Abstract
For more than a decade, fluorescence resonance energy transfer (FRET) imaging methods have been developed to study dynamic interactions between molecules at the nanometer scale in live cells. Here, we describe a protocol to measure FRET by the acceptor-sensitized emission method as detected by total internal reflection fluorescence (TIRF) imaging to study the interaction of appropriately labeled plasma membrane-associated molecules that regulate the earliest stages of antigen-mediated signaling in live B lymphocytes. This protocol can be adapted and applied to many cell types where there is an interest in understanding signal transduction mechanisms in live cells.
Keywords: B lymphocyte, fluorescence resonance energy transfer (FRET), total internal reflection fluorescence (TIRF) imaging, B cell receptor (BCR) signaling, planar lipid bilayer, immune synapse
1. Introduction
B lymphocytes play a pivotal role in the adaptive immune system by producing antibodies against pathogens (1). Binding of antigens to B cell receptors (BCRs) leads to BCR clustering that triggers the recruitment of Lyn, the first kinase in the BCR signaling cascade (2). Current evidence indicates that B cells contact antigen on the surface of antigen presenting cells (APCs). At the interface of the contact between B cell and APC, an immune synapse (IS), a structure associated with B cell activation, is organized. Within the IS, various receptor-ligand interactions take place, including the association of the BCR with the antigen and the lymphocyte function-associated antigen-1 (LFA-1) with the intercellular cell adhesion molecule-1 (ICAM-1) (3, 4). Live cell imaging techniques provide both the temporal and spatial information concerning the dynamic association of the BCR with signaling molecules over the time and length scale that are critical to decipher the earliest events that occur during the contact of live B cells with APCs bearing antigen. The advent of fluorescent protein technologies has made such observations possible by standard microscopy, and the development of sensitive imaging techniques, such as TIRF imaging, has enabled the study of signaling proteins at a single molecule level (5, 6).
TIRF imaging is a spatially limited technique that is particularly well suited to the study of molecules near the plasma membrane. In TIRF imaging, fluorophores in specimens are excited with collimated light, typically from a laser, that is introduced at a specific angle. The incident beam of excitation light, brought either through a prism or a high numerical aperture, oil-immersion objective lens, is totally internally reflected to form an evanescent field at the interface of the coverslip and aqueous media bathing the cells. In the case of through-lens systems, an evanescent field is produced along the optical axis by virtue of the difference in refractive index between oil/glass and aqueous medium of the cell. The evanescent field exponentially decays as it enters the cell, penetrating to a depth of ~100 nm, with depth being dependent on the angle of incidence and the wavelength of excitation light. As a result, TIRF imaging greatly reduces background fluorescence outside the plane of focus to the point where fluorescent emission from single fluorophores can be detected.
To study the interaction of membrane-associated proteins between B cells and APCs by TIRF imaging, planar lipid bilayers bearing mobile ligands have been developed to mimic APC (7). This system provides a means to the study the spatial and temporal dynamics of receptor-signaling molecule interaction in live B cells introduced to the lipid bilayer-coated chamber coverslip. In contrast to standard confocal microscopy, TIRF imaging enables the entire surface area of the cell in contact with the planar lipid bilayer to be imaged as interfering fluorescent signals from the interior of the cell are eliminated. In addition, because highly sensitive CCD cameras are used to capture images, frame rates can greatly exceed (>30 fps) those obtained on a typical scanning confocal microscope.
We have taken advantage of TIRF microscope technology and combined it with FRET imaging to study dynamic signal transduction events in live B cells (8, 9). FRET is the non-radioactive transfer of energy from a donor fluorophore to an acceptor fluorophore by dipole–dipole coupling. The result is a quenching of donor fluorescence and a concomitant increase (sensitization) in acceptor emission. Energy transfer can take place only over a limited distance (within 10 nm); thus, the closer the donor and acceptor fluorophores, the more efficient is the energy transfer. FRET efficiency (E) is exquisitely sensitive to the distance separating donor and acceptor fluorophores, being inversely dependent by the 6th power (10). This dependency provides a nanometer-scale resolution that can be measured via standard light microscopes. Several techniques have been developed to detect FRET, which include acceptor photobleaching, fluorescence lifetime imaging (FLIM) of the donor fluorophore, and acceptor-sensitized emission (also called three cube FRET) (11–13). While the sensitized emission approach requires the most careful controls and corrections of all the methods, it allows FRET measurements to be performed on relatively inexpensive equipment (compared to FLIM) and allows for dynamic measurements in live cells (compared to acceptor photobleaching, which is an end point method that destroys the fluorescence of the cell). To measure FRET by the sensitized emission, images from three channels are acquired: donor channel (D), acquired with the donor laser excitation wavelength and donor emission filter; FRET channel (F), acquired with donor laser excitation wavelength and acceptor emission filter; and acceptor channel (A), acquired with acceptor laser excitation wavelength and acceptor emission filter. Acquired images are processed to calculate FRET efficiency and FRET efficiency can be expressed as either FRET efficiency for the acceptor (Ea) or FRET efficiency for the donor (Ed) according to the following equations (14):
| [1] |
| [2] |
where FRa and FRd are determined by
| [3] |
| [4] |
In these equations, D, F and A are background-subtracted fluorescence intensities in the donor, FRET, and acceptor channels, respectively. β is correction factor for donor spectral bleed-through into FRET channel (F/D) when excited by 442 nm laser and is determined from cells expressing only the donor fluorophore. γ and δ are correction factors obtained from cells expressing only the acceptor fluorophore for acceptor crosstalk captured in FRET channel (F/A) and spectral bleed-through of acceptor emission back into donor channel (D/F), respectively. Ka and Kd are conversion constants between FRET ratios and FRET efficiencies determined from cells expressing a FRET positive control. The positive control is a fusion of the donor and the acceptor fluorophore at distance close enough to produce non-zero FRET at a 1:1 molar ratio. In the protocol below, we monitor the association of the BCR with Lyn in B cells contacting antigen-bound membranes by FRET/TIRF imaging, using CH27 cells which express Lyn-CFP (donor) and Igα-YFP (acceptor), and biotinylated antigens which are bound on the biotin-lipid membrane through a streptavidin bridge, to reveal the initiation of BCR signaling in response to the membrane-bound antigens in a spatial and temporal way.
2. Materials
2.1. Cell Culture
CH27, a mouse B lymphoma cell line (see Note 1).
Cell culture medium: Iscove’s modified Dulbecco medium (IMDM) supplemented with 15% fetal bovine serum (FBS) (Gibco/BRL, Bethesda, MD), 10 mM of MEM non-essential amino acids solution (GIBCO), 50 mM β-mercaptoethanol, 10,000 units/mL of penicillin and 10,000 μg/mL of streptomycin solution (Pen Strep, 100×) (GIBCO).
Plasmids: Lyn-CFP (the FRET donor), Igα-YFP (the FRET acceptor), and Lyn16-CFP-YFP (the FRET positive control). These plasmids can be obtained from authors’ laboratory (see Note 2). It is recommended to purify plasmid DNAs with an endotoxin-free column prior to transfections (QIAGEN, Valencia, CA).
Cell line Nucleofector kit V and NuclefectorII machine (Amaxa Inc., USA). Keep the reagent at 4°C and use within 90 days after mixing the two components.
12-well culture plates (Corning, Corning, NY).
2.2. Preparation of Planar Lipid Bilayers Bearing Antigen
2.2.1. Cleaning of All Glass Items
Fresh KOH/EtOH cleaning solution: 2.1 M KOH, 85% EtOH (see Note 3). Caution: this solution is corrosive and KOH liberates toxic gas when it contacts water; avoid exposure.
Rinsing solution: 95% EtOH, 100% EtOH.
NextGen™ V 5 mL clear glass vials with a V-shaped bottom (Wheaton Science Products, Millville, NJ).
Coplin jars with lids.
Roll & Grow™ Plating Glass Beads (MP Biomedicals, Solon, OH).
Deionized ultrapure water.
2.2.2. Preparation of Small Unilamellar Vesicles (SUVs)
25 mM 1,2-Dioleoyl-sn-Glycero-3-phosphocholine (DOPC) (Avanti Polar Lipids, Alabaster, AL). This is provided in chloroform as a 20 mg/mL solution.
10 mM 1,2-Dioleoyl-sn-Glycero-3-phosphoethanolamine-cap-biotin (DOPE-cap-biotin) (Avanti Polar Lipids). This is provided in chloroform as a 10 mg/mL solution. Keep both DOPC and DOPE-cap-biotin at −20°C (see Note 4).
200 μL and 1 mL size Hamilton syringes.
High purity chloroform. Caution: this is toxic, flammable, and can easily evaporate. Store at room temperature in a safety storage cabinet.
Compressed argon gas with gas regulator (Model: 10-575-110, Fisher Scientific, Pittsburg, PA). Insert a Pasteur pipette into the 0.45-μm syringe-type filter (Corning) connected to the gas regulator via tubing for drying lipid mixtures.
Water-bath-type sonicator (Model: G112SP1G, Laboratory Supplies, Hicksville, NY).
Phosphate-buffered saline (PBS), pH7.4: Prepare 10×stock with 1.55 M NaCl, 16.9 mM KH2PO4, and 26.5 mM Na2HPO4, pH7.4 (adjusted with HCl), and dilute with fresh ultrapure deionized water and filter with a disposable bottle-top vacuum filter.
Beckman rotor, SW55Ti and Ultra-Clear 13×51 mm ultracentrifuge tubes (Beckman Instruments Inc., Palo Alto, CA).
Individually wrapped serological glass pipettes and Pasteur pipettes (Kimble Glass, Owens, IL).
2.2.3. Preparation of Antigen-Tethered Planar Lipid Bilayer
SUV working solution (0.1 mM): Dilute 20 μL of 5 mM SUV stock solution with 1 mL of freshly prepared PBS. The SUV stock solution is stable for several months in a refrigerator, but planar lipid bilayers must be prepared freshly on the same day of image acquisition from diluted SUV working solution.
24×50 mm #1.5 coverglasses.
Nanostrip solution (OM Group Ultra Pure Chemicals, Derbyshire, UK). This is a strong acid and will react with carbohydrate. Protect skin and eyes when using. Store at room temperature in the safety storage cabinet.
Lab-Tek chamber #1.0 Borosilicate coverglasses (Nalge Nunc International, Rochester, NY) (see Note 5).
Sylgard 164 Silicone Elastomer adhesive (Dow Corning, Midland, MI).
Biotinylated goat Fab anti-mouse IgM and streptavidin (Jackson Immuno Research Lab, West Grove, PA): Prepare for 20 μM and 5 mg/mL stock solution, respectively, in 50% glycerol and keep at −20°C. Both are stable for at least 1 year in this condition.
2.3. FRET/TIRF Imaging
2.3.1. Imaging Reagents
Imaging buffer: 1×PBS or 1×Hank’s balanced salt solution (HBSS) containing 0.1% FBS (Gibco/BRL). Freshly prepare with ultrapure deionized water and filter it with disposable bottle-top filter system.
FluoSpheres polystyrene microspheres, 1.0 μm (430/465 nm) (Molecular Probes, Eugene, OR). Store at 2–6°C and protect from light.
2.3.2. Imaging Equipment
Olympus IX-81 microscope equipped with a TIR illumination port and an IX2 ZDC autofocus laser system (Olympus USA, Center Valley, PA).
Olympus TIRF microscope objective lenses: 60×, 1.45 NA PlanApo and 100×, 1.45 NA PlanApo.
442/514 nm polychroic mirror for Olympus filter cube (Chroma Technology, Rockingham, VT).
442 and 514 nm laser-line (notch) blocking filters (Semrock, Rochester, NY). Required for additional laser blocking in the emission path.
FC/APC single-mode fiber optic cable (OZ Optics, Ottawa, ON, Canada).
FW1000 excitation and emission filter wheels (ASI, Eugene, OR).
MS2000 automated X-Y, piezo Z stage (ASI, Eugene, OR).
Two channel optical splitter, DualView (MAG Biosystems, Tucson, AZ) equipped with a 505 nm dichroic filter and HQ485/30 nm and HQ560/50 nm emission filters (Chroma Technology, Rockingham, VT) to collect CFP and YFP signals, respectively.
40 mW, 442 nm diode laser (BlueSky Research, Milpitas, CA).
300 mW argon gas 488/514 nm laser (Dynamic Laser, Salt Lake City, UT).
Cascade 512II EM-CCD, −70°C cooled camera (Photometrics, Tucson, AZ).
Acousto-optical tunable filter and multichannel controller (AOTF) (NEOS, Melbourne, FL).
Computer equipped with: Intel Core Duo, 2.66 GHz processor, 4 GB RAM, Windows XP, 500 GB hard drive, four or more 9 pin com ports, three or more open PCI slots.
MetaMorph system control software (Molecular Devices, Downingtown, PA).
Image-Pro Plus Software 6.3 (MediaCybernetics, Bethesda, MD) and Microsoft Office Excel for image analysis.
3. Methods
To obtain robust and reproducible FRET data, accurate determination of the correction factors, β, γ , and δ is necessary. The FRET channel contains not only the sensitized acceptor emission by FRET but also spectral contaminants from the direct excitation and emission of CFP and YFP that the correction factors eliminate. Moreover, an accurate determination of the conversion constants, Ka or Kd, are required to allow FRET data to be compared between different experiments. Therefore, image acquisitions of control and experimental samples must be performed for each imaging session and the constants calculated from control cells for each experiment.
3.1. Cell Culture and Preparation of Cells Expressing Lyn-CFP and/or Igα-YFP, and Lyn16-CFP-YFP
Maintain CH27 cells in 15% IMDM culture media in T-25 culture flasks at 37°C under 7% CO2 in a humidified incubator. Passage cells by 1:15 dilution of cells into fresh media when confluent (see Note 6).
Prepare purified plasmids, Lyn-CFP, Igα-YFP, Lyn16-CFP-YFP, so that they are ready for transient transfection using Amaxa Nucleofection 1 day before imaging (see Note 7 for other transfection options).
Passage cells 3 days before imaging by diluting cells 1:15 in fresh media in T-75 flask.
One day before imaging, when cells reach a density of ~1.5–2.0×106 cells/mL, transfer 3.0×106 cells to a 15-mL centrifuge tube for each transfection. Pellet cells at 90×g at room temperature for 10 min (see Note 8).
While centrifuging, set Amaxa Nucleofector II device to program A-23. For CH27 cells, best transfection efficiencies are obtained with Nucleofector Kit V reagent and program A-23, which is recommended for the control GFP plasmid in Amaxa protocol.
Aspirate medium completely from the cell pellet with a 2-mL pipette connected to a vacuum waste tank (see Note 9).
Add 100 μL of reagent V pre-mixed with supplement and resuspend the cell pellet completely by gently shaking and pipetting once or twice using micropipette. Perform steps 7–10 for each sample separately.
Use 4 μg of plasmid DNA for each transfection. Use 2 μg of DNA per each plasmid if transfecting two plasmids. Mix DNA with the resuspended cells by brief, gentle shaking.
Carefully transfer the sample to an Amaxa-certified cuvette. Avoid air bubbles while pipetting, and close with blue cap.
Insert the cuvette into the holder of Nucleofector II device, and press ≫X≪ button to start program A-23.
Immediately after completion, remove the cuvette from holder and gently add about 1 mL of warm culture media to the bottom of the cuvette using plastic pipette provided in the kit, and transfer to a 12-well culture plate containing 1 mL of warm media per well.
Incubate overnight at 37°C under 7% CO2 in a humidified incubator.
Check cells the next day for viability and transfection efficiency using inverted epi-fluorescence microscope. Confirm expression levels and distribution of Lyn-CFP and Igα-YFP (see Note 10).
3.2. Preparation of Planar Lipid Bilayers Bearing Antigen
3.2.1. Cleaning of All Glass Items in KOH/EtOH Cleaning Solution
Separate glass vials from caps and immerse ten vials in freshly prepared KOH/EtOH cleaning solution for 10 min. Make sure that the vials are completely filled with the cleaning solution; remove air bubbles if required.
Remove the cleaning solution and transfer the vials to a 2-L beaker containing 1 L of 95% EtOH. Rinse briefly by moderately swirling the beaker several times.
With vigorous shaking, rinse each vial thoroughly under a flow of ultrapure deionized water.
Bake vials at 160°C for 1 h.
Place all caps in 100% EtOH in a beaker and vigorously shake for a few minutes. Then discard the solution.
Rinse three times with a copious amount of ultrapure deionized water to remove EtOH.
Dry caps at 60°C.
Likewise, clean glass beads and three Coplin jars with KOH/EtOH solution, rinse, and oven-dry.
3.2.2. Preparation of SUVs
Fill three cleaned vials with 4 mL of 100% EtOH and another three vials with 4 mL of chloroform in a fume hood.
Place the tip of a Hamilton syringe in the first EtOH vial and move the piston up and down several times to clean. Repeat the process sequentially in the next two EtOH vials followed by three chloroform vials. Air-dry the syringe on Kimwipes.
Using two syringes, transfer to a cleaned vial 1 mL of 25 mM DOPC and 25 μL of 10 mM DOPE-cap-biotin in chloroform, resulting in a 100:1 ratio of DOPC:DOPE-cap-biotin.
Tighten the cap and mix briefly by tapping. For vials containing stock lipid solutions, immediately fill them with argon gas. Store lipid stocks at −20°C.
Dry the lipid mixture in a glass vial under a stream of argon gas. Slowly rotate the vial until the solution is dried up and a thin film of lipid is formed on the walls (see Note 11).
Completely evaporate residual chloroform by putting the vial under high vacuum at room temperature overnight (see Note 12).
For lipid hydration, degas ~20 mL of fresh 1×PBS in a nanostrip-cleaned side-arm flask (see Note 13). Degassing is complete when solution stops releasing bubbles when shaken.
Hydrate the lipid film with 5 mL of degassed PBS at room temperature. Fill the vial with argon gas and cap tightly.
Vortex for about 30 s until the lipid film is completely dissociated from the wall and makes an opaque solution. Keep on ice.
Fill the sonicator with a mixture of ice and water to achieve bath temperature of <4°C. Continuously sonicate the lipid mixture in 10-min rounds until it becomes clear (see Note 14). This indicates the formation of SUVs.
Transfer the SUVs into a 5-mL ultracentrifuge tube to fit Beckman SW55Ti rotor and clarify the SUV solution by ultracentrifugation at 4°C for 1 h at 46,800 g (see Note 15).
Transfer supernatant containing the SUVs into a new 5-mL tube using a 5-mL sterile serological pipette and continue clarifying with ultracentrifugation at 4°C for 8 h at 54,700 g using the same rotor. Carefully transfer supernatant into a new 15-mL plastic tube.
Fill the tube with argon gas; cap and seal it with parafilm. The SUVs are ready for use (see Note 16).
3.2.3. Preparation of Antigen-Tethered Planar Lipid Bilayers
Fill each of three Coplin jars with Nanostrip, deionized ultrapure water, and 100% EtOH, respectively.
Place 24×50 #1.5 coverslips into the jar containing Nanostrip for at least 1 h.
Transfer the coverslips with forceps into the jar containing deionized ultrapure water and agitate several times. Discard, fill with fresh ultrapure water, and repeat the process ten times.
Transfer the coverslips into the jar containing 100% EtOH for several minutes.
Take out a coverslip with forceps and blow-dry it completely with a stream of argon gas. Place the coverslip on top of a monolayer of cleaned glass beads in a sterile 100-mm round culture dish.
Tear off the bottom coverslip of an 8-well Lab-Tek chamber and attach the Nanostrip-cleaned coverslip to chamber with Sylgard 164 (see Note 17).
Dilute 20 μL of 5 mM SUVs into 1 mL of PBS for a 0.1 mM final concentration of SUVs. Dispense 200 μL of diluted SUVs into each of the wells. Wait for 10 min.
Rinse planar lipid bilayer in each well with about 20 mL of 1×PBS, keeping it in solution at all times. After the last rinse, fill the chamber to the top with PBS (see Note 18).
Remove 400 μL of PBS from the filled well and add 250 μL of 5 μg/mL solution of streptavidin in PBS. Mix twice by gently pipetting up and down with a micropipette (see Note 19).
Incubate for 10 min and wash excess streptavidin by repeating step 8.
Again, remove 400 μL and add 250 μL of 10 nM biotinylated goat Fab anti-mouse IgM (antigen) and incubate 20 min (see Note 20). Save some chambers for no antigen controls.
Wash unbound excess antigen by repeating step 8.
Immediately before imaging, exchange PBS with imaging buffer. Follow step 8 only with 10 mL of imaging buffer (see Note 21).
3.3. Image Acquisition Using Total Internal Reflection Fluorescence (TIRF) Microscope
For FRET imaging in our experiments, images in the CFP, FRET, and YFP channels are captured as a set from control cells expressing Lyn-CFP-only, Igα-YFP-only, Lyn16-CFP-YFP, a positive FRET control, and from experimental cells expressing both fluorophores. To study the dynamic association of two molecules using FRET imaging, time-lapse imaging is used and three images are obtained at each time interval. The procedure for acquiring images for FRET analysis is adapted to the equipment in our facility: an inverted Olympus IX-81, through-lens TIRF microscope that is equipped with Olympus 60×1.45 N.A. and 100×1.45 N.A. objective lenses, an optical splitter (DualView) for simultaneous imaging of the CFP and FRET channels, and a Cascade 512 II EM-CCD camera. Three-channel image set for each time interval is acquired under the following excitation (laser) and emission wavelengths:
Under identical conditions, all control cells are imaged each day. Lyn16-CFP-YFP expressing cells are also subjected to complete YFP photobleaching. CFP intensity is recorded before and after photobleaching to determine FRET efficiency (explained in detail below).
At least 1 h prior to the preparation of lipid bilayers and biological specimens, turn on power on all equipment including lasers. It is important to equilibrate the stage and lens heaters to 37°C and to stabilize gas laser.
Choose 442 or 514 nm laser lines (see Note 22).
Set 442/514 nm excitation filter (clean-up) in front of the AOTF (see Note 23).
Choose 442/514 nm polychroic mirror to reflect excitation light to the specimen.
Select the emission path to the camera port and ensure that appropriate emission filters are selected in the DualView image splitter (see Note 24).
Choose Olympus 100×1.45 N.A. oil-immersion objective lens.
Select the following camera settings as a starting point: 50 ms exposure time; full chip (512 ×512 pixel image size); 16bit gray scale; no pixel binning; no frame averaging; EM-mode; 5 MHz transfer speed; gain 2 (standard conversion gain); EM-gain 3000; overlap mode (clear pre-sequence: clears CCD charge buildup twice before image acquisition) (see Note 25).
Focus at specimen on the coverslip in a live acquisition mode. Remove the specimen and wipe away the oil (see Note 26).
Focus 442 and 514 laser lines to the back focal plane of selected objective lens. First, ensure that field diaphragm is open on TIR illumination port and tilt back bright field illumination swing arm to free a path for laser beam. Micrometer on TIR illumination port controls the beam angle. Return the micrometer (rightward) to its neutral setting so the beam is projected on the ceiling. Loosen the locking bolt and slide the fiber optic coupler until a small, focused spot followed by a series of diffraction rings is observed on the ceiling (see Note 27).
-
Align dual image splitter (DualView; MAGS Biosystems, Tuscon, AZ) onto CCD chip. A step-by-step alignment protocol is provided by the manufacturer. In steps 10a–c below, we provide an overview and commentary. It is important to align the apparatus before every imaging session. While alignment is usually stable, it is best to leave filter cassette in place once the alignment is complete. For FRET imaging, DualView is equipped with a filter cassette containing a 505 dichroic beamsplitter, and an HQ485/30 (470–500 nm), and HQ560/50 (535–585 nm) emission filters for simultaneous acquisition of CFP (left) and FRET (right) image, respectively.
Choose open position in the emission path filter wheel. Emission filters required for the experiment are contained in the DualView (see Note 28).
Place the alignment grid provided by the manufacturer in specimen holder and focus grid lines in bright field in a live acquisition mode. Follow the procedure outlined by the manufacturer to align the X, Y position. The grid is used for major adjustments and is typically needed when filter cassette is changed; otherwise, fine adjustments between experiments are made with alignment beads (see step 10c). For final adjustments with the grid, display live, false-colored, overlapped image of left and right sides of the CCD. We use red and green color combination to display yellow overlap (alignment). If further alignment is required, split the distance of adjustment by moving left/right or up/down knobs equally (see Note 29).
Perform fine adjustments with FluoSpheres (430/465 nm); these beads fluoresce in both CFP and FRET channels. The beads can be mounted or used in solution after settling to the bottom of a Lab-Tek chamber. Image beads in a live acquisition mode. Overlap left and right image in the software and adjust the DualView so that beads exactly overlap near the center of the image. Digitally magnifying the image so that individual pixels are viewed is helpful. Due to optical aberration, the beads at the periphery will not overlap (see Note 30).
Return to the standard acquisition mode and capture images of several fields of beads. These images will be used for alignment during image processing. Adjust laser power appropriately, so as not to saturate the image (see Note 31).
Allow time for stage heater to re-equilibrate (see Note 32).
Place the chamber containing the antigen-tethered planar lipid bilayer freshly prepared in imaging buffer onto the oil-mounted lens and allow for 10 min to reach 37°C.
Set imaging software to acquire two images for each time interval. For the first image, specimen is excited with the 442 nm laser and emission in the CFP and FRET channels is captured simultaneously. For the second image, specimen is excited with the 514 nm laser to capture YFP channel. Set interval time for 2 s and capture 500 frames.
Prepare CFP-only and YFP-only control cells at 1×104 cells/mL in the imaging buffer, mix at 1:1 ratio, and drop 20 μL of the mixture into a control chamber with no antigen positioned over the 100×objective lens. These control cells will be imaged with Lyn16-CFP-YFP FRET positive control cells (see steps 20–22). Find cells under transmitted light using a live acquisition mode (see Note 33).
Turn off transmitted light illumination source, excite the specimen with both 442 and 514 nm laser lines, and find a field that includes both CFP and YFP single-positive cells (see Note 34).
Illuminate the specimen by TIR. Turn the micrometer to the left on the TIR illumination port while using fine focus adjustment to image the cells proximal to the coverslip. As TIR angle is increased, a point will be reached where image intensity increases relative to the initial 0° TIR angle setting. Move past this point, again while adjusting focus, until image intensity is lost. Then slowly turn back the micrometer to the right to enter TIR-mode. Note micrometer setting for TIR angle (see Note 35).
Adjust laser power so that image is not saturated (see Note 36).
Perform test time-lapse acquisition for the calculation of correction factors and bleaching rate.
Prepare Lyn16-CFP-YFP FRET positive control cells at 2.0×106 cells/mL in imaging buffer and slowly drop 50 μL of cells into the same chamber containing single positive control cells.
View cells introduced to planar lipid bilayer in a live acquisition mode. Adjust focus and wait until they firmly settle down on the bilayer.
Acquire a set of CFP, FRET, and YFP channel images (pre-bleach) and then completely bleach YFP with 514 nm line at full power with TIR angle 0°. Return the micrometer to TIR angle noted in step 17 and acquire another three sequential image sets (post-bleach) under the same acquisition conditions to obtain Ka and Ebleaching.
Move chamber coverglass so that the center of the chamber containing antigens-tethered PLB can be placed over the lens and repeat steps 15–17 for image acquisition of experimental cells expressing both Lyn-CFP and Igα-YFP.
Prepare experimental cells expressing both Lyn-CFP and Igα-YFP cells at 2×106 cells/mL in imaging buffer and slowly drop 50 μL of cells into the chamber containing single positive control cells on antigen-tethered lipid bilayer.
View progress of introduced cells to the planar lipid bilayer in a live acquisition mode. Adjust the focus. Begin time-lapse image acquisition when blurred fluorescence images start to appear on the screen under a live image mode (see Note 37).
3.4. Image Processing for FRET Imaging
The principles of FRET calculations have been described elsewhere (12, 14, 15). For our protocol using B cells expressing Igα-YFP and Lyn-CFP, the use of FRET efficiency for the acceptor (Ea), that is FRET efficiency normalized for fluorescence intensity of YFP, is advantageous because YFP rather than CFP is limiting in the FRET pair, Lyn-CFP and Igα-YFP. For our microscope system, the δ factor is typically negligible (δ ≈0), so the equation for Ea can be simplified to the following:
| [5] |
To calculate FRET, raw images in each channel must be processed by background subtraction, image filtering, and masking of background for each channel, followed by image arithmetic on a pixel-by-pixel basis according to the equation (10.5). These processing steps are necessary to obtain the final FRET image and avoid false-positive or false-negative results. Processing of images can be performed using image processing software that is capable of image alignment and arithmetic, such as the Image Pro Plus (Media Cybernetics).
Open image that contains the CFP-FRET dual view of fluorescence beads.
Separate it into CFP and FRET channel images by creating new area of interests (AOIs) for the left and right half of the image.
Align separated CFP and FRET images using alignment function in image processing software. Alternatively, find the best alignment manually by overlaying the two images and moving one with respect to the other. Record the amount of horizontal and vertical shift, rotation, and scaling that was applied to obtain complete alignment of beads in the two images.
If necessary, merge individual image files of a sequence of CFP-FRET and YFP images at each time frame by stacking them in series.
Separate all DualView images based on the alignment determined in step 3. This will produce three sequences of images: CFP, FRET, and YFP.
For each image, set AOI to background (an area without cells) and determine average background value. Subtract that value from respective image using arithmetic operations.
Smoothen images by applying a low-pass (averaging) filter to reduce the noise of individual pixels. The size, passes, and strength of the filter are determined empirically to smoothen the image, but not to blur cellular details completely (see Note 38).
Determine the threshold value in each channel image to distinguish the foreground of the cell from background in that particular channel, using the mean plus three standard deviations of pixel intensity of the background. Mask the background by thresholding the image at that intensity. As a result, background pixels in all three (CFP, FRET, and YFP) channels should have zero intensity.
-
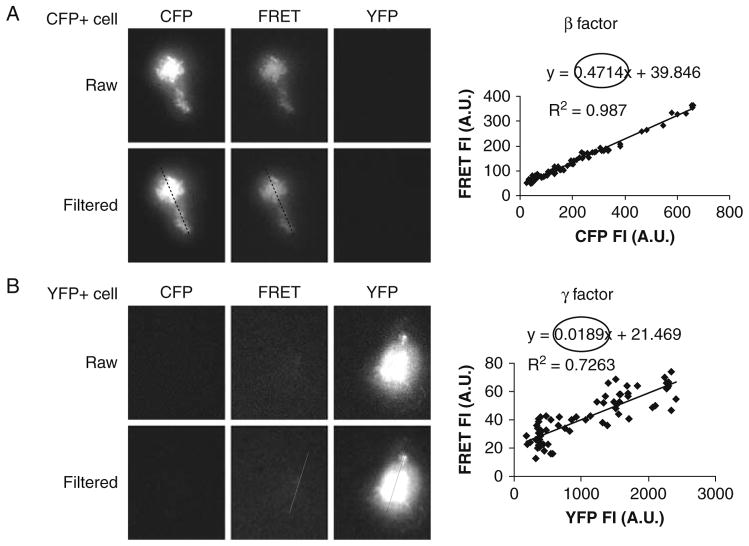
Determine β from cells expressing CFP only.
, where FRET and CFP are background-subtracted intensity values in AOIs from the cells in FRET and CFP channels as excited by the 442 nm laser (Fig. 10.1a).
-
Likewise, determine γ from cells expressing YFP only.
, where FRET and YFP are background-subtracted intensity values in AOIs from the cells in FRET and YFP channels as excited by the 442 and 514 nm laser, respectively (Fig. 10.1b).
-
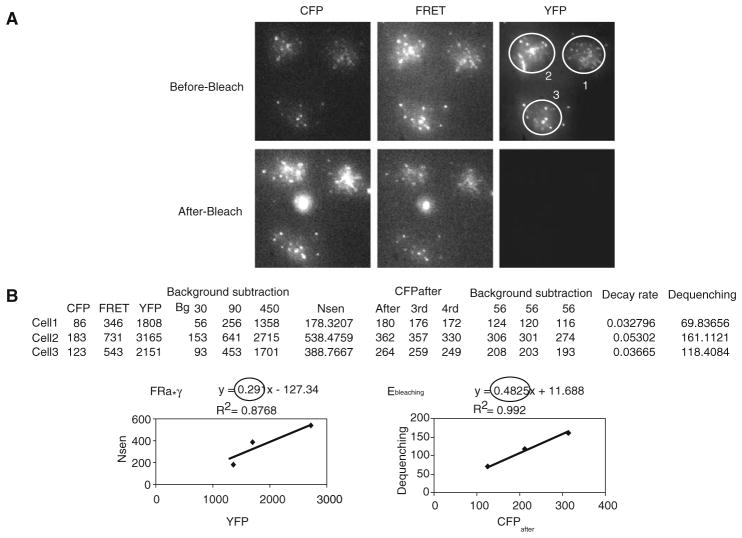
Determine FRa, FRET efficiency (Ebleaching), and Ka from images before and after YFP photobleaching of cells expressing Lyn16-CFP-YFP using the following series of equations:
[6] where Nsen is the sensitized acceptor emission by FRET:[7] [8] where dequenching is difference of fluorescence intensities of CFP before (CFPbefore) and after acceptor photobleaching (CFPafter) (see Note 39) (see Fig. 10.2):[9] [10] First, record in a MS Excel sheet the mean fluorescence intensities of CFP, FRET, and YFP from the same AOIs drawn over the cell contact areas from each channel image before YFP photo-bleaching (before-bleach), and subtract background value from each. An example table of raw and background subtracted intensity values, which was obtained from images is provided in Fig. 10.2b.
Calculate Nsen using equation [7] from background-subtracted FRET, CFP, and YFP mean intensities.
Calculate dequenching by equation [9].
Make intensity plots for Nsen vs YFP and dequenching vs CFPafter from the values calculated in step 12c. Determine FRa and Ebleaching from the slopes of the relationships of Nsen vs YFP and dequenching vs CFPafter, respectively (Fig. 10.2b).
Obtain Ka using equation [10] (see Note 40).
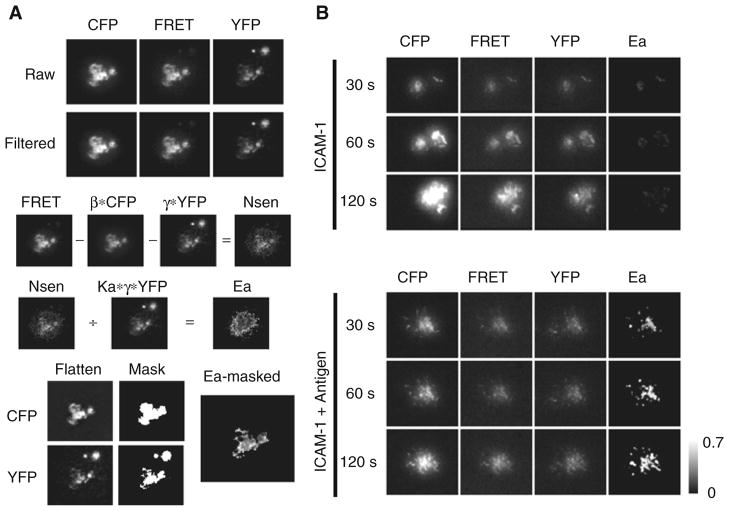
Finally, produce FRET efficiency (Ea) images from sample cells by pixel-by-pixel arithmetic image calculation using equation [5] (see Fig. 10.3a).
-
Because the resulting Ea image from step 12 shows artificial FRET in the region outside the cell contact zone (TIRF imaging plane) due to background noise, mask the Ea image by following steps:
Apply a flatten enhancing filter to both CFP and YFP images to augment the borders along contact zones.
Set the threshold for the enhanced Flatten images as done in step 8 and create mask image for both.
Then, apply the combined CFP and YFP masks over the Ea image resulting in the masked Ea image. An sample result is shown in Fig. 10.3b.
Fig. 10.1.
Determination of correction factors: (a) for CFP bleed-through (β) and (b) for YFP crosstalk (γ) from direct excitation of CFP and YFP by 442 nm laser, respectively. Cells expressing either Lyn-CFP alone (CFP+ cell) or Igα-YFP alone (YFP+ cell) were settled on planar lipid bilayer. Cells were excited sequentially with 442 and 514 nm laser light by TIRF microscope. Fluorescent emission was split through DualView to acquire CFP and FRET images (442 nm excitation) and the YFP image (514 nm excitation). Images were aligned, split to obtain raw CFP, FRET, and YFP images from the control cells, and then background subtracted and a Lo-pass filter applied (Filtered) as described in Section 3. To calculate β, intensity data was obtained from a line drawn over the same relative position in the cell in CFP and FRET filtered image and was plotted as FRET vs CFP (a) for each pixel in the line. β was determined from the slope of the line in the plot. γ (B) was calculated similarly using YFP and FRET filtered images. Similar results are obtained by averaging fluorescence intensities from several control cells.
Fig. 10.2.
Determination of Ebleaching and Ka from Lyn16-CFP-YFP FRET positive control. (a) CFP, FRET, and YFP images before and after YFP photobleaching were acquired from cells expressing Lyn16-CFP-YFP on planar lipid bilayer and processed as described in Section 3. AOIs from three cells were drawn over cell contact areas from each image before (Pre-Bleach) and after bleaching (Post-Bleach). Note that images are not saturated on the 16 bit gray scale. (b) Procedure to obtain Ka from the CFP, FRET, and YFP images in a Microsoft Excel sheet is shown. Shown are a table containing the values that include background-subtracted YFP, Nsen, CFPafter and Dequenching, calculated from the mean fluorescence intensities of AOIs drawn in (a), and intensity plots for Nsen vs YFP and Dequenching vs CFPafter from the values of the table. FRa and Ebleaching were determined from the slope of the line of Nsen vs YFP and Dequenching vs CFPafter, respectively. Finally Ka was obtained from the equation (10).
Fig. 10.3.
Membrane-bound-antigen-induced association of Lyn with BCR. Cells were dropped onto planar lipid bilayer with or without antigen and time-lapse images were acquired at 37°C for 15 min. (a) Procedure for image processing to obtain the FRET efficiency (Ea) image from experimental cells expressing both Lyn-CFP and Igα-YFP is shown. Images taken 1 min after cell contact to the antigen-bound membrane were processed and shown are the representative images in each step described in Section 3. β*CFP, γ *YFP, Ka* γ*YFP, N4sen and Ea images were obtained from pixel-by-pixel image calculation according to the appropriate equations. Raw; background-subtracted images, Filtered: Lopass-filtered images, Flatten: flatten-filtered images, Mask: masked images from flattened images, Ea-masked: masked image of Ea with both CFP and YFP masks. (b) CFP, FRET, YFP and Ea images at each indicated time after cell contact in the absence (ICAM-1) or presence of antigen (ICAM-1 + Antigen) on a planar lipid bilayer are shown. Images were processed as in (a) and shown are the filtered CFP, FRET, and YFP images and the masked Ea image. Ea is shown as gray scale is from 0 to 0.7. The fluorescence intensities of CFP, FRET, and YFP images are not comparable.
Acknowledgments
This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Footnotes
CH27 cells, which express on their surface a phosphorylcholine (PC) specific BCR, can be obtained from authors’ lab.
Although specific design of chimeric constructs needs to be considered for each gene, Lyn and Igα were fused to the N-terminus of CFP or YFP using pECFP-N1 or pEYFP-N1 vector (BD Biosciences Clontech) by inserting these genes in-frame at the Bam HI site upstream of the CFP or YFP vector. In both cases, there is a 6 amino acid spacer between the gene of interest and the tag which gave us the best expression of these fusion proteins at plasma membrane. Full length Igα and mouse Lyn cDNA clones are available commercially (Open Biosystems, Huntsville, AL) for PCR templates. The membrane targeted FRET positive control plasmid, Lyn16-CFP-YFP, has been previously described (15).
To make cleaning solution, dissolve 120 g KOH into 120 mL of deionized water in a beaker. Add about 1 L of 95% EtOH to the KOH solution and mix thoroughly using a stirrer. Wear suitable protective clothing, gloves, and eye/face protection when making the cleaning solution. Do not keep it in a glass bottle with glue-jointed bottom for an extended period of time.
After each use, fill the bottle with argon and wrap the cap with parafilm to avoid evaporation of chloroform. The lipids are easily oxidized when exposed to the air.
This particular Lab-Tek chamber is used because the coverslip can be easily removed by hand.
CH27 cells grow better in 7% CO2 than 5%. When first thawed from liquid nitrogen, incubate them at 7%. Once cells are growing, they can be cultured at either condition. When confluent after about 2–3 days, the media turns from pink to orange-yellow color. Otherwise, keep track of cell density by counting cells. When cells reach about 1.5×106 cells/mL, dilute cells into fresh media. Cells are usually passaged every 3 days.
Plasmid transfection conditions must be determined empirically for each particular B cell line. We found that creating stable transfectants with a ratio of ~1:1 of CFP to YFP worked best in FRET experiments. Retroviral infection system using MSCV vector and PT67 packaging cell line (Clontech) followed by FACS sorting for positives worked best for CH27 cells. Stable cell lines expressing both CFP and YFP can be established by sequential or simultaneous infection followed by repetitive sorting for CFP and/or YFP positive cells. Usually, we use “spin infection” by centrifugation at 1844 g for 90 min followed by concentration of the viral supernatant by centrifugation at 2300 g for 20 min using a centrifugal filter device (Amicon Ultra-15 with 10 k MW cut-off, Millipore). Stable cell lines for positive clones can be established by drug selection or sorting by FACS.
To achieve maximal transfection efficiency, it is important to maintain cells with good viability (over 90%) during passage. CH27 cells show maximum transfection efficiency when cell density reaches ~1.5–2.0×106 cells/mL on the day of transfection.
It is very important to remove any residual liquid from the pellet to obtain good transfection efficiency. A sterile 200 μL micropipette tip at the end of aspiration pipette works well to remove any remaining medium. Cell pellets become loose with time, which can lead to accidental aspiration of cells. We recommend a maximum of four samples to be spun at a time.
If transfection efficiencies are less than 10%, chances are low to capture cells expressing both CFP and YFP in the same imaging field. Sort for positive cells by FACS if available or use stable cell transfectants.
It is important to keep the pressure low enough as to not to splash the mixture on the walls of the vial.
Cover the vial with a Kimwipe to inhibit dust from getting inside.
To clean side arm flask, swirl about 50 mL of Nanostrip in the flask for a few minutes, discard, and then rinse with a copious amount of deionized ultrapure water. Dry the flask in an oven.
To avoid overheating the sonicator bath, do not sonicate for more than 10 min. Pause sonication, place the vial on ice, and add more ice to the bath to adjust water temperature.
An opaque pellet will be seen at the bottom of the tube after the first round of centrifugation. Pellet size depends on the extent of sonication.
SUVs are stable for several months, when stored at 4°C under argon. To minimize repeated exposure of SUV stock to air and to prolong stability, store in 1 mL aliquots in 15-mL sterile plastic tubes under argon sealed with parafilm. The SUV mixture can be filtered through a 0.2-μm syringe-type Whatman polysulfone filter for further purification; resulting concentration of lipid is about half of the original determined by measurement of OD at 234 nm using UV–vis spectrophotometer.
Mix Sylgard components ~1:1 on a clean surface with a 200-μL micropipette tip and use quickly before hardening. With the chamber upside down, fill the channels at the base of each well. Syringe with a short, thick (18 G) needle can be used. Avoid applying too much silicone. Using forceps or with gloved hands holding only the edges, place the coverslip over the bottom of the chamber and apply gentle pressure along the channels. Place chambers upright on a Kimwipe, cover with about 0.5 kg weight, and leave for 30 min before use. Prepare chambers on the day of use as dust will re-accumulate and prevent the formation of a lipid bilayer.
To avoid exposing the lipid bilayer to air during washing, remove solution with a 200-μL micropipette tip attached to aspiration pipette while simultaneously adding 1×PBS with a slow, consistent flow rate using a 10-mL serological pipette attached to auto-pipetter.
To monitor mobility/integrity of the lipid bilayer, mix 1:9 Alexa488-labeled streptavidin with unlabeled streptavidin and add 250 μL to one well.
Dilute antigen with 1×PBS to 10 nM and microcentrifuge at maximum speed for about 10 min to eliminate aggregates.
Planar lipid bilayers are less stable in the presence of serum. It lasts at most for 1–2 h. Its mobility can be tested by a simple FRAP experiment with fluorescently labeled planar lipid bilayer in a chamber.
We recommend MetaMorph software to control imaging and laser (via AOTF) systems.
Despite the 104 blocking power of AOTF, we found it necessary to clean-up (block) non-selected laser lines emanating from the gas laser as this light was detected by our EM-CCD. Filters work best when located on the laser table. Ours are held in a filter wheel placed after the AOTF; filter position on the table is flexible and is only defined by what laser lines require for blocking. Interestingly, unwanted reflections detected by the EM-CCD were seen when excitation filters were placed in the TIR illumination port or filter cube holder of the microscope.
We place 442 nm notch filter in the microscope filter cube to further block escaping excitation light from the camera to decrease background signal and spurious reflection patterns.
Once optimized for cells, it is important to keep all settings the same. Maximal useful EM-gain is idiosyncratic for each specific camera type and needs to be determined empirically. Best EM-gain is reached when the ratio of signal to background no longer improves with further increase of gain.
The goal is to place objective lens at correct working distance before focusing laser lines. Changing the height of objective lens will change the distance of objective’s back focal plane relative to laser focusing lens in TIR illumination port. Laser focusing can be achieved directly through the specimen as long as the cell number is low, which minimizes light scatter.
Laser light will exit the objective lens as collimated beam, which can be observed through a concentrated solution of fluorophore if so desired. We replaced the original Olympus TIR illumination port and tube lens with a 100 and 200 mm focal length achromatic lens, respectively, from JML Optical. These second-party lenses offer superior color correction, allowing both beams to co-focus. Be aware that this only improves laser co-focusing and does not affect the differences in penetration depth of specific excitation wavelengths.
Laser-line blocking filters (notch filters) can be placed in this filter wheel, polychroic filter cube of the microscope, or in DualView filter cassette.
For a CCD camera with a 512×512 pixel dimension, most image acquisition software, including MetaMorph, designates the first pixel position in the upper left corner as 0,0 and the bottom right as 511,511; therefore, we divide horizontal axis from 0 to 255 for the left side and 256 to 511 for the right.
To our knowledge, diffraction-limited beads (<300 nm) that fluoresce well from CFP channel into the FRET channel when mounted on coverslips are not available commercially. We therefore use 1-μm-sized beads.
Most imaging software can be set to display saturated pixels in red.
We occasionally monitor well temperature with YSI 4600 thermometer equipped with a 400 series probe.
Adjust tungsten lamp intensity appropriately.
For finding cells, TIR illumination angle can be kept at 0°.
Ideally, it is better to work with subregions because even illumination of the entire specimen plane (512×512 region) at an optimal TIR angle is not possible.
For our experiments, we use ~2 and ~1.0 mW for 442 and 514 nm laser lines, respectively (measured with a power meter through 100×lens). It is important to minimize photobleaching during time lapse by keeping laser power to a minimum while still achieving adequate signal.
To compensate for focus drift, our system is equipped with the Olympus laser autofocusing device, which works well for long time-lapse experiments.
Typically within small regions of interest where FRET is measured, we found that flat field correction was unnecessary. To correct for uneven illumination of the field of view, each pixel value of the image is divided by the mean intensity of normalized flat field image, which is pixel-by-pixel ratio image between CFP and FRET channels obtained using a fluorophore solution in a chamber that emits in both CFP and FRET channels.
With some exception such as degradation of fusion protein in certain cells, FRET positive control cells show empirically about 55±10% Ebleaching regardless of imaging conditions due to the fact that the fusion protein with 2 amino acid linker between CFP and YFP maintains the fixed 1:1 molar ratio between donor and acceptor and constant FRET.
| [11] |
| [12] |
References
- 1.Fearon DT, Carroll MC. Regulation of B lymphocyte responses to foreign and self-antigens by the CD19/CD21 complex. Annu Rev Immunol. 2000;18:393–422. doi: 10.1146/annurev.immunol.18.1.393. [DOI] [PubMed] [Google Scholar]
- 2.Dal Porto JM, Gauld SB, Merrell KT, Mills D, Pugh-Bernard AE, Cambier J. B cell antigen receptor signaling 101. Mol Immunol. 2004;41:599–613. doi: 10.1016/j.molimm.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 3.Pierce SK. Lipid rafts and B-cell activation. Nat Rev Immunol. 2002;2:96–105. doi: 10.1038/nri726. [DOI] [PubMed] [Google Scholar]
- 4.Tolar P, Sohn HW, Pierce SK. Viewing the antigen-induced initiation of B-cell activation in living cells. Immunol Rev. 2008;221:64–76. doi: 10.1111/j.1600-065X.2008.00583.x. [DOI] [PubMed] [Google Scholar]
- 5.Axelrod D. Cell-substrate contacts illuminated by total internal reflection fluorescence. J Cell Biol. 1981;89:141–145. doi: 10.1083/jcb.89.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kandere-Grzybowska K, Campbell C, Komarova Y, Grzybowski BA, Borisy GG. Molecular dynamics imaging in micropatterned living cells. Nat Methods. 2005;2:739–741. doi: 10.1038/nmeth796. [DOI] [PubMed] [Google Scholar]
- 7.Brian AA, McConnell HM. Allogeneic stimulation of cytotoxic T cells by supported planar membranes. Proc Natl Acad Sci U S A. 1984;81:6159–6163. doi: 10.1073/pnas.81.19.6159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evans J, Yue DT. New turf for CFP/YFP FRET imaging of membrane signaling molecules. Neuron. 2003;38:145–147. doi: 10.1016/s0896-6273(03)00234-4. [DOI] [PubMed] [Google Scholar]
- 9.Sohn HW, Tolar P, Pierce SK. Membrane heterogeneities in the formation of B cell receptor-Lyn kinase microclusters and the immune synapse. J Cell Biol. 2008;182:367–379. doi: 10.1083/jcb.200802007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Truong K, Ikura M. The use of FRET imaging microscopy to detect protein-protein interactions and protein conformational changes in vivo. Curr Opin Struct Biol. 2001;11:573–578. doi: 10.1016/s0959-440x(00)00249-9. [DOI] [PubMed] [Google Scholar]
- 11.Ciruela F. Fluorescence-based methods in the study of protein-protein interactions in living cells. Curr Opin Biotechnol. 2008;19:338–343. doi: 10.1016/j.copbio.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 12.van Rheenen J, Langeslag M, Jalink K. Correcting confocal acquisition to optimize imaging of fluorescence resonance energy transfer by sensitized emission. Biophys J. 2004;86:2517–2529. doi: 10.1016/S0006-3495(04)74307-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zal T, Gascoigne NR. Photobleaching-corrected FRET efficiency imaging of live cells. Biophys J. 2004;86:3923–3939. doi: 10.1529/biophysj.103.022087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tolar P, Sohn HW, Pierce SK. The initiation of antigen-induced B cell antigen receptor signaling viewed in living cells by fluorescence resonance energy transfer. Nat Immunol. 2005;6:1168–1176. doi: 10.1038/ni1262. [DOI] [PubMed] [Google Scholar]
- 15.Sohn HW, Tolar P, Jin T, Pierce SK. Fluorescence resonance energy transfer in living cells reveals dynamic membrane changes in the initiation of B cell signaling. Proc Natl Acad Sci USA. 2006;103:8143–8148. doi: 10.1073/pnas.0509858103. [DOI] [PMC free article] [PubMed] [Google Scholar]