of CAS indicates a catalytic role for Lys-95 in the second half reaction. Based on the structure of Gm-CAS with PLP-Cys in the active site (Figure 4), the amino acid side chain is oriented toward the active site entrance and is ideally positioned for nucleophilic attack by CN− in the second half reaction. Subsequent protonation of the α-carbon of β-cyanoalanine by Lys-95 leads to product release and regeneration of the enzyme-bound Schiff base form of PLP in the second half reaction (Figure 6A), which resets the active site for the next round of catalysis.
Figure 6.
Summary of Reactions Catalyzed in the BSAS Enzyme Family of Plants.
(A) Reaction mechanism of CAS.
(B) Overview of enzymatic activities in the BSAS family, showing first half reaction substrates leading to the α-aminoacrylate intermediate and second half reaction substrates and products.
Given the chemical parallels between CAS and OASS, what governs the ability of each enzyme to preferentially form the same reactive intermediate from either Cys or O-acetylserine? Comparison of the Gm-CAS active site structure and sequence with other CAS and OASS proteins from a variety of plant species identified three amino acids near the PLP-Cys ligand that covaried between the two enzymes (Figures 5A and 5B). Single point mutations of these residues in Gm-OASS (T81M, S181M, and T185S) were introduced to test their potential effect on enhancing the CAS activity of this enzyme (Figure 5C, Table 3). Although the Gm-OASS mutants displayed altered ratios of CAS:OASS activity, the individual changes failed to convert the substrate preference of OASS into that of a CAS. Nonetheless, the T185S mutant was sufficient to generate a more stable α-aminoacrylate intermediate when incubated with Cys, suggesting this residue in OASS sets the reaction equilibrium so that it is preferable for Cys biosynthesis (see Supplemental Figure 2 online). Ultimately, combining all three changes into Gm-OASS successfully altered the reaction chemistry to that of a CAS and allowed formation of a stable α-aminoacrylate intermediate from Cys (Figure 5C, Table 3). These results imply a dynamic interplay between active site structure, the substrate, and the subtle conformational changes that occur during catalysis (Austin et al., 2008). The effect of combining the three mutations suggests that multiple structural changes likely alter the environment around the PLP molecule, which in turn affects how the cofactor can move during the reaction sequence to shape the chemical landscape and allow Cys to form the reactive α-aminoacrylate instead of the nonreactive external aldimine (Eliot and Kirsch, 2004). Thus, active site structure in CAS and OASS guides the chemical reaction sequence to tailor their respective biological activities.
Although the Gm-OASS T81M/S181M/T185S mutant functions as a CAS, its catalytic efficiency does not match that of the authentic Gm-CAS (Table 1). This suggests that additional changes, inside and outside the active sites, are required for optimization of function between CAS and OASS. Considering the dynamic features of CAS, such as the ligand-induced movement of the N-terminal domain and likely localized structural changes of PLP during the reaction sequence, multiple amino acid differences will contribute to the evolutionary changes that distinguish CAS and the efficient detoxification of CN− from OASS and its role in Cys biosynthesis.
The structure of Gm-CAS also has implications for how the plant BSAS family evolved multiple enzymatic activities (Figure 6B). In addition to CAS and OASS, other members of the BSAS enzyme family catalyze yet other specialized reactions. Cys desulfhydrase (DES) is a minor cytosolic isoform that regulates Cys homeostasis by catalyzing the elimination of sulfide from Cys to yield pyruvate in late developmental stages and under biotic/abiotic stress conditions (Alvarez et al., 2010; Álvarez et al., 2012). S-sulfocysteine synthase (SSCS) is a chloroplast-localized enzyme with a role in long-day light-dependent redox control in this organelle and catalyzes the addition of thiosulfate to O-acetylserine (Bermúdez et al., 2010). At the biochemical level, all four types of BSAS (CAS, OASS, DES, and SSCS) share similar substrates and reaction chemistry. CAS and DES both use Cys as a substrate and release sulfide to form the α-aminoacrylate intermediate. These enzymes differ in the second half reaction with CAS using CN− to complete the reaction sequence and DES preventing any nucleophilic addition. Similarly, both OASS and SSCS accept O-acetylserine and sulfide, but larger nucleophiles (i.e., thiosulfate) are used by SSCS for the second half reaction.
The evolution of BSAS enzyme function needs to satisfy three requirements: binding of the first substrate, formation of a reactive intermediate from that substrate, and nucleophilic attack by the second substrate (Figure 6B). Using the structure of Gm-CAS as a guide, we have demonstrated how combination of three mutations in the active site of an OASS leads to formation of the reactive α-aminoacrylate intermediate from Cys and improvement in CAS activity (Figure 5, Table 3). The DES from Arabidopsis also uses Cys as a substrate (Alvarez et al., 2010), but differs from both CAS and OASS at the three active site residues identified here (Met, Met, and Ser in CAS; Thr, Ser, and Thr in OASS; and Ile, Ala, and Thr in DES) (Figure 5). It remains to be determined whether these changes alter Cys versus O-acetylserine preference in DES. The second half reaction also provides an added level of substrate discrimination, which leads to biochemical diversity in the BSAS enzyme family. Changes in the size of the active site entrance in the closed form of the enzyme would provide a steric gate to control nucleophile access to the α-aminoacrylate intermediate. The biosynthesis of Cys in bacteria by two OASS isoforms (CysK and CysM) provides an example of this. CysK is specific for sulfide, whereas CysM accepts sulfide, thiosulfate, and a diverse range of larger nucleophiles (Maier, 2003; Claus et al., 2005). In this case, differences in surface loops were proposed to determine second substrate access to the active site; however, additional functional studies are required to demonstrate this plausible model in plant BSAS enzymes that display different reaction product specificity. The evolutionary diversification of biochemical function in the plant BSAS family to span essential reactions for CN− detoxification (CAS), Cys synthesis (OASS) and homeostasis (DES), and metabolism of other thiol-containing compounds (SSCS) likely results from the accumulation of multiple changes that tailor enzymatic properties.
METHODS
Protein Expression, Purification, and Mutagenesis
The pET-28a-Gm-CAS (Glyma09g39390; GmBSAS3;1) bacterial expression construct encodes for an N-terminally hexahistidine-tagged protein lacking the first 51 amino acids of a putative mitochondrial localization sequence. The Gm-OASS (Glyma11g00810; GmBSAS1;1) expression construct was previously described (Chronis and Krishnan, 2003). Site-directed mutagenesis to generate the Gm-CAS K95A mutant used the QuikChange PCR mutagenesis method (Agilent Technologies) (H. Yi and J.M. Jez, unpublished data).
Expression and purification of Gm-OASS used protocols similar to those used for Arabidopsis thaliana OASS (Bonner et al., 2005; Francois et al., 2006; Kumaran and Jez, 2007; Kumaran et al., 2009). For bacterial expression of wild-type and mutant Gm-CAS, Escherichia coli BL21(DE3) cells were transformed with each expression construct and grown in Terrific broth containing 50 μg L−1 kanamycin at 37°C, until A600 nm reached ∼1.0. Addition of isopropyl-β-d-1-thiogalactopyranoside (1 mM final) was used to induce expression of N-terminal His-tagged protein, with the cells grown at 20°C for 18 h. Cells were harvested by centrifugation (3000g for 15 min at 4°C), and then resuspended in lysis buffer (50 mM Tris-HCl, pH 8.0, 20 mM imidazole, 500 mM NaCl, 10% glycerol, and 1% Tween-20). Sonication was used for cell lysis. Cell debris was removed by centrifugation (12,000g for 30 min at 4°C), and the supernatant was loaded on a Ni2+-nitrilotriacetic acid agarose (Qiagen). After extensive washing (50 mM Tris-HCl, pH 8.0, 20 mM imidazole, 500 mM NaCl, and 10% glycerol), the His-tagged proteins were eluted (50 mM Tris-HCl, pH 8.0, 250 mM imidazole, 500 mM NaCl, and 10% glycerol). Eluted proteins were further purified by size-exclusion chromatography on a HiLoad 26/60 Superdex-200 fast protein liquid chromatography column (GE Healthcare Life Sciences) using a buffer containing 25 mM HEPES (pH 7.5) and 100 mM NaCl. Fractions of purified protein with the expected size were pooled and stored at −80°C for enzyme assay after addition of final 10% glycerol or were concentrated to ∼10 to 12 mg mL−1 for protein crystallization. Protein concentration was determined by the Bradford method (Protein Assay; Bio-Rad) with BSA as standard.
Enzyme Assays
Assays of OASS activity were determined by measuring the amount of Cys produced, as previously described (Bonner et al., 2005). Analysis of CAS activity was performed in 0.5 mL reaction mix containing 100 mM Tris-HCl, pH 9.0, Cys (0 to 1 mM), and KCN (0 to 2 mM) (Han et al., 2008). All reactions were initiated by addition of protein (∼200 ng for CAS or ∼1 μg for OASS). The CAS reaction was terminated after 10 min of incubation at 25°C by addition of 50 μL of 30 mM FeCl3 in 1.2 N HCl and 50 μL of 20 mM N,N-dimethyl-p-phenylenediamine dihydrochloride in 7.2 N HCl. The amount of methylene blue produced from one molecule of sulfide and two molecules of N,N-dimethyl-p-phenylenediamine dihydrochloride was colorimetrically determined at 670 nm. Steady state kinetic parameters were determined by nonlinear fitting of initial velocity versus substrate concentration to the Michaelis-Menten equation in SigmaPlot.
Spectroscopic Analysis
Binding of Cys, O-acetylserine, and β-cyanoalanine to Gm-CAS and Gm-OASS was monitored by recording the change in PLP signal using a Beckman DU800 UV/visible spectrophotometer (Bonner et al., 2005). Wavelength spectra (350 to 600 nm) of protein (200 to 500 μg) in the absence or presence of different ligands were obtained in 25 mM HEPES, pH 7.5, 100 mM NaCl.
Protein Crystallization and Structure Determination
Crystals of Gm-CAS were obtained by the vapor diffusion method in 4 μL hanging drops of a 1:1 mixture of protein and crystallization buffer (10% polyethylene glycol 8000, 0.2 M NaCl, and 0.1 sodium/potassium phosphate, pH 7, and 0.2 M MgSO4) at 4°C over a 0.7-mL reservoir. The K95A mutant was crystallized in similar drops from a solution containing 20% polyethylene glycol 3350, 0.2 M potassium citrate, pH 7.5. All crystals were stabilized in cryoprotectant (crystallization solution with 30% glycerol) before flash freezing in liquid nitrogen. Diffraction data were collected at beamline 19ID of the Argonne National Lab Advanced Photon Source, with indexing, integration, and scaling performed with HKL3000 (Minor et al., 2006). The structure of Gm-CAS was solved by molecular replacement implemented in PHASER (McCoy et al., 2007) using the structure of Arabidopsis OASS (Bonner et al., 2005; Protein Data Bank [PDB]: 1Z7W) as a search model. Four molecules were present in the asymmetric unit corresponding to two noncrystallographic dimers. Model building of the Gm-CAS structure was performed in COOT (Emsley and Cowtan, 2004), and all refinements were performed with PHENIX (Adams et al., 2010). The final model includes residues 51 to 367, 52 to 372, 44 to 372, and 51 to 367 in chains A to D, respectively, four PLPs, and 233 water molecules. The crystal structure of the Gm-CAS K95A mutant was solved by molecular replacement using the refined wild-type structure as a search model. Six molecules were present in the asymmetric unit, with two noncrystallographic dimers formed by chains A (residues 51 to 372) and B (residues 51 to 372) and chains C (residues 51 to 372) and D (residues 50 to 372). Chains E (residues 51 to 372) and F (residues 51 to 372) each form a crystallographic dimer. After initial placement of each monomer, portions of the model corresponding to α1 to α4, β3 to β5, and β9 were removed and manually rebuilt, because these features changed in position. The final model also contains six copies of PLP linked to a Cys in an external aldimine linkage and 1731 water molecules. Building and refinement of the Gm-CAS K95A mutant was performed as described above. Crystal parameters, data collection statistics, and refinement statistics for the structures are summarized in Table 2.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: XP_003534555 (soybean CAS), ABF13209.1 (apple CAS1), ABF13210.1 (apple CAS2), NP_191703.1 (Arabidopsis CAS), BAA07177.1 (spinach CAS), AAV48452.1 (rice CAS), AAL66291.1 (soybean OASS1), NP_001238392.1 (soybean OASS2), XP_003554689.1 (soybean OASS3), Q00834 (spinach OASS), and CAA56593 (Arabidopsis OASS).Coordinates and structure factors for wild-type Gm-CAS (PDB: 3VBE) and the Gm-CAS K95A mutant (PDB: 3VC3) have been deposited in the Research Collaboratory for Structural Bioinformatics PDB.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Ligand Interactions in Gm-CAS.
Supplemental Figure 2. Spectroscopic Analysis of Gm-OASS Mutants.
Acknowledgments
This article was funded by a grant from the U.S. Department of Agriculture (NRI-2005-02518) to J.M.J. M.J. received an American Society of Plant Biologists Summer Undergraduate Research Fellowship. Portions of this research were carried out at the Argonne National Laboratory Structural Biology Center of the Advanced Photon Source, a national user facility operated by the University of Chicago for the Department of Energy Office of Biological and Environmental Research (DE-AC02-06CH11357).
AUTHOR CONTRIBUTIONS
H.Y., M.J., and J.M.J. designed the research; H.Y., M.J., and J.M.J. performed research; H.Y., M.J., and J.M.J. analyzed data; and H.Y. and J.M.J. wrote the article.
Glossary
- CAS
β-cyanoalanine synthase
- BSAS
β-substituted alanine synthase
- OASS
O-acetylserine sulfhydrylase
- PLP
pyridoxal phosphate
- Amax
photosynthetic capacity
- TSGN
to be defined
- DES
desulfhydrase
- SSCS
S-sulfocysteine synthase
- PDB
Protein Data Bank
References
- Adams P.D., et al. (2010). PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez C., Bermúdez M.A., Romero L.C., Gotor C., García I. (2012). Cysteine homeostasis plays an essential role in plant immunity. New Phytol. 193: 165–177 [DOI] [PubMed] [Google Scholar]
- Alvarez C., Calo L., Romero L.C., García I., Gotor C. (2010). An O-acetylserine(thiol)lyase homolog with L-cysteine desulfhydrase activity regulates cysteine homeostasis in Arabidopsis. Plant Physiol. 152: 656–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akopyan T.N., Braunstein A.E., Goryachenkova E.V. (1975). Beta-cyanoalanine synthase: Purification and characterization. Proc. Natl. Acad. Sci. USA 72: 1617–1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin M.B., O’Maille P.E., Noel J.P. (2008). Evolving biosynthetic tangos negotiate mechanistic landscapes. Nat. Chem. Biol. 4: 217–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermúdez M.A., Páez-Ochoa M.A., Gotor C., Romero L.C. (2010). Arabidopsis S-sulfocysteine synthase activity is essential for chloroplast function and long-day light-dependent redox control. Plant Cell 22: 403–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleecker A.B., Kende H. (2000). Ethylene: A gaseous signal molecule in plants. Annu. Rev. Cell Dev. Biol. 16: 1–18 [DOI] [PubMed] [Google Scholar]
- Blumenthal S.G., Hendrickson H.R., Abrol Y.P., Conn E.E. (1968). Cyanide metabolism in higher plants. 3. The biosynthesis of β-cyanolanine. J. Biol. Chem. 243: 5302–5307 [PubMed] [Google Scholar]
- Bonner E.R., Cahoon R.E., Knapke S.M., Jez J.M. (2005). Molecular basis of cysteine biosynthesis in plants: Structural and functional analysis of O-acetylserine sulfhydrylase from Arabidopsis thaliana. J. Biol. Chem. 280: 38803–38813 [DOI] [PubMed] [Google Scholar]
- Burkhard P., Tai C.H., Ristroph C.M., Cook P.F., Jansonius J.N. (1999). Ligand binding induces a large conformational change in O-acetylserine sulfhydrylase from Salmonella typhimurium. J. Mol. Biol. 291: 941–953 [DOI] [PubMed] [Google Scholar]
- Castric P.A., Farnden K.J., Conn E.E. (1972). Cyanide metabolism in higher plants. V. The formation of asparagine from -cyanoalanine. Arch. Biochem. Biophys. 152: 62–69 [DOI] [PubMed] [Google Scholar]
- Chronis D., Krishnan H.B. (2003). Sulfur assimilation in soybean: Molecular cloning and characterization of O-acetylserine(thiol)lyase (cysteine synthase). Crop Sci. 43: 1819–1827 [Google Scholar]
- Claus M.T., Zocher G.E., Maier T.H.P., Schulz G.E. (2005). Structure of the O-acetylserine sulfhydrylase isoenzyme CysM from Escherichia coli. Biochemistry 44: 8620–8626 [DOI] [PubMed] [Google Scholar]
- Elias M., Sudhakaran P.R., Nambisan B. (1997). Purification and characterization of β-cyanoalanine synthase from cassava tissues. Phytochemistry 46: 469–472 [Google Scholar]
- Eliot A.C., Kirsch J.F. (2004). Pyridoxal phosphate enzymes: Mechanistic, structural, and evolutionary considerations. Annu. Rev. Biochem. 73: 383–415 [DOI] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. (2004). Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Francois J.A., Kumaran S., Jez J.M. (2006). Structural basis for interaction of O-acetylserine sulfhydrylase and serine acetyltransferase in the Arabidopsis cysteine synthase complex. Plant Cell 18: 3647–3655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García I., Castellano J.M., Vioque B., Solano R., Gotor C., Romero L.C. (2010). Mitochondrial beta-cyanoalanine synthase is essential for root hair formation in Arabidopsis thaliana. Plant Cell 22: 3268–3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudey J.S., Tittle F.L., Spencer M.S. (1989). A role for ethylene in the metabolism of cyanide by higher plants. Plant Physiol. 89: 1306–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S.E., Seo Y.S., Heo S., Kim D., Sung S.K., Kim W.T. (2008). Structure and expression of MdFBCP1, encoding an F-box-containing protein 1, during Fuji apple (Malus domestica Borkh.) fruit ripening. Plant Cell Rep. 27: 1291–1301 [DOI] [PubMed] [Google Scholar]
- Hatzfeld Y., Maruyama A., Schmidt A., Noji M., Ishizawa K., Saito K. (2000). beta-Cyanoalanine synthase is a mitochondrial cysteine synthase-like protein in spinach and Arabidopsis. Plant Physiol. 123: 1163–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami F., Takayama K., Murakoshi I. (1988a). Purification and properties of β-cyano-L-alanine synthase from Lathyrus latifolius. Phytochemistry 27: 3385–3389 [Google Scholar]
- Ikegami F., Takayama K., Tajima C., Murakoshi I. (1988b). Purification and properties of β-cyano-L-alanine synthase from Spinacia oleracea. Phytochemistry 27: 2011–2016 [Google Scholar]
- Kumaran S., Jez J.M. (2007). Thermodynamics of the interaction between O-acetylserine sulfhydrylase and the C-terminus of serine acetyltransferase. Biochemistry 46: 5586–5594 [DOI] [PubMed] [Google Scholar]
- Kumaran S., Yi H., Krishnan H.B., Jez J.M. (2009). Assembly of the cysteine synthase complex and the regulatory role of protein-protein interactions. J. Biol. Chem. 284: 10268–10275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai K.W., Yau C.P., Tse Y.C., Jiang L., Yip W.K. (2009). Heterologous expression analyses of rice OsCAS in Arabidopsis and in yeast provide evidence for its roles in cyanide detoxification rather than in cysteine synthesis in vivo. J. Exp. Bot. 60: 993–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier T.H.P. (2003). Semisynthetic production of unnatural L-α-amino acids by metabolic engineering of the cysteine-biosynthetic pathway. Nat. Biotechnol. 21: 422–427 [DOI] [PubMed] [Google Scholar]
- Maruyama A., Ishizawa K., Takagi T. (2000). Purification and characterization of beta-cyanoalanine synthase and cysteine synthases from potato tubers: Are beta-cyanoalanine synthase and mitochondrial cysteine synthase same enzyme? Plant Cell Physiol. 41: 200–208 [DOI] [PubMed] [Google Scholar]
- Maruyama A., Ishizawa K., Takagi T., Esashi Y. (1998). Cytosolic beta-cyanoalanine synthase activity attributed to cysteine synthases in cocklebur seeds. Purification and characterization of cytosolic cysteine synthases. Plant Cell Physiol. 39: 671–680 [DOI] [PubMed] [Google Scholar]
- Maruyama A., Saito K., Ishizawa K. (2001). Beta-cyanoalanine synthase and cysteine synthase from potato: Molecular cloning, biochemical characterization, and spatial and hormonal regulation. Plant Mol. Biol. 46: 749–760 [DOI] [PubMed] [Google Scholar]
- McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. (2007). Phaser crystallographic software. J. Appl. Cryst. 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier M., Janosik M., Kery V., Kraus J.P., Burkhard P. (2001). Structure of human cystathionine beta-synthase: A unique pyridoxal 5′-phosphate-dependent heme protein. EMBO J. 20: 3910–3916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.M., Conn E.E. (1980). Metabolism of hydrogen cyanide by higher plants. Plant Physiol. 65: 1199–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minor W., Cymborowski M., Otwinowski Z., Chruszcz M. (2006). HKL-3000: The integration of data reduction and structure solution—from diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr. 62: 859–866 [DOI] [PubMed] [Google Scholar]
- Møller B.L. (2010). Functional diversifications of cyanogenic glucosides. Curr. Opin. Plant Biol. 13: 338–347 [DOI] [PubMed] [Google Scholar]
- Ngo H., Harris R., Kimmich N., Casino P., Niks D., Blumenstein L., Barends T.R., Kulik V., Weyand M., Schlichting I., Dunn M.F. (2007). Synthesis and characterization of allosteric probes of substrate channeling in the tryptophan synthase bienzyme complex. Biochemistry 46: 7713–7727 [DOI] [PubMed] [Google Scholar]
- Palmer G. (1993). Current issues in the chemistry of cytochrome c oxidase. J. Bioenerg. Biomembr. 25: 145–151 [DOI] [PubMed] [Google Scholar]
- Peiser G.D., Wang T.T., Hoffman N.E., Yang S.F., Liu H.W., Walsh C.T. (1984). Formation of cyanide from carbon 1 of 1-aminocyclopropane-1-carboxylic acid during its conversion to ethylene. Proc. Natl. Acad. Sci. USA 81: 3059–3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravilious G.E., Jez J.M. (2012). Structural biology of plant sulfur metabolism: From assimilation to biosynthesis. Nat. Prod. Rep., in press [DOI] [PubMed] [Google Scholar]
- Simanshu D.K., Savithri H.S., Murthy M.R. (2006). Crystal structures of Salmonella typhimurium biodegradative threonine deaminase and its complex with CMP provide structural insights into ligand-induced oligomerization and enzyme activation. J. Biol. Chem. 281: 39630–39641 [DOI] [PubMed] [Google Scholar]
- Tai C.H., Nalabolu S.R., Simmons J.W., III, Jacobson T.M., Cook P.F. (1995). Acid-base chemical mechanism of O-acetylserine sulfhydrylases-A and -B from pH studies. Biochemistry 34: 12311–12322 [DOI] [PubMed] [Google Scholar]
- Tittle F.L., Goudey J.S., Spencer M.S. (1990). Effect of 2,4-dichlorophenoxyacetic acid on endogenous cyanide, beta-cyanoalanine synthase activity, and ethylene evolution in seedlings of soybean and barley. Plant Physiol. 94: 1143–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrilow A.G., Hawkesford M.J. (2000). Cysteine synthase (O-acetylserine (thiol) lyase) substrate specificities classify the mitochondrial isoform as a cyanoalanine synthase. J. Exp. Bot. 51: 985–993 [DOI] [PubMed] [Google Scholar]
- Warrilow A.G., Hawkesford M.J. (2002). Modulation of cyanoalanine synthase and O-acetylserine (thiol) lyases A and B activity by beta-substituted alanyl and anion inhibitors. J. Exp. Bot. 53: 439–445 [DOI] [PubMed] [Google Scholar]
- Watanabe M., Kusano M., Oikawa A., Fukushima A., Noji M., Saito K. (2008). Physiological roles of the β-substituted alanine synthase gene family in Arabidopsis. Plant Physiol. 146: 310–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtele E.S., Nikolau B.J., Conn E.E. (1984). Tissue distribution of beta-cyanoalanine synthase in leaves. Plant Physiol. 75: 979–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtele E.S., Nikolau B.J., Conn E.E. (1985). Subcellular and developmental distribution of beta-cyanoalanine synthase in barley leaves. Plant Physiol. 78: 285–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y., Nakamura T., Kusano T., Sano H. (2000). Three Arabidopsis genes encoding proteins with differential activities for cysteine synthase and beta-cyanoalanine synthase. Plant Cell Physiol. 41: 465–476 [DOI] [PubMed] [Google Scholar]
- Yi H., Galant A., Ravilious G.E., Preuss M.L., Jez J.M. (2010a). Sensing sulfur conditions: Simple to complex protein regulatory mechanisms in plant thiol metabolism. Mol. Plant 3: 269–279 [DOI] [PubMed] [Google Scholar]
- Yi H., Ravilious G.E., Galant A., Krishnan H.B., Jez J.M. (2010b). From sulfur to homoglutathione: Thiol metabolism in soybean. Amino Acids 39: 963–978 [DOI] [PubMed] [Google Scholar]
- Yip W.K., Yang S.F. (1988). Cyanide metabolism in relation to ethylene production in plant tissues. Plant Physiol. 88: 473–476 [DOI] [PMC free article] [PubMed] [Google Scholar]