Abstract
HIV-1 protease is one of the main therapeutic targets in HIV. However, a major problem in treatment of HIV is the rapid emergence of drug-resistant strains. It should be particularly helpful to clinical therapy of AIDS if one method can be used to predict antivirus capability of compounds for different variants. In our study, proteochemometric (PCM) models were created to study the bioactivity spectra of 92 chemical compounds with 47 unique HIV-1 protease variants. In contrast to other PCM models, which used Multiplication of Ligands and Proteins Descriptors (MLPD) as cross-term, one new cross-term, i.e. Protein-Ligand Interaction Fingerprint (PLIF) was introduced in our modeling. With different combinations of ligand descriptors, protein descriptors and cross-terms, nine PCM models were obtained, and six of them achieved good predictive abilities (Q2 test>0.7). These results showed that the performance of PCM models could be improved when ligand and protein descriptors were complemented by the newly introduced cross-term PLIF. Compared with the conventional cross-term MLPD, the newly introduced PLIF had a better predictive ability. Furthermore, our best model (GD & P & PLIF: Q2test = 0.8271) could select out those inhibitors which have a broad antiviral activity. As a conclusion, our study indicates that proteochemometric modeling with PLIF as cross-term is a potential useful way to solve the HIV-1 drug-resistant problem.
Introduction
Acquired immunodeficiency syndrome (AIDS), caused by human immunodeficiency virus (HIV), is one of the most fatal diseases to threat human life for its infectivity and high mortality. Since its recognition in 1981, more than 60 million people have been infected with HIV around the world, and approximately 25 million people have died of AIDS. Nowadays, more than 34 million are living with HIV infection [1], [2]. Currently, the main strategies for treating AIDS are through disrupting one or several key steps of HIV life cycle to control the replication rate of HIV virus.
HIV-1 protease is one of the main therapeutic targets in HIV and it is a dimeric protein composed of two identical 99-residue chains. The protease cleaves the Gag-Pol polyprotein into structure proteins and enzymes, which is a necessary step for the generation of new infectious virus particles, and nine of the twenty-eight FDA-approved anti-HIV drugs in current use target the HIV-1 protease. However, mutations were found in the protease soon after the HIV protease inhibitors were introduced, and the high mutation rate of HIV-1 protease allows the virus to escape from the antiviral therapy [3]. So it is necessary to acquire a reasonable method to predict antivirus capability of compounds for a wide spectrum of HIV.
To date, for experimental methods, high-throughput screen is mostly used to filter novel compounds against all kinds of targets as well as HIV mutated variants; for in silico methods, molecular docking [4], [5], [6], pharmacophore models [7], [8], quantitative structure-activity relationship (QSAR) [6], [9], [10], [11] etc are widely used to virtually screen antiviral compounds against HIV mutated variants. However, these methods are limited to the study of the molecular recognition of one series of ligands interacting with single target. In addition, the experimental assays are not only cost-consuming but also limited by the repertoire of compounds [12]. What the previous methods obtained are only suitable for single variant rather than an overall bioactivity profile of compounds' activity against series of variants. Although several methods have been proposed on multi-target, like Liu et.al [13], [14]_ENREF_13_ENREF_13 applied multi-task learning in QSAR to analyze and design the novel multi-target HIV-1 inhibitors as well as HIV-HCV co-inhibitors; Ragno et.al [5], De Martino et.al [15] and Sotriffer et.al [16] used cross-docking to gain insight on the mode of action of new anti-HIV agents against both wild-type and resistant strains, in such multi-target QSAR models, there are no explicit descriptions for targets, especially for the interaction information of target-ligand pairs [13], [14]. On the other hand, it is well known that docking is time-consuming, and the accuracy and versatility of the scoring functions are the main issues for the current docking algorithms [17], [18], [19], [20], [21].
More recently, proteochemometric modeling has been widely used to study the mechanisms for molecular recognition of series of proteins, and widely applied in multiple variants- [22], [23], [24], superfamily- [25], [26], kinome- [27], as well as proteome-wide interaction [28], [29], [30]. This method combines both the ligand and target descriptors, and then correlates them to the activity data. Therefore, PCM models can be considered as an extension of the QSAR models, which are only based on the ligand information. So far proteochemometrics have been successfully applied to HIV-1 protease [23], [24] and reverse transcriptase [22] to analyze drug resistance over the mutational space for multiple variants and multiple inhibitors.
However, in most of previous proteochemometric modeling, cross-terms were derived from Multiplication of Ligand and Protein Descriptors (MLPD) [23], [24], [25], [26], [31]. Cross-term is an additional introduced term. Although it was introduced to account for the complementarity of the properties of the interacting entities and it can describe the two entities simultaneously, the significance is not easy to understand. In addition, a lot of descriptors will be generated by MLPD so that it is computationally time-costive and with much redundancy. To address this issue, here we presented a new cross-term protein-ligand interaction fingerprint (PLIF) [32], [33], [34], [35], which describes the interaction of a protein's residues with its ligand. In our study, we used PLIF to construct PCM models to analyze bioactivity profiles of series of inhibitors against series of HIV-1 protease variants comprehensively.
Results and Discussion
Kernel Selection
Our PCM modeling was performed based on support vector regression (SVR). To select an effective kernel function for SVR, 10-fold cross-validation was first performed based on all the data set with all the four kernel functions in choices. The results of Q2 CV of each model with different combinations of descriptor blocks were listed in Table 1 . From the table, the results show that most of the models run with Normalized Poly Kernel function obtained better predictive ability than those with the other three kernel functions. The paired t-test also showed that Normalized Poly Kernel function was more suitable for this dataset in PCM modeling (p-values are 0.0006827, 8.652e-06, 0.0301, compared with Poly Kernel function, Puk function and RBF Kernel function respectively). Therefore, Normalized Poly Kernel function was selected here.
Table 1. Q2 CV of each model with different combinations of descriptor blocks.
| Models with different descriptor combinations | Normalized Poly Kernel | Poly Kernel | Puk | RBF Kernel |
| GD×P | 0.6429 | 0.3643 | 0.1586 | 0.2988 |
| DLI×P | 0.6327 | 0.2054 | 0.2511 | 0.4221 |
| PLIF | 0.5572 | 0.5727 | 0.1627 | 0.5475 |
| GD & P & GD×P | 0.6476 | 0.3615 | 0.1581 | 0.2916 |
| GD & P & PLIF | 0.7022 | 0.3572 | −0.0214 | 0.6988 |
| GD & P | 0.6623 | 0.5702 | 0.2759 | 0.6731 |
| DLI & P & DLI×P | 0.6273 | 0.2243 | 0.2509 | 0.4155 |
| DLI & P & PLIF | 0.6731 | 0.3475 | 0.0306 | 0.6880 |
| DLI & P | 0.6095 | 0.5195 | 0.3831 | 0.6544 |
Support vector regression has a number of advantages over the conventional linear regressions, especially for its robustness to avoid overfitting [28], [36], [37]. By the use of the non-linear kernel, SVM projects the data into a high-dimensional feature space and correlation is then performed in this hyperspace. The selection of the kernel function for SVR is very important because we may construct learning machines based on how this inner-product kernel is generated. The four kernels (summarized in Table 2 ) are implemented in SMOreg of Weka and commonly used in support vector machine. In previous SVM classification studies, experiments were carried out using two to four of these kernels for comparison. In different studies, different kernel was adapted [38], [39], [40]. Therefore, a kernel that performs well on one dataset does not necessarily perform well on another one. In our regression analysis, Normalized Poly Kernel indicated the best predictive ability among others.
Table 2. Summary of Kernels.
| Type of Kernels | Functions |
| Normalized Poly Kernel |
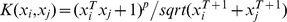
|
| Poly Kernel |
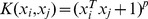
|
| Puk |
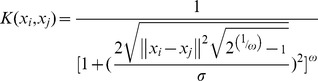
|
| RBF Kernel |
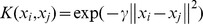
|
Development and evaluation of the PCM models
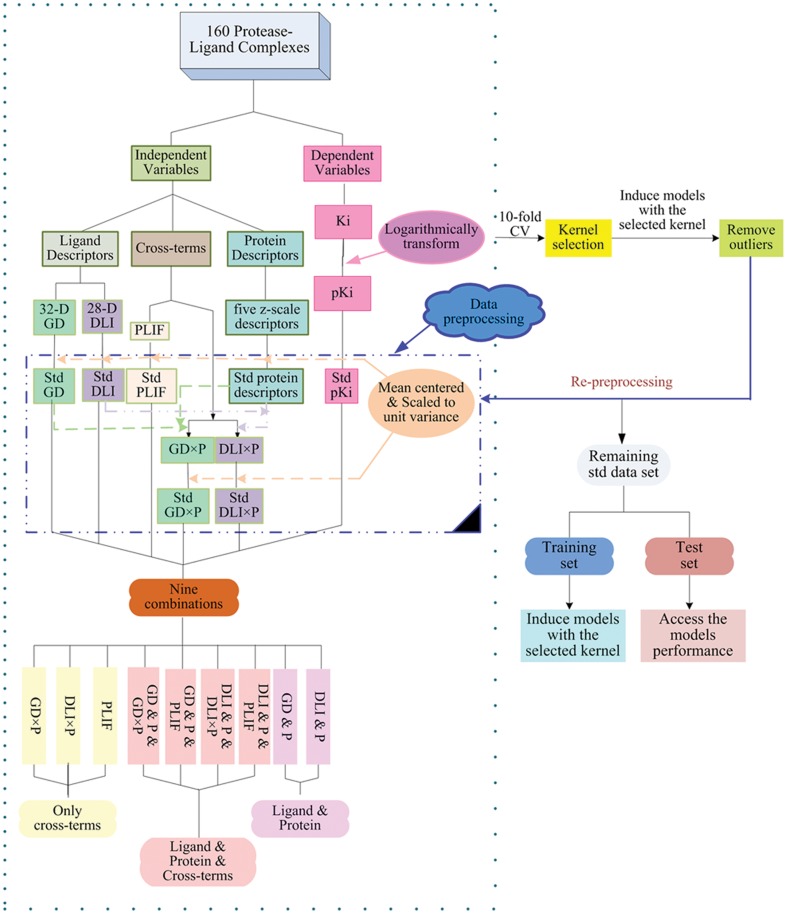
With the selected Normalized Poly Kernel function, nine PCM models with different descriptor combinations were created from all the datasets. 20 ligand-protein pairs behaved as outliers (Z-score> = 2.0 in no less than five of these nine models) , thus they were removed (see Table S3).
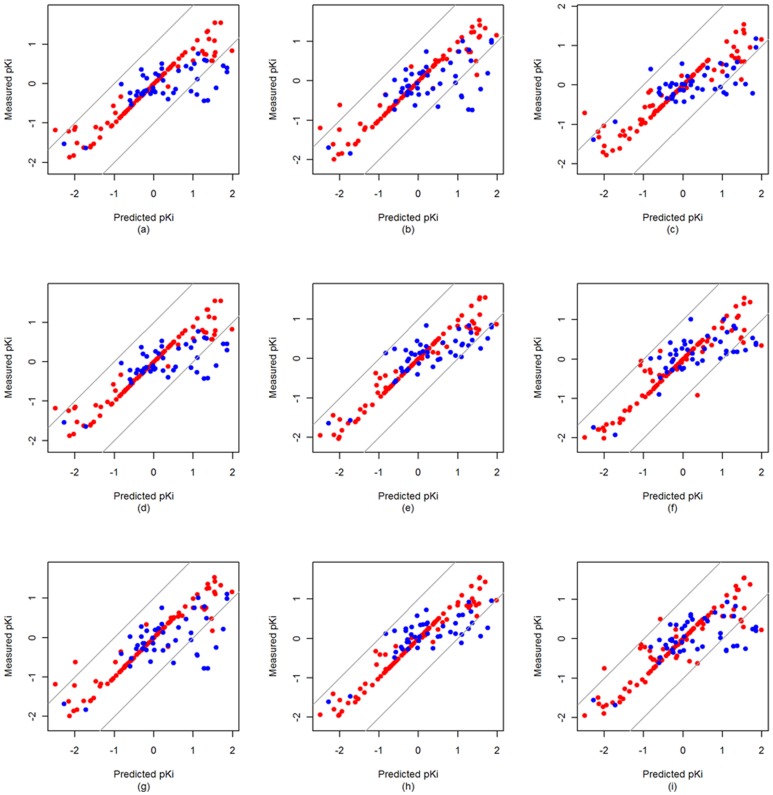
Diverse Subset method was used to split the remaining dataset into a training set (95 inhibitor-protease pairs) (see Table S1) and a test set (45 inhibitor-protease pairs) (see Table S2). The training set was used to create models and the test set was used to evaluate the performance of different models with different descriptor combinations. The obtained goodness-of-fit (R2) and predictive ability (Q2 test) of models were illustrated in Table 3 and Figure 1 . As a result, nine new PCM models were obtained, and six of them achieved reasonably good predictive ability (Q2 test>0.7). The results indicate that the SVR with the selected kernel, as well as the data partition strategy etc are all suitable for the present study.
Table 3. Goodness-of-fit (R2) and predictive ability (Q2 test) of the obtained models.
| Models with different descriptor combinations | GD | DLI | ||
| R2 | Q2 test | R2 | Q2 test | |
| PLIFa | 0.9621 | 0.7470 | 0.9621 | 0.7470 |
| MLPDa | 0.9700 | 0.7101 | 0.9722 | 0.6702 |
| L & P & PLIFb | 0.9716 | 0.8271 | 0.9731 | 0.7929 |
| L & P &MLPDb | 0.9696 | 0.7129 | 0.9727 | 0.6612 |
| L&Pc | 0.9350 | 0.7298 | 0.9241 | 0.6134 |
Models created using only cross-terms.
Models created using ligand and protein descriptors with cross-terms.
Models created using ligand and protein descriptors.
Figure 1. Graphical illustrations of the goodness-of-fit and predictive ability of the obtained models with the selected kernel.
Goodness-of-fit is shown as red solid circles, and predictive ability is shown as blue solid circles. The predicted versus measured activity values using different combinations of descriptor blocks, i.e. GD×P (a), DLI×P (b), PLIF (c), GD & P & GD×P (d), GD & P & PLIF (e), GD & P (f), DLI & P & DLI×P (g), DLI & P & PLIF (h), DLI & P (i) are shown in the figure.
Performance of PLIF as Cross-terms in Proteochemometric Modeling
From Table 3 , we found that when including the PLIF cross-terms, the models obtained better predictive abilities than that of the models without PLIF. For the comparison purpose, we also used the conventional cross-terms MLPD to build PCM models, which is commonly used in previous proteochemometric modeling studies [22], [23], [24], [25], [26]. Obviously, for each kind of ligand descriptors, the newly introduced cross-terms PLIF outperformed the conventional MLPD whether we used only the cross-terms or the combinations of ligand, protein descriptors and cross-terms blocks to create models (see Table 3).
Cross-terms are influenced by both the ligand and the target part [31]. They are intended to describe the properties of the interface between ligand and protein. PLIF is a kind of interaction fingerprint which is calculated from the ligand-target complexes and directly describes the interaction of ligand with protein from hydrogen bonds, ionic interactions, and surface interactions [33]. Therefore, PLIF is inherent to be a suitable cross-term with no surprising that the model performance would be improved by using PLIF as cross-terms. In contrast, MLPD is derived by multiplying ligand and protein descriptors, which is not an essential reflection of the ligand-protein binding. In addition, our results also displayed that the use of MLPD as cross-terms could not improve the model performance significantly as PLIF did, and sometimes even deteriorate the predictive ability. Such result is probably explained by that the PCM models in this study were created using support vector machine, which is a non-linear machine learning method, which is actually expected to fulfill the same purpose as the MLPD does. Therefore, we may conclude that only when a suitable cross-term such as PLIF is used in proteochemometric modeling, the model performance can be improved significantly.
Bioactivity Spectra of HIV-1 Protease Inhibitors
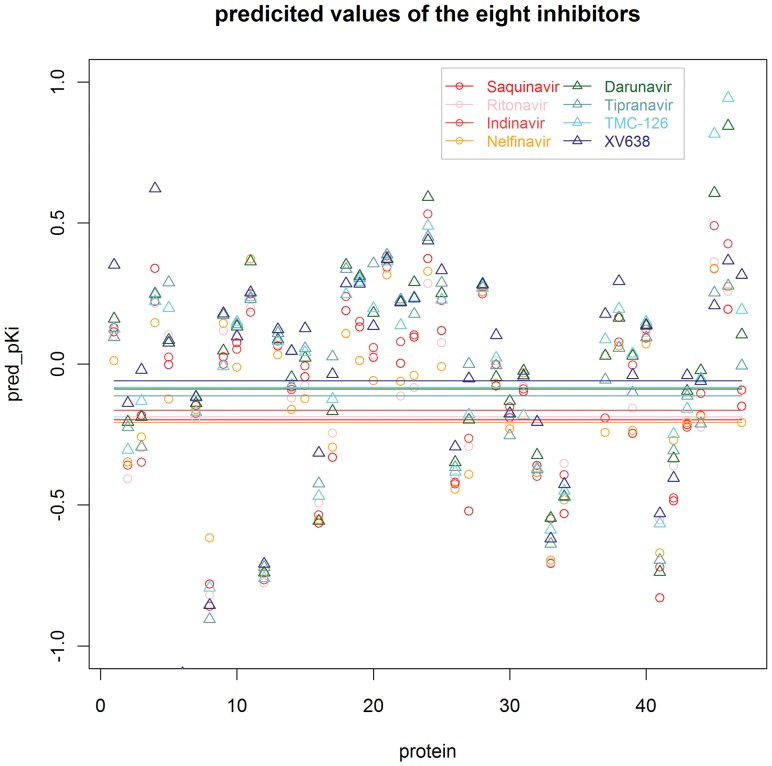
Bioactivities of the four first-generation and four second-generation inhibitors against the 47 protease variants were predicted using our selected best PCM model. The results (shown in Figure 2 ) display that the predicted activities of the second-generation inhibitors are higher than the first-generation ones for most variants. The average predicted values of the second-generation ones are also higher than that of the first-generation ones. Furthermore, the number of proteins for the eight inhibitors whose predicted activities are higher than zero is 10 for Saquinavir, 15 for Ritonavir, 15 for Indinavir, 12 for Nelfinavir, 22 for Darunavir, 23 for Tipranavir, 21 for TMC-126 and 25 for XV638 respectively.
Figure 2. Predicted inhibitory activity (pKi) of the selected eight compounds against 47 proteases.
Red, pink, brown, orange circles stand for the first-generation inhibitors, i.e. Saquinavir, Ritonavir, Indinavir, Nelfinavir respectively; Darkgreen, cadetblue, cyan, blue triangles stand for the second-generation ones, i.e. Darunavir, Tipranavir, TMC-126, XV638 respectively. The lines indicate the average values for each of them.
As we all know, the arrival of the early HIV-1 protease inhibitors was a pivotal moment in the development of antiretroviral therapy. However, the rapid emerging resistance to the first-generation of protease inhibitors occurred, which brought a substantial and persistent problem in the treatment of AIDS. Hence, to inhibit these drug-resistant HIV protease variants, second-generation approaches have been developed. As a result, the second-generation inhibitors should have a broader antiviral activity. Meanwhile, all the above-mentioned results also indicate that the second-generation inhibitors are potent against a wider spectrum of protease variants. Thus it can be seen that our derived model provides a useful way to discovery novel inhibitors which have a broad antiviral activity.
Conclusions
To sum up, we have successfully applied proteochemometric modeling in the study of the bioactivity spectra of HIV-1 protease inhibitors and introduced a new cross-term PLIF into proteochemometrics. Our results showed that when cross-terms were introduced into proteochemometric modeling, the newly introduced cross-term PLIF could always improve the model performance significantly. In addition, we also found that PLIF had a better predictive ability than that of the conventional MLPD. Furthermore, our best derived model shows the ability to discover novel inhibitors with broad antiviral activity. Our study indicates that PLIF could improve the resolution and predictive ability of the PCM model and consequently have potential application to solve the HIV-1 drug-resistant problem.
Materials and Methods
Data set
To create PCM models with PLIF, protein-ligand complexes of HIV-1 protease variants with their inhibitors and the corresponding activity values were collected. Activity is described with K i value which is an inhibition constant and less susceptible by the experiment circumstances than the others such as IC50, EC50 and etc. As a result, 160 protease-ligand complexes with inhibition constants (K i) were retrieved from PDB database, including 92 chemical compounds and 47 HIV-1 protease variants. Inhibition constants of the 160 unique inhibitor-protease pairs were collected from the literatures (see Table S1, S2 and S3). More recent studies suggest that not only the active-site mutations but also distant mutations may influence the drugs' ability to inhibit the protease [41], thus all the mutations in the variants should be taken into consideration in the design of the inhibitors. The fragment 501–599 of P03366 protein was considered as a wild-type protease, and all of the proteases in the 160 complexes were aligned to the wild-type. As a result, the number of the sequences of mutated proteases differing from the wild-type sequence ranges from one to twenty-one (5.17 on average). In all the data set, 69 compounds are collected with K i value for only one type of HIV-1 protease variant, and there are 22 protease variants which only have one ligand with collected K i value for each of them.
The data set was divided into a training set (67%) and an external test set (33%) according to the Diverse Subset partition strategy built in MOE [33], which is used to rank entries in a database based on the distance of their sequences from each other. The general framework for our proteochemometric modeling is presented in Figure 3 .
Figure 3. General framework for our proteochemometric modeling.
Numerical Descriptions for Proteochemometric Modeling
Description of Proteases
Of the 99 amino acids in each protease monomer, 48 positions were found to be mutated in the data set. Mutated positions were coded using the five z-scale descriptors, z1–z5, of amino acids derived by Sandberg et al [42]. The five z-scales are the principal components of 26 computed and measured physicochemical properties of amino acids, and represent essential hydrophobicity/hydrophilicity (z1), steric bulk properties and polarizability (z2), polarity (z3), and electronic effects (z4 and z5) of amino acids. In this way, the varying parts of protease sequences were represented by 48×5 = 240 protease descriptors.
Description of Protease Inhibitors
The inhibitors were represented with two typical kinds of feature space respectively, i.e. 32-dimensional general descriptors (GD) and 28-dimensional Drug-Like Index (DLI). The two sets of descriptors are widely applied in describing organic compounds, and they describe compounds from the views of intrinsic characteristics and drug-like properties respectively. GD includes atomic contributions to logP, molar refractivity, and atomic partial charge [43]. These descriptors characterize physical properties of compounds. They were successfully used to build reasonably good QSAR/QSPR models of boiling point, vapor pressure, free energy of salvation in water, water solubility, receptor class, activity against thrombin/trypsin/factor Xa, blood-brain barrier permeability and compound classification etc. On the other hand, DLI characterizes the hierarchy of drug structures in terms of rings, links, and molecular frameworks [44]. DLI was initially used to rank compounds in a library to select drug-like compounds. In contrast to GD, DLI characterizes simple topological indices of compounds. Therefore, the obtained models will be validated with the two sets of features respectively.
Protease-Inhibitor Cross-terms
Interaction fingerprints have been developed to enhance the representation and analysis of three-dimensional protein-ligand interactions, such as SIFt (structure interaction fingerprint) [45], APIF (atom-pairs-based interaction fingerprint) [46], Pharm-IF (pharmacophore-based interaction fingerprint) [32], PLIF (protein-ligand interaction fingerprint) [32], [33] etc. Additionally, MM-PBSA/GBSA [47], [48], [49], [50] can also generate protein-ligand interaction spectra, which is based on the binding energy. Here, protein-ligand interaction fingerprints were calculated as protease-inhibitor cross-terms using the functions built in MOE [33]. PLIF summarizes the interactions between ligands and proteins using a fingerprint scheme. The interactions are classified into six types: sidechain hydrogen bonds (donor or acceptor), backbone hydrogen bonds (donor or acceptor), ionic interactions, and surface interactions in which a residue may participate. Setting the “Maximum # Bits” as 1000, and using the other default settings, raw protein-ligand interaction data were calculated, and then fingerprint bits were generated. Finally, totally 46 descriptors were extracted whose values represented the strength of the corresponding interaction with the ligand (these 46 descriptors were listed in Table S4). In order to assess the efficiency of introduction of PLIF, MLPD (GD: 32×240 = 7680 cross-terms or DLI: 28×240 = 6720 cross-terms) was also adopted for a complementary comparison.
Preprocessing of data
Prior to the calculation of MLPD and further building PCM models, all descriptors were mean centered and scaled to unit variance. The dependent variable (K i) was logarithmically transformed and also mean centered and scaled to unit variance prior to the use in the computations..
Proteochemometrics Modeling
Selection of Kernel of Support Vector Regression
All models were created using support vector regression (SVR) built in the Weka [36] suit (Weka implementation “SMOreg”), which is a collection of machine learning algorithms for data mining tasks. The kernel of SMOreg implemented in Weka consists of Normalized Poly Kernel (normalized polynomial kernel), Poly Kernel (polynomial kernel), Precomputed Kernel Matrix Kernel, Puk (Pearson VII function-based universal kernel), RBF Kernel (Radial Basis Function kernel), String Kernel. Since Precomputed Kernel Matrix Kernel is based on a static kernel matrix that is read from a file, and String Kernel can't handle multi-valued nominal attributes, the kernel was selected from the left four nonlinear functions. The SMOreg algorithm was run with no normalization/standardization on each of the four kernels. The efficacy of the four kernels was assessed by Q2 (predictive ability) with 10-fold cross-validation.
Model Induction and Validation
We used nine different combinations of descriptor blocks, i.e. three kinds of cross-terms (PLIF, MLPD of GD×P, MLDP of DLI×P), two combinations of ligand and protein descriptors without cross-term (GD & P, DLI & P), and four combinations of ligand, protein descriptors with cross-terms (GD & P & PLIF, GP & P & GP×P, DLI & P & PLIF, DLI & P& DLI×P) to create models from all the datasets with the selected kernel. The Z score method was adopted for the detection of outliers [51], [52], [53]. Any pair is considered as an outlier with removing, if it shows a value of Z-score no lower than 2.0 in no less than five of these nine models. Then the left datasets were split into a training set and a test set. We created nine new models with the training set and assessed the models performance with the external test set. At last, the derived models were quantified by the goodness-of-fit (R2) and predictive ability (Q2 test).
Finally, we selected four first-generation inhibitors, which are the first four drugs (Saquinavir, Ritonavir, Indinavir and Nelfinavir) [54] approved by Food and Drug Administration (FDA), and four second-generation ones, of which two (Darunavir [55] and Tipranavir [56]) are the most recently approved drugs [54] and the other two (TMC-126 [57] and XV638 [58]) are reported to be extremely potent against a wide spectrum of HIV. If there were no experimental complexes for the eight inhibitors against the 47 protease variants, these inhibitors were docked into the protease to derive complexes and generate PLIFs using MOE (presented in Table S5). Subsequently, the best derived model was used to predict their bioactivity spectra.
Supporting Information
Training set used for construction of the proteochemometric models.
(DOCX)
Test set used for assessment of the proteochemometric models.
(DOCX)
Removed outliers.
(DOCX)
PLIFs for all the experimental complexes.
(XLSX)
PLIFs for the eight inhibitors against all the proteases.
(XLSX)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This study was supported in part by grants from National Natural Science Foundation of China (20903058, 30976611), Research Fund for the Doctoral Program of Higher Education of China (20100072120050), Natural Science Foundation of Ningbo (2010A610025) and TCM modernization of Shanghai (09dZ1972800). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Dieffenbach CW, Fauci AS. Thirty years of HIV and AIDS: future challenges and opportunities. Ann Intern Med. 2011;154:766–771. doi: 10.7326/0003-4819-154-11-201106070-00345. [DOI] [PubMed] [Google Scholar]
- 2.Cohen J. HIV prevention. Halting HIV/AIDS epidemics. Science. 2011;334:1338–1340. doi: 10.1126/science.334.6061.1338. [DOI] [PubMed] [Google Scholar]
- 3.Clavel F, Hance AJ. HIV Drug Resistance. New England Journal of Medicine. 2004;350:1023–1035. doi: 10.1056/NEJMra025195. [DOI] [PubMed] [Google Scholar]
- 4.Ragno R, Mai A, Sbardella G, Artico M, Massa S, et al. Computer-aided design, synthesis, and anti-HIV-1 activity in vitro of 2-alkylamino-6-[1-(2,6-difluorophenyl)alkyl]-3,4-dihydro-5-alkylpyrimidin- 4(3H)-ones as novel potent non-nucleoside reverse transcriptase inhibitors, also active against the Y181C variant. J Med Chem. 2004;47:928–934. doi: 10.1021/jm0309856. [DOI] [PubMed] [Google Scholar]
- 5.Ragno R, Frasca S, Manetti F, Brizzi A, Massa S. HIV-reverse transcriptase inhibition: inclusion of ligand-induced fit by cross-docking studies. J Med Chem. 2005;48:200–212. doi: 10.1021/jm0493921. [DOI] [PubMed] [Google Scholar]
- 6.Cichero E, Buffa L, Fossa P. 3,4,5-Trisubstituted-1,2,4-4H-triazoles as WT and Y188L mutant HIV-1 non-nucleoside reverse transcriptase inhibitors: docking-based CoMFA and CoMSIA analyses. J Mol Model. 2011;17:1537–1550. doi: 10.1007/s00894-010-0857-7. [DOI] [PubMed] [Google Scholar]
- 7.Wang RR, Gao YD, Ma CH, Zhang XJ, Huang CG, et al. Mangiferin, an anti-HIV-1 agent targeting protease and effective against resistant strains. Molecules. 2011;16:4264–4277. doi: 10.3390/molecules16054264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Namba AM, Da Silva VBAR, Borges Ramos TMIT, Vermelho RBON, Baptista NZAN, et al. Virtual Screening and Toxicology Prediction of Novel Potential Non- Nucleoside Reverse Transcriptase Inhibitors. Current Bioactive Compounds. 2009;5:128–136. [Google Scholar]
- 9.Jayatilleke PR, Nair AC, Zauhar R, Welsh WJ. Computational studies on HIV-1 protease inhibitors: influence of calculated inhibitor-enzyme binding affinities on the statistical quality of 3D-QSAR CoMFA models. J Med Chem. 2000;43:4446–4451. doi: 10.1021/jm9905357. [DOI] [PubMed] [Google Scholar]
- 10.Hu R, Doucet JP, Delamar M, Zhang R. QSAR models for 2-amino-6-arylsulfonylbenzonitriles and congeners HIV-1 reverse transcriptase inhibitors based on linear and nonlinear regression methods. Eur J Med Chem. 2009;44:2158–2171. doi: 10.1016/j.ejmech.2008.10.021. [DOI] [PubMed] [Google Scholar]
- 11.Zhu R, Wang F, Liu Q, Kang T. Quantitative Structure-Activity Relationship of IOPY/ISPY Analogues as HIV-1 Non-Nucleoside Reverse Transcriptase Inhibitors. Acta Chim Sinica. 2011;69:1731–1736. [Google Scholar]
- 12.Stahura FL, Bajorath J. Virtual screening methods that complement HTS. Comb Chem High Throughput Screen. 2004;7:259–269. doi: 10.2174/1386207043328706. [DOI] [PubMed] [Google Scholar]
- 13.Liu Q, Che D, Huang Q, Cao Z, Zhu R. Multi-target QSAR Study in the Analysis and Design of HIV-1 Inhibitors. Chin J Chem. 2010;28:1587–1592. [Google Scholar]
- 14.Liu Q, Zhou H, Liu L, Chen X, Zhu R, et al. Multi-target QSAR modelling in the analysis and design of HIV-HCV co-inhibitors: an in-silico study. BMC Bioinformatics. 2011;12:294. doi: 10.1186/1471-2105-12-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Martino G, La Regina G, Ragno R, Coluccia A, Bergamini A, et al. Indolyl aryl sulphones as HIV-1 non-nucleoside reverse transcriptase inhibitors: synthesis, biological evaluation and binding mode studies of new derivatives at indole-2-carboxamide. Antivir Chem Chemother. 2006;17:59–77. doi: 10.1177/095632020601700202. [DOI] [PubMed] [Google Scholar]
- 16.Sotriffer CA, Dramburg I. “In situ cross-docking” to simultaneously address multiple targets. J Med Chem. 2005;48:3122–3125. doi: 10.1021/jm050075j. [DOI] [PubMed] [Google Scholar]
- 17.Warren GL, Andrews CW, Capelli AM, Clarke B, LaLonde J, et al. A critical assessment of docking programs and scoring functions. J Med Chem. 2006;49:5912–5931. doi: 10.1021/jm050362n. [DOI] [PubMed] [Google Scholar]
- 18.Leach AR, Shoichet BK, Peishoff CE. Prediction of protein-ligand interactions. Docking and scoring: successes and gaps. J Med Chem. 2006;49:5851–5855. doi: 10.1021/jm060999m. [DOI] [PubMed] [Google Scholar]
- 19.Kitchen DB, Decornez H, Furr JR, Bajorath J. Docking and scoring in virtual screening for drug discovery: methods and applications. Nat Rev Drug Discov. 2004;3:935–949. doi: 10.1038/nrd1549. [DOI] [PubMed] [Google Scholar]
- 20.Perola E, Walters WP, Charifson PS. A detailed comparison of current docking and scoring methods on systems of pharmaceutical relevance. Proteins. 2004;56:235–249. doi: 10.1002/prot.20088. [DOI] [PubMed] [Google Scholar]
- 21.Moitessier N, Englebienne P, Lee D, Lawandi J, Corbeil CR. Towards the development of universal, fast and highly accurate docking/scoring methods: a long way to go. Br J Pharmacol. 2008;153(Suppl 1):S7–26. doi: 10.1038/sj.bjp.0707515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Junaid M, Lapins M, Eklund M, Spjuth O, Wikberg JE. Proteochemometric modeling of the susceptibility of mutated variants of the HIV-1 virus to reverse transcriptase inhibitors. PLoS One. 2010;5:e14353. doi: 10.1371/journal.pone.0014353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lapins M, Wikberg JE. Proteochemometric modeling of drug resistance over the mutational space for multiple HIV protease variants and multiple protease inhibitors. J Chem Inf Model. 2009;49:1202–1210. doi: 10.1021/ci800453k. [DOI] [PubMed] [Google Scholar]
- 24.Lapins M, Eklund M, Spjuth O, Prusis P, Wikberg JE. Proteochemometric modeling of HIV protease susceptibility. BMC Bioinformatics. 2008;9:181. doi: 10.1186/1471-2105-9-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lapinsh M, Prusis P, Lundstedt T, Wikberg JE. Proteochemometrics modeling of the interaction of amine G-protein coupled receptors with a diverse set of ligands. Mol Pharmacol. 2002;61:1465–1475. doi: 10.1124/mol.61.6.1465. [DOI] [PubMed] [Google Scholar]
- 26.Lapinsh M, Prusis P, Uhlen S, Wikberg JE. Improved approach for proteochemometrics modeling: application to organic compound–amine G protein-coupled receptor interactions. Bioinformatics. 2005;21:4289–4296. doi: 10.1093/bioinformatics/bti703. [DOI] [PubMed] [Google Scholar]
- 27.Lapins M, Wikberg JE. Kinome-wide interaction modelling using alignment-based and alignment-independent approaches for kinase description and linear and non-linear data analysis techniques. BMC Bioinformatics. 2010;11:339. doi: 10.1186/1471-2105-11-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strombergsson H, Daniluk P, Kryshtafovych A, Fidelis K, Wikberg JE, et al. Interaction model based on local protein substructures generalizes to the entire structural enzyme-ligand space. J Chem Inf Model. 2008;48:2278–2288. doi: 10.1021/ci800200e. [DOI] [PubMed] [Google Scholar]
- 29.Strombergsson H, Kryshtafovych A, Prusis P, Fidelis K, Wikberg JE, et al. Generalized modeling of enzyme-ligand interactions using proteochemometrics and local protein substructures. Proteins. 2006;65:568–579. doi: 10.1002/prot.21163. [DOI] [PubMed] [Google Scholar]
- 30.Strömbergsson H, Lapins M, Kleywegt GJ, Wikberg JES. Towards Proteome–Wide Interaction Models Using the Proteochemometrics Approach. Mol Inf. 2010;29:499–508. doi: 10.1002/minf.201000052. [DOI] [PubMed] [Google Scholar]
- 31.van Westen GJP, Wegner JK, Ijzerman AP, van Vlijmen HWT, Bender A. Proteochemometric modeling as a tool to design selective compounds and for extrapolating to novel targets. MedChemComm. 2011;2:16–30. [Google Scholar]
- 32.Sato T, Honma T, Yokoyama S. Combining machine learning and pharmacophore-based interaction fingerprint for in silico screening. J Chem Inf Model. 2010;50:170–185. doi: 10.1021/ci900382e. [DOI] [PubMed] [Google Scholar]
- 33.Molecular Operation Environment. 2009.10 ed. Montreal, Quebec, Canada: Chemical Computing Group Inc; 2009. [Google Scholar]
- 34.Huang D, Kang H, Zhang DF, Sheng Z, Liu Q, et al. Comparison of Ligand-, Target Structure-, and Protein-Ligand Interaction Fingerprint-based Virtual Screening Methods. ACTA CHIMICA SINICA. 2011;69:515–522. [Google Scholar]
- 35.Kang H, Sheng Z, Zhu R, Huang Q, Liu Q, et al. Virtual Drug Screen Schema Based on Multiview Similarity Integration and Ranking Aggregation. J Chem Inf Model. 2012 doi: 10.1021/ci200481c. [DOI] [PubMed] [Google Scholar]
- 36.Witten IH, Frank E, Hall MA. Data Mining: Practical Machine Learning Tools and Techniques (Third Edition) San Francisco: Morgan Kaufmann; 2011. pp. 217–223. [Google Scholar]
- 37.Yao XJ, Panaye A, Doucet JP, Zhang RS, Chen HF, et al. Comparative study of QSAR/QSPR correlations using support vector machines, radial basis function neural networks, and multiple linear regression. J Chem Inf Comput Sci. 2004;44:1257–1266. doi: 10.1021/ci049965i. [DOI] [PubMed] [Google Scholar]
- 38.Isa D, Blanchfield P, Chen ZY. Intellectual Property Management System for the Super-Capacitor Pilot Plant. the proceddings of IC-AIProceedings of the 2009 International Conference on Artificial Intelligence, ICAI 2009. Las Vegas Nevada, USA: CSREA Press; 2009. pp. 708–714. [Google Scholar]
- 39.Liangpei Z, Xin H, Bo H, Pingxiang L. A pixel shape index coupled with spectral information for classification of high spatial resolution remotely sensed imagery. Geoscience and Remote Sensing, IEEE Transactions on. 2006;44:2950–2961. [Google Scholar]
- 40.Muller KR, Ratsch G, Sonnenburg S, Mika S, Grimm M, et al. Classifying ‘drug-likeness’ with kernel-based learning methods. J Chem Inf Model. 2005;45:249–253. doi: 10.1021/ci049737o. [DOI] [PubMed] [Google Scholar]
- 41.Muzammil S, Ross P, Freire E. A major role for a set of non-active site mutations in the development of HIV-1 protease drug resistance. Biochemistry. 2003;42:631–638. doi: 10.1021/bi027019u. [DOI] [PubMed] [Google Scholar]
- 42.Sandberg M, Eriksson L, Jonsson J, Sjostrom M, Wold S. New chemical descriptors relevant for the design of biologically active peptides. A multivariate characterization of 87 amino acids. J Med Chem. 1998;41:2481–2491. doi: 10.1021/jm9700575. [DOI] [PubMed] [Google Scholar]
- 43.Labute P. A widely applicable set of descriptors. J Mol Graph Model. 2000;18:464–477. doi: 10.1016/s1093-3263(00)00068-1. [DOI] [PubMed] [Google Scholar]
- 44.Xu J, Stevenson J. Drug-like index: a new approach to measure drug-like compounds and their diversity. J Chem Inf Comput Sci. 2000;40:1177–1187. doi: 10.1021/ci000026+. [DOI] [PubMed] [Google Scholar]
- 45.Deng Z, Chuaqui C, Singh J. Structural interaction fingerprint (SIFt): A novel method for analyzing three-dimensional protein-ligand binding interactions. Journal of Medicinal Chemistry. 2004;47:337–344. doi: 10.1021/jm030331x. [DOI] [PubMed] [Google Scholar]
- 46.Perez-Nueno VI, Rabal O, Borrell JI, Teixido J. APIF: A New Interaction Fingerprint Based on Atom Pairs and Its Application to Virtual Screening. Journal of Chemical Information and Modeling. 2009;49:1245–1260. doi: 10.1021/ci900043r. [DOI] [PubMed] [Google Scholar]
- 47.Kar P, Knecht V. Energetic basis for drug resistance of HIV-1 protease mutants against amprenavir. Journal of Computer-Aided Molecular Design. 2012;26:215–232. doi: 10.1007/s10822-012-9550-5. [DOI] [PubMed] [Google Scholar]
- 48.Kar P, Knecht V. Origin of Decrease in Potency of Darunavir and Two Related Antiviral Inhibitors against HIV-2 Compared to HIV-1 Protease. Journal of Physical Chemistry B. 2012;116:2605–2614. doi: 10.1021/jp211768n. [DOI] [PubMed] [Google Scholar]
- 49.Cai YF, Schiffer CA. Decomposing the Energetic Impact of Drug Resistant Mutations in HIV-1 Protease on Binding DRV. Journal of Chemical Theory and Computation. 2010;6:1358–1368. doi: 10.1021/ct9004678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Swanson JMJ, Henchman RH, McCammon JA. Revisiting free energy calculations: A theoretical connection to MM/PBSA and direct calculation of the association free energy. Biophysical Journal. 2004;86:67–74. doi: 10.1016/S0006-3495(04)74084-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vays VK, Jain A, Ghate M, Maliwal D. QSAR modeling of some substituted alkylidenepyridazin-3-one as a non-cAMP-based antiplatelet agent. Medicinal Chemistry Research. 2011;20:355–363. [Google Scholar]
- 52.Gupta L, Patel A, Karthikeyan C, Trivedi P. QSAR studies on dihydro-alkoxy-benzyl-oxopyrimidines (DABOs) derivatives, a new series of potent, broad-spectrum non-nucleoside reverse transcriptase inhibitors. Journal of Current Pharmaceutical Research. 2010;01:19–25. [Google Scholar]
- 53.Jamloki A, Karthikeyan C, Moorthy NSHN, Trivedi P. QSAR analysis of some 5-amino-2-mercapto-1,3,4-thiadiazole based inhibitors of matrix metalloproteinases and bacterial collagenase. Bioorganic & Medicinal Chemistry Letters. 2006;16:3847–3854. doi: 10.1016/j.bmcl.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 54.Wensing AMJ, van Maarseveen NM, Nijhuis M. Fifteen years of HIV Protease Inhibitors: raising the barrier to resistance. Antiviral Research. 2010;85:59–74. doi: 10.1016/j.antiviral.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 55.Ghosh AK, Dawson ZL, Mitsuya H. Darunavir, a conceptually new HIV-1 protease inhibitor for the treatment of drug-resistant HIV. Bioorganic & Medicinal Chemistry. 2007;15:7576–7580. doi: 10.1016/j.bmc.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Doyon L, Tremblay S, Bourgon L, Wardrop E, Cordingley MG. Selection and characterization of HIV-1 showing reduced susceptibility to the non-peptidic protease inhibitor tipranavir. Antiviral Research. 2005;68:27–35. doi: 10.1016/j.antiviral.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 57.Yoshimura K, Kato R, Kavlick MF, Nguyen A, Maroun V, et al. A potent human immunodeficiency virus type 1 protease inhibitor, UIC-94003 (TMC-126), and selection of a novel (A28S) mutation in the protease active site. Journal of Virology. 2002;76:1349–1358. doi: 10.1128/JVI.76.3.1349-1358.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ala PJ, Huston EE, Klabe RM, Jadhav PK, Lam PY, et al. Counteracting HIV-1 protease drug resistance: structural analysis of mutant proteases complexed with XV638 and SD146, cyclic urea amides with broad specificities. Biochemistry. 1998;37:15042–15049. doi: 10.1021/bi980386e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Training set used for construction of the proteochemometric models.
(DOCX)
Test set used for assessment of the proteochemometric models.
(DOCX)
Removed outliers.
(DOCX)
PLIFs for all the experimental complexes.
(XLSX)
PLIFs for the eight inhibitors against all the proteases.
(XLSX)