SUMMARY
We investigated the potential of in-depth quantitative proteomics to reveal plasma protein signatures that reflect lung tumor biology. We compared plasma protein profiles of four mouse models of lung cancer with profiles of models of pancreatic, ovarian, colon, prostate, and breast cancer and two models of inflammation. A protein signature for Titf1/Nkx2-1, a known lineage-survival oncogene in lung cancer, was found in plasmas of mouse models of lung adenocarcinoma. An EGFR signature was found in plasma of an EGFR mutant model, and a distinct plasma signature related to neuroendocrine development was uncovered in the small-cell lung cancer model. We demonstrate relevance to human lung cancer of the protein signatures identified on the basis of mouse models.
INTRODUCTION
Mouse models of cancer, in particular genetically engineered mouse models (GEMs) in which genetic alterations in oncogenes and tumor suppressors known to be mutated in human cancer are introduced, recapitulate many features of the human disease on a more uniform genetic background. For example, mouse models have been used to study the role of mutant KRAS and EGFR in the genesis of lung adenocarcinomas. As in humans with EGFR mutant lung cancer, mice with this disease respond to treatment with a tyrosine kinase inhibitor (Politi et al., 2006). Similarly, the deletion of Rb and Trp53, genes that are inactivated in the vast proportion of human small-cell lung cancers (SCLCs), in the murine lung leads to the development of tumors that recapitulate the features of this histological subtype (Meuwissen et al., 2003; Schaffer et al., 2010).
To determine whether we could identify plasma protein signatures that are common to lung adenocarcinomas or that reflect pathways driving tumor development, we examined the plasma proteomes of several mouse lung tumor models, in comparison with the plasma proteomes of models of other tumor types. We also extended these findings to human patients with lung cancer.
RESULTS
A Plasma Protein Signature for Lung Adenocarcinoma
Plasma proteins from tumor-bearing mice and age-matched littermate controls were subjected to quantitative profiling by mass spectrometry as previously described (Faca et al., 2008) (see Table S1, which is available with this article online). Plasma was collected from three well-characterized models of lung adenocarcinoma: TetO-EGFRL858R/CCSP-rtTA (Lung-EGFR) (Politi et al., 2006), TetO-Kras4bG12D/CCSP-rtTA (Lung-Kras) (Fisher et al., 2001), and urethane treated (Lung-Urethane) (Horio et al., 1996). Plasma was collected 5–6 weeks (Lung-EGFR) and 5–6 months (Lung-Kras) after starting doxycycline treatment, representing early and intermediate stages of tumorigenesis, respectively (Fisher et al., 2001; Politi et al., 2006). For the urethane mouse model, plasma was collected at 38–42 weeks of age from mice bearing multiple adenomas and adenocarcinomas that harbor frequent Kras and Trp53 mutation (Horio et al., 1996). Plasma was also collected from a mouse model of small-cell lung cancer (Lung-SCLC) (Meuwissen et al., 2003), aged for 6–9 months following AdCre infection. In the SCLC model, AdCre-infected Trp53lox/lox; Rblox/lox mice develop neuroendocrine tumors that resemble SCLCs histologically (Meuwissen et al., 2003). The resulting proteomic data from tumor-bearing mice and matched controls for all four lung cancer models were compared with corresponding data from other models for which plasma proteome data was available to delineate lung cancer signatures. They consisted of two mouse models of breast cancer (MMTV-rtTA/TetO-NeuNT; (Breast-HER2) [Moody et al., 2002; Pitteri et al., 2011] and Tg(MMTV-PyMT)634Mul; (Breast-PyMT 0.5 cm) and (Breast-PyMT 1.0 cm), representing early and late stages of tumor development, respectively [Pitteri et al., 2008]), two prostate cancer models (Ptenpc−/− mice bearing nonmetastatic tumors compared with Ptenpc−/−;Smad4pc−/− mice bearing metastatic tumors; (Prostate-Strain Comparison) [Ding et al., 2011]), and one model each of colon (ApcΔ580/+; (Colon-Apc) [Hung et al., 2009)], ovarian (LSL-KrasG12D/+;PtenloxP/loxP; (Ovary-Kras/Pten) [Dinulescu et al., 2005; Pitteri et al., 2009)] and pancreatic ductal adenocarcinoma (PDAC) (Pdx1-Cre;LSL-KrasG12D;Ink4a/Arflox/lox; (Pancreas-PanIN [pancreatic intraepithelial neoplasia]) and (Pancreas-PDAC), representing early and late stage of tumor development, respectively [Faca et al., 2008]). Additionally, the plasma proteomes of two mouse models of inflammatory disease were also assessed to ascertain whether alterations observed in the plasma of the tumor models were related to general inflammatory processes. These included a mouse model bearing necrotic granulomatous tissue induced by carra-geenan-sponge implantation (Confounder-Acute Inflammation) and a model consisting of type II collagen-induced arthritis (Confounder-Chronic Inflammation) (Kelly-Spratt et al., 2011).
For each model, we quantified the relative concentrations of proteins in case and control plasmas by differential cysteine alkylation in intact proteins using isotopically labeled acrylamide (Faca et al., 2006). We identified protein products from 5361 unique genes with <5% false discovery rate from ~13 million spectra in 14 proteomic profiling experiments with plasmas from individual mouse models (Table S2). Case:control protein concentration ratios were calculated according to identification of cysteine-containing peptides from the protein products of 2261 unique genes (Table S3).
Unsupervised hierarchical clustering analysis of quantified plasma proteins revealed clustering of models by organ type, with all four lung cancer models clustering together as well as the two breast cancer models, suggestive of organ-type specific plasma protein signatures (Figure 1). To further explore protein signatures associated specifically with lung adenocarcinoma, the plasma proteome data was analyzed for proteins that were significantly elevated (case:control ratio > 1.25; p < 0.05, t test) in mice bearing lung adenocarcinoma tumors or only identified in tumor bearing mice from at least two of the three lung adenocarcinoma models, but not in other mouse models. This filtering strategy yielded 13 proteins (Table 1; Lung adeno subgroup in Table S4) including the known lung surfactant proteins (Sftpb and Sftpd) (Pérez-Gil, 2008). Three additional proteins (Igsf4a, Ppbp, and Prtg) exhibited increased levels in mice bearing adenocarcinomas as well as in mice bearing SCLC tumors but not in nonlung tumor models, thus representing proteins more broadly associated with lung cancer (Table 1; Lung cancer subgroup in Table S4). Independent evidence of PPBP as a lung cancer marker has been reported on the basis of human studies (Yee et al., 2009). Proteins with significantly decreased concentrations (case:control ratio < 0.8; p < 0.05, t test) were also identified in the lung adenocarcinoma models (Table S4). Of potential broader cancer relevance, we identified 16 additional proteins that were altered across multiple cancer models, but not in confounder mouse models, and thus represent potential broad epithelial tumor signatures (Common subgroup in Table S4). One such protein is Wfdc2, which has been associated with ovarian cancer (Hellström et al., 2003). Of interest, WFDC2 was found to be expressed in human lung tumor tissue, although not previously known to be elevated in lung cancer plasmas (Bingle et al., 2006; Galgano et al., 2006). Another subset of seven proteins was elevated in plasmas from both cancer models and the inflammatory models (Inflammation subgroup in Table S4). This subset included platelet-derived chemokine Cxcl4/Pf4, a cleaved form of which has been recently reported as associated with breast ductal carcinoma in situ (Solassol et al., 2010) and Angptl3, which has not been previously associated with lung cancer. Angptl3 plays a role in lipid metabolism (Koishi et al., 2002), induces angiogenesis (Camenisch et al., 2002), and has been found to be associated with the sera of ovarian cancer (Lin et al., 2009).
Figure 1. Clustering of Plasma Proteome Profiles of Mouse Models of Lung Cancer.

Unsupervised hierarchical clustering was performed for proteins quantified in plasmas from mouse models based on differential cysteine labeling. Plasma protein profiles of all four lung cancer mouse models yielded a distinct cluster. A close-up view of distinct clusters of proteins with elevated and decreased levels in plasma from lung cancer mouse models is shown in the lower panels.
Table 1.
Plasma Proteins with Significantly Elevated Levels in Tumor-Bearing Mice from the Lung Adenocarcinoma Models
| Gene | Lung Mouse Models
|
Cell Lines
|
|||||
|---|---|---|---|---|---|---|---|
| EGFR | Kras | Urethane | SCLC | Total Cell Extract | Cell Surface | Media | |
| Sftpb | 4.83 | 3.41 | 1.99 | — | 0 | 1 | 2 |
|
| |||||||
| Morc3 | * | 10 | 10 | — | 0 | 0 | 0 |
|
| |||||||
| Fgfr2 | 9.8 | * | — | — | 2 | 1 | 0 |
|
| |||||||
| Adam10 | 7.85 | 1.5 | — | — | 3 | 12 | 11 |
|
| |||||||
| Man2b2 | 4.22 | 1.46 | — | — | 1 | 0 | 7 |
|
| |||||||
| Tfpi2 | 10 | * | — | — | 5 | 5 | 8 |
|
| |||||||
| Son | * | 10 | — | — | 1 | 1 | 6 |
|
| |||||||
| Eif2ak3 | * | 9.03 | — | — | 1 | 0 | 0 |
|
| |||||||
| Hamp2 | * | 3.07 | — | — | NA | NA | NA |
|
| |||||||
| Ncan | * | 1.45 | — | — | 2 | 1 | 0 |
|
| |||||||
| Sftpd | 10 | — | 10 | — | 2 | 1 | 3 |
|
| |||||||
| Mocs1 | 1.71 | — | 1.3 | — | 0 | 0 | 1 |
|
| |||||||
| Nup188 | — | * | 10 | — | 0 | 1 | 1 |
|
| |||||||
| Igsf4a | 3.55 | 1.48 | 1.55 | 3.32 | 0 | 0 | 0 |
|
| |||||||
| Ppbp | 1.63 | 1.7 | 5.12 | 1.39 | 0 | 0 | 0 |
|
| |||||||
| Prtg | 10 | 10 | — | 10 | 0 | 0 | 0 |
Numbers for each mouse model indicate the case:control ratio based on the identification of peptides containing cysteine residues labeled with heavy and light acrylamide isotopes. Dashes (—) indicate that the quantitative ratio was not calculated. Asterisks (*) indicate proteins that were only identified in plasma from tumor-bearing mice. Thirteen proteins were significantly elevated (case:control ratio > 1.25; p < 0.05, t test) in mice bearing lung adenocarcinoma tumors or only identified in tumor bearing mice from at least two of the three lung adenocarcinoma models, but not in any other mouse models. Three proteins (Igsf4a, Ppbp, and Prtg) were increased in mice bearing adenocarcinomas and SCLC tumors but not in the other models. The numbers in the columns for cell lines indicate the number of cell lines in which a protein was identified in any of the whole cell extract, the cell surface, or the conditioned media. The entire dataset is available in Table S4 and Table S5.
Next, we examined whether proteins with increased levels in plasmas from lung tumor-bearing mice are expressed in lung cancer cells and may be released into the extracellular space (Faça and Hanash, 2009). To this end, the proteomes of 21 human lung adenocarcinoma cell lines were analyzed by mass spectrometry to identify proteins in whole cell extracts, and proteins specifically on the cell surface or released into the medium. These comparisons revealed that 25 of 39 proteins found at increased levels in plasmas from mice with lung adenocarcinoma (Table S5) were identified in conditioned media, and an overlapping set of 26 proteins were found in the cell surface compartment of lung adenocarcinoma cell lines. Twenty one of the 39 proteins were enriched more than 5-fold in the conditioned media compared to their abundance in whole cell extracts (Table 1 and Table S5). Together, these profile comparisons suggest that lung cancer cells are a likely contributing source for the increased levels of these proteins in plasmas from mice with lung adenocarcinomas.
A Titf1/Nkx2-1 Signature in the Plasma of Mice with Lung Adenocarcinomas
Sftpb and Sftpd, which were elevated in lung adenocarcinoma plasma profiles are secreted by alveolar cells and play an essential role in lung function by reducing surface tension (Pérez-Gil, 2008). These proteins are encoded by genes that are known targets of Titf1/Nkx2-1, a master transcription factor of peripheral airway cells and a known lineage-specific determinant of survival in lung cancer cells (Tanaka et al., 2007; Weir et al., 2007). Expression of Titf1/Nkx2-1 protein in lung tumors was confirmed by immunohistochemistry (Figure 2A). On this basis, we speculated that additional targets of Titf1/Nkx2-1 might be represented at increased levels in the plasma proteomes of the lung adenocarcinoma models. To further delineate a Titf1/Nkx2-1 plasma proteome signature, we examined mRNA micro-array data from a set of 111 human NSCLC tumors, including both adenocarcinomas and squamous cell carcinomas (accession number, GSE3141; Bild et al., 2006). Orthologous genes encoding four proteins with increased levels in the murine lung adenocarcinoma plasmas (SFTPB, SFTPD, NPC2, and WFDC2) exhibited a strong positive correlation with TITF1/NKX2-1 mRNA levels (Spearman correlation coefficient > 0.6; p < 0.00001) (Table S6). NPC2, a cholesterol transfer protein implicated in Niemann-Pick disease (Vanier and Millat, 2004), was not previously known to be elevated in lung cancer plasma. Expression of the NPC2 gene showed a strong correlation with TITF1/NKX2-1 mRNA across the set of human lung tumors (Spearman correlation coefficient = 0.64) providing supportive evidence for NPC2 as a potential target of this transcription factor in lung cancer cells. Immunohistochemical staining for Npc2 and Sftpb in mouse lung cancer tissue (Figure 2A) indicated that these proteins as well as Titf1/Nkx2-1 are coexpressed in lung tumor cells. Additional data associating NPC2 with lung cancer included identification of NPC2 in media of all 21 lung adenocarcinoma cell lines (Table S5) and occurrence of NPC2 among 300 proteins found in pleural effusions associated with lung cancer (Pernemalm et al., 2009).
Figure 2. A Titf1/Nkx2-1 Signature in the Plasma of Lung Adenocarcinoma Tumor-Bearing Mice.
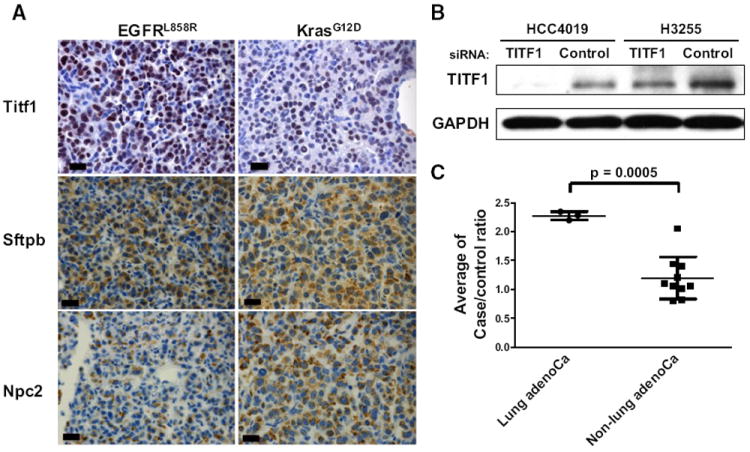
(A) Immunohistochemical staining of mouse lung cancer tissue for Titf1, Sftpb, and Npc2 in the EGFR and Kras lung cancer models. Scale bars represent 20 μm.
(B) Knockdown of TITF1 with siRNA in two lung cancer cell lines, HCC4019 (KRAS mutant) and H3255 (EGFR mutant), was verified by western blotting. GAPDH served as a loading control.
(C) The average ratios of potential Titf1-regulated proteins in plasma from mice with disease relative to controls for the different mouse models. Bars indicate mean and standard deviation of ratios for proteins quantified in each mouse model. p value was calculated by t test.
To further determine the role of Titf1/Nkx2-1 in the expression of genes encoding proteins found at elevated levels in plasmas from lung cancer models, we examined the effects of inhibiting Titf1/Nkx2-1 in two lung adenocarcinoma cell lines with high expression of TITF1/NKX2-1: HCC4019 (KRAS mutant) and H3255 (EGFR L858R mutant) (Figure 2B). TITF1/NKX2-1 knockdown experiments were performed twice with each cell line. Gene expression analysis yielded a total of 964 genes with average case:control RNA ratios of less than 0.75 in one or both cell lines following knockdown. Plasma data were available for protein products of 34 genes among the 964 potential TITF1/NKX2-1-regulated genes, including Sftpa1, Sftpb, Sftpd, and Npc2 (Table S7). The average plasma ratios of these proteins in tumor-bearing mice from lung adenocarcinoma mouse models relative to controls were significantly higher than plasma ratios for other mouse models (mean ± standard deviation: 2.28 ± 0.07 versus 1.20 ± 0.37; p = 0.0005, t test) (Figure 2C), providing further supporting evidence for a Titf1/Nkx2-1 regulated protein set in plasmas of mice with lung adenocarcinoma. We also observed a strong correlation between expression of genes in this set and TITF1/NKX2-1 mRNA levels in NSCLC tumors according to expression array analysis (Table S7), further supporting their regulation by TITF1/NKX2-1 transcription factor.
Dominant Networks in Plasmas from Mice with Genetically Engineered Lung Cancers
We searched for dominant networks among proteins with significantly altered levels in plasmas from genetically engineered lung cancer mouse models based on Ingenuity Pathway Analysis (http://www.ingenuity.com/). We observed 21, 15, and 17 networks with statistical scores of more than 10 (score = −log10 [p value] based on the right-tailed Fisher’s exact test) in EGFR, Kras, and SCLC lung cancer mouse models, respectively. Protein networks involving TGFβ were observed in plasma with high significance among all lung cancer models (network #4 in the Lung-EGFR model; #5 in the Lung-Kras model; #2 in the Lung-SCLC model) (Table S8). NFκB was also found in networks with high significance among all three models. Networks involving TGFβ or NFκB may reflect a host response to the tumors based on the associated proteins and/or may reflect functional roles for these pathways in lung tumor formation. NFκB signaling has been shown to be required for mouse lung adenocarcinoma development (Meylan et al., 2009). Interestingly a network involving Egfr was the most statistically significant network (score = 38) in the EGFR mutant mouse model (Figure 3A), whereas it was observed as network #8 (score = 23) in the Kras mutant mouse model. A network involving Egfr was also highly significant (score = 41) in the SCLC model (Figure S1A). Messenger RNA expression of Egfr was found to be elevated in tumor tissue compared to normal lung tissue in this SCLC model (accession number, GSE18534; Schaffer et al., 2010). Thus, this SCLC model may recapitulate EGFR-positive human SCLC, which occurs in ~40% of SCLC (Schmid et al., 2010). The plasma Egfr network observed in the EGFR mutant mouse model includes proteins that bind directly with EGFR (Met, Cd44, Cdh1, Ndn, Sh3bgrl, and Rin1) and proteins that interact indirectly with EGFR (Adam10 and Trak2/Als2cr3). Among proteins that bind directly with EGFR, Met, Cdh1, and Sh3bgrl were predominantly elevated in tumor bearing mice from the EGFR mutant mouse model (Table S3). Moreover CDH1 occurred at higher levels in EGFR mutant lung adenocarcinoma cell lines than in KRAS mutant lung adenocarcinoma cell lines (Figure 3B). Interestingly, one form of Cd44 (IPI00265503) was only identified in the EGFR mutant mouse model. This form represents a longer Cd44 variant (Figure 3C). Proteomic analysis of 21 lung adenocarcinoma cell lines revealed that peptides encoding the CD44 variant region were identified in 4 of 10 EGFR mutant cell lines, but none of 11 KRAS mutant cell lines (p = 0.0351, Fisher’s exact test) (Figure S1B). Concordantly, alternative splicing RNA variants of CD44 were associated with EGFR mutant compared with KRAS mutant lung adenocarcinoma cell lines (p = 0.0237, Fisher’s exact test) (Figure 3D).
Figure 3. An Egfr Protein Network in the Plasma of Mice with Tumors Induced by Mutant EGFR.

(A) A protein network involving EGFR was found in the plasma from mice with EGFR mutant lung adenocarcinoma. Proteins highlighted in red were significantly elevated or uniquely identified, and proteins highlighted in green were significantly decreased in plasma from tumor-bearing mice. Solid lines and dashed lines indicate direct and indirect interactions, respectively.
(B) Normalized spectral counts, indicative of protein abundance, of CDH1 in conditioned media and cell surface compartments of 10 EGFR mutant lung cancer cell lines compared with 11 KRAS mutant lung cancer cell lines. Columns indicate average and error bars indicate standard deviation.
(C) Cd44 peptides identified in lung cancer mouse models. Black bars indicate sequence location of peptides.
(D) Alternative splicing variants of CD44 in 21 lung adenocarcinoma cell lines. The locations of the common and the representative variant form of CD44 observed in EGFR mutant cell lines are indicated with an asterisk and an arrowhead, respectively. The no template control is indicated as NTC.
(E) Ratios of plasma Egfr levels in (1) lung adenocarcinoma tumor-bearing mice compared to controls, (2) in mice following injection of IL-6 relative to baseline, and (3) in tumor-bearing mice compared to controls for other mouse models investigated. Two replicates of IL-6 injection experiment (#1 and #2) were independently analyzed by mass spectrometry. The error bar indicates standard deviation.
(F) Return toward baseline following tumor regression of proteins with altered levels in tumor-bearing mice in the EGFR mutant mouse model. p value was calculated by paired t test.
Interestingly, although proteins that form an Egfr network were observed in plasma from tumor-bearing mice in the EGFR mouse model, levels of endogenous mouse Egfr in plasma from tumor-bearing mice in this model as well as the two other lung adenocarcinoma models were reduced (Table S4) compared to other models (p = 0.0018, t test) (Figure 3E). We hypothesized that soluble factors produced by lung tumors and/or through a host response may induce down-regulation of circulating levels of soluble Egfr, a major source of which in the mouse is the liver. Indeed, Egfr mRNA expression levels in liver from tumor-bearing EGFR mutant mouse model were decreased compared to controls (data not shown). In view of reports that IL-6 is induced at high levels in some primary human lung adenocarcinomas (Gao et al., 2007) and that its levels are increased in the plasma of patients with lung cancer (De Vita et al., 1998; Kaminska et al., 2006), although IL-6 was not detected by mass spectrometry in plasmas from mouse models possibly because of assay sensitivity, we examined the systemic effect of IL-6 administration on plasma levels of soluble Egfr. A similar reduction in plasma Egfr levels was observed with IL-6 treatment as in plasmas from lung tumor-bearing mice (Figure 3E), consistent with our hypothesis that soluble factors may induce down-regulation of Egfr in host tissues with lung tumor development.
Erlotinib-Treatment of Mice with EGFR Mutant Lung Tumors Restores Levels of Plasma Proteins toward Baseline
To assess protein levels in the context of treatment-induced changes in tumor volume, we compared plasma protein profiles of tumor-bearing mice carrying the EGFRL858R transgene before and after erlotinib treatment (Lung-Regression model; Politi et al., 2006). CCSP-rtTA; EGFRL858R mice were fed doxycycline for 5–6 weeks to induce tumors and were subsequently treated with 25 mg/kg/day of erlotinib for two weeks, after which plasma was collected. Plasma samples from mice in which tumor regression was documented were analyzed and compared to their control erlotinib-treated littermates. In total, levels of 91 of 164 proteins that were either elevated or decreased in plasmas from tumor-bearing mice in the EGFR model and that were quantified in the regression model, returned toward baseline (Figure 3F and Table S9). Thus, using this approach, we have identified proteins (e.g., Npc2 and Adam10) whose abundance reflects tumor progression and regression status in the EGFR model.
Plasma from SCLC Tumor-Bearing Mice Exhibits a Distinct Signature
SCLC exhibits distinct molecular characteristics compared to lung adenocarcinoma. Analysis of the plasma proteome of the SCLC mouse model yielded 116 proteins that were significantly elevated (case:control ratio > 1.25; p < 0.05, t test) in tumor-bearing mice but not in either the EGFR or Kras model (Figure 4A and Table S10). Ingenuity Pathway Analysis of these 116 proteins revealed that 33 (28.4%) proteins were associated with Neurological Disease or Nervous System Development and Function pathways (Table S10), pointing to a neural signature in the plasma protein profile of the SCLC mouse model, consistent with the neuroendocrine feature of SCLC. A significant network in the SCLC model consisted of proteins in the proteasome-ubiquitin pathway (Figure S1A and Table S8). Anti-apoptotic Bcl-2 overexpression, induced by proteasome-mediated NFκB activation, is observed in up to 90% of SCLCs, and inhibition of the proteasome pathway represents a potential therapeutic modality (Ben-Ezra et al., 1994; Lara et al., 2006),. A subset of the proteins with increased plasma levels (case:control ratios > 1.5) in tumor-bearing mice in the SCLC model also exhibited increased expression (case:control ratio > 1.25) of their corresponding genes in tumor tissue compared to normal lung tissue from the SCLC mouse model (accession number, GSE18534; (Schaffer et al., 2010) (Figure 4B), including Ncam1/Cd56, which has been previously associated with SCLC (Ledermann et al., 1994; Vangsted et al., 1994).
Figure 4. A Distinct Neuroendocrine Signature in Plasma of Mice with SCLC.
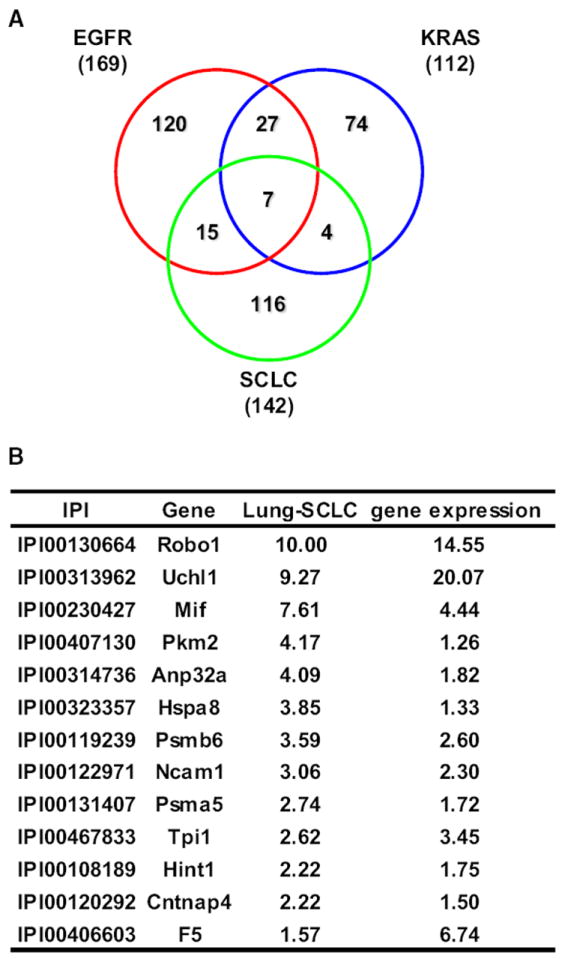
(A) Venn diagrams of elevated (case:control ratio > 1.25; p < 0.05, t test) proteins that were in common or unique to the EGFR, Kras, and SCLC mouse models.
(B) Plasma protein case:control ratios (>1.5) and corresponding gene expression ratios (>1.25) in tumor tissue from the SCLC mouse model compared to controls.
Concordance of Plasma Protein Changes Between Murine and Human Lung Cancer
To determine the potential relevance to human lung cancer of findings from mouse models, we performed assays in human blood samples of a set of proteins consisting of SFTPB, WFDC2, ANGPTL3, and EGFR for NSCLC and ROBO1 for SCLC, for which ELISAs were available. One source of human plasmas was from newly diagnosed smokers with operable NSCLC (n = 28) and control subjects matched for age, sex, smoking status, ethnicity, and plasma collection protocol (n = 39) (newly diagnosed set). A second set consisted of sera collected from subjects 0–11 months before they were diagnosed with NSCLC (n = 26) and matched control subjects who remained cancer free over a 4-year follow-up period (n = 26) (prediagnostic set) that were part of the Carotene and Retinol Efficacy Trial cohort study (Goodman et al., 2004). Demographics of subjects in these two sample sets are summarized in Table 2. For the group of newly diagnosed subjects, EGFR levels were decreased in subjects with lung cancer, and SFTPB, WFDC2, and ANGPTL3 levels were elevated with statistical significance, concordant with our findings in mouse models (p < 0.05 in t test or Mann-Whitney test) (Figure 5A). A receiver operating characteristic (ROC) analysis of the combined panel of EGFR, SFTPB, WFDC2, and ANGPTL3 using a linear combination rule yielded an AUC of 0.882 for the newly diagnosed set (Figure 5B). To further assess the potential contribution of the marker panel to early detection of lung cancer, we compared performance of the marker panel in the prediagnostic sample set with the performance of a previously validated panel (Qiu et al., 2008) consisting of autoantibodies to ANXA1, YWHAQ, and LAMR1 (autoantibody panel) in the same sample set (Figures 5C and 5D). For the group of subjects with sera collected before a diagnosis of NSCLC, an AUC of 0.808 was observed for the EGFR, SFTPB, WFDC2, and ANGPTL3 panel compared with 0.828 for the autoantibody panel and an AUC of 0.898 for the two panels combined.
Table 2.
Demographics of Subjects and Histology of Tumors
| Newly Diagnosed Set
|
Prediagnostic Set
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| NSCLC
|
SCLC
|
Control
|
NSCLC
|
Control
|
||||||
| No. | % | No. | % | No. | % | No. | % | No. | % | |
| Total | 28 | 100 | 10 | 100 | 39 | 100 | 26 | 100 | 26 | 100 |
|
| ||||||||||
| Age (years) | ||||||||||
|
| ||||||||||
| Mean | 63.3 | 64.1 | 61.7 | 65.2 | 64.5 | |||||
|
| ||||||||||
| SD | 10.7 | 7.9 | 9.6 | 6.4 | 6.1 | |||||
|
| ||||||||||
| Sex | ||||||||||
|
| ||||||||||
| Female | 13 | 46 | 8 | 80 | 22 | 56 | 6 | 23 | 6 | 23 |
|
| ||||||||||
| Male | 15 | 54 | 2 | 20 | 17 | 44 | 20 | 77 | 20 | 77 |
|
| ||||||||||
| Histology | ||||||||||
|
| ||||||||||
| Adenocarcinoma | 17 | 61 | — | — | 11 | 42 | — | |||
|
| ||||||||||
| Squamous | 11 | 39 | — | — | 10 | 38 | — | |||
|
| ||||||||||
| Other NSCLC | — | — | — | 5 | 19 | — | ||||
|
| ||||||||||
| SCLC | — | 10 | 100 | — | — | — | ||||
|
| ||||||||||
| Time span to diagnosis (months)
| ||||||||||
| Mean | 6.2 | |||||||||
|
| ||||||||||
| Range | 0–11 | |||||||||
Figure 5. Concordance of Plasma Protein Changes Between Murine and Human NSCLC.

(A–D) Plasma levels of EGFR, SFTPB, WFDC2, and ANGPTL3 were determined by ELISA in samples from newly diagnosed subjects with NSCLC (n = 28) relative to matched control subjects (n = 39) (newly diagnosed set) (A) and in prediagnostic sera samples (n = 26) relative to matched controls (n = 26) (prediagnostic set) (C). Data are log2-transformed, and the average of control samples is set to 1.0. Mean ± standard error (SE) for controls and subjects with NSCLC are shown. Receiver operating characteristic (ROC) curves for each ELISA assay and combined panel were analyzed in the newly diagnosed set (B) and in prediagnostic set (D). ROC curves for a previously validated panel of autoantibodies to ANXA1, YWHAQ, and LAMR1 (Qiu et al., 2008) alone and in combination with the ELISA panel are also shown (D).
(E) Levels of ROBO1 in plasmas from subjects with newly diagnosed SCLC (n = 10) relative to control subjects without cancer (n = 39). Data are log2-transformed, and the average of control samples is set to 1.0. Boxes indicate 25th and 75th percentiles, and horizontal lines inside the boxes indicate median. Bars indicate 10th and 90th percentiles, and black circles indicate data outside the 10th and 90th percentiles.
Given the finding of substantially increased levels of the neural protein Robo1 in tumor-bearing mouse plasma from the SCLC mouse model (Figure 4B), we examined the levels of this protein in human plasmas. An ELISA assay targeting the ectodomain of ROBO1 yielded significantly increased levels in plasma from patients with SCLC in comparison with levels in plasmas from control subjects without cancer (n = 10 and 39, respectively; Figure 5E), On the other hand, the levels of ROBO1 were not significantly elevated in plasmas from subjects with NSCLC (n = 28) in the newly diagnosed set (data not shown), suggesting that ROBO1 is a SCLC-specific biomarker, concordant with the findings from the SCLC mouse model.
DISCUSSION
Genetically engineered mouse models of human cancer have generated remarkably concordant histopathology, genetic profiles, and response to therapeutics when compared to their human counterparts. However, the use of such models to identify blood-based cancer signatures has been limited, and most published studies have focused on single models (Faca et al., 2008; Pitteri et al., 2011). The present study compared plasma proteomes of a relatively large number of mouse models on the basis of quantitative mass spectrometry. Analysis of the plasma proteome profiles resulted in a clustering of the four lung tumor models together, separate from other tumor models. Among 39 proteins present at increased levels in plasmas from mice with lung adenocarcinoma (Table S5), 17 were extracellular proteins, seven were localized to the plasma membrane, and 11 were intracellular, as determined by Ingenuity Pathway analysis. Furthermore, 21 of the proteins were demonstrated to be enriched in conditioned media of lung adenocarcinoma cell lines.
Of note, some previously described protein associations with lung cancer (e.g., CEA, CYFRA21-1, and serum amyloid A) were not revealed in our proteomic analysis of plasmas from lung adenocarcinoma mouse models. CEA (Ceacam5) was neither quantified nor identified in any mouse models, and CYFRA21-1 (fragment of Krt19) was only identified in the acute inflammation model. On the other hand, CEACAM5 and KRT19 were identified in 13 and 16 of 21 human lung adenocarcinoma cell lines, respectively (data not shown). These discrepancies may be due to the limitations of assay sensitivity of mass spectrometry or the lack of significant elevations of these proteins at early stages of tumor development (Van’t Westeinde and van Klaveren, 2011). Serum amyloid A (Saa1) was identified in all mouse models used in this study, but was not quantified (Tables S2 and S3), because none of the identified Saa1 peptides contained cysteine (data not shown). Therefore, Saa1 still could be a potential biomarker of lung cancer.
Strong evidence was obtained for a Titf1/Nkx2-1-related signature in plasmas from mice with lung adenocarcinomas. The evidence included expression of Titf1/Nkx2-1 and several of its targets in mouse lung tumors, a positive correlation between mRNA levels of protein signature genes and TITF1/NKX2-1 expression in human lung cancers and cell lines, and reduced expression of protein signature genes following knockdown of TITF1/NKX2-1 with short inhibitory RNA. The finding that a master regulator specific to a tissue lineage during airway development, as in the case of TITF1/NKX2-1, regulates production of proteins that are released into circulation with tumor development provides a new avenue to search for cancer protein signatures in plasma based on protein products of genes under the control of master developmental regulators that are also expressed during tumor development.
The discovery of an Egfr network in plasma from the mutant EGFR lung cancer model with the highest statistical significance among networks observed in that model suggests the occurrence in plasma of proteins that inform about genes and pathways driving tumor development. The Egfr signature included Adam10, which is known to be induced by Egfr signaling (McCulloch et al., 2004; Yan et al., 2002). Interestingly, Cdh1 and Cd44, which were also part of the Egfr containing network, are substrates of Adam10 (Maretzky et al., 2005; Murai et al., 2004). CD44 isoforms have been reported to occur in cancer cells (Ponta et al., 2003). Although inclusion of variant exons is stimulated by both RAS/MAPK pathway and EGF signaling (Huot et al., 2009; Weg-Remers et al., 2001), alternatively spliced isoforms of CD44 were found to occur predominantly in EGFR mutant lung adenocarcinoma cell lines concordant with our proteomic finding of peptides encoding Cd44 variant exon sequences only in the EGFR mutant mouse model (Figure 3D). Alternative splicing of CD44 has been linked to epithelial-mesenchymal transition triggers (Warzecha et al., 2009). Our CD44 peptide findings in the EGFR mutant mouse model and EGFR mutant lung adenocarcinoma cell lines may be related to an epithelial phenotype (Deng et al., 2009).
In our study, we observed distinct plasma protein signatures in SCLC compared to NSCLC. They included proteins that reflected a neural lineage consistent with neuroendocrine features of SCLC. Mouse Robo1 was the neural protein most elevated in plasma from the SCLC mouse model compared to control mice. Robo1 is a highly conserved axon guidance receptor and a member of the NCAM family. Targeted deletion of the Robo1 gene affects lung development and results in bronchial hyperplasia (Xian et al., 2001), and Slit-Robo1 signaling induces angiogenesis (Wang et al., 2003). Assay of ROBO1 plasma levels in human SCLC cases yielded concordant findings with the mouse model with statistically significantly increased levels in cases compared to healthy controls.
We obtained further evidence for concordance between findings in lung cancer mouse models plasmas and human blood samples with observations of reduced levels of circulating EGFR in our study and in a prior study (Lemos-González et al., 2007) and increased levels of circulating SFTPB, WFDC2, and ANGPTL3 in our study. Of particular relevance to lung cancer detection are results of assays using prediagnostic sera, which also yielded concordant results indicative of a potential utility of findings from mouse models for blood-based early detection strategies of lung cancer and for monitoring of subjects with the disease for tumor status.
EXPERIMENTAL PROCEDURES
Mouse Models and Mass Spectrometry Analysis for Mouse Plasma Samples
All animal experiments were conducted in accordance with institutional and national guidelines and regulations, under approval by the IACUC at Fred Hutchinson Cancer Research Center. Plasma samples from mouse models were obtained from tumor-bearing mice and control littermates. For each mouse model, an independent pool of case and control plasma was created. Pools of case and control plasma were formed by combining aliquots of plasma from 4–10 mice (Table S1). Details on mouse models, plasma sample preparation, mass spectrometry analysis, and statistical analysis are provided in Supplemental Experimental Procedures.
Cell Lines
Twenty-one lung adenocarcinoma cell lines were profiled (Table S5). Detailed analysis procedures and methods for cell culture, collection of whole cell extracts, conditioned media and cell surface proteins, and mass spectrometry analysis are given in Supplemental Experimental Procedures.
Informatic Analysis
For unsupervised hierarchical clustering of mouse plasma protein profile, the Cluster program (http://rana.lbl.gov/EisenSoftware.htm) was used to perform complete linkage hierarchical clustering of plasma proteins with filtering at least 40% presence and log2 transformation, and the result was displayed with the aid of TreeView software (http://rana.lbl.gov/EisenSoftware.htm) (Eisen et al., 1998).
For pathway analysis, Ingenuity Pathway Analysis (IPA) Software (Ingenuity Systems, Mountain View, CA) was used to perform pathway analysis of elevated/decreased and uniquely identified proteins from plasma of genetically engineered lung mouse models.
Histology and Immunohistochemistory
Tissues were fixed in 4% paraformaldehyde overnight at room temperature, placed in 70% ethanol, and sent for paraffin embedding and sectioning (Histoserv). The primary antibodies used for immunohistochemistry were anti-NPC2 (used at a dilution of 1:500, Sigma Cat. No. HPA000835), anti-surfactant protein B (used at a 1:250 dilution; Abcam Cat. No. ab40876), and anti-thyroid transcription factor 1 (used at a dilution of 1:200; Dako Cat. No. M3575).
Gene Expression and Western Blot Analysis
Microarray data for TITF1/NKX2-1 knockdown experiments was generated with the Illumina Human HT-12 array platform. Details are found in Supplemental Experimental Procedures.
Human Plasma Samples
Plasma samples were obtained at the time of diagnosis from subjects with operable NSCLC (n = 28), subjects with SCLC (n = 10), and healthy control subjects (n = 39) matched for age (±5 years), sex, and smoking history and plasma collection protocol. Twenty-six pairs of prediagnostic NSCLC sera samples and matched controls were collected 0–11 months prior to diagnosis as part of the Carotene and Retinol Efficacy Trial (CARET) at the time when they were completely asymptomatic. Control subjects were matched for age, sex, and smoking history and were not diagnosed with cancer over a 4-year follow-up period, irrespective of their state of general health otherwise. All human plasma samples were obtained with the informed consent and the approval of IRB at Fred Hutchinson Cancer Research Center.
ELISA Assays and Statistical Analysis
Levels of circulating EGFR (R& D Systems, dilution 1:300), SFTPB (USCN life, dilution 1:2000), WFDC2 (IBL-America, dilution 1:10), and ANGPTL3 (IBL-America, dilution 1:50) were measured according to the manufacturer’s protocols. The resulting data were normalized according to the mean of control subjects in each assay, and p values were calculated by use of the t test and Mann-Whitney test. To avoid biasing issues in ROC analysis, panels were generated using a linear combination of all assayed proteins. The level of ROBO1 was determined by in-house sandwich ELISA for ectodomain of ROBO1. Details on the ROBO1 ELISA assay are provided in Supplemental Experimental Procedures.
Supplementary Material
Significance.
We have applied a comparative strategy of mouse models of cancer to uncover protein signatures in plasma that reflect cell lineages of lung cancer, or that reflect pathways driving tumor development. Proteins not previously associated with lung cancer were identified in plasmas from lung cancer models compared with plasmas from other cancer models or models of inflammation. We also obtained evidence for concordant findings in human lung cancer cell lines and in plasmas collected from subjects with lung cancer at the time of diagnosis as well as in sera collected from asymptomatic subjects prior to diagnosis supporting the merits of comparative profiling of mouse models of cancer for the discovery of proteomic signatures relevant to humans.
Acknowledgments
Funding support was provided by the National Cancer Institute’s Mouse Models of Human Cancer program; the Canary, Labrecque, and Uniting Against Lung Cancer Foundations; and the Department of Defense (DOD) Congressionally Mandated Lung Cancer Research program. K.S.P. is a Parker B. Francis Fellow. D.M.D. is supported by the DOD (grant no. W81XWH-10-1-0263) and by the Burroughs Wellcome, V Foundation, Mary Kay Ash, and Rivkin Foundations.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes one figure, 10 tables, supplemental references, and supplemental Experimental Procedures and can be found with this article online at doi:10.1016/j.ccr.2011.08.007.
The authors declare no competing financial interests.
ACCESSION NUMBERS
Microarray data sets were deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo) with the accession number GSE28480.
References
- Ben-Ezra JM, Kornstein MJ, Grimes MM, Krystal G. Small cell carcinomas of the lung express the Bcl-2 protein. Am J Pathol. 1994;145:1036–1040. [PMC free article] [PubMed] [Google Scholar]
- Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, Joshi MB, Harpole D, Lancaster JM, Berchuck A, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- Bingle L, Cross SS, High AS, Wallace WA, Rassl D, Yuan G, Hellstrom I, Campos MA, Bingle CD. WFDC2 (HE4): a potential role in the innate immunity of the oral cavity and respiratory tract and the development of adenocarcinomas of the lung. Respir Res. 2006;7:61. doi: 10.1186/1465-9921-7-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camenisch G, Pisabarro MT, Sherman D, Kowalski J, Nagel M, Hass P, Xie MH, Gurney A, Bodary S, Liang XH, et al. ANGPTL3 stimulates endothelial cell adhesion and migration via integrin alpha vbeta 3 and induces blood vessel formation in vivo. J Biol Chem. 2002;277:17281–17290. doi: 10.1074/jbc.M109768200. [DOI] [PubMed] [Google Scholar]
- De Vita F, Orditura M, Auriemma A, Infusino S, Catalano G. Serum concentrations of proinflammatory cytokines in advanced non small cell lung cancer patients. J Exp Clin Cancer Res. 1998;17:413–417. [PubMed] [Google Scholar]
- Deng QF, Zhou CC, Su CX. Clinicopathological features and epidermal growth factor receptor mutations associated with epithelial-mesenchymal transition in non-small cell lung cancer. Respirology. 2009;14:371–376. doi: 10.1111/j.1440-1843.2009.01496.x. [DOI] [PubMed] [Google Scholar]
- Ding Z, Wu CJ, Chu GC, Xiao Y, Ho D, Zhang J, Perry SR, Labrot ES, Wu X, Lis R, et al. SMAD4-dependent barrier constrains prostate cancer growth and metastatic progression. Nature. 2011;470:269–273. doi: 10.1038/nature09677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinulescu DM, Ince TA, Quade BJ, Shafer SA, Crowley D, Jacks T. Role of K-ras and Pten in the development of mouse models of endometriosis and endometrioid ovarian cancer. Nat Med. 2005;11:63–70. doi: 10.1038/nm1173. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faca V, Coram M, Phanstiel D, Glukhova V, Zhang Q, Fitzgibbon M, McIntosh M, Hanash S. Quantitative analysis of acrylamide labeled serum proteins by LC-MS/MS. J Proteome Res. 2006;5:2009–2018. doi: 10.1021/pr060102+. [DOI] [PubMed] [Google Scholar]
- Faça VM, Hanash SM. In-depth proteomics to define the cell surface and secretome of ovarian cancer cells and processes of protein shedding. Cancer Res. 2009;69:728–730. doi: 10.1158/0008-5472.CAN-08-3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faca VM, Song KS, Wang H, Zhang Q, Krasnoselsky AL, Newcomb LF, Plentz RR, Gurumurthy S, Redston MS, Pitteri SJ, et al. A mouse to human search for plasma proteome changes associated with pancreatic tumor development. PLoS Med. 2008;5:e123. doi: 10.1371/journal.pmed.0050123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher GH, Wellen SL, Klimstra D, Lenczowski JM, Tichelaar JW, Lizak MJ, Whitsett JA, Koretsky A, Varmus HE. Induction and apoptotic regression of lung adenocarcinomas by regulation of a K-Ras transgene in the presence and absence of tumor suppressor genes. Genes Dev. 2001;15:3249–3262. doi: 10.1101/gad.947701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galgano MT, Hampton GM, Frierson HF., Jr Comprehensive analysis of HE4 expression in normal and malignant human tissues. Mod Pathol. 2006;19:847–853. doi: 10.1038/modpathol.3800612. [DOI] [PubMed] [Google Scholar]
- Gao SP, Mark KG, Leslie K, Pao W, Motoi N, Gerald WL, Travis WD, Bornmann W, Veach D, Clarkson B, Bromberg JF. Mutations in the EGFR kinase domain mediate STAT3 activation via IL-6 production in human lung adenocarcinomas. J Clin Invest. 2007;117:3846–3856. doi: 10.1172/JCI31871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman GE, Thornquist MD, Balmes J, Cullen MR, Meyskens FL, Jr, Omenn GS, Valanis B, Williams JH., Jr The Beta-Carotene and Retinol Efficacy Trial: incidence of lung cancer and cardiovascular disease mortality during 6-year follow-up after stopping beta-carotene and retinol supplements. J Natl Cancer Inst. 2004;96:1743–1750. doi: 10.1093/jnci/djh320. [DOI] [PubMed] [Google Scholar]
- Hellström I, Raycraft J, Hayden-Ledbetter M, Ledbetter JA, Schummer M, McIntosh M, Drescher C, Urban N, Hellström KE. The HE4 (WFDC2) protein is a biomarker for ovarian carcinoma. Cancer Res. 2003;63:3695–3700. [PubMed] [Google Scholar]
- Horio Y, Chen A, Rice P, Roth JA, Malkinson AM, Schrump DS. Ki-ras and p53 mutations are early and late events, respectively, in urethane-induced pulmonary carcinogenesis in A/J mice. Mol Carcinog. 1996;17:217–223. doi: 10.1002/(SICI)1098-2744(199612)17:4<217::AID-MC5>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Hung KE, Faca V, Song K, Sarracino DA, Richard LG, Krastins B, Forrester S, Porter A, Kunin A, Mahmood U, et al. Comprehensive proteome analysis of an Apc mouse model uncovers proteins associated with intestinal tumorigenesis. Cancer Prev Res (Phila) 2009;2:224–233. doi: 10.1158/1940-6207.CAPR-08-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huot ME, Vogel G, Richard S. Identification of a Sam68 ribonu-cleoprotein complex regulated by epidermal growth factor. J Biol Chem. 2009;284:31903–31913. doi: 10.1074/jbc.M109.018465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminska J, Kowalska M, Kotowicz B, Fuksiewicz M, Glogowski M, Wojcik E, Chechlinska M, Steffen J. Pretreatment serum levels of cytokines and cytokine receptors in patients with non-small cell lung cancer, and correlations with clinicopathological features and prognosis: M-CSF—an independent prognostic factor. Oncology. 2006;70:115–125. doi: 10.1159/000093002. [DOI] [PubMed] [Google Scholar]
- Kelly-Spratt KS, Pitteri SJ, Gurley KE, Liggitt D, Chin A, Kennedy J, Wong CH, Zhang Q, Buson TB, Wang H, et al. Plasma proteome profiles associated with inflammation, angiogenesis, and cancer. PLoS ONE. 2011;6:e19721. doi: 10.1371/journal.pone.0019721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koishi R, Ando Y, Ono M, Shimamura M, Yasumo H, Fujiwara T, Horikoshi H, Furukawa H. Angptl3 regulates lipid metabolism in mice. Nat Genet. 2002;30:151–157. doi: 10.1038/ng814. [DOI] [PubMed] [Google Scholar]
- Lara PN, Jr, Chansky K, Davies AM, Franklin WA, Gumerlock PH, Guaglianone PP, Atkins JN, Farneth N, Mack PC, Crowley JJ, Gandara DR. Bortezomib (PS-341) in relapsed or refractory extensive stage small cell lung cancer: a Southwest Oncology Group phase II trial (S0327) J Thorac Oncol. 2006;1:996–1001. [PubMed] [Google Scholar]
- Ledermann JA, Pasini F, Olabiran Y, Pelosi G. Detection of the neural cell adhesion molecule (NCAM) in serum of patients with small-cell lung cancer (SCLC) with “limited” or “extensive” disease, and bone-marrow infiltration. Int J Cancer Suppl. 1994;8:49–52. doi: 10.1002/ijc.2910570710. [DOI] [PubMed] [Google Scholar]
- Lemos-González Y, Rodriíguez-Berrocal FJ, Cordero OJ, Gómez C, Páez de la Cadena M. Alteration of the serum levels of the epidermal growth factor receptor and its ligands in patients with non-small cell lung cancer and head and neck carcinoma. Br J Cancer. 2007;96:1569–1578. doi: 10.1038/sj.bjc.6603770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin B, White JT, Wu J, Lele S, Old LJ, Hood L, Odunsi K. Deep depletion of abundant serum proteins reveals low-abundant proteins as potential biomarkers for human ovarian cancer. Proteomics Clin Appl. 2009;3:853–861. doi: 10.1002/prca.200800141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maretzky T, Reiss K, Ludwig A, Buchholz J, Scholz F, Proksch E, de Strooper B, Hartmann D, Saftig P. ADAM10 mediates E-cadherin shedding and regulates epithelial cell-cell adhesion, migration, and beta-catenin translocation. Proc Natl Acad Sci USA. 2005;102:9182–9187. doi: 10.1073/pnas.0500918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulloch DR, Akl P, Samaratunga H, Herington AC, Odorico DM. Expression of the disintegrin metalloprotease, ADAM-10, in prostate cancer and its regulation by dihydrotestosterone, insulin-like growth factor I, and epidermal growth factor in the prostate cancer cell model LNCaP. Clin Cancer Res. 2004;10:314–323. doi: 10.1158/1078-0432.ccr-0846-3. [DOI] [PubMed] [Google Scholar]
- Meuwissen R, Linn SC, Linnoila RI, Zevenhoven J, Mooi WJ, Berns A. Induction of small cell lung cancer by somatic inactivation of both Trp53 and Rb1 in a conditional mouse model. Cancer Cell. 2003;4:181–189. doi: 10.1016/s1535-6108(03)00220-4. [DOI] [PubMed] [Google Scholar]
- Meylan E, Dooley AL, Feldser DM, Shen L, Turk E, Ouyang C, Jacks T. Requirement for NF-kappaB signalling in a mouse model of lung adenocarcinoma. Nature. 2009;462:104–107. doi: 10.1038/nature08462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody SE, Sarkisian CJ, Hahn KT, Gunther EJ, Pickup S, Dugan KD, Innocent N, Cardiff RD, Schnall MD, Chodosh LA. Conditional activation of Neu in the mammary epithelium of transgenic mice results in reversible pulmonary metastasis. Cancer Cell. 2002;2:451–461. doi: 10.1016/s1535-6108(02)00212-x. [DOI] [PubMed] [Google Scholar]
- Murai T, Miyazaki Y, Nishinakamura H, Sugahara KN, Miyauchi T, Sako Y, Yanagida T, Miyasaka M. Engagement of CD44 promotes Rac activation and CD44 cleavage during tumor cell migration. J Biol Chem. 2004;279:4541–4550. doi: 10.1074/jbc.M307356200. [DOI] [PubMed] [Google Scholar]
- Pérez-Gil J. Structure of pulmonary surfactant membranes and films: the role of proteins and lipid-protein interactions. Biochim Biophys Acta. 2008;1778:1676–1695. doi: 10.1016/j.bbamem.2008.05.003. [DOI] [PubMed] [Google Scholar]
- Pernemalm M, De Petris L, Eriksson H, Brandén E, Koyi H, Kanter L, Lewensohn R, Lehtioö J. Use of narrow-range peptide IEF to improve detection of lung adenocarcinoma markers in plasma and pleural effusion. Proteomics. 2009;9:3414–3424. doi: 10.1002/pmic.200800814. [DOI] [PubMed] [Google Scholar]
- Pitteri SJ, Faca VM, Kelly-Spratt KS, Kasarda AE, Wang H, Zhang Q, Newcomb L, Krasnoselsky A, Paczesny S, Choi G, et al. Plasma proteome profiling of a mouse model of breast cancer identifies a set of up-regulated proteins in common with human breast cancer cells. J Proteome Res. 2008;7:1481–1489. doi: 10.1021/pr7007994. [DOI] [PubMed] [Google Scholar]
- Pitteri SJ, JeBailey L, Faça VM, Thorpe JD, Silva MA, Ireton RC, Horton MB, Wang H, Pruitt LC, Zhang Q, et al. Integrated proteomic analysis of human cancer cells and plasma from tumor bearing mice for ovarian cancer biomarker discovery. PLoS ONE. 2009;4:e7916. doi: 10.1371/journal.pone.0007916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitteri SJ, Kelly-Spratt KS, Gurley KE, Kennedy J, Buson TB, Chin A, Wang H, Zhang Q, Wong CH, Chodosh LA, et al. Tumor micro-environment-derived proteins dominate the plasma proteome response during breast cancer induction and progression. Cancer Res. 2011;71:5090–5100. doi: 10.1158/0008-5472.CAN-11-0568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politi K, Zakowski MF, Fan PD, Schonfeld EA, Pao W, Varmus HE. Lung adenocarcinomas induced in mice by mutant EGF receptors found in human lung cancers respond to a tyrosine kinase inhibitor or to down-regulation of the receptors. Genes Dev. 2006;20:1496–1510. doi: 10.1101/gad.1417406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponta H, Sherman L, Herrlich PA. CD44: from adhesion molecules to signalling regulators. Nat Rev Mol Cell Biol. 2003;4:33–45. doi: 10.1038/nrm1004. [DOI] [PubMed] [Google Scholar]
- Qiu J, Choi G, Li L, Wang H, Pitteri SJ, Pereira-Faca SR, Krasnoselsky AL, Randolph TW, Omenn GS, Edelstein C, et al. Occurrence of autoantibodies to annexin I, 14-3-3 theta and LAMR1 in prediagnostic lung cancer sera. J Clin Oncol. 2008;26:5060–5066. doi: 10.1200/JCO.2008.16.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer BE, Park KS, Yiu G, Conklin JF, Lin C, Burkhart DL, Karnezis AN, Sweet-Cordero EA, Sage J. Loss of p130 accelerates tumor development in a mouse model for human small-cell lung carcinoma. Cancer Res. 2010;70:3877–3883. doi: 10.1158/0008-5472.CAN-09-4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid K, Bago-Horvath Z, Berger W, Haitel A, Cejka D, Werzowa J, Filipits M, Herberger B, Hayden H, Sieghart W. Dual inhibition of EGFR and mTOR pathways in small cell lung cancer. Br J Cancer. 2010;103:622–628. doi: 10.1038/sj.bjc.6605761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solassol J, Rouanet P, Lamy PJ, Allal C, Favre G, Maudelonde T, Mangé A. Serum protein signature may improve detection of ductal carcinoma in situ of the breast. Oncogene. 2010;29:550–560. doi: 10.1038/onc.2009.341. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Yanagisawa K, Shinjo K, Taguchi A, Maeno K, Tomida S, Shimada Y, Osada H, Kosaka T, Matsubara H, et al. Lineage-specific dependency of lung adenocarcinomas on the lung development regulator TTF-1. Cancer Res. 2007;67:6007–6011. doi: 10.1158/0008-5472.CAN-06-4774. [DOI] [PubMed] [Google Scholar]
- Van’t Westeinde SC, van Klaveren RJ. Screening and early detection of lung cancer. Cancer J. 2011;17:3–10. doi: 10.1097/PPO.0b013e3182099319. [DOI] [PubMed] [Google Scholar]
- Vangsted A, Drivsholm L, Andersen E, Bock E. New serum markers for small-cell lung cancer. II. The neural cell adhesion molecule, NCAM. Cancer Detect Prev. 1994;18:291–298. [PubMed] [Google Scholar]
- Vanier MT, Millat G. Structure and function of the NPC2 protein. Biochim Biophys Acta. 2004;1685:14–21. doi: 10.1016/j.bbalip.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Wang B, Xiao Y, Ding BB, Zhang N, Yuan X, Gui L, Qian KX, Duan S, Chen Z, Rao Y, Geng JG. Induction of tumor angiogenesis by Slit-Robo signaling and inhibition of cancer growth by blocking Robo activity. Cancer Cell. 2003;4:19–29. doi: 10.1016/s1535-6108(03)00164-8. [DOI] [PubMed] [Google Scholar]
- Warzecha CC, Sato TK, Nabet B, Hogenesch JB, Carstens RP. ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol Cell. 2009;33:591–601. doi: 10.1016/j.molcel.2009.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weg-Remers S, Ponta H, Herrlich P, König H. Regulation of alternative pre-mRNA splicing by the ERK MAP-kinase pathway. EMBO J. 2001;20:4194–4203. doi: 10.1093/emboj/20.15.4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BA, Woo MS, Getz G, Perner S, Ding L, Beroukhim R, Lin WM, Province MA, Kraja A, Johnson LA, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xian J, Clark KJ, Fordham R, Pannell R, Rabbitts TH, Rabbitts PH. Inadequate lung development and bronchial hyperplasia in mice with a targeted deletion in the Dutt1/Robo1 gene. Proc Natl Acad Sci USA. 2001;98:15062–15066. doi: 10.1073/pnas.251407098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Shirakabe K, Werb Z. The metalloprotease Kuzbanian (ADAM10) mediates the transactivation of EGF receptor by G protein-coupled receptors. J Cell Biol. 2002;158:221–226. doi: 10.1083/jcb.200112026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee J, Sadar MD, Sin DD, Kuzyk M, Xing L, Kondra J, McWilliams A, Man SF, Lam S. Connective tissue-activating peptide III: a novel blood biomarker for early lung cancer detection. J Clin Oncol. 2009;27:2787–2792. doi: 10.1200/JCO.2008.19.4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


