Abstract
Introduction
Humoral immune responses play a pivotal role in naturally acquired immunity to malaria. Understanding which humoral responses are impaired among individuals at higher risk for malaria may improve our understanding of malaria immune control and contribute to vaccine development.
Methods
We compared humoral responses with 483 Plasmodium falciparum antigens between adults in, Kisumu (high, year-long malaria transmission leading to partial immunity), and adults in Kisii (low, seasonal malaria transmission). Then within each site, we compared malaria-specific humoral responses between those at higher risk for malaria (CD4+ ≤ 500) and those at lower risk for malaria (CD4+>500). A protein microarray chip containing 483 P. falciparum antigens and 71 HIV antigens was used. Benjamini–Hochberg adjustments were made to control for multiple comparisons.
Results
Fifty-seven antigens including CSP, MSP1, LSA1 and AMA1 were identified as significantly more reactive in Kisumu than in Kisii. Ten of these antigens had been identified as protective in an earlier study. CD4+ T-cell count did not significantly impact humoral responses.
Conclusion
Protein microarrays are a useful method to screen multiple humoral responses simultaneously. This study provides useful clues for potential vaccine candidates. Modest decreases in CD4 counts may not significantly impact malaria-specific humoral immunity.
Keywords: HIV-1, Humoral Immunity, Malaria, Vaccine
1 Introduction
Malaria continues to be a significant cause of morbidity and mortality worldwide [1] (www.who.int/malaria/wmr2008). Adults living in malaria endemic areas develop some level of partial immunity to malaria as a result of repeated exposures to parasite antigens [2]. This naturally acquired partial immunity involves both humoral and cellular immune responses and appears protective against symptomatic disease, but not against malaria infection [2–9]. A greater understanding of immune responses among individuals with partial immunity may lead to the identification of vaccine targets with the potential to dramatically reduce malaria-related morbidity and mortality.
The humoral arm of the immune system is believed to play a key role in naturally acquired partial immunity to malaria [10, 11]. Early studies showed that transfer of serum from partially immune individuals to non-immune individuals conferred some protection from severe outcomes related to malaria [12, 13]. Immuno-epidemiological studies have also shown associations between high levels of some malaria antibodies with protection [14, 15]. HIV-1 infection may affect some but not all malaria-specific antibody responses [16–20]. The specific mechanism explaining how this happens has not been well studied. HIV-1 infection is known to decrease the number and function of memory B cells [21–23]. HIV-1 infection may also impair B-cell proliferation and antibody response to some antigens [24].
Evaluating the specific defects in humoral immunity that result in higher risk of malaria disease among a cohort of HIV-1-infected adults offers a unique model for identifying which Plasmodium falciparum (Pf) proteins may be important for mediating protection. Acquired partial immunity to malaria is negatively impacted by decreased parasite exposure [2] and increasing HIV-1-associated immunodeficiency [25–27]. HIV-1 infection has been associated with impairments to some but not all malaria-specific antibodies [16–20]. By identifying which antibody responses are lost or impaired among those with limited parasite exposure or lower CD4 count, we may be able to identify antigen-specific antibodies that are protective. These proteins may prove essential for the development and/or characterization of an effective malaria vaccine.
Protein microarrays allow for the evaluation of humoral responses to hundreds of malaria antigens [28, 29]. Microarrays can also profile the antibody repertoire from a large sample size providing statistically meaningful results not possible by other methods [28, 29]. Using this technology, we previously constructed a protein microarray containing ~23% of the Pf proteome. These antigens were selected according to specific sets of criteria, including pattern of stage-specific gene or protein expression deduced from genomic or proteomic data sets, subcellular localization, secondary structure and known immunogenicity or antigenicity in human and animal models. Using this protein microarray, we profiled the antibody repertoire among Malian children between the ages of 8 and 10 [30] and identified 491 immunoreactive proteins. Of these 491 immunoreactive proteins, we found that the humoral responses to four leading malaria vaccine candidate antigens (CSP, MSP1, LSA1 and AMA1) were equally reactive in both protected and unprotected children [30]. Furthermore, we identified an additional 49 proteins that were associated with protection from clinical malaria among Malian children.
Here, we extend this approach and compare the Pf antibody repertoires of two geographically distinct locations with differing levels of endemicity and immunity to malaria. Our aim was to identify potential vaccine candidates by defining which antibody responses are more reactive among adults with presumed partial immunity to malaria compared with those without. Additionally, within each location we compared the antibody repertoire of those with high CD4 counts to those with low CD4 counts. In this way, we evaluated the effect of increasing HIV-1-associated immunodeficiency on humoral immunity to malaria in two populations with different baseline malaria immunity.
2 Methods
2.1 Study design
We performed a cross-sectional analysis of stored samples gathered from 150 antiretroviral naïve HIV-1 sero-positive adults participating in a large randomized controlled trial evaluating the effect of deworming on markers of HIV-1 disease progression in Kenya. Samples were collected between May, 2008 to May, 2009. All individuals provided written informed consent to participate in this study. Both the parent trial and this study were independently approved by the IRB of the University of Washington and the Ethical Review Board of the Kenya Medical Research Institute. The parent trial has been registered as NCT00507221 at http://clinicaltrials.gov.
2.2 Population
Totally, 150 stored plasma samples from individuals recruited in an ongoing randomized clinical trial (RCT) were used for this study. To be enrolled in the parent study participants had to be older than 18, non-pregnant, antiretroviral naïve, have a CD4+ count >350 and be willing and able to give informed consent. For this study, we restricted participants to those in the 20–40 age group. Seventy-five samples meeting the above criteria were randomly selected from Kisii and another 75 were randomly selected from Kisumu.
2.3 Study sites
Kisumu and Kisii represent two areas of differing malaria endemicity. The entomologic inoculation rates (EIR) is 31.1 infectious bites per person per year in Kisumu district as compared with 0.4 in Kisii [31]. Malaria transmission is relatively low and seasonal in Kisii (during rainy season), while Kisumu experiences high-intensity malaria transmission throughout the year. Levels of protective immunity differ between individuals at each of these sites. Kisii is prone to malaria epidemics because adults in Kisii do not have partial immunity to malaria [32, 33]. Adults in areas of high malaria transmission such as Kisumu typically acquire partial immunity to malaria which protects them from clinical malaria disease and death [34].
2.4 Microarray construction
A detailed description of the Pf ORF cloning, in vitro expression, array printing method and probing methods has been published elsewhere [30, 35]. Previous antigens were selected based on specific sets of criteria, including pattern of stage-specific gene or protein expression deduced from genomic or proteomic datasets, subcellular localization, secondary structure, and known immunogenicity or antigenicity in human and animal models. Proteins printed on the array were selected based on seroreactivity in our previous results of individuals from malaria endemic regions [30] or patients vaccinated with irradiated sporozites [35]. As such, a total of 499 Pf proteins from 382 Pf ORFs were selected to be printed on the arrays used here. Pf proteins containing multiple exons and sequences larger than 3000 bp were cloned as overlapping segments. An additional 72 proteins from four HIV Clades (A1, A2, B and D) were printed on the array. Microarrays containing all 571 of these proteins, expressed from in vitro transcription/translation (IVTT) reactions were fabricated and probed as previously described [30, 35]. In brief, ORFs cloned into the pXT7 vector were expressed under the T7 promoter in a 5 h Escherichia coli-based cell-free IVTT reaction according to the manufacturer’s instructions. Proteins were printed using an Omni Grid 100 microarray printer (Genomic Solutions) and analyzed for fluorescence on a Perkin Elmer ScanArray Express HT microarray scanner. IVTT Expression efficiency was determined by probing against the carboxy-terminal HA tags for each spot. Ninety-seven percent of the spotted proteins had signals greater than the average of “No DNA” control reactions plus 2.5 times the standard deviation, and were considered positively expressed. In this manner, a protein microarray of 571 spotted proteins was fabricated, composed of 71 HIV and 483 Pf proteins, with positive and negative controls.
2.5 Data and statistical analysis
Intensities were quantified using QuantArray software, utilizing automatic background subtraction for each spot. Proteins were considered to be expressed if either tag’s signal intensity was greater than the average signal intensity of the IVTT reaction without plasmid, plus 2.5 times the standard deviation. “No DNA” controls, consisting of IVTT reactions without addition of plasmid, were averaged and used to subtract background reactivity from the unmanipulated raw data. Proteins with average signal intensities greater than the average “No DNA” controls plus 2.5 times the standard deviation were considered to be seroreactive. All results presented are expressed as signal intensity. As previously reported [36], the “vsn” package in the Bioconductor suite (http://Bioconductor.org/) in the R statistical environment (http://www.R-project.org) was used to calculate seroreactivity [37]. In addition to the variance correction, this method calculates maximum likelihood shifting and scaling calibration parameters for different arrays, using known non-differentially expressed spots. This calibration has been shown to minimize experimental effects [37]. We used raw values for the positive and negative controls to calibrate and then normalize the entire data set using the vsn package. Differential analysis of the normalized signals was performed using a Bayes-regularized t-test adapted from Cyber-T for protein arrays [38–41]. All p-values herein are Benjamini–Hochberg p-values adjusted for false-discovery, unless stated otherwise [42].
We compared each study site to determine differences in malaria antibody profile. Within each site, study participants were then divided into two groups based on CD4 count. Individuals with CD4 counts of 200–500 were compared to individuals with CD4 counts >500. The choice of CD4 cutoff was influenced by prior studies that have made comparisons between those with CD4 <200, 200–500 and >500 [25–27]. These studies have suggested an increase in malaria incidence with decreasing CD4 count. Because there were no individuals with CD4 counts <200, we are only able to make comparisons between those with CD4 counts between 200 and 500 and those with CD4 counts >500.
3 Results
3.1 Study participant characteristics
Plasma samples from 150 HIV-1-infected adults from two locations of different malaria endemicity were used for this study. Seventy-five of the samples were from individuals living in Kisii and 75 were from Kisumu. The demographic characteristics of the population, stratified by site of enrollment, are presented in Table 1. The mean age of the selected participants was 29.9 years, with a range of 20–40. Individuals from the Kisumu site were significantly younger than those from Kisii (28.7 versus 31.2 years, p-value<0.01). Overall, 78.7% of all participants were women, with a significantly higher proportion of women at the Kisii site (85.3%) compared to the Kisumu site (72%) (p<0.05). Individuals in Kisumu had higher rates of bednet usage (80.6 versus 28.4%, p<0.01). The differences in age and gender were not felt to adversely affect our outcomes of interest (humoral responses to malaria). Bednet use was higher in Kisumu. This could potentially decrease parasite exposure in Kisumu affecting our ability to detect differences between sites. The median CD4+ count of the individuals was 560 (range: 275–1612). There was no significant difference in the number of individuals with CD4+ counts <500 between the sites.
Table 1.
Basic demographic information
| Variable | Kisumu N=75 | Kisii N=75 | p-Value |
|---|---|---|---|
| Age – mean (SD) | 28.7 (5.5) | 31.2 (5.7) | N/A |
| Male gender – no. (%) | 21 (28.0) | 11 (14.7) | N/A |
| Use of bed net – no. (%) | 58 (80.6) | 21 (28.4) | <0.01a) |
| CD4 count ≤ 500 – no. (%) | 29 (38.7) | 26 (34.7) | 0.6 |
Significant p-value.
3.2 Identification of Pf-specific Ab profile differences between regions of high and low malaria endemicity
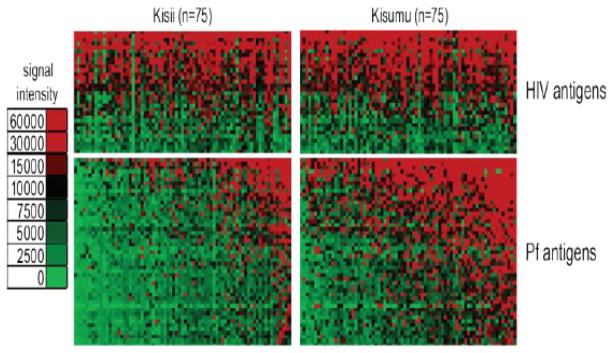
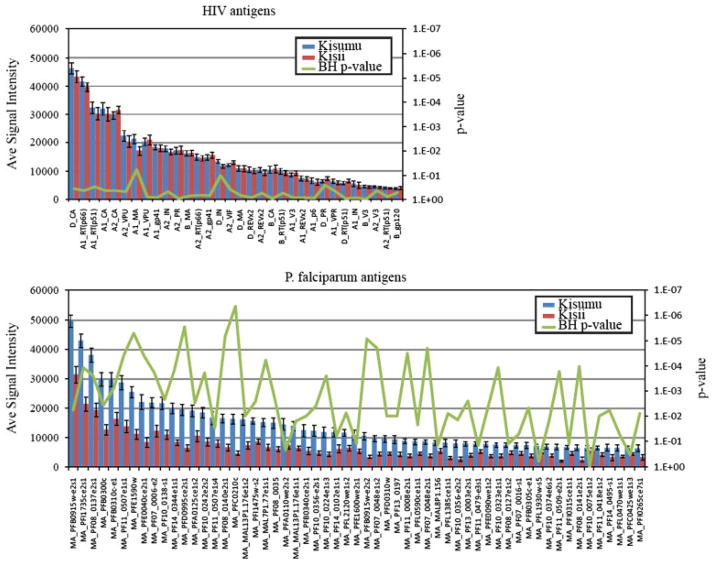
We profiled the humoral immune response to Pf antigens between sites. As shown in Fig. 1, individuals from Kisumu (presumed partial immunity) had higher and broader reactivity to malaria antigens as compared to individuals from Kisii. We identified 57 antigens with significantly different seroreactivity between Kisumu and Kisii (p-values <0.05) after adjustment for false discovery (Fig. 2B). All 57 antigens were more seroreactive in Kisumu as compared with Kisii. The 57 differentially reactive proteins are listed in Supporting Information Table 1. Among these 57 are four of the leading vaccine candidates; CSP, MSP2, LSA1 and AMA1 (Table 2). There were no significant differences in seroreactivity to HIV antigens between sites (Fig. 2A).
Figure 1.
Profiling the antibody repertoire of a collection of malaria unprotected and partially protected individuals. Arrays containing 499 P. falciparum proteins were probed with 150 patient samples. The normalized antibody signal intensity to the 64 seroreactive antigens is shown according to the colorized scale with red strongest, green weakest, and black in between. The antigens are in rows and sorted by decreasing average signal intensity. Patient samples are in columns and are sorted left to right by increasing average intensity to the 64 seroreactive antigens.
Figure 2.
Identification of differentially reactive antigens. The mean seroreactivity of each antigen was compared between unprotected and partially protected groups plotted red and blue, respectively, with SEM. “e” and “s” at the end of the genes refers to exon number and segment number. Corresponding p-values corrected for false discovery for each antigen are shown as a green line on the secondary axis. (A) The 64 seroreactive antigens are compared to the control HIV seroreactive ORFs and indicate that HIV seroreactivity was equal between groups. (B) Only the 64 seroreactive antigens are displayed and the corresponding locus tags are labeled on the x-axis.
Table 2.
Humoral reactivity of vaccine candidates by site
| Antigen | Average intensity for Kisumu (N=75) | Average intensity for Kisii (N=75) | p-Value | p-Value (Benjamini– Hochberg adjusted)a) |
|---|---|---|---|---|
| Circumsporozoite Protein (CSP) | 16 690.5 | 4715.3 | <0.01b) | <0.01b) |
| Merozoite Surface Protein 1 (MSP-1) | 15 668.7 | 8907.7 | <0.01b) | <0.01b) |
| Liver Stage Antigen 1 (LSA-1) | 12 471.7 | 4775.3 | <0.01b) | <0.01b) |
| Apical Membrane Antigen 1 (AMA-1) | 5803.1 | 2933.3 | <0.01b) | <0.01b) |
Benjamini–Hochberg adjusts for multiple comparisons were used to minimize the likelihood of false discovery.
Significant p-value.
3.3 Proteomic features of differentially reactive antigens
The 57 differentially seroreactive antigens were compared with our previous reported results by Crompton et al. in which we identified 49 highly seroreactive antigens that correlate with protection [30]. Ten of the 49 previously identified antigens were also identified here as differentially reactive between the two locations. Furthermore, an additional 12 of the 49 previously identified antigens were identified here as seroreactive, but were not significantly higher in the Kisumu group. The remaining 32 were below the seroreactive threshold. A list of the 10 antigens that were identified as possible markers of immunity in both studies is outlined in Table 3. All 10 proteins were predicted to be probable antigens utilizing Vaxijen software (Table 3) (http://www.ddg-pharmfac.net/vaxijen/VaxiJen/Vaxi-Jen.html).
Table 3.
Antigens reported as protective by both Crompton et al and this study
| Gene ID | Protein | Parasite stage that protein is maximally expressed | BH p-value | Total AA length | VaxiJen | Predicted CD4 T-cell epitopes | # of AA predicted to be part of a B-cell linear epitope |
|---|---|---|---|---|---|---|---|
| PF07_0006 | Starp antigen | Early ring | 1.76E–04 | 594 | 0.9644 (probable antigen) | 2 | 416 |
| PF14_0344 | Hypothetical protein | Merozoite | 1.43E–04 | 993 | 0.7561 (probable antigen) | 8 | 722 |
| PF11_0507 | Antigen 332, putative | Early trophozoite | 3.45E–05 | 6093 | 0.6278 (probable antigen) | 14 | 4323 |
| PFL0590c | p-type ATPase, putative | Early trophozoite | 2.04E–02 | 1209 | 0.5079 (probable antigen) | 28 | 423 |
| PFE0090w | Hypothetical protein | Early schizogony | 3.55E–03 | 1076 | 0.7821 (probable antigen) | 12 | 514 |
| PF13_0190 | Hypothetical protein | Early trophozoite | 2.52E–02 | 524 | 0.5532 (probable antigen) | 0 | 251 |
| PFE0060w | Hypothetical protein | Early trophozoite | 3.25E–03 | 408 | 0.6228 (probable antigen) | 7 | 189 |
| PF11_0008 | Erythrocyte membrane protein 1 (PfEMP1) | Gametocyte | 3.02E–05 | 2994 | 0.6532 (probable antigen) | 19 | 1574 |
| PF13_0003 | Erythrocyte membrane protein 1 (PfEMP1) | Early ring | 2.30E–03 | 3346 | 0.6463 (probable antigen) | 18 | 1774 |
| PF10_0356 | Liver stage antigen, putative | Merozoite | 3.82E–03 | 1162 | 0.7935 (probable antigen) | 1 | 827 |
Immuno-informatics software was used to determine the probability of CD4 T-cell and B-cell epitopes on the 10 antigens identified as being associated with protection in both this study and Crompton et al. Multipred2 available at http://cvc.dfci.harvard.edu/multipred2/index.php, was used to predict CD4 T-cell epitopes of the HLA DR1 supertype. Nine out of the 10 antigens were predicted to have CD4 T-cell epitopes of the HLA DR1 supertype. Only PF13_0190 was not predicted to bind to HLA DR1 supertype allele. Bepipred software available at http://www.cbs.dtu.dk/services/BepiPred was used to predict the presence of linear B-cell epitopes (Table 3). All ten antigens were predicted to have B-cell epitopes.
A detailed list comparing all P. falciparum antigens used on the array is presented in Supporting Information Table 1. The 483 antigens that were printed on the array were down-selected for being seroreactive on an array published earlier containing a diversified set of 2320 Pf proteins. Twenty-five of the 142 (17%) seroreactive proteins on the array presented here contained a predicted signal peptide sequence, compared with 9.7% of the whole Pf proteome, and fifty of the seroreactive proteins (35%) contain one or more predicted transmembrane domains compared with 31% in the Pf proteome.
3.4 Identification of conserved antibody profile in patients with moderately diminished CD4 count
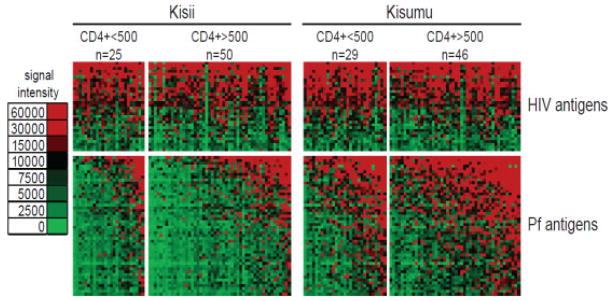
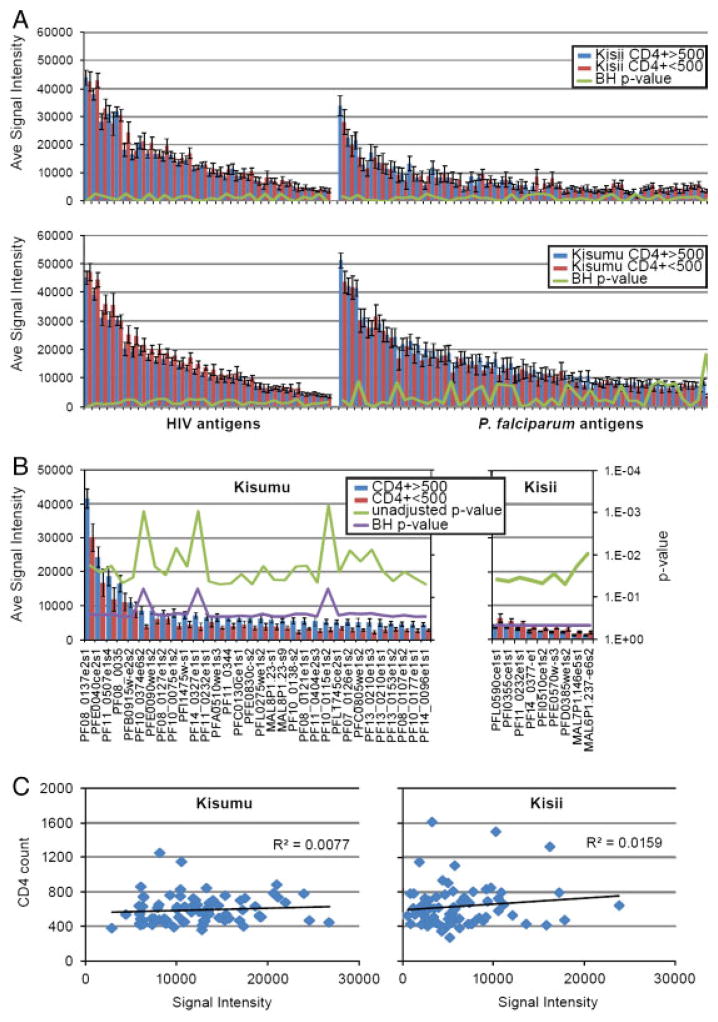
Humoral responses were compared between individuals with high CD4 counts (CD4>500) and individuals with lower CD4 counts (CD4 ≤ 500) in the high transmission site (Kisumu) and low transmission site (Kisii). A comparison of the antibody profiles of individuals with CD4 count >500 to those with CD4 count ≤ 500 is presented in Fig. 3. The mean seroreactivity for HIV and Pf antigens are displayed as bar graphs in Fig. 4A. Individuals with higher CD4 counts did not have significantly higher antibody response to either HIV or Pf antigens after adjustment for false discovery for either site. Furthermore, CD4 cell count did not correlate with average signal intensity of the seroreactive antigens for either Kisumu or Kisii (R2=0.008 and R2=0.016, respectively) (Fig. 4C).
Figure 3.
Comparison of the seroreactivity between low and high CD4 counts within each group. Normalized antibody signal intensity against the 161 most seroreactive antigens are shown according to the colorized scale. (A) Individuals from Kisii (unprotected) and Kisumu (partial protection) are organized into groups of CD4 count below 500 and above. (B) 32 antigens were found to be differentially reactive (unadjusted for false discovery). These antigens are rows and are grouped according to differentially reactive and cross-reactive.
Figure 4.
Differential seroreactivity of P. falciparum in patients with low and high CD4 count. Mean seroreactivity of “low” and “high” CD4 counts are plotted in red and blue, respectively. Corresponding unadjusted p-values for each antigen are shown on the secondary axis. “e” and “s” at the end of the genes refers to exon number and segment number. (A) Comparison of the mean seroreactivity of HIV and P. falciparum antigens between low and high CD4 count individuals within Kisumu and Kisii. (B) Identification of differentially reactive antigens is displayed and the corresponding locus tags are labeled on the x-axis. (C) A scatter plot comparison of the CD4 count (y-axis) and the mean seroreactivity to the 161 seroreactive antigens (x-axis). A linear regression trend line and R2 is displayed for Kisii and Kisumu.
Although we did not find significant differences between CD4 count groups after adjustment for multiple comparisons, we observed a possible trend in decreasing seroreactivity to Pf antigens in lower CD4 count patients. Thirty-two proteins were more reactive among those with higher CD4 counts when not adjusted for multiple comparisons (Fig. 4B). A complete list of these 32 antigens is provided in Supporting Information Table 2. Of the four leading vaccine candidates (CSP, MSP1, LSA1 and AMA1) printed on the array, only AMA1 was more reactive among the high CD4 count group in Kisumu in unadjusted analysis (Table 4). In Kisii, nine antigens were found to be differentially reactive between high CD4 and low CD4 groups without adjustment for false discovery. Paradoxically, all nine were more reactive in the low CD4 group as compared with the high CD4 group (Fig. 4B). However, unlike the differences found in Kisumu, all nine of these proteins were relatively weakly immunoreactive (<5000 signal intensity) in high and low CD4 group, and may represent artifacts. None of the four leading vaccine candidates were differentially reactive in unadjusted analysis when comparing CD4 groups in Kisii (Table 5).
Table 4.
Humoral reactivity of vaccine candidates by CD4 count within Kisumu
| Antigen | Average intensity CD4>500 (N=46) | Average intensity CD4 ≤ 500 (N=29) | p-Value | p-Value (Benjamini– Hochberg adjusted) |
|---|---|---|---|---|
| Circumsporozoite Protein (CSP) | 17 293.3 | 14646.5 | 0.4 | 0.8 |
| Merozoite Surface Protein 1 (MSP-1) | 17 142.2 | 12734.6 | 0.08 | 0.3 |
| Liver Stage Antigen 1 (LSA-1) | 12 396.1 | 11583.5 | 0.8 | 0.9 |
| Apical Membrane Antigen 1 (AMA-1) | 6507.9 | 4191.4 | 0.05a) | 0.3 |
Significant p-value.
Table 5.
Humoral reactivity of vaccine candidates by CD4 count within Kisii
| Antigen | Average intensity CD4>500 (N=46) | Average intensity CD4 ≤ 500 (N=29) | p-Value | p-Value (Benjamini– Hochberg adjusted) |
|---|---|---|---|---|
| Circumsporozoite Protein (CSP) | 4538.7 | 5295.3 | 0.5 | 0.8 |
| Merozoite Surface Protein 1 (MSP-1) | 4780.5 | 5734.1 | 0.4 | 0.8 |
| Liver Stage Antigen 1 (LSA-1) | 5789.9 | 3192.6 | 0.2 | 0.7 |
| Apical Membrane Antigen 1 (AMA-1) | 3100.1 | 2792.6 | 1.0 | 1.0 |
4 Discussion
Understanding which Pf proteins are immune targets of acquired partial immunity could provide crucial insights into malaria immunology and vaccine development. Fifty-seven antigens were significantly more reactive among individuals with presumed partial immunity as compared with individuals without. Further studies need to be performed on these 57 antigens to determine if they are important in immune control of malaria.
The 57 antigens include 4 antigens that have been studied as potential vaccine candidates. The 4 antigens are CSP, AMA1, LSA1 and MSP1. Previous immuno-epidemiological and vaccine studies have suggested a potential role for antibodies against CSP [6, 43], AMA1 [44, 45], LSA1 [6] and MSP1 [46] in mediating protection from malaria. Our findings differ from those reported in Crompton et al. In that study, these four antigens were not found to correlate with protection. One potential rationale for the differences is that all participants in that previous study were exposed to high levels of malaria parasites (an EIR as high as 60 bites/month during the rainy season) and may have induced a maximum or near maximum antibody response to these four antigens [30]. In this study, individuals from a high transmission area were compared with the individuals from a much lower level of transmission, which may have allowed for the arrays to identify these differences.
Ten of the 57 antigens identified as significantly more reactive among individuals with presumed partial immunity to malaria were identified as protective in Crompton et al. The 10 antigens are STARP, two PfEMP1 proteins, LSA1, antigen 332, p-type ATPase and 4 hypothetical proteins. All 10 antigens were predicted to possess B-cell epitopes. All antigens also appeared to possess CD4 T-cell epitopes with the exception of 1 of the hypothetical proteins. It is however possible that this antigen possessed CD4 T-cell epitopes recognized by an HLA class II supertype other than HLA DR1.
STARP antigen is expressed on the surface of the sporozoite stage [47]. Although this antigen has not been studied extensively as a vaccine candidate, earlier studies suggested that humoral responses to this protein may play a role in immune protection [47]. In one study, monoclonal antibodies to STARP antigen prevented sporozoite invasion into hepatocytes [47]. PfEMP1 is an antigen that is thought to play a role in parasite adhesion to capillaries [48]. The process of parasite adhesion is believed to be a crucial part of the pathogenesis of cerebral malaria [48]. PfEMP1 is highly polymorphic [49] making it a challenging vaccine candidate. Population level studies have however suggested that antibodies to a broad array of PfEMP1 variants may play a role in acquired immunity to malaria in endemic areas [50]. LSA-1 is an antigen that has been studied as a vaccine candidate [51]. Cellular immune responses to LSA-1 are thought to be important in malaria immune control [52].
We found significant individual-to-individual variation within each site. These variations were not explained by differences in CD4 cell count, and are likely due to small differences in parasite exposure within each site. Factors such as consistent bed net use, use of other mosquito repelling techniques, possible travel between areas of differing rates of malaria transmission, and other factors may also contribute to intra-site differences.
CD4 count did not significantly affect humoral responses to malaria in this cohort. CD4 cells play a critical role in malaria immune control [53]. Earlier studies have shown that CD4 cells are important in the generation of antibodies to pathogens [54, 55]. It is possible that the increased malaria susceptibility seen in HIV-1-infected adults with lower CD4 counts is primarily related to impairments in cellular immunity and not humoral immunity. Cellular immune control of malaria parasites is mediated by the release of cytokines such as IFN-γ and HIV-1 infection may lead to impaired cytokine responses to malaria parasites [53, 56]. Another possible explanation for the lack of association between CD4 count and humoral responses to malaria is that the low CD4 count group in this cohort may not have been sufficiently immunocompromised to detect differences. The median CD4 count in the low CD4 groups in Kisii and Kisumu were 432 and 449 respectively. It is possible that malaria-specific humoral immunity is impaired in severely immunocompromised HIV-1-infected adults with lower CD4 counts contributing to higher risk of malaria in that group.
Although this study provides useful new insights on malaria immune responses, there are a number of weaknesses in our study design. Our basic assumption in this study is that intensity of malaria transmission correlates with partial immunity. However, this does not take into account the fact that individuals may travel between sites potentially causing a bias towards the null hypothesis. To truly determine who has partial immunity to malaria, it would have been important to obtain longitudinal data on clinical malaria incidence within this cohort. Alternatively, an antibody functional assay could have been used. Second, our sample size may have limited our ability to detect differentially reactive antibody responses associated with loss of CD4+ cells. The samples collected for this study had relatively high CD4 counts in this cohort (median=560, range=275–1612). Hence we were unable to analyze which responses are lost as CD4 counts drop below 200. Finally, partial immunity to malaria involves both cellular and humoral arms of the immune system. Our study is only able to evaluate humoral immunity.
In summary, this study utilizes a relatively new technique to study humoral immune responses to malaria among individuals of differing susceptibility to malaria. We have identified 57 antigens that were significantly more reactive among a group of adults with presumed partial immunity to malaria as compared with adults prone to clinical malaria. Given the limitations of this study, it will be important for more studies to be conducted to determine the role of these 57 antigens in acquired partial immunity to malaria.
Supplementary Material
Clinical Relevance.
Naturally acquired immunity to malaria develops among adults living in malaria endemic areas after years of exposure to malaria. This immunity protects individuals from severe disease and death but not from malaria infection. Studies suggest an important role for humoral immune responses. Despite years of research there is still no clear understanding of the targets of naturally acquired immunity to malaria. In this study, we utilize protein microarray techniques to define which malaria specific immune responses are impaired among individuals at higher risk for malaria. The immune responses identified by this study may play an important role in naturally acquired malaria immunity. The results of this study may provide new insights into malaria immunology and vaccine development.
Acknowledgments
The authors acknowledge the participants at the two sites in Kenya as well as the staff of the UW/KEMRI collaboration. They acknowledge Xiaolin Tan for her assistance with data analysis. They thank Vladimir Diaz-Ochoa and Vivian E. Chen for assistance with cloning of the HIV antigens. This work was supported by the Division of Intramural Research of the National Institute of Allergy and Infectious Diseases, National Institutes of Health, and by National Institute of Allergy and Infectious Diseases Grant number 1R43AI066791 (P.L.F.). This study was also funded in part by Antigen Discovery Inc. through funds from two National Institutes of Health small business innovation and research (SBIR) grants, 1R43AI066791 (X.L.) and 1R43AI075692 (X.L.).
This company provided the study with the Plasmodium expression plasmids that were used for the study. The company used funds from the following NIH grants to produce the plasmids, (1) 1R43AI066791 “Scanning the P. falciparum proteome for vaccine antigens”, and (2) 1R43AI075692 “Antibodies to malaria blood stages in single and mixed-species infections”.
Footnotes
The authors have declared the following potential conflict of interest: One of the authors (Xiaowu Liang) is a manager at Antigen Discovery Inc. (ADi).
References
- 1.World Malaria Report. Published by the World Health Organization; 2008. www.who.int/malaria/wmr2008. [Google Scholar]
- 2.Langhorne J, Ndungu FM, Sponaas AM, Marsh K. Immunity to malaria: more questions than answers. Nat Immunol. 2008;9:725–732. doi: 10.1038/ni.f.205. [DOI] [PubMed] [Google Scholar]
- 3.Cohen S, Mc GI, Carrington S. Gamma-globulin and acquired immunity to human malaria. Nature. 1961;192:733–737. doi: 10.1038/192733a0. [DOI] [PubMed] [Google Scholar]
- 4.McGregor IA. The passive transfer of human malarial immunity. Am J Trop Med Hyg. 1964;13:237–239. doi: 10.4269/ajtmh.1964.13.237. [DOI] [PubMed] [Google Scholar]
- 5.Sabchareon A, Burnouf T, Ouattara D, Attanath P, et al. Parasitologic and clinical human response to immunoglobulin administration in falciparum malaria. Am J Trop Med Hyg. 1991;45:297–308. doi: 10.4269/ajtmh.1991.45.297. [DOI] [PubMed] [Google Scholar]
- 6.Chelimo K, Ofulla AV, Narum DL, Kazura JW, et al. Antibodies to Plasmodium falciparum antigens vary by age and antigen in children in a malaria-holoendemic area of Kenya. Pediatr Infect Dis J. 2005;24:680–684. doi: 10.1097/01.inf.0000172151.28851.fd. [DOI] [PubMed] [Google Scholar]
- 7.John CC, Zickafoose JS, Sumba PO, King CL, Kazura JW. Antibodies to the Plasmodium falciparum antigens circumsporozoite protein, thrombospondin-related adhesive protein, and liver-stage antigen 1 vary by ages of subjects and by season in a highland area of Kenya. Infect Immun. 2003;71:4320–4325. doi: 10.1128/IAI.71.8.4320-4325.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.John CC, Tande AJ, Moormann AM, Sumba PO, et al. Antibodies to pre-erythrocytic Plasmodium falciparum antigens and risk of clinical malaria in Kenyan children. J Infect Dis. 2008;197:519–526. doi: 10.1086/526787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gray JC, Corran PH, Mangia E, Gaunt MW, et al. Profiling the antibody immune response against blood stage malaria vaccine candidates. Clin Chem. 2007;53:1244–1253. doi: 10.1373/clinchem.2006.081695. [DOI] [PubMed] [Google Scholar]
- 10.Bull PC, Marsh K. The role of antibodies to Plasmodium falciparum-infected-erythrocyte surface antigens in naturally acquired immunity to malaria. Trends Microbiol. 2002;10:55–58. doi: 10.1016/s0966-842x(01)02278-8. [DOI] [PubMed] [Google Scholar]
- 11.Osier FH, Fegan G, Polley SD, Murungi L, et al. Breadth and magnitude of antibody responses to multiple Plasmodium falciparum merozoite antigens are associated with protection from clinical malaria. Infect Immun. 2008;76:2240–2248. doi: 10.1128/IAI.01585-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen S, McGregor IA, Carrington S. Gamma-globulin and acquired immunity to malaria. Nature. 1961;192:733–737. doi: 10.1038/192733a0. [DOI] [PubMed] [Google Scholar]
- 13.Sabchareon A, Burnouf T, Ouatara D, Attanath P, et al. Parasitologic and clinical human response to immunoglobulin administration in falciparum malaria. Am J Trop Med Hyg. 1991;45:297–308. doi: 10.4269/ajtmh.1991.45.297. [DOI] [PubMed] [Google Scholar]
- 14.Perraut R, Marrama L, Diouf B, et al. Antibodies to the conserved C-terminal domain of the Plasmodium falciparum merozoite surface protein 1 and to the merozoite extract and their relationship with in vitro inhibitory antibodies and protection against clinical malaria in a Senegalese village. J Infect Dis. 2005;191:264–271. doi: 10.1086/426398. [DOI] [PubMed] [Google Scholar]
- 15.Roussilhon C, Oeuvray C, Müller-Graf C, et al. Long-term clinical protection from falciparum malaria is strongly associated with IgG3 antibodies to merozoite surface protein 3. PLoS Med. 2007;4:e320. doi: 10.1371/journal.pmed.0040320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ayisi JG, Branch OH, Rafi-Janajreh A, et al. Does infection with human immunodeficiency virus affect the antibody responses to Plasmodium falciparum antigenic determinants in asymptomatic pregnant women? J Infect. 2003;46:164–172. doi: 10.1053/jinf.2002.1088. [DOI] [PubMed] [Google Scholar]
- 17.Migot F, Ouedraogo JB, Diallo J, et al. Selected P. falciparum responses are maintained in AIDS adults in Burkina Faso. Parasite Immunol. 1996;18:333–339. doi: 10.1046/j.1365-3024.1996.d01-116.x. [DOI] [PubMed] [Google Scholar]
- 18.Mount AM, Mwapasa V, Elliott SR, et al. Impairment of humoral immunity to Plasmodium falciparum malaria in pregnancy by HIV infection. Lancet. 2004;363:1860–1867. doi: 10.1016/S0140-6736(04)16354-X. [DOI] [PubMed] [Google Scholar]
- 19.Kashamuka M, Nzila A, Mussey L, et al. Short report: analysis of anti-malaria immune response during human immunodeficiency virus infection in adults in Kinshasa, Democratic Republic of Congo. Am J Trop Med Hyg. 2003;68:376–378. [PubMed] [Google Scholar]
- 20.Ned RM, Moore JM, Chaisavaneeyakorn S, Vdhayakumar V, et al. Modulation of immune responses during HIV-malaria co-infection in pregnancy. Trends Parasitol. 2005;21:284–291. doi: 10.1016/j.pt.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 21.De Milito A. B lymphocyte dysphunctions in HIV infection. Curr HIV Res. 2004;2:11–21. doi: 10.2174/1570162043485068. [DOI] [PubMed] [Google Scholar]
- 22.Cagigi A, Nilsson A, De Milito A, Chiodi F, et al. B cell immunopathology during HIV-1 infection: lessons to learn for HIV-1 vaccine design. Vaccine. 2008;26:3016–3025. doi: 10.1016/j.vaccine.2007.11.063. [DOI] [PubMed] [Google Scholar]
- 23.Longwe H, Gordon S, Malamba S, et al. Characterising B cell numbers and memory B cells in HIV infected and uninfected Malawian adults. BMC Infect Dis. 2010;10:280–285. doi: 10.1186/1471-2334-10-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Madhi SA, Kuwanda L, Cutland C, Kayhty H, et al. Quantitative and qualitative antibody response to pneumococcal conjugate vaccine among African human immunodeficiency virus-infected and uninfected children. Pediatr Infect Dis J. 2005;24:410–416. doi: 10.1097/01.inf.0000160942.84169.14. [DOI] [PubMed] [Google Scholar]
- 25.Laufer MK, van Oosterhout JJ, Thesing PC, Thumba F, et al. Impact of HIV-associated immunosuppression on malaria infection and disease in Malawi. J Infect Dis. 2006;193:872–878. doi: 10.1086/500245. [DOI] [PubMed] [Google Scholar]
- 26.French N, Nakiyingi J, Lugada E, Watera C, et al. Increasing rates of malarial fever with deteriorating immune status in HIV-1-infected Ugandan adults. AIDS. 2001;15:899–906. doi: 10.1097/00002030-200105040-00010. [DOI] [PubMed] [Google Scholar]
- 27.Whitworth J, Morgan D, Quigley M, Smith A, et al. Effect of HIV-1 and increasing immunosuppression on malaria parasitaemia and clinical episodes in adults in rural Uganda: a cohort study. Lancet. 2000;356:1051–1056. doi: 10.1016/S0140-6736(00)02727-6. [DOI] [PubMed] [Google Scholar]
- 28.Vigil A, Davies DH, Felgner PL. Defining the humoral immune response to infectious agents using high-density protein microarrays. Future Microbiol. 2010;5:241–251. doi: 10.2217/fmb.09.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davies DH, Liang X, Hernandez JE, Randall A, et al. Profiling the humoral immune response to infection by using proteome microarrays: high-throughput vaccine and diagnostic antigen discovery. Proc Natl Acad Sci USA. 2005;102:547–552. doi: 10.1073/pnas.0408782102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crompton PD, Kayala MA, Traore B, Kayentao K, et al. A prospective analysis of the Ab response to Plasmodium falciparum before and after a malaria season by protein microarray. Proc Natl Acad Sci USA. 2010;107:6958–6963. doi: 10.1073/pnas.1001323107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ndenga B, Githeko A, Omukunda E, Munyekenye G, et al. Population dynamics of malaria vectors in western Kenya highlands. J Med Entomol. 2006;43:200–206. doi: 10.1603/0022-2585(2006)043[0200:pdomvi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 32.Hay SI, Noor AM, Simba M, Busalo M, et al. Clinical epidemiology in the highlands of western Kenya. Emerg Infect Dis. 2002;8:543–548. doi: 10.3201/eid0806.010309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lindsay SW, Maartens WJM. Malaria in the African Highlands: past, present and future. Bull World Health Organ. 1998;76:33–45. [PMC free article] [PubMed] [Google Scholar]
- 34.Snow RW, Omumbo JA, Lowe B, Molyneux CS, et al. Relationship between severe malaria morbidity in children and level of Plasmodium falciparum transmission in Africa. Lancet. 1997;349:1650–1654. doi: 10.1016/S0140-6736(97)02038-2. [DOI] [PubMed] [Google Scholar]
- 35.Doolan DL, Mu Y, Unal B, Sundaresh S, et al. Profiling humoral immune responses to P. falciparum infection with protein microarrays. Proteomics. 2008;8:4680–4694. doi: 10.1002/pmic.200800194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sundaresh S, Randall A, Unal B, Petersen JM, et al. From protein microarrays to diagnostic antigen discovery: a study of the pathogen Francisella tularensis. Bioinformatics. 2007;23:i508–i518. doi: 10.1093/bioinformatics/btm207. [DOI] [PubMed] [Google Scholar]
- 37.Kreil DP, Karp NA, Lilley KS. DNA microarray normalization methods can remove bias from differential protein expression analysis of 2D difference gel electrophoresis results. Bioinformatics. 2004;20:2026–2034. doi: 10.1093/bioinformatics/bth193. [DOI] [PubMed] [Google Scholar]
- 38.Sundaresh S, Doolan DL, Hirst S, Mu Y, et al. Identification of humoral immune responses in protein microarrays using DNA microarray data analysis techniques. Bioinformatics. 2006;22:1760–1766. doi: 10.1093/bioinformatics/btl162. [DOI] [PubMed] [Google Scholar]
- 39.Long AD, Mangalam HJ, Chan BY, Tolleri L, et al. Improved statistical inference from DNA microarray data using analysis of variance and a Bayesian statistical framework. Analysis of global gene expression in Escherichia coli K12. J Biol Chem. 2001;276:19937–19944. doi: 10.1074/jbc.M010192200. [DOI] [PubMed] [Google Scholar]
- 40.Baldi P, Long AD. A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics. 2001;17:509–519. doi: 10.1093/bioinformatics/17.6.509. [DOI] [PubMed] [Google Scholar]
- 41.Sundaresh S, Randall A, Unal B, Petersen JM, et al. From protein microarrays to diagnostic antigen discovery: A study of the pathogen Francisella tularensis. Bioinformatics. 2007;23:i508–i518. doi: 10.1093/bioinformatics/btm207. [DOI] [PubMed] [Google Scholar]
- 42.Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9:811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- 43.Abdulla S, Oberholzer R, Juma O, Kubhoja S, et al. Safety and immunogenicity of RTS,S/AS02D malaria vaccine in infants. N Engl J Med. 2008;359:2533–2544. doi: 10.1056/NEJMoa0807773. [DOI] [PubMed] [Google Scholar]
- 44.Remarque EJ, Faber BW, Kocken CH, Thomas AW. Apical membrane antigen 1: a malaria vaccine candidate in review. Trends Parasitol. 2008;24:74–84. doi: 10.1016/j.pt.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 45.Thera MA, Doumbo OK, Coulibaly D, Diallo DA, et al. Safety and immunogenicity of an AMA-1 malaria vaccine in Malian adults: results of a phase 1 randomized controlled trial. PLoS One. 2008;3:e1465. doi: 10.1371/journal.pone.0001465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perraut R, Marrama L, Diouf B, Sokhna C, et al. Antibodies to the conserved C-terminal domain of the Plasmodium falciparum merozoite surface protein 1 and to the merozoite extract and their relationship with in vitro inhibitory antibodies and protection against clinical malaria in a Senegalese village. J Infect Dis. 2005;191:264–271. doi: 10.1086/426398. [DOI] [PubMed] [Google Scholar]
- 47.Pasquetto V, Fidock DA, Gras H, Badell E, et al. Plasmodium falciparum sporozoite invasion is inhibited by naturally acquired or experimentally induced polyclonal antibodies to the STARP antigen. Eur J Immunol. 1997;27:2502–2513. doi: 10.1002/eji.1830271007. [DOI] [PubMed] [Google Scholar]
- 48.Rao A, Kumar MK, Joseph T, Bulusu G. Cerebral malaria: insights from host–parasite protein–protein interactions. Malar J. 2010;9:155. doi: 10.1186/1475-2875-9-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mackintosh CL, Christodoulou Z, Mwangi TW, Kortok M, et al. Acquisition of naturally occurring antibody responses to recombinant protein domains of Plasmodium falciparum erythrocyte membrane protein 1. Malar J. 2008;7:155. doi: 10.1186/1475-2875-7-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bull PC, Lowe BS, Kortok M, Molyneux CS, et al. Parasite antigens on the infected red cell surface are targets for naturally acquired immunity to malaria. Nat Med. 1998;4:358–360. doi: 10.1038/nm0398-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pichyangkul S, Kum-Arb U, Youngvanitchit A, Limsalakpetch A, et al. Pre-clinical evaluation safety and immunogenicity of Plasmodium falciparum LSA1/AS01B when administered alone or in concurrently with RTS,S/AS01B in rhesus primates. Infect Immun. 2008;76:229–238. doi: 10.1128/IAI.00977-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Speak C, Duffy PE. Antigens for pre-erythrocytic vaccines: building on success. Parasite Immunol. 2009;31:539–546. doi: 10.1111/j.1365-3024.2009.01139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stevenson MM, Riley EM. Innate immunity to malaria. Nat Rev Immunol. 2004;4:169–180. doi: 10.1038/nri1311. [DOI] [PubMed] [Google Scholar]
- 54.Sette A, Moutaftsi M, Moyron-Quiroz J, McCausland MM, et al. Selective CD4+ T cell help for antibody responses to a large viral pathogen: deterministic linkage of specificities. Immunity. 2008;28:847–858. doi: 10.1016/j.immuni.2008.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jing L, Davies DH, Chong TM, Chun S, et al. An extremely diverse CD4 response to vaccinia virus in humans is revealed by proteome-wide T-cell profiling. J Virol. 2008;82:7120–7134. doi: 10.1128/JVI.00453-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moore JM, Ayisi J, Nahlen BL, Misore A, et al. Immunity to placental malaria. II. Placental antigen-specific cytokine responses are impaired in human immunodeficiency virus-infected women. J Infect Dis. 2000;182:960–964. doi: 10.1086/315755. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.