Abstract
Due to its ability to form biofilms on medical devices, Staphylococcus epidermidis has emerged as a major pathogen of nosocomial infections. In this study, we investigated the role of the two-component signal transduction system ArlRS in regulating S. epidermidis biofilm formation. An ArlRS-deficient mutant, WW06, was constructed using S. epidermidis strain 1457 as a parental strain. Although the growth curve of WW06 was similar to that of SE1457, the mutant strain was unable to form biofilms in vitro. In a rabbit subcutaneous infection model, sterile disks made of polymeric materials were implanted subcutaneously followed with inoculation of WW06 or SE1457. The viable bacteria cells of WW06 recovered from biofilms on the embedded disks were much lower than that of SE1457. Complementation of arlRS genes expression from plasmid in WW06 restored biofilm-forming phenotype both in vivo and in vitro. WW06 maintained the ability to undergo initial attachment. Transcription levels of several genes involved in biofilm formation, including icaADBC, sigB, and sarA, were decreased in WW06, compared to SE1457; and icaR expression was increased in WW06, detected by real-time reverse-transcription PCR. The biofilm-forming phenotype was restored by overexpressing icaADBC in WW06 but not by overexpressing sigB, indicating that ArlRS regulates biofilm formation through the regulation of icaADBC. Gel shift assay showed that ArlR can bind to the promoter region of the ica operon. In conclusion, ArlRS regulates S. epidermidis biofilm formation in an ica-dependent manner, distinct from its role in S. aureus.
Introduction
Staphylococcus epidermidis is an opportunistic pathogen that normally colonizes human skin and mucosal surfaces. Over the past two decades, S. epidermidis has emerged as a major pathogen of nosocomial infections, particularly infections involving indwelling medical device [1]–[3]. S. epidermidis pathogenesis is associated with its ability to colonize polymer surfaces to form multilayered biofilms, which impair the efficacy of antibiotic treatments and serve to protect the bacteria from the host immune system [4], [5].
Staphylococcal biofilm formation is a complicated process that is regulated by multiple regulatory factors including SigB, agr, SarA, and two-component signal transduction systems (TCSs) [6]–[10]. TCSs mediate a diverse range of adaptive responses to environmental stresses and play a vital role in bacterial pathogenesis [11]–[14]. In S. epidermidis, whereas the TCSs Agr, LytSR and SaeRS are known to be involved in biofilm formation [15]–[17], the role of the ArlRS TCS remains unclear.
The ArlRS TCS was first identified in Staphylococcus aureus [18], [19]. ArlRS strikingly modifies the extracellular proteolytic activity of S. aureus and is a regulator of virulence gene expression. Mutations in either arlR or arlS increase the production of Protein A, α-toxin, β-hemolysin, lipase, coagulase and serine protease. It was suggested that ArlRS may interact with both agr and sarA regulatory loci to modulate the virulence regulation network. A recent study found ArlRS positively regulate S. aureus capsule formation in a sigma B dependent manner [20]. Transcriptional profile analysis showed that in S. aureus, ArlRS positively regulated virulence factor genes such as sdrC, sdrD and sdrE, and functioned as a repressor of several toxin genes including lukD, lukE, phlC and hlgC [21]. Furthermore, ArlRS was found to be involved in S. aureus biofilm formation. An arlS gene transposition mutant of S. aureus clinical strain MT23142 exhibited an increased ability to form biofilms on polymer surfaces, resulting from altered bacterial autolysis and peptidoglycan hydrolase activity [18]. Deletion of the arlRS locus in S. aureus clinical strain 15981 enhanced initial cell attachment and biofilm formation when cultured in Hussain-Hastings-White-modified medium (HHWm) [22].
Staphylococcal biofilm formation occurs in two steps: bacterial cells attach to the material surface; intercellular adhesion between bacterial cells forms multi-layered structures. In the second step, poly-N-acetylglucosamine (PNAG) in S. aureus or polysaccharide intercellular adhesin (PIA) in S. epidermidis synthesized by ica operon-encoded enzymes play a key role [5], [23].
Toledo-Arana’s study demonstrated that although the accumulation of PNAG was increased in the arlRS mutant of S. aureus, deletion of icaADBC did not result in biofilm abolishment. This finding indicates ArlRS may be involved in S. aureus biofilm formation in an ica-independent manner [22].
Here, we demonstrate for the first time that the ArlRS plays an important role in the regulation of S. epidermidis biofilm formation, and acts in an ica-dependent manner distinct from the role of ArlRS in S. aureus biofilm formation.
Materials and Methods
Ethics Statement
All procedures performed on rabbits were conducted according to relevant national and international guidelines (the Regulations for the Administration of Affairs Concerning Experimental Animals, China, and the NIH Guide for the Care and Use of Laboratory Animals) and were approved by the Institutional Animal Care and Use Committee (IACUC) of Shanghai Medical College of Fudan University (IACUC Animal Project Number: 20110307-066).
Bacterial Strains, Plasmids and Growth Media
Bacterial strains and plasmids used in this study are listed in Table 1. B-Medium and Tryptic soy broth (TSB, Oxoid, Cambridge, UK) were used for S. epidermidis cultivation and biofilm formation. Media were supplemented with erythromycin (10 µg/ml), ampicillin (100 µg/ml), tetracycline (10 µg/ml) or chloramphenicol (10 µg/ml) when appropriate for purposes of selection.
Table 1. Plasmids and bacterial strains.
| Discription | |
| Plasmids | |
| pBT2 | temperature-sensitive E.coli-Staphylococcus shuttle vector |
| pTXicaADBC | icaADBC cloned in pTX15, a xylose inducible plasmid |
| pTXsigB | sigB cloned in pTX15 |
| pCNarlS | arlS cloned in pCN51, a Cd2+ inducible plasmid |
| pCNarlRS | arlRS cloned in pCN51 |
| Bacterial strains | |
| RP62A | a standard strain of S. epidermidis, biofilm positive |
| ATCC12228 | a standard strain of S. epidermidis, biofilm negative |
| SE1457 | a clinical strain of S. epidermidis, biofilm positive |
| WW06 | a arlS gene deletion mutant of SE1457, in which ArlR cannot be translated |
| ParlS | WW06 complemented with the plasmid pCNarlS |
| ParlRS | WW06 complemented with the plasmid pCNarlRS |
| PsigB | WW06 complemented with the plasmid pTXsigB |
| PicaADBC | WW06 complemented with the plasmid pTXicaADBC |
| ΔicaC | icaC gene deletion mutant of SE1457 |
| ΔatlE | atlE gene deletion mutant of SE1457 |
| icaCpRS | ΔicaC complemented with the plasmid pCNarlRS |
Construction of arlS Gene Knockout Mutant and Complementation Strains
The arlS gene in S. epidermidis SE1457 was deleted using the temperature-sensitive vector pBT2 [24]. Briefly, an erythromycin-resistance cassette (ermB) was inserted into the pBT2 plasmid. Then the regions flanking arlS gene were amplified by PCR and inserted into pBT2-ermB. Primers for PCR were designed according to the genomic sequence of S. epidermidis RP62A (GenBank accession number CP000029). Primer sequences are listed in Table 2. The recombinant plasmid, designated pBT2-arlS, was transformed by electroporation into S. aureus strain RN4220 then into SE1457. A procedure for allelic displacement of the arlS gene was performed as previous described [24], [25]. The mutant, designated WW06, was verified by PCR, RT-PCR and direct sequencing. Complementation studies were performed using a vector pCN51 [26] with a shine-dalgarno sequence plus either the arlS gene alone or arlRS genes. The resulting plasmids, pCN-arlS and pCN-arlRS, were transformed by electroporation into WW06, forming two complementary strains, ParlS and ParlRS, respectively.
Table 2. Primers used in this study.
| Name | Primers | Applications | |
| ICAP-S | 5′-CTTTCAATTCTAAAATCTCCCCCTT-3′ | icaADBC promoter | |
| ICAP-AS | 5′-TTTTTCACCTACCTTTCGTTAGTTA-3′ | icaADBC promoter | EMSA |
| ICAA-S | 5′-GAGGGAATCAAACAAGCA-3′ | icaA fragment | |
| ICAA-AS | 5′-AGGCACTAACATCCAGCA-3′ | icaA fragment | |
| U-S | 5′-CCGGAATTCTGGAACCGTATCGACAC-3′ | arlS (upstream) | |
| U-A | 5′-CGCGGATCCGGTGGTATGATTCAGGTTG-3′ | arlS (upstream) | Gene knockout |
| D-S | 5′-AACTGCAGCGAATCACATACCCTACG-3′ | arlS (downstream) | |
| D-A | 5′-CTAGCTAGCCTTGTATGTGGGGGGAAT-3′ | arlS (downstream) | |
| U-S’ | 5′-CGCTAAAGAAATACGTTGT-3′ | arlS (upstream) | |
| U-A’ | 5′-GCGCTAGGGACCTCTTTA-3′ | arlS (upstream) | |
| D-S’ | 5′-CTATTGTGAGTTATTAGTGG-3′ | arlS (downstream) | |
| D-A’ | 5′-AAGTGTCAAAGGTCTACTG-3′ | arlS (downstream) | Mutant validation |
| erm-S | 5′-CTATTGTGAGTTATTAGTGG-3′ | ermB cassette | |
| erm-A | 5′-GCGCTAGGGACCTCTTTA-3′ | ermB cassette | |
| arlS-S' | 5′-TCAACCTGAATCATACCACC-3′ | arlS amplification | |
| arlS-A' | 5′-GATGCTTATTACGACGCTCAT-3′ | arlS amplification | |
| sarA-S | 5′-TTCAAAAATCAATGACTGCT-3′ | sarA amplification | |
| sarA-AS | 5′-TTCCTCTTCTTTATTCTCAC-3′ | sarA amplification | |
| sigB-S | 5′-TCACCTGAACAAATTAACCAATG-3′ | sigB amplification | |
| sigB-AS | 5′-CACCTATTAGACCAACCATACC-3′ | sigB amplification | |
| arlS-S | 5′-ATTATTCAAGGTCATCTCAA-3′ | arlS amplification | |
| arlS-A | 5′-ATCTCGCTATTTATGTCTAC-3′ | arlS amplification | |
| icaA-S | 5′-TTATTGGTTGTATCAAGCGAAGTC-3′ | icaA amplification | Real-time PCR |
| icaA-AS | 5′-TCCTCAGTAATCATGTCAGTATCC-3′ | icaA amplification | |
| luxS-S | 5′-GAGCAGATGATTATATTGT-3′ | luxS amplification | |
| luxS-AS | 5′-TGTTAGATTCACTTGTTG-3′ | luxS amplification | |
| rsbU-S | 5′-TCTCTTCATACAGTCCAT-3′ | rsbU amplification | |
| rsbU-AS | 5′-ATAGGTTCAGGTATTCCA-3′ | rsbU amplification | |
| icaR-S | 5′-GGAGCACTAGATAATTGAACAT-3′ | icaR amplification | |
| icaR-S | 5′-CATTGACGGACTTTACCAG-3′ | icaR amplification | |
| gyrB-S | 5′-TAGTATTGACGAGGCATTAGCA-3′ | gyrB amplification | |
| gyrB-AS | 5′-TATCCGCCACCTCCGA-3′ | gyrB amplification | |
Expression of Recombinant ArlR (rArlR) and Preparation of the anti-rArlR Antiserum
The arlR gene was amplified from the genomic DNA of SE1457 and inserted into the vector pET-28a(+) to obtain the recombinant plasmid pET-arlR. The recombinant plasmid was then transformed into Escherichia coli BL21 (DE3). The expressed His-tagged ArlR protein was purified using the ProBond™ Purification System (Invitrogen, Carlsbad, USA) according to the manufacturer’s instructions.
Five Balb/c mice were immunized with 5 μg purified rArlR coupled with aluminum hydroxide adjuvant by intraperitoneal injection, and boosted after four weeks. The antiserum was collected and anti-rArlR antibodies were detected with an immune dot-blot.
Detection of ArlR Expression in SE1457 and WW06 by Western Blot
Overnight cultures of S. epidermidis strains SE1457 and WW06 were inoculated in 100 ml TSB and incubated at 37°C for 4 h. After centrifugation, pellets were washed three times with distilled water and resuspended in 2 ml phosphate buffered saline. After incubation with 100 µg lysostaphin at 37°C for 1 h, cells were ultrasonicated and centrifuged. Supernatants were assayed for total protein concentration by Bradford method. One microgram of bacterial cell extracts were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis and then transferred to polyvinylidene fluoride membranes (GE-Whatman, Shanghai, China); purified rArlR (200 ng) was used as a control. Blots were probed with anti-rArlR antiserum (diluted 1∶1000) followed by horseradish peroxidase-labeled goat anti-mouse IgG (diluted 1∶2000) (Sangon, Shanghai, China). HRP activity was visualized via chemiluminescence detection using CSPD (Roche, Mannheim, Germany).
Bacterial Initial Attachment Assay
The initial attachment ability of S. epidermidis cells was tested as described by Heilmann et al. [27].
Semi-quantitative Detection of Biofilms
S. epidermidis biofilm formation was detected as previously described [28]. Overnight bacterial cultures grown in TSB were diluted 1∶200, then transferred to 96-well polystyrene microtiter plates (200 µl per well). After incubation at 37°C for 24 h, wells were washed gently three times with PBS and stained with 2% crystal violet for 5 min. Next, the plate was rinsed under running tap water, air-dried, and ethanol was used to dissolve the crystal violet; finally, absorbance was determined at 570 nm.
Scanning Electron Microscope Observations of Biofilms
Overnight cultures of S. epidermidis strains were diluted in 1∶200 in TSB and added in 96-well plates containing fragments of a central vein catheter (ABLE, Guangdong, China). After incubation at 37°C for 24 h, catheter fragments were washed and fixed with 2.5% glutaric dialdehyde (pH 7.4), followed by a secondary fixation with 1% osmium tetroxide. Catheter fragments were then washed twice with distilled water, dehydrated in 70%, 95% and 100% ethanol, dried, fixed onto cylindrical metal plates and gilded with an ion sputter (HCP-2, Hitachi, Tokyo, Japan). Catheter fragments were observed for biofilm formation using a scanning electron microscope (XL-30, Philips, Eindhoven, Netherlands). SEM micrographs were taken at ×3000 and ×10000 magnifications.
Biofilm Formation of the arlRS Mutant in vivo
Three New Zealand White female rabbits (2.0–2.5 kg) were anesthetized by intravascular injection with sodium pentobarbital (35 mg/kg of body weight). After the back of rabbit was denuded of fur, sterile polyethylene disks (1.0 cm diameter, 0.1 cm thickness, with 2 mm chimb) were implanted subcutaneously at four sites bilateral to the spine, as reported previously [29]–[31], three disks per site. About 108 S. epidermidis cells from overnight cultures of SE1457, WW06 and ParlRS were collected, resuspended in TSB and injected subcutaneously into different implantation site respectively, using fresh TSB as a negative control. Three days after, the rabbits were euthanized and the implants were removed. The biofilms were scraped from the disks and the viable bacteria (colony forming units, CFUs) were determined. Differences in log10 CFU counts among groups were assessed with one-way analysis of variance (ANOVA) tests using software SPSS version 11.5 (SPSS Inc., Chicago, IL, USA).
Semi-quantitative Detection of Polysaccharide Intercellular Adhesin (PIA)
PIA detection was performed as previously described [32] using wheat germ agglutinin (WGA) as a surrogate for anti-PIA antibody, using an icaC deletion mutant of SE1457 as a PIA negative control, and strain RP62A as a PIA positive control. Overnight cultures of SE1457, WW06, RP62A and ΔicaC in TSB supplemented with 0.5% glucose were diluted to an OD578 of 0.07 with fresh media, added into sterile 6-well cell cultures plates (6 ml/well) (Nunc, Roskilde, Denmark), and incubated at 37°C under static conditions. After 24 h incubation, cultures and the attached biofilms were transferred into 2 ml tubes and centrifuged. To prepare cell surface extracts for PIA detection, pellets were resuspended in 0.5 M ethylenediaminetetraacetic acid, pH 8.0 (3 ml per gram wet weight), followed by heating at 100°C for 5 min. After centrifugation, 40 µl of the supernatant was mixed with 10 µl of proteinase K (20 mg/ml) and incubated at 37°C for 3 h, followed by heating at 100°C for 10 min. Different dilutions of the extracts were transferred to a nitrocellulose membrane, using a 96-well dot blot system (Biometra, Goettingen, Germany). Membranes were blocked with 3% bovine serum albumin and incubated with 2 µg/ml HRP-labeled wheat germ agglutinin (WGA-HRP conjugate, Lectinotest Laboratory, Ukraine) for 1 h. HRP activity was visualized by chromogenic detection using 4-Chloro-1-Naphthol.
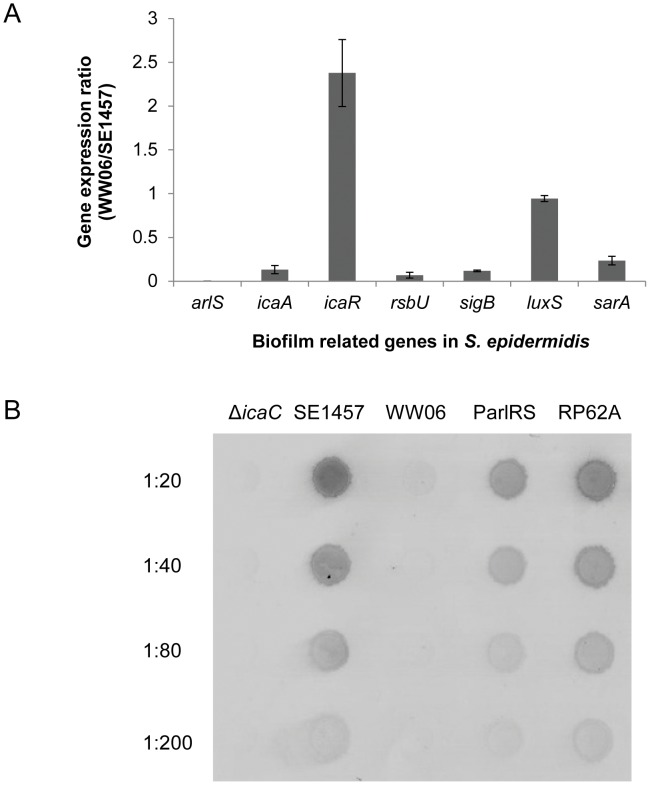
Analysis of Transcriptional Levels of Biofilm-related Genes in WW06
To define the arlRS regulon, transcriptional levels of biofilm-related genes, icaADBC, icaR, rsbU, sigB, luxS and sarA, in the arlRS-deficient strain WW06 and the wild-type parent strain SE1457 were analyzed by RT-qPCR analysis using the ABI 7500 real-time PCR system. Gene-specific primers (Table 2) were designed to yield approximately 100 bp specific products; the housekeeping gene gyrB was used as an endogenous control. All samples were analyzed in triplicate and normalized against gyrB gene expression.
Electrophoretic Mobility Shift Assay (EMSA)
Interaction of the recombinant ArlR and the icaADBC promoter was analyzed by EMSA using the DIG Gel Shift Kit (Roche). rArlR was phosphorylated prior to gel shift reaction by incubating rArlR with 50 mM acetylphosphate for 1 h. The DNA fragment upstream icaADBC (ICAP, 216 bp) was amplified and linked with digoxin-labeled dd-UTP. The resulting DNA fragment, Dig-ICAP, was used for EMSA in native PAGE. Lanes 1–2 were loaded with 0.8 ng Dig-ICAP alone and 0.8 ng Dig-ICAP mixed with 2 µg rArlR, respectively. In lanes 3–5, increasing amounts of unlabeled ICAP (12.5, 62.5, 125 fold increase in the amount of Dig-ICAP in lane 2, respectively) was added as a specific competitor to the mixture of Dig-ICAP and rArlR. In lane 6, 100 ng of a 405 bp icaA fragment was added as a nonspecific competitor. Primers used for to amplify partial icaA gene and ICAP are listed in Table 2. The DNA fragments were transferred to positively charged nylon membranes (Roche) by electro-blotting and visualized by an enzyme immunoassay following the manufacturer’s instruction.
Cell Autolysis Assay
Autolysis assays for S. epidermidis strains were performed as previously described [33]. Strains SE1457, WW06, ParlRS and ΔatlE were cultured in TSB containing 1 M NaCl at 37°C to exponential phase (OD580 = 0.7). Additionally, WW06 and ParlRS were cultured in TSB containing 1 M NaCl and 2 µM CdCl2. Bacterial cells in 50 ml cultures were harvested by centrifugation, washed twice with ice-cold water and resuspended in 50 ml of 0.05 M Tris/HCl (pH 7.2) containing 0.05% (v/v) Triton X-100. Cells were then incubated at 30°C with shaking (200 rpm), and OD580 was measured at 30 min intervals.
Results
Construction of an arlRS Gene-deficient Mutant Strain of S. epidermidis
In S. epidermidis strain SE1457, the TCS arlRS consists of two genes, arlS and arlR, which share the same promoter and overlap by 4 bp. To investigate the function of arlRS in S. epidermidis biofilm formation, an ArlRS deficient mutant was constructed by replacing the arlS gene with an erythromycin-resistant cassette in the biofilm-forming SE1457 strain. The mutant was designated WW06. Western blot analysis of the whole cell extracts from WW06 using antiserum against the recombinant ArlR (rArlR, Figure S1) showed that no ArlR was expressed in WW06 (Figure S2). Thus, WW06 was an ArlRS-deficient mutant.
ArlRS Deficiency in S. epidermidis Abolishing Biofilm Formation in vitro
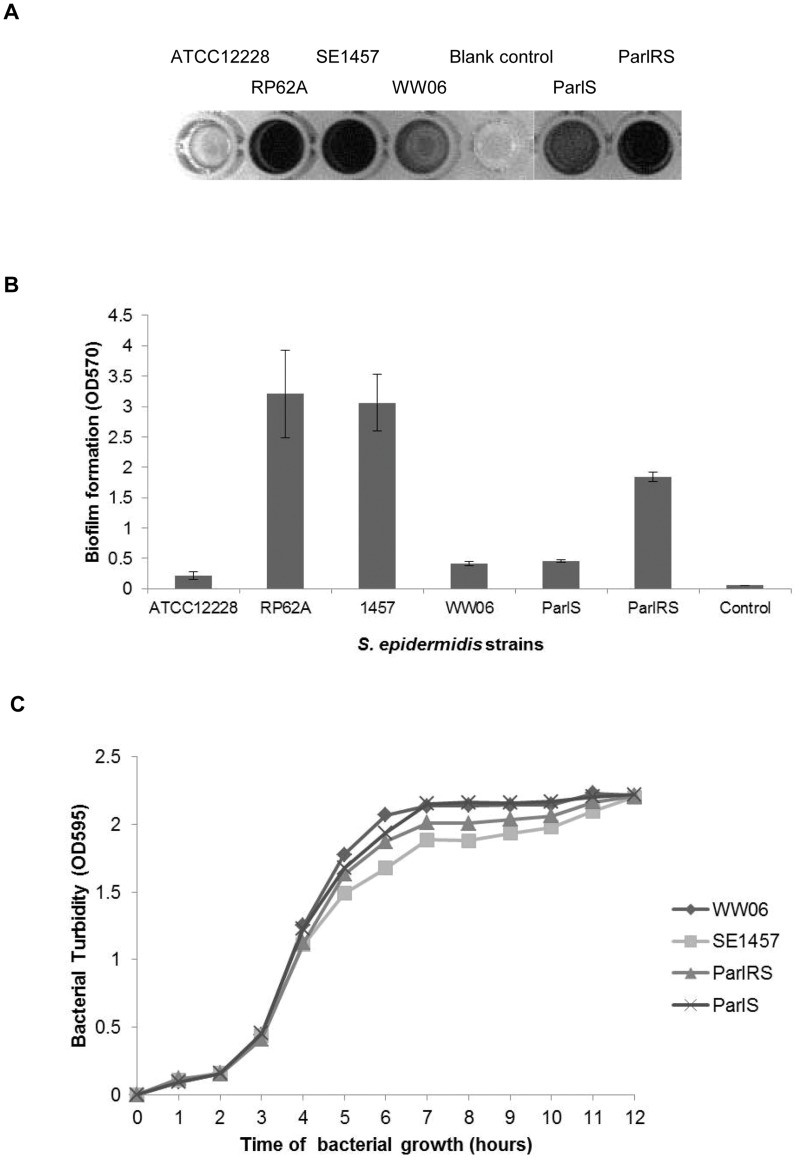
The impact of ArlRS mutation on biofilm formation was investigated using both polystyrene microtiter plates and intravenous catheters. In the microtiter plates, biofilm formation in WW06 was dramatically decreased (OD570 = 0.41±0.04), compared to its wild-type counterpart (OD570 = 3.06±0.47) (Figure 1 A, B). To rule out the possibility of a polar effect, two complementation strains, ParlS and ParlRS, were constructed using plasmids expressing arlS gene and arlRS genes respectively. While ParlRS partially restored biofilm-forming ability (OD570 = 1.84±0.08), the biofilm-forming ability of ParlS (OD570 = 0.45±0.02) was similar to that of WW06.
Figure 1. Effect of ArlRS deficiency on S.epidermidis growth and biofilm formation in vitro.
(A)Semi-quantitative detection of biofilms formed by SE1457, WW06, ParlS and ParlRS over a 24 h period in 96-well polystyrene microtiter plates: The biofilms were stained with 2% crystal violet. (B) The crystal violet was dissolved in ethanol and the absorbance was determined at 570 nm. The mean value ± standard deviation for each strain is shown. (C) Growth curves of S. epidermidis strains: Overnight cultures of SE1457, WW06, ParlS and ParlRS were diluted to OD595 = 0.08 in 150 ml fresh TSB and incubated at 37°C with shaking.The value of OD595 was monitored hourly. The curves represent the means of three independent experiments with standard deviation (SD). Staphylococcus epidermidis strain ATCC12228, a non-biofilm forming control; RP62A, a biofilm positive control.
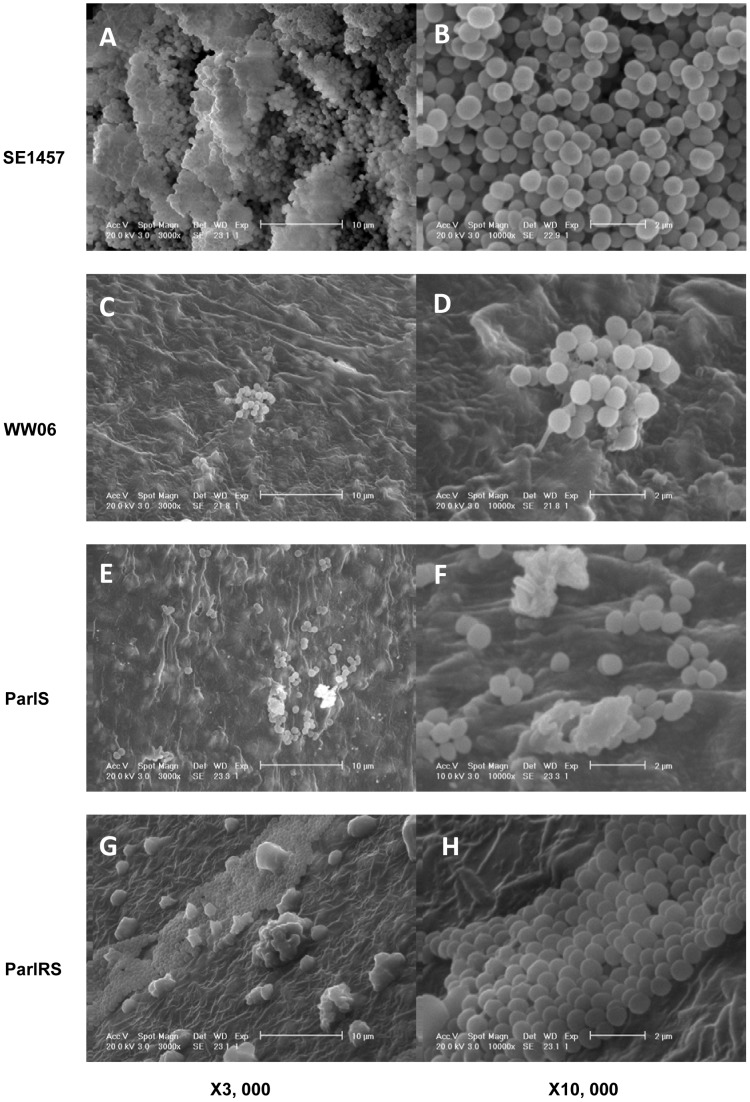
To further investigate the ability of S. epidermidis strains WW06, ParlS, ParlRS, and SE1457 to form biofilms, these strains were incubated with catheter fragments in 96-well plates at 37°C for 24 h and biofilm formation morphology was assessed by SEM. The wild-type strain SE1457 generated a compact, thick biofilm on the catheter surface, whereas the mutant WW06 formed only a few bacterial cell clusters. ParlRS formed a flat biofilm on the catheter, whereas ParlS formed only a small number of microcolonies similar to WW06 (Figure 2).
Figure 2. Biofilms formed on catheter fragments surfaces observed by scanning electron microscope (SEM).
S. epidermidis strains WW06, ParlS, ParlRS and SE1457 were incubated with central vein catheter fragments in 96-well plates. After incubation at 37°C for 24 h, biofilm formed on the surface of the catheter fragments were observed under a transmission electron microscope (XL-30, Philips). Images were obtained at different magnifications (x3000, x10000) for biofilms formed by SE1457 (a, b), WW06 (c, d), ParlS (e, f) and ParlRS(g, h).
In terms of bacterial growth, the growth curves of SE1457, WW06, ParlS and ParlRS were similar (Figure 1C). Additionally, in an assay measuring initial adherence, the numbers of attached cells of SE1457 and WW06 were 3.64×104/cm2 and 3.58×104/cm2, respectively, which showed that mutation of arlRS exhibited little effect on bacterial growth and primary attachment.
Influence of ArlRS Deficiency on S.epidermidis Biofilm Formation in vivo
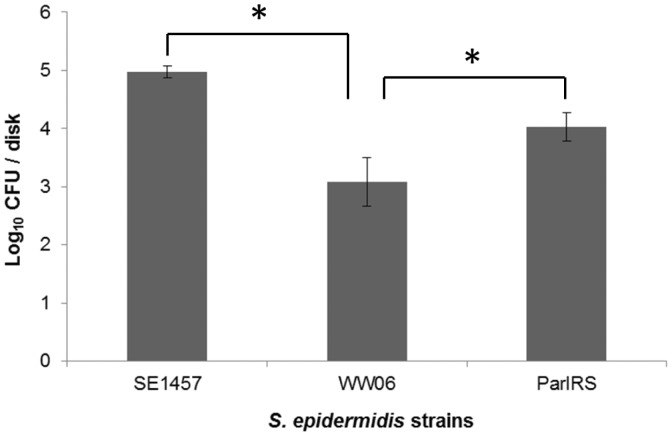
To investigate the effect of ArlRS deficiency on S. epidermidis biofilm formation in vivo, a biofilm infected rabbit model was established. The biofilm formation of SE1457, WW06 and ParlRS on the implanted disks in vivo was observed under an optical microscope and evaluated by determination of viable cells recovered from biofilms. SE1457 formed much thicker biofilms than WW06 on the implanted disks. The log10CFU/disk number of viable cells recovered from the biofilms of the arlRS mutant WW06 was significantly lower than that of SE1457 (3.08±0.42 vs 4.97±0.11, P<0.001) (Figure 3). The log10 number of viable CFU in biofilm of the complementation strain ParlRS was partially restored, which is higher than that of WW06 (4.02±0.25 vs 3.08±0.42, P<0.001).
Figure 3. Influence of ArlRS deficiency on viable S. epidermidis cells recovery from the implanted disks in the rabbit model.
Biofilms of SE1457, WW06 and ParlRS were formed in vivo on sterile polyethylene disks that were implanted subcutaneously in three New Zealand White female rabbits. Three days after infection, the implants were removed. Biofilms were scraped from the disks and CFUs of the viable bacteria recovered from the biofilms were determined and expressed as mean ± standard deviation. Asterisks denote statistically significant difference, P<0.001.
ArlRS Mutation Affecting Bacterial Autolysis
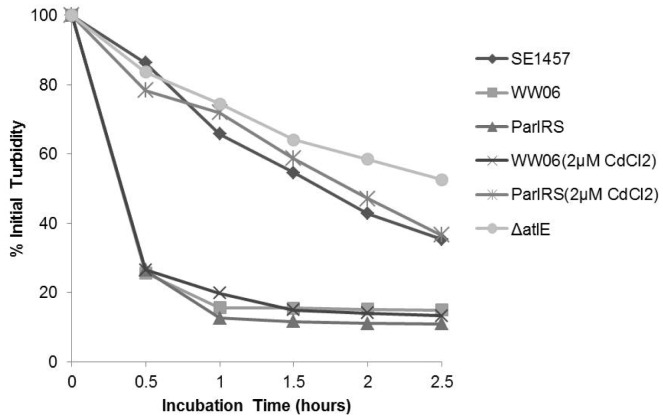
To investigate the effect of ArlRS on bacterial autolysis, SE1457, WW06 and ParlRS were incubated with 0.1% Triton X-100 for 2.5 h and the percent autolysis was calculated. The lytic percentage of WW06 reached 97.83%, which was significantly higher than the 55.38% lytic percentage for SE1457. The autolysis rate of ParlRS was similar to that of WW06 when cultured in media without cadmium chloride (CdCl2); however, when cultured with 2 µM CdCl2 to induce arlRS expression, the lytic percentage of ParlRS was similar to wild-type (Figure 4).
Figure 4. Effect of arlRS mutation on S. epidermidis Triton X-100-induced autolysis.
SE1457, WW06, ParlRS and ΔatlE were cultured in TSB containing 1 M NaCl at 37°C to exponential phase (OD580 = 0.7). WW06 and ParlRS were also cultured in TSB containing 1 M NaCl and 2 µM CdCl2. Triton X-100 induced autolysis of the strains was determined by measuring the change in the value of OD595 at 30 min intervals; results are expressed as lysis percentages. Percent lysis was calculated as follows: [(OD0-ODat time t/OD0)] X 100%]. Experiments were carried out three times independently.
Transcriptional Analysis of Biofilm-related Genes in WW06
To define the arlRS regulon and the role of ArlRS in S. epidermidis biofilm formation, total RNAs from the arlRS-deficient strain WW06 and the wild-type parent strain SE1457 in mid-log growth phase (4 h) were extracted, and transcriptional levels of several biofilm-related genes, rsbU, sigB, sarA, icaR, luxS and icaA, were analyzed by RT-qPCR, using the housekeeping gene gyrB as an internal control. The time point was selected because arlRS transcription in SE1457 reached peak levels at 4 h [34].
Transcription of the biofilm-related genes rsbU, sigB, sarA and icaA in WW06 were found to be down-regulated 14.5 fold, 8.4 fold, 4.3 fold and 7.5 fold, as compared to SE1457, whereas icaR transcription was upregulated 2.4 fold and expression of the quorum sensing-related gene luxS was similar (Figure 5A).
Figure 5. Regulation of expression of biofilm-related genes by ArlRS.
(A) Effect of ArlRS deficiency on transcription levels of the biofilm-related genes. Gene expression profiles of the arlRS-deficient strain WW06 and the wild-type parent strain SE1457 in the mid-log growth phase were analyzed using RT-qPCR. The experiment was performed in triplicate and the expression ratios of the biofilm-related genes in WW06 and SE1457 are represented as mean ± standard deviation. (B) Effect of arlRS mutation on PIA synthesis. PIA was extracted from cells grown for 24 h under biofilm conditions in 6-well cell culture plates. PIA was detected by dot blot analysis using wheat germ agglutinin coupled to horseradish peroxidase. The PIA-deficient S. epidermidis mutant ΔicaC was used as a negative control, and RP62A was used as a positive control.
ArlRS Involving in S. epidermidis Biofilm Formation in an ica-dependent Manner
In contrast to S. aureus, where the ArlRS TCSs has been implicated in ica-independent biofilm formation, RT-qPCR analysis of the ArlRS-deficient mutant of S. epidermidis showed down-regulation of icaADBC transcription. To determine whether the down-regulation of icaADBC expression in WW06 functionally impacted biofilm formation, PIA formation was examined. SE1457 and WW06 were grown under conditions favoring biofilm formation and PIA concentrations were analyzed by a semi-quantitative dot blot assay; using a non-biofilm forming strain ΔicaC as a negative control and strong biofilm producing strain RP62A as a positive control. As expected, no surface-located PIA was detected from either WW06 or ΔicaC, whereas large amount of PIA production was observed in both SE1457 and RP62A. PIA synthesis was restored in the complementation strain ParlRS (Figure 5B).
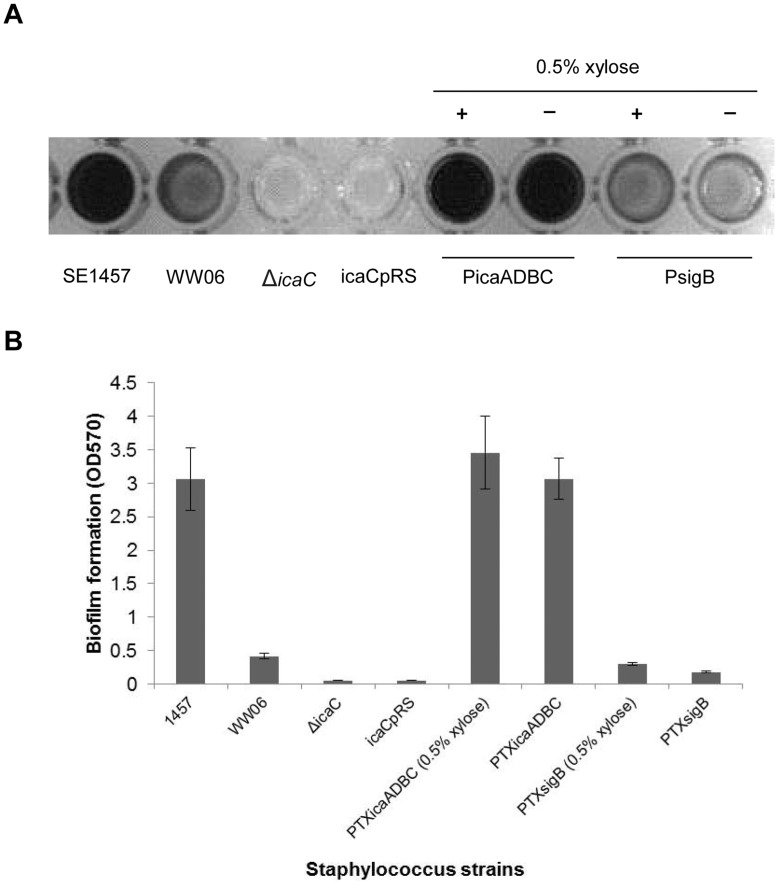
To confirm that ArlRS is an upstream regulator of ica-dependent biofilm formation in S. epidermidis, the arlRS mutant was complemented with icaADBC on a plasmid under the control of xylose inducible promoter (pTX-icaADBC). This complementation restored biofilm-forming ability. In the contrast, overexpressing arlRS in the ΔicaC mutant did not recover biofilm formation (Figure 6). Although transcription of the sigB gene was down-regulated in WW06, when the mutant was complemented with a plasmid-encoding sigB gene (pTX-sigB), no obvious differences in biofilm formation were observed (Figure 6).
Figure 6. Biofilm formation in complementation strains PicaADBC, icaCpRS and PsigB in 96-well microtiter plates.
(A) SE1457, WW06, ΔicaC and icaCpRS were cultivated in TSB; PicaADBC and PsigB were cultivated in TSB with or without 0.5% (g/ml) xylose. Biofilms formed over 24 h in 96-well microtiter plates were stained with 2% crystal violet. (B) The crystal violet was dissolved in ethanol and the absorbance was determined at 570 nm. The mean value ± standard deviation for each strain is shown.
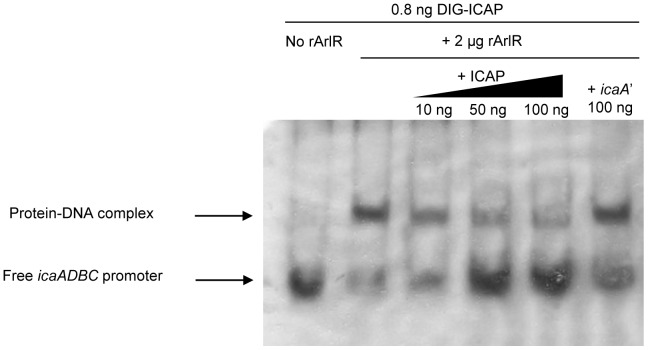
As ArlR is a DNA binding protein, we hypothesized that ArlR may bind directly to the promoter region of icaADBC to modulate transcription of the operon. Thus, EMSA were performed with digoxin-labeled DNA directly upstream of icaADBC (ICAP, 216 bp) and recombinant ArlR (rArlR). As shown in Figure 7, rArlR bound to Dig-ICAP and formed a DNA-protein complex, shifting Dig-ICAP behind (lane 2) compared to Dig-ICAP alone (lane 1). When increasing amounts of unlabeled ICAP was added as a specific competitor, more free Dig-ICAP was observed (lanes 3–5), while when a nonspecific competitor, icaA fragments, were added, no Dig-ICAP was shifted back (lane 6). It indicated a potential mechanism for ArlRS regulation of ica expression.
Figure 7. Binding of ArlR to the icaADBC promoter.
Lanes were loaded as follows: lane 1, 0.8 ng Dig-ICAP alone; lane 2, 0.8 ng Dig-ICAP and 2 µg rArlR; lanes 3–5, Dig-ICAP, rArlR and increasing amounts of unlabeled ICAP (12.5, 62.5,125 fold increase in Dig-ICAP, respectively); and lane 6, Dig-ICAP, rArlR and 100 ng of a 405 bp icaA fragment. The DIG-labeled DNA fragments were transferred to positively charged nylon membranes and visualized by an enzyme immunoassay using anti-Digoxigenin-AP, Fab-fragments and the chemiluminescent substrate CSPD. Chemiluminescent signals were recorded on X-ray film. DIG-ICAP, digoxin-labeled icaADBC promoter region; ICAP, unlabeled icaADBC promoter region; icaA’, icaA gene fragment.
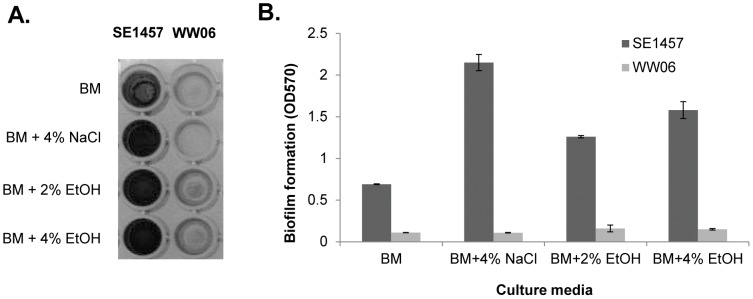
To further investigate the role of IcaR in regulating WW06 biofilm formation, effect of ethanol or sodium chloride on biofilm production of WW06 and SE1457 was detected. Addition of 4% NaCl to BM media increased SE1457 biofilm formation (approximately 3.1 fold), while had no effect on that of WW06, shown in Figure 8. SE1457 showed enhanced biofilm production in BM media supplemented with either 2% or 4% ethanol (1.8 fold or 2.3 fold, respectively), whereas ethanol showed very limited effect on WW06 biofilm formation (1.3 fold −1.4 fold increase).
Figure 8. Effect of ethanol or sodium chloride on biofilm formation of SE1457 and WW06.
(A) Semi-quantitative detection of biofilms formed by SE1457, WW06 in BM supplemented with 4% NaCl, 2% EtOH or 4% EtOH in 96-well polystyrene microtiter plates: The biofilms were stained with 2% crystal violet. (B) The crystal violet was dissolved in ethanol and the absorbance was determined at 570 nm. The mean value ± standard deviation for each strain is shown.
Discussion
The two-component signal transductionsystem ArlRS is a global regulator of virulence genes in S. aureus [19], modulating the extracellular proteolytic activity, capsule formation, biofilm formation and production of Protein A, α-toxin, β-hemolysin, lipase, coagulase and serine protease. However, the function of ArlRS in S. epidermidis was unclear. Thus, we investigated the role of ArlRS in S. epidermidis, an opportunistic pathogen that lacks many virulence genes [35] but can form biofilms as a major pathogenic factor. First, an ArlRS deficient mutant WW06 was constructed. WW06 showed dramatically decreased biofilms formation in vitro and in vivo. No obvious difference was found in either growth curves or initial attachment of WW06 and SE1457. Extracellular DNA has been reported to play an important role in biofilm formation [36]–[38]. In this study, however, the quantity of eDNA in the unwashed biofilms of the two strains was similar (data not shown), although the arlRS mutant strain WW06 exhibited a significantly higher autolysis rate than SE1457. Thus, we hypothesize that ArlRS modulates S. epidermidis biofilm formation by regulating the expression of the biofilm-related genes involved in the step of intercellular adhesion.
Many factors are involved in the second step of biofilm formation, e.g. polysaccharide intercellular adhesin (PIA) [39], accumulation associated protein (Aap) [40], [41], Extracellular matrix binding protein (Embp) [42], etc. The ica (intercellular adhesin) operon plays a crucial role by synthesizing PIA that is a main component of the extracellular polymeric substances (EPS). The ica operon is composed of the icaR (regulatory) gene and icaADBC (biosynthesis) genes. The icaR gene is located upstream of the icaADBC and is transcribed divergently, encoding a transcriptional repressor that negatively regulates icaADBC transcription in both S. aureus and S. epidermidis. Staphylococcal biofilms formation involves ica-dependent and ica-independent pathways [43]. In the present study, we have demonstrated that ArlRS activates icaADBC transcription, leading to PIA production and S. epidermidis biofilm formation. First, RT-qPCR analysis showed that icaADBC expression was significantly reduced in the ArlRS-deficient mutant WW06. Secondly, whereas semi-quantitative PIA detection revealed strong PIA production in the wild-type strain SE1457, little or no PIA expression was detected in WW06 mutant. Furthermore, complementation of icaADBC in WW06 restored its ability to form biofilms, whereas overexpression of arlRS in the icaC deletion mutant of SE1457 (ΔicaC) did not recover biofilm formation. Together, these results indicate that icaADBC is the downstream biofilm-related effector in the ArlRS regulation pathway. Moreover, EMSA demonstrated that recombinant ArlR binds to the 216-bp region located upstream of icaADBC. Overall, it reveals that ArlRS regulates S. epidermidis biofilm formation in an ica-dependent manner.
Previous studies have showed that ArlRS promotes biofilm formation in S. aureus strain ISP794 and strain 15981 which is capable of producing strong biofilms in TSB while unable to produce a biofilm in HHWm [18], [22]. Fournier, B. et, al. have found that an arlS gene transposition mutant (BF16) of S. aureus clinical strain ISP794 exhibited an increased ability to form biofilms on polymer surfaces, resulting from enhanced bacterial autolysis and altered peptidoglycan hydrolase activity. In the present study, arlRS mutation in S. epidermidis resulted in increased Triton X-100 induced bacterial autolysis, but decreased biofilm formation. The difference of altered biofilm formation between arlS mutant of S. aureus (BF16) and arlRS mutant of S. epidermidis (WW06) could be explained that due to different methods used for gene mutation (transposon insertion in the former and gene knockout in the latter), ArlR is absent in WW06 while present and may have function in BF16. Toledo-Arana, A. et, al. have reported that deletion of the arlRS locus in S. aureus clinical strain 15981 enhanced initial cell attachment and provoked accumulation of PNAG and biofilm formation when cultured in Hussain-Hastings-White-modified medium (HHWm). However, in HHWm the biofilm formation of the arlRS and icaADBC operon double mutant increased at the same level as the arlRS mutant, suggesting ArlRS is involved in early stage of strain 15981’s biofilm development in HHWm in an ica-independent manner. Interestingly, mutation of arlRS in S. epidermidis resulted in decreased biofilm formation in TSB in an ica-dependent manner. Bioinformatics analysis showed that arlR and arlS genes are conserved in S. epidermidis (over 95% nucleotide sequence identities among strains RP62A, SE1457 and ATCC12228) while S. epidermidis strain RP62A and S. aureus strain COL [44] share relatively high nucleotide sequence identities of arlR and arlS genes (74.8% and 70%, respectively). It appears ArlRS is involved in different mechanisms that regulate biofilm formation in the closely related species.
Current findings indicate that icaADBC expression in S. epidermidis is regulated by multiple regulatory factors including RsbU, SigB, IcaR, SarA and LuxS[8], [45]–[48]. Knobloch, J.K. et, al. reported that inactivation of rsbU gene (a positive regulator of sigB) in S. epidermidis was associated with both a reduction in PIA levels and biofilm formation, and the regulation of biofilm formation by rsbU is SigB dependent. Handke, L.D. et, al. reported that icaADBC transcription is down-regulated in the sigB mutant [45]. The regulation of icaADBC transcription by SigB was found to be mediated through the upregulation of IcaR expression. IcaR is a transcriptional repressor of icaADBC [49]. The icaR gene deletion in S. epidermidis strain O-47 [50] led to increased PIA synthesis. In the present study, decreased transcription of rsbU, sigB and icaADBC, and increased icaR expression are observed in the arlRS mutant WW06, compared with SE1457. The results are consistent with the previous findings, indicating a potential mechanism that the regulation of icaADBC expression by ArlRS may be mediated by IcaR. The global transcriptional regulator SarA plays an important role in staphylococcal biofilm formation [8], [51]–[54]. Transcription of sarA was decreased by about 4.3 fold in the WW06 mutant, which is consistent with previous findings that SarA positively regulates ica operon expression in an icaR-independent manner; its role in ArlRS regulation pathway requires further investigation. As a quorum sensing system, LuxS repressed S. epidermidis biofilm formation through a cell-cell signaling mechanism based on autoinducer 2 secretions. LuxS affects biofilm formation by altering production of PIA via transcriptional regulation of the ica operon. In the present study, transcriptional levels of luxS in the SE1457 and WW06 were similar, indicating luxS may not play a role in the ArlRS regulation pathway in S. epidermidis.
Taken together, it reveals that in S. epidermidis, the ArlRS two-component signal transduction system is involved in the modulation of bacterial autolysis and in the regulation of biofilm formation an ica-dependent manner, which is distinct from the role of ArlRS in S. aureus and deserves further study.
Supporting Information
Purification of the recombinant ArlR by affinity chromatography. The arlR gene was cloned in the expression vector pET28a(+) to form pET-arlR, which was transformed in the E. coli BL21(DE3+). After induction with 0.4 mM IPTG for 12 h, the recombinant ArlR was purified by affinity chromatography. M: Protein molecular weight marker; rArlR: the purified recombinant ArlR (about 27 kDa).
(TIF)
Detection of ArlR expression in SE1457 and WW06 by Western blot. The lanes were loaded with 200 ng purified recombinant ArlR (rArlR), 1 µg bacterial cells extract of WW06, and 1 µg bacterial cells extract of SE1457, respectively. Antiserum from the mouse immunized with 5 µg recombinant ArlR was diluted by 1∶1000. An unspecific protein band with a lower molecular mass was present in each lane.
(TIF)
Acknowledgments
We thank Prof. J.M. van Dijl (University of Groningen, Netherlands) for the gift of the plasmid pCN51.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the National Science and Technology Major Project of China [2012ZX09301002-005, 2012ZX10003008-010; http://www.nmp.gov.cn], the National Natural Science Foundation of China [81101214, 30800036, J0730860; http://www.nsfc.gov.cn/Portal0/default106.htm], the Scientific Technology Development Foundation of Shanghai [10410700600, 09DZ1908602; http://www.stcsm.gov.cn], the Program of Ministry of Science and Technology of China [2010DFA32100; http://www.nmp.gov.cn], and the Specialized Research Fund for the Doctoral Program of Higher Education [20100071120049; http://www.cutech.edu.cn]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Otto M. Staphylococcus epidermidis–the ‘accidental’ pathogen. Nat Rev Microbiol. 2009;7:555–567. doi: 10.1038/nrmicro2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ziebuhr W, Hennig S, Eckart M, Kranzler H, Batzilla C, et al. Nosocomial infections by Staphylococcus epidermidis: how a commensal bacterium turns into a pathogen. Int J Antimicrob Agents. 2006;28:S14–20. doi: 10.1016/j.ijantimicag.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 3.von Eiff C, Proctor RA, Peters G. Coagulase-negative staphylococci. Pathogens have major role in nosocomial infections. Postgrad Med 110: 63–64, 69–70, 73–66. 2001. [PubMed]
- 4.Vuong C, Otto M. Staphylococcus epidermidis infections. Microbes Infect. 2002;4:481–489. doi: 10.1016/s1286-4579(02)01563-0. [DOI] [PubMed] [Google Scholar]
- 5.Otto M. Staphylococcal biofilms. Curr Top Microbiol Immunol. 2008;322:207–228. doi: 10.1007/978-3-540-75418-3_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knobloch JK, Jager S, Horstkotte MA, Rohde H, Mack D. RsbU-dependent regulation of Staphylococcus epidermidis biofilm formation is mediated via the alternative sigma factor sigmaB by repression of the negative regulator gene icaR. Infect Immun. 2004;72:3838–3848. doi: 10.1128/IAI.72.7.3838-3848.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lauderdale KJ, Boles BR, Cheung AL, Horswill AR. Interconnections between Sigma B, agr, and proteolytic activity in Staphylococcus aureus biofilm maturation. Infect Immun. 2009;77:1623–1635. doi: 10.1128/IAI.01036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tormo MA, Marti M, Valle J, Manna AC, Cheung AL, et al. SarA is an essential positive regulator of Staphylococcus epidermidis biofilm development. J Bacteriol. 2005;187:2348–2356. doi: 10.1128/JB.187.7.2348-2356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharma-Kuinkel BK, Mann EE, Ahn JS, Kuechenmeister LJ, Dunman PM, et al. The Staphylococcus aureus LytSR two-component regulatory system affects biofilm formation. J Bacteriol. 2009;191:4767–4775. doi: 10.1128/JB.00348-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dubrac S, Boneca IG, Poupel O, Msadek T. New insights into the WalK/WalR (YycG/YycF) essential signal transduction pathway reveal a major role in controlling cell wall metabolism and biofilm formation in Staphylococcus aureus. J Bacteriol. 2007;189:8257–8269. doi: 10.1128/JB.00645-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skerker JM, Prasol MS, Perchuk BS, Biondi EG, Laub MT. Two-component signal transduction pathways regulating growth and cell cycle progression in a bacterium: a system-level analysis. PLoS Biol. 2005;3:e334. doi: 10.1371/journal.pbio.0030334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bader MW, Sanowar S, Daley ME, Schneider AR, Cho U, et al. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell. 2005;122:461–472. doi: 10.1016/j.cell.2005.05.030. [DOI] [PubMed] [Google Scholar]
- 13.Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 14.Hoch JA. Two-component and phosphorelay signal transduction. Curr Opin Microbiol. 2000;3:165–170. doi: 10.1016/s1369-5274(00)00070-9. [DOI] [PubMed] [Google Scholar]
- 15.Zhu T, Lou Q, Wu Y, Hu J, Yu F, et al. Impact of the Staphylococcus epidermidis LytSR two-component regulatory system on murein hydrolase activity, pyruvate utilization and global transcriptional profile. BMC Microbiol. 2010;10:287. doi: 10.1186/1471-2180-10-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vuong C, Gerke C, Somerville GA, Fischer ER, Otto M. Quorum-sensing control of biofilm factors in Staphylococcus epidermidis. . J Infect Dis. 2003;188:706–718. doi: 10.1086/377239. [DOI] [PubMed] [Google Scholar]
- 17.Lou Q, Zhu T, Hu J, Ben H, Yang J, et al. Role of the SaeRS two-component regulatory system in Staphylococcus epidermidis autolysis and biofilm formation. BMC Microbiol. 2011;11:146. doi: 10.1186/1471-2180-11-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fournier B, Hooper DC. A new two-component regulatory system involved in adhesion, autolysis, and extracellular proteolytic activity of Staphylococcus aureus. J Bacteriol. 2000;182:3955–3964. doi: 10.1128/jb.182.14.3955-3964.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fournier B, Klier A, Rapoport G. The two-component system ArlS-ArlR is a regulator of virulence gene expression in Staphylococcus aureus. Mol Microbiol. 2001;41:247–261. doi: 10.1046/j.1365-2958.2001.02515.x. [DOI] [PubMed] [Google Scholar]
- 20.Meier S, Goerke C, Wolz C, Seidl K, Homerova D, et al. sigmaB and the sigmaB-dependent arlRS and yabJ-spoVG loci affect capsule formation in Staphylococcus aureus. Infect Immun. 2007;75:4562–4571. doi: 10.1128/IAI.00392-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang X, Zheng L, Landwehr C, Lunsford D, Holmes D, et al. Global regulation of gene expression by ArlRS, a two-component signal transduction regulatory system of Staphylococcus aureus. J Bacteriol. 2005;187:5486–5492. doi: 10.1128/JB.187.15.5486-5492.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Toledo-Arana A, Merino N, Vergara-Irigaray M, Debarbouille M, Penades JR, et al. Staphylococcus aureus develops an alternative, ica-independent biofilm in the absence of the arlRS two-component system. J Bacteriol. 2005;187:5318–5329. doi: 10.1128/JB.187.15.5318-5329.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cramton SE, Gerke C, Schnell NF, Nichols WW, Gotz F. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect Immun. 1999;67:5427–5433. doi: 10.1128/iai.67.10.5427-5433.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bruckner R. Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol Lett. 1997;151:1–8. doi: 10.1111/j.1574-6968.1997.tb10387.x. [DOI] [PubMed] [Google Scholar]
- 25.Vuong C, Gotz F, Otto M. Construction and characterization of an agr deletion mutant of Staphylococcus epidermidis. Infect Immun. 2000;68:1048–1053. doi: 10.1128/iai.68.3.1048-1053.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Charpentier E, Anton AI, Barry P, Alfonso B, Fang Y, et al. Novel cassette-based shuttle vector system for gram-positive bacteria. Appl Environ Microbiol. 2004;70:6076–6085. doi: 10.1128/AEM.70.10.6076-6085.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heilmann C, Hussain M, Peters G, Gotz F. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol Microbiol. 1997;24:1013–1024. doi: 10.1046/j.1365-2958.1997.4101774.x. [DOI] [PubMed] [Google Scholar]
- 28.Christensen GD, Simpson WA, Younger JJ, Baddour LM, Barrett FF, et al. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J Clin Microbiol. 1985;22:996–1006. doi: 10.1128/jcm.22.6.996-1006.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carmen JC, Roeder BL, Nelson JL, Beckstead BL, Runyan CM, et al. Ultrasonically enhanced vancomycin activity against Staphylococcus epidermidis biofilms in vivo. J Biomater Appl. 2004;18:237–245. doi: 10.1177/0885328204040540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rediske AM, Roeder BL, Nelson JL, Robison RL, Schaalje GB, et al. Pulsed ultrasound enhances the killing of Escherichia coli biofilms by aminoglycoside antibiotics in vivo. Antimicrob Agents Chemother. 2000;44:771–772. doi: 10.1128/aac.44.3.771-772.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rediske AM, Roeder BL, Brown MK, Nelson JL, Robison RL, et al. Ultrasonic enhancement of antibiotic action on Escherichia coli biofilms: an in vivo model. Antimicrob Agents Chemother. 1999;43:1211–1214. doi: 10.1128/aac.43.5.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gerke C, Kraft A, Sussmuth R, Schweitzer O, Gotz F. Characterization of the N-acetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesin. J Biol Chem. 1998;273:18586–18593. doi: 10.1074/jbc.273.29.18586. [DOI] [PubMed] [Google Scholar]
- 33.Brunskill EW, Bayles KW. Identification and molecular characterization of a putative regulatory locus that affects autolysis in Staphylococcus aureus. J Bacteriol. 1996;178:611–618. doi: 10.1128/jb.178.3.611-618.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu JR, Sun ZP, Xu T, Wu Y, Li DW, et al. Detection of ArlR expression in different growth phases of Staphylococcus epidermidis. Journal of Microbes and Infection. 2009;4:92–96. [Google Scholar]
- 35.Zhang YQ, Ren SX, Li HL, Wang YX, Fu G, et al. Genome-based analysis of virulence genes in a non-biofilm-forming Staphylococcus epidermidis strain (ATCC 12228). Mol Microbiol. 2003;49:1577–1593. doi: 10.1046/j.1365-2958.2003.03671.x. [DOI] [PubMed] [Google Scholar]
- 36.Qin Z, Ou Y, Yang L, Zhu Y, Tolker-Nielsen T, et al. Role of autolysin-mediated DNA release in biofilm formation of Staphylococcus epidermidis. Microbiology. 2007;153:2083–2092. doi: 10.1099/mic.0.2007/006031-0. [DOI] [PubMed] [Google Scholar]
- 37.Allesen-Holm M, Barken KB, Yang L, Klausen M, Webb JS, et al. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol Microbiol. 2006;59:1114–1128. doi: 10.1111/j.1365-2958.2005.05008.x. [DOI] [PubMed] [Google Scholar]
- 38.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- 39.Mack D, Fischer W, Krokotsch A, Leopold K, Hartmann R, et al. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear beta-1,6-linked glucosaminoglycan: purification and structural analysis. J Bacteriol. 1996;178:175–183. doi: 10.1128/jb.178.1.175-183.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hussain M, Herrmann M, von Eiff C, Perdreau-Remington F, Peters G. A 140-kilodalton extracellular protein is essential for the accumulation of Staphylococcus epidermidis strains on surfaces. Infect Immun. 1997;65:519–524. doi: 10.1128/iai.65.2.519-524.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rohde H, Burdelski C, Bartscht K, Hussain M, Buck F, et al. Induction of Staphylococcus epidermidis biofilm formation via proteolytic processing of the accumulation-associated protein by staphylococcal and host proteases. Mol Microbiol. 2005;55:1883–1895. doi: 10.1111/j.1365-2958.2005.04515.x. [DOI] [PubMed] [Google Scholar]
- 42.Christner M, Franke GC, Schommer NN, Wendt U, Wegert K, et al. The giant extracellular matrix-binding protein of Staphylococcus epidermidis mediates biofilm accumulation and attachment to fibronectin. Mol Microbiol. 2010;75:187–207. doi: 10.1111/j.1365-2958.2009.06981.x. [DOI] [PubMed] [Google Scholar]
- 43.O’Gara JP. ica and beyond: biofilm mechanisms and regulation in Staphylococcus epidermidis and Staphylococcus aureus. FEMS Microbiol Lett. 2007;270:179–188. doi: 10.1111/j.1574-6968.2007.00688.x. [DOI] [PubMed] [Google Scholar]
- 44.Gill SR, Fouts DE, Archer GL, Mongodin EF, Deboy RT, et al. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J Bacteriol. 2005;187:2426–2438. doi: 10.1128/JB.187.7.2426-2438.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Handke LD, Slater SR, Conlon KM, O’Donnell ST, Olson ME, et al. SigmaB and SarA independently regulate polysaccharide intercellular adhesin production in Staphylococcus epidermidis. Can J Microbiol. 2007;53:82–91. doi: 10.1139/w06-108. [DOI] [PubMed] [Google Scholar]
- 46.Conlon KM, Humphreys H, O’Gara JP. icaR encodes a transcriptional repressor involved in environmental regulation of ica operon expression and biofilm formation in Staphylococcus epidermidis. J Bacteriol. 2002;184:4400–4408. doi: 10.1128/JB.184.16.4400-4408.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu L, Li H, Vuong C, Vadyvaloo V, Wang J, et al. Role of the luxS quorum-sensing system in biofilm formation and virulence of Staphylococcus epidermidis. Infect Immun. 2006;74:488–496. doi: 10.1128/IAI.74.1.488-496.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Knobloch JK, Bartscht K, Sabottke A, Rohde H, Feucht HH, et al. Biofilm formation by Staphylococcus epidermidis depends on functional RsbU, an activator of the sigB operon: differential activation mechanisms due to ethanol and salt stress. J Bacteriol. 2001;183:2624–2633. doi: 10.1128/JB.183.8.2624-2633.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jeng WY, Ko TP, Liu CI, Guo RT, Liu CL, et al. Crystal structure of IcaR, a repressor of the TetR family implicated in biofilm formation in Staphylococcus epidermidis. Nucleic Acids Res. 2008;36:1567–1577. doi: 10.1093/nar/gkm1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gotz F. Staphylococcus and biofilms. Mol Microbiol. 2002;43:1367–1378. doi: 10.1046/j.1365-2958.2002.02827.x. [DOI] [PubMed] [Google Scholar]
- 51.Beenken KE, Mrak LN, Griffin LM, Zielinska AK, Shaw LN, et al. Epistatic relationships between sarA and agr in Staphylococcus aureus biofilm formation. PLoS One. 2010;5:e10790. doi: 10.1371/journal.pone.0010790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trotonda MP, Manna AC, Cheung AL, Lasa I, Penades JR. SarA positively controls bap-dependent biofilm formation in Staphylococcus aureus. J Bacteriol. 2005;187:5790–5798. doi: 10.1128/JB.187.16.5790-5798.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Valle J, Toledo-Arana A, Berasain C, Ghigo JM, Amorena B, et al. SarA and not sigmaB is essential for biofilm development by Staphylococcus aureus. Mol Microbiol. 2003;48:1075–1087. doi: 10.1046/j.1365-2958.2003.03493.x. [DOI] [PubMed] [Google Scholar]
- 54.Beenken KE, Blevins JS, Smeltzer MS. Mutation of sarA in Staphylococcus aureus limits biofilm formation. Infect Immun. 2003;71:4206–4211. doi: 10.1128/IAI.71.7.4206-4211.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Purification of the recombinant ArlR by affinity chromatography. The arlR gene was cloned in the expression vector pET28a(+) to form pET-arlR, which was transformed in the E. coli BL21(DE3+). After induction with 0.4 mM IPTG for 12 h, the recombinant ArlR was purified by affinity chromatography. M: Protein molecular weight marker; rArlR: the purified recombinant ArlR (about 27 kDa).
(TIF)
Detection of ArlR expression in SE1457 and WW06 by Western blot. The lanes were loaded with 200 ng purified recombinant ArlR (rArlR), 1 µg bacterial cells extract of WW06, and 1 µg bacterial cells extract of SE1457, respectively. Antiserum from the mouse immunized with 5 µg recombinant ArlR was diluted by 1∶1000. An unspecific protein band with a lower molecular mass was present in each lane.
(TIF)