Abstract
Non-human primates have emerged as an important resource for the study of human disease and evolution. The characterization of genomic variation between and within non-human primate species could advance the development of genetically defined non-human primate disease models. However, non-human primate specific reagents that would expedite such research, such as exon-capture tools, are lacking. We evaluated the efficiency of using a human exome capture design for the selective enrichment of exonic regions of non-human primates. We compared the exon sequence recovery in nine chimpanzees, two crab-eating macaques and eight Japanese macaques. Over 91% of the target regions were captured in the non-human primate samples, although the specificity of the capture decreased as evolutionary divergence from humans increased. Both intra-specific and inter-specific DNA variants were identified; Sanger-based resequencing validated 85.4% of 41 randomly selected SNPs. Among the short indels identified, a majority (54.6%–77.3%) of the variants resulted in a change of 3 base pairs, consistent with expectations for a selection against frame shift mutations. Taken together, these findings indicate that use of a human design exon-capture array can provide efficient enrichment of non-human primate gene regions. Accordingly, use of the human exon-capture methods provides an attractive, cost-effective approach for the comparative analysis of non-human primate genomes, including gene-based DNA variant discovery.
Introduction
Non-human primates are increasingly studied as highly relevant animal models for human biomedical diseases and disorders. Members of the Macaca genus are among the most commonly studied non-human primates, due to their close evolutionary relationship to humans, analogous disease susceptibilities, and wide-spread commercial availability. The rhesus macaque (Macaca mulatta), estimated to have shared a common ancestor with humans approximately 25 million years ago (MYA) [1], is one of the most widely studied macaques. Genetic studies have shown the rhesus macaque to have common genetic risk factors with humans for age-related macular degeneration [2] behavioral disorders [3], [4]_ENREF_4 and reproductive disorders such as amennorhea [5]. A close relative of the rhesus macaque, the Japanese macaque (M. fuscata) has served as a model for multiple sclerosis [6] and ischemia [7], [8]_ENREF_8. The crab-eating or cynomolgus macaque (M. fascicularis) is widely used in studies of amyotrophic lateral sclerosis [9], and depression [10], among other disorders.
The chimpanzee (Pan troglodytes), is more closely related to humans than the macaques, sharing a common ancestor approximately 5–7 MYA [1]. The more recent divergence between humans and the chimpanzee has been of particular importance to the study of human evolution and speciation [11], [12]_ENREF_12. In the field of comparative genomics, the chimpanzee genome provides a critical insight into studies of positive selection in primate genomes [13]. The chimpanzee has also served as a important model for neuroscience research, including studies of cognition [14], neurobiology [15], and behavior [16].
With the recent advance in genomic technologies, interest in comparative analysis of non-human primates, particularly as they relate to biomedical and evolutionary studies, has been rapidly expanding [17], [18]. However, such studies are limited by the financial costs, computational requirements and effort required to generate genome-wide variant data on a large scale. Although improvements in next-generation sequencing (NGS) technology have already sharply reduced the cost of sequencing, the non-human primate still significantly lags behind in the comprehensive characterization of genome variation.
Exome sequencing has proven to be a powerful and efficient approach in human genetics studies [19], as it allows an unbiased investigation of almost all protein-coding regions in a large sample of individuals, at a fraction of the cost of whole genome sequencing. The method has been successfully applied to causative gene identification of several rare monogenic diseases such as Miller syndrome [20] spinocerebellar ataxias [21] and retinitis pigementosa [22]. A study of 50 Tibetan exomes uncovered a number of high-altitude adaptation related genes [23]. If the human exome-capture tools can be applied to the closely related non-human primate species, it could provide an opportunity to efficiently advance the pace of discovery of non-human primate sequence variants.
The human and chimpanzee genomes are about 99% identical, while macaques and human genomes are an estimated 93% conserved [17], [18]. Given the high level of sequence conservation for coding regions among primates, we considered whether it would be feasible to efficiently enrich the exonic sequences of primate species using human-based exon capture designs. Applying exon-capture technology to non-human primate research would not only minimize cost, but it would also reduce the computational effort required for deep sequence analysis. Importantly, exome-sequencing approaches would expedite the discovery sequence variants of greatest interest to many investigators, those located in gene coding regions.
Similar efforts have been used to successfully enrich and sequence target regions of the Neanderthal genome [24]. More than a megabase of captured sequence was recovered from Neanderthal DNA, despite DNA degradation and the presence of significant microbial DNA contamination. This achievement provides support for the use of human exon-capture reagents for the study of more distantly related human ancestors.
Here we report an effort to use human based exome capture to analyze chimpanzee and macaque exomes. Nineteen non-human primates, involving 3 species, were evaluated. We report the utility of the human exon array tool for exon enrichment, DNA variant discovery, and for comparative genomic analysis.
Results and Discussion
Capture and Sequencing
We sequenced the exomes of nine chimpanzees (CM), two crab-eating macaques (CE) and eight Japanese macaques (JP). Exonic sequences were enriched with the Agilent SureSelect all exon capture array (Human All Exon V1 for Human, CM and CE and Human All Exon V2 for JP)(Santa Clara, CA), targeting ∼38 Mb (∼46 Mb for JP) of DNA in nearly ∼18,000 human consensus coding DNA sequences (CCDS). Sequencing was performed on an Illumina Hiseq2000 sequencer (San Diego, CA). The two human individuals (HM) were sequenced using the same workflow. The human genome [hg19, UCSC] was used as the reference for alignment of all sequenced individuals, since it enabled consistent comparisons for all individuals using the same coordinate system. All reads were aligned using SOAPaligner [25] with a gap-free model and for SNP calling and coverage calculations. BWA [26] gap tolerant alignments were also used to detect short indels. For the non-human primates, we also mapped reads to their own or nearest reference genome (chimpanzees to panTro2, crab-eating macaques and Japanese macaque to rheMac2) to evaluate sequencing quality (Table S2).
Evaluation and Comparison of Capture Performance
In order to perform unbiased evaluation, we compared the capture performance and sequencing quality in different species using equivalent metrics. Low quality reads and sequencing adaptor reads were filtered. Reads that mapped to the same chromosomal location and also had the same orientation were identified as duplicated reads. The one with highest mean quality score was retained and used in the analysis. The exomes of all 21 individuals were sequenced with a mean depth ≥28 fold (Table 1) on the array design target region (TR). Coverage of the TR ranged from 91.06% to 97.73%, with 67.59% to 81.33% of the sites in the TR covered by more than 10 reads. A gene-by-gene coding region coverage statistic was implemented for both theoretical TR and by actual sequence data. The theoretical analysis found that 83.68% (15559/18594) of CCDS genes’ coding regions should have ≥90% covered by TR (Table 2). The recovered sequence data showed that 80.40%–80.65% of the genes were covered by ≥90% of sequence reads for humans and chimpanzees, and by 77.76% for macaques (Figure 1). Since coverage of chimpanzee CCDS was nearly equivalent to that of humans, we primarily focused on the macaques for our additional analysis. When mapping to the closest reference genome, the mapping rate was ≥84% in all 21 individuals, indicating a high quality sequencing data.
Table 1. Data production.
| CE1 | CE2 | JP (mean ± s.d.) | CM (mean ± s.d.) | HM1 | HM2 | |
| Target region (Mb) | 37.63 | 37.63 | 45.88 | 37.63 | 37.63 | 37.63 |
| # of clean reads(Mb) | 31.71 | 51.12 | 98.43±8.24 | 27.55±2.75 | 25.56 | 25.56 |
| Reads mapped to humangenome(Mb) | 18.12 | 28.05 | 55.68±6.07 | 23.54±2.30 | 23.58 | 23.35 |
| (fraction of clean reads%) | 57.13 | 54.86 | 56.48±2.06 | 85.46±1.06 | 92.26 | 91.37 |
| Reads mapped to human genomeafter filter duplication(Mb) | 17.66 | 27.30 | 48.44±4.93 | 22.22±2.23 | 22.74 | 22.42 |
| (fraction of clean reads%) | 55.69 | 53.40 | 49.19±2.56 | 80.67±1.68 | 88.97 | 87.72 |
| Reads mapped to own genome(Mb) | 27.02 | 45.92 | 88.22±8.09 | 24.22±2.15 | 23.58 | 23.35 |
| (fraction of clean reads%) | 85.20 | 89.83 | 89.58±1.19 | 88.01±1.79 | 92.26 | 91.37 |
| Reads mapped to target region(Mb) | 14.26 | 21.04 | 88.22±8.09 | 16.67±1.71 | 17.08 | 17.35 |
| (fraction of clean reads%) | 44.96 | 41.17 | 40.66±2.95 | 60.51±1.60 | 66.82 | 67.89 |
| Mean depth of target region | 30.72 | 45.77 | 70±8.52 | 35.03±3.40 | 35.41 | 36.14 |
| Coverage of target region(%) | 91.06 | 92.70 | 93.29±0.14 | 97.16±0.18 | 97.08 | 97.73 |
| Capture specificity | 52.77 | 45.83 | 45.36±2.84 | 68.78±2.21 | 72.43 | 74.30 |
| Fraction of target covered > = 4X(%) | 81.14 | 83.67 | 87.03±0.40 | 91.06±0.83 | 91.30 | 92.27 |
| Fraction of target covered> = 10X(%) | 67.59 | 72.32 | 78.80±0.86 | 78.41±2.38 | 79.09 | 79.91 |
| Rate of nucleotide mismatch (%) | 2.90 | 2.76 | 2.76±0.07 | 1.03±0.05 | 0.44 | 0.48 |
Summary of captured target sequence coverage for each non-human primate exome and two human exomes. The total size of the captured target is 37,627,322 bp for CE and 45,880,359 bp for JP. Each exome was compared to the human reference genome. Listed for each exome are the details of captured data including alignment, depth and coverage of target region. The exomes of all twenty one individuals were sequenced with a mean depth ≥28 fold on array designed target region (TR). Coverage of target region ranged from 91.06% to 97.73%, and 67.59% to 81.33% of sites in the target region were covered by more than 10 reads. Non-human primates were aligned to their own or closest genomes (rheMac2 was used as reference genome for crab-eating cynomolgus and Japanese macaque) to evaluate the capture efficiency.
Table 2. Comparison of different scanning regions and theoretical coverage of genes.
| Target Region | Target Orthologous Region | Target Orthologous Region &depth≥10 | |
| Length(bp) | 37,627,322 | 35,163,761 | 17,067,287 |
| Fraction of raw Target Region | 100.00% | 93.45% | 45.36% |
| Coverage of CCDS genes | 86.21% | 80.63% | 44.93% |
Summary of theoretical analysis for target region, target orthologous region and target orthologous region & depth≥10. Target orthologous region was the region we used to assess for capture efficiency and the target orthologous region & depth≥10 was used for SNP detection. 18,594 CCDS genes were used to analyze the theoretical coverage.
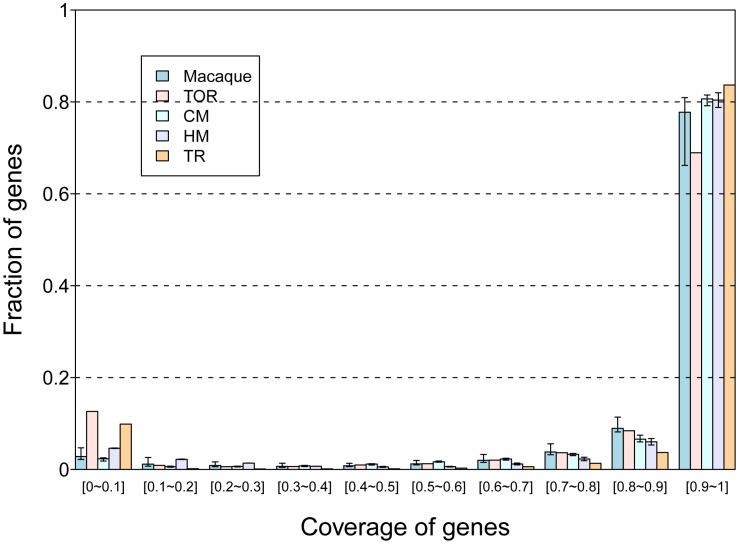
Figure 1. The profile of gene coverage.
The distribution of coverage of 18,594 gene coding regions of human CCDS by theoretical target region, target orthologous region and sequencing data from 3 species is shown. The sequencing coverage of each gene was inferred from one individual in each species. Sequencing data showed that 80.40%–80.45% of genes were covered ≥90% for both humans and chimpanzees, and 77.76% for macaques. Coverage of macaque CCDSs was higher than target orthologous region. The coverage of human and chimpanzee genes was close when concerns target region, and higher for macaque compared to target orthologous region.
To further consider the exon sequence recovery, we used pairwise blastz alignment result of hg19/rheMac2 (download from UCSC genome browser) to evaluate the orthologous level between human and rhesus macaque in the TR. Evaluating a fragment from the target region of the human reference genome alignment to the Indian macaque reference genome, we calculated an orthologous score (OS) for every position and separated them into three levels: OS = 0, no alignment, unable to be captured theoretically; OS = 1, only one hit, high level of 1∶1 orthologous coverage; OS = 2, multiple hits at different locations in the macaque reference, suggesting misalignment and potential for calling false polymorphisms (Table 3). We defined the region where OS = 1 as target orthologous region (TOR) for macaques and only used data in those regions in the following analysis. The region contains 93.5% of the initial target region. In TOR, 12,817 genes theoretically could be ≥90% covered (Table 2) and the actual sequence data showed that 88.86% (11389/12817) of them were ≥90% covered (Table S1). Thus based upon the TOR analysis, the coverage was close to the expected.
Table 3. Distribution of orthologous score (OS) for macaques.
| OS = 0 | OS = 1 | OS = 2 | |
| Length | 1.78 M | 35.16 M | 0.68 M |
| Percent of target region | 4.73% | 93.45% | 1.82% |
A fragment from the target region of the human reference genome was aligned to the Indian macaque reference genome. The OS was calculated for every site and classified into three possible levels: OS = 0, no alignment hit, unable to be captured theoretically; OS = 1, only one hit, high level of 1∶1 orthologous, well covered; OS = 2, multiple hits at different locations in the macaque reference, would give rise to misalignment and potentially false polymorphisms.
In assessing capture specificity, we defined the ratio of the reads aligned to a target region to the reads mapped to the species’ closest reference genome. We found that capture specificity ranged from 40% to 74% (Table 1) and decreased as a function of sequence divergence. The percentage of clean reads that mapped to the TR for macaque were much less than that of human (∼35% vs. ∼68%).
In order to further investigate the influence of specific genomic features on capture efficiency, we evaluated how targets that failed to be captured differed from targets that were successfully captured (more than 50% of bases covered at least by one read) (Table 4A and 4B). We found that failed targets generally had more nucleotide differences from human (6.95% vs. 3.29%), higher percentage of indels (1.93% vs. 0.64%) and a higher GC content (57% vs. 47%) in macaques. More detailed inspection of GC content revealed that the total GC content influence capture rate over a broad range (Figure 2 A and 2B). Target regions with moderate GC content (30%–50%) yielded higher coverage rates than regions with either high GC or low GC content (Figure 3A). This finding is likely the result of the methods employed for the exon-capture, which included use of annealing temperatures that were optimize for the binding of target sequences with a moderate GC content.
Table 4. Genomic features of captured and not captured targets.
| (A) Genomic features of captured targets | |||||
| # of targets | # of bases | mismatch | Indel bases | GC | |
| CE1 | 155,610 | 35,682,723 | 3.07% | 0.55% | 47.13% |
| CE2 | 157,062 | 36,002,143 | 3.10% | 0.56% | 47.16% |
| JP1 | 185,636 | 44,284,360 | 3.28% | 0.63% | 47.26% |
| JP2 | 186,122 | 44,395,960 | 3.29% | 0.64% | 47.30% |
| JP3 | 185,628 | 44,295,992 | 3.28% | 0.63% | 47.26% |
| JP4 | 185,584 | 44,280,953 | 3.28% | 0.63% | 47.24% |
| JP5 | 185,481 | 44,251,493 | 3.28% | 0.63% | 47.24% |
| JP6 | 185,540 | 44,256,348 | 3.28% | 0.63% | 47.23% |
| JP7 | 186,066 | 44,364,842 | 3.29% | 0.64% | 47.28% |
| JP8 | 185,812 | 44,305,013 | 3.28% | 0.64% | 47.26% |
| (B) Genomic features of not captured targets | |||||
| # of targets | # of bases | mismatch | Indel bases | GC | |
| CE1 | 9,961 | 1,944,599 | 5.85% | 1.47% | 56.29% |
| CE2 | 8,509 | 1,625,179 | 5.90% | 1.47% | 57.15% |
| JP1 | 8,381 | 1,595,999 | 6.85% | 1.87% | 53.84% |
| JP2 | 7,895 | 1,484,399 | 6.86% | 1.89% | 53.27% |
| JP3 | 8,389 | 1,584,367 | 6.83% | 1.88% | 53.84% |
| JP4 | 8,433 | 1,599,406 | 6.82% | 1.88% | 54.26% |
| JP5 | 8,536 | 1,628,866 | 6.80% | 1.85% | 54.13% |
| JP6 | 8,477 | 1,624,011 | 6.86% | 1.87% | 54.27% |
| JP7 | 7,951 | 1,515,517 | 6.95% | 1.93% | 53.70% |
| JP8 | 8,205 | 1,575,346 | 6.92% | 1.88% | 53.99% |
We examined the following genomic features for the 165,571 (for CE) and 194,017 (for JP) human targets with best reciprocal orthologs in the Indian macaque genome: the number of nucleotide differences between human and macaque, the number of indel bases between human and macaque and the GC content. (A) A target was considered “captured” if more than half of the human targeted bases were covered by at least one sequence read. (B) Otherwise, the target was not captured.
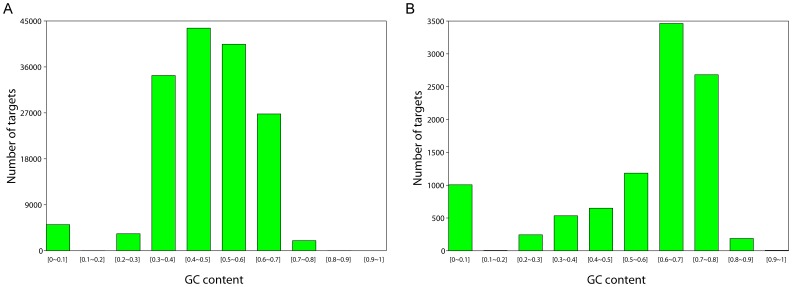
Figure 2. GC distribution of captured and un-captured targets by CE1.
Sequenced CE1 exome was compared with its closest reference genome (rheMac2) to examine the influence of GC content on coverage. (A) Captured targets were defined as targets with more than 50% of bases covered at least by one read. The captured targets were mainly of moderate GC content. (B) Uncaptured targets were less than a half of the bases coverd by at least one read. The uncaptured targets were dominated by high GC content, except for a small portion with low GC content.
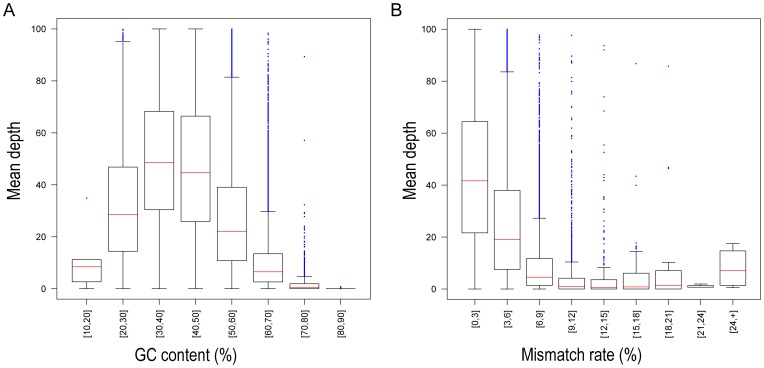
Figure 3. Average sequencing depth of captured targets versus human reference and rhesus macaque reference mismatch.
Mismatch rate of each target region was calculated by comparing human reference and rhesus macaque references respectively. (A) The X-axis identifies the mismatch rate region, and the Y-axis identifies the mean depth of target region, as calculated from CE2. As the mismatch rate increases, the mean depth decreases. (B) Average sequencing depth of captured targets versus each target GC content. Targets with extreme GC content were poorly captured; the mean depth was lower compared with moderate GC content.
Finally, we calculated the mismatch rate with the human and rhesus macaque reference genomes and correlated that with the average sequencing depth for all 158,852 autosomal targets. We found that sequencing depth decreased as sequence divergence increased (Figure 3B).
The remaining unrecovered TOR in macaques totaled 1.74 Mb, accounted for by three potential factors: 1) there were ∼0.22 Mb sites absent from both HM and CE in this capture array design; 2) the rhesus macaque genome was used to identify the TOR, and sequence divergence of the CE and JP genomes could have limited exon capture potential; 3) the amount of sequence, in combination with the reduced efficiency of capture, may have resulted in a biased representation of exon coverage.
We conclude that we can predict the capture performance using available pairwise alignment information for those species that have their own/related reference genome. We observed TOR regions for Orangutan [ponAbe2] and Marmoset [calJac3] had a percentage of 92.26% and 88.80% separately in TR, using the same analysis with the IR genome. This demonstrates a potential utility of human based chip among these species. On the other hand for species without reference genomes, but which are very closely related to humans, exon capture should be sufficiently efficient to make sequencing the majority of the exome, as well as genotyping each individual, feasible.
Variation Discovery and Analysis
The use of exome sequencing for mutation discovery in human studies is critically dependent on the accurate identification of DNA polymorphisms and genotypes. We were interested in whether human exon capture technology would also enable the accurate detection of variants in non-human primates. Using the reference genomes of humans and non-human primates, it was possible to identify both variations between species and within species.
We used SOAPsnp [27] for calling genotypes. In order to directly compare different individuals, we only called genotypes in TORs where every individual had a read depth of ≥10X and had high quality reads. The qualifying regions totaled 17.07 Mb, or 45.36% of the original TR (Table 2). In total, 79,704 ∼311,477 single base variants were detected within non-human primates (compared with human reference) and 15,013∼27,391 of them were candidate intra-species polymorphisms (Table 5A).
Table 5. (A) Comparison of sequencing based point variants and variants between references. (B) Coding SNP summary; piN refers to X and piS refers to Y. dN indicates X and dS indicates Y.
| (A) Comparison of sequencing based point variants and variants between references. | ||||||||
| Total SNPs | consistent | inconsistent | non-overlap | candidate intra-species polymorphism | consistent rate | |||
| hom | het | hom | het | |||||
| JP1 | 311,252 | 287,901 | 4,067 | 532 | 37 | 18,715 | 23,351 | 99.81% |
| JP2 | 310,564 | 287,724 | 3,935 | 536 | 42 | 18,327 | 22,840 | 99.80% |
| JP3 | 310,948 | 288,188 | 3,927 | 524 | 45 | 18,264 | 22,760 | 99.81% |
| JP4 | 310,935 | 287,036 | 4,115 | 521 | 47 | 19,216 | 23,899 | 99.81% |
| JP5 | 311,477 | 287,427 | 4,195 | 519 | 40 | 19,296 | 24,050 | 99.81% |
| JP6 | 310,847 | 287,096 | 4,131 | 525 | 36 | 19,059 | 23,751 | 99.81% |
| JP7 | 310,132 | 285,929 | 4,181 | 528 | 45 | 19,449 | 24,203 | 99.80% |
| JP8 | 311,362 | 287,570 | 4,131 | 513 | 40 | 19,108 | 23,792 | 99.81% |
| CE1 | 306,534 | 279,641 | 5,929 | 543 | 49 | 20,372 | 26,893 | 99.79% |
| CE2 | 303,992 | 276,601 | 5,910 | 517 | 44 | 20,920 | 27,391 | 99.80% |
| CM1 | 80,882 | 64,328 | 2,398 | 67 | 18 | 14,071 | 16,554 | 99.87% |
| CM2 | 80,571 | 64,042 | 2,446 | 71 | 21 | 13,991 | 16,529 | 99.86% |
| CM3 | 80,721 | 64,433 | 2,379 | 68 | 19 | 13,822 | 16,288 | 99.87% |
| CM4 | 80,061 | 63,718 | 2,392 | 70 | 15 | 13,866 | 16,343 | 99.87% |
| CM5 | 80,895 | 64,498 | 2,271 | 74 | 18 | 14,034 | 16,397 | 99.86% |
| CM6 | 80,737 | 64,230 | 2,391 | 68 | 16 | 14,032 | 16,507 | 99.87% |
| CM7 | 80,381 | 64,034 | 2,407 | 77 | 16 | 13,847 | 16,347 | 99.86% |
| CM8 | 79,704 | 64,691 | 2,047 | 71 | 16 | 12,879 | 15,013 | 99.87% |
| CM9 | 80,500 | 64,228 | 2,346 | 70 | 17 | 13,839 | 16,272 | 99.87% |
| (B) Coding SNP summary | ||||||||
| Non synonymous SNPs | Synonymous SNPs | Heterozygotes | piN/piS | Non synonymous divergence | Synonymous divergence | dn/ds | Neutrality Index | |
| JP1 | 8,172 | 10,757 | 13,096 | 0.7597 | 60,162 | 155,835 | 0.3861 | 1.9678 |
| JP2 | 7,948 | 10,503 | 12,651 | 0.7567 | 60,089 | 155,702 | 0.3859 | 1.9608 |
| JP3 | 7,896 | 10,513 | 12,638 | 0.7511 | 60,224 | 156,046 | 0.3859 | 1.9461 |
| JP4 | 8,429 | 10,922 | 13,662 | 0.7717 | 59,936 | 155,274 | 0.386 | 1.9993 |
| JP5 | 8,502 | 10,991 | 13,827 | 0.7735 | 59,995 | 155,564 | 0.3857 | 2.0058 |
| JP6 | 8,382 | 10,876 | 13,563 | 0.7707 | 60,022 | 155,398 | 0.3862 | 1.9953 |
| JP7 | 8,603 | 11,053 | 14,014 | 0.7783 | 59,738 | 154,756 | 0.386 | 2.0164 |
| JP8 | 8,348 | 10,913 | 13,643 | 0.765 | 60,104 | 155,724 | 0.386 | 1.9819 |
| CE1 | 7,477 | 13,753 | 16,904 | 0.5437 | 58,382 | 151,782 | 0.3846 | 1.4134 |
| CE2 | 7,850 | 13,719 | 17,330 | 0.5722 | 57,596 | 149,784 | 0.3845 | 1.4881 |
| CM1 | 5,207 | 7,449 | 9,975 | 0.699 | 16,421 | 31,766 | 0.5169 | 1.3522 |
| CM2 | 5,292 | 7,492 | 10,240 | 0.7064 | 16,301 | 31,622 | 0.5155 | 1.3702 |
| CM3 | 5,119 | 7,305 | 9,714 | 0.7008 | 16,399 | 31,852 | 0.5148 | 1.3611 |
| CM4 | 5,191 | 7,477 | 10,044 | 0.6943 | 16,246 | 31,466 | 0.5163 | 1.3447 |
| CM5 | 5,211 | 7,325 | 9,698 | 0.7114 | 16,461 | 31,879 | 0.5164 | 1.3777 |
| CM6 | 5,179 | 7,389 | 9,885 | 0.7009 | 16,359 | 31,759 | 0.5151 | 1.3607 |
| CM7 | 5,176 | 7,417 | 9,833 | 0.6979 | 16,296 | 31,670 | 0.5146 | 1.3562 |
| CM8 | 4,763 | 6,789 | 8,287 | 0.7016 | 16,511 | 31,975 | 0.5164 | 1.3587 |
| CM9 | 5,272 | 7,329 | 9,934 | 0.7193 | 16,342 | 31,719 | 0.5152 | 1.3962 |
| HM1 | 2,944 | 4,024 | 4,282 | 0.7316 | ||||
| HM2 | 2,957 | 4,092 | 4,310 | 0.7226 | ||||
We considered the possibility of cross DNA contamination contributing to the SNPs called. Though we used care in the DNA extraction, capture, library construction and sequencing procedures, we nonetheless evaluated whether the derived exome sequence contained evidence of human DNA contamination. We compared all exome sequencing data to the closest reference sequence. We found that the exome sequences and closest reference sequences exceeded a 99% match in all 19 non-human primate individuals (Table 5A). These results indicate that the sequence data used for genotype calling aligned most closely with the reference genome and did not suggest the presence of contaminating human DNA.
The abundance of polymorphisms discovered in coding and flanking regions enabled us to explore the potential evolutionary history of these species on a population scale. We found the macaques had the highest level of heterozygosity, followed by the chimpanzee. These data suggest that the macaques have a higher effective population size than chimpanzees, which in turn have a higher effective size than humans (Table 5B). In all species the ratio of nonsynonymous to synonymous SNPs was larger than the ratio of nonsynonymous to synonymous differences (Neutrality index, Table 5B) This is consistent with previous studies of effective population sizes in these species, [13], [17] suggesting that nonsynonymous deleterious alleles are the subject of selection pressure within each species.
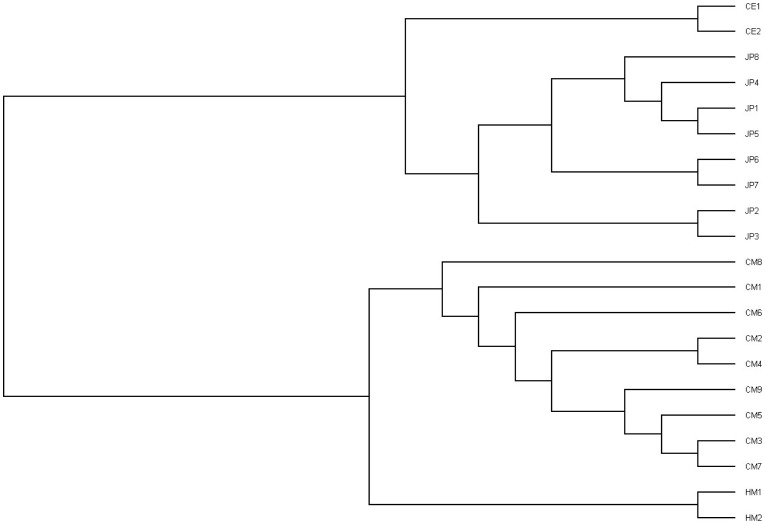
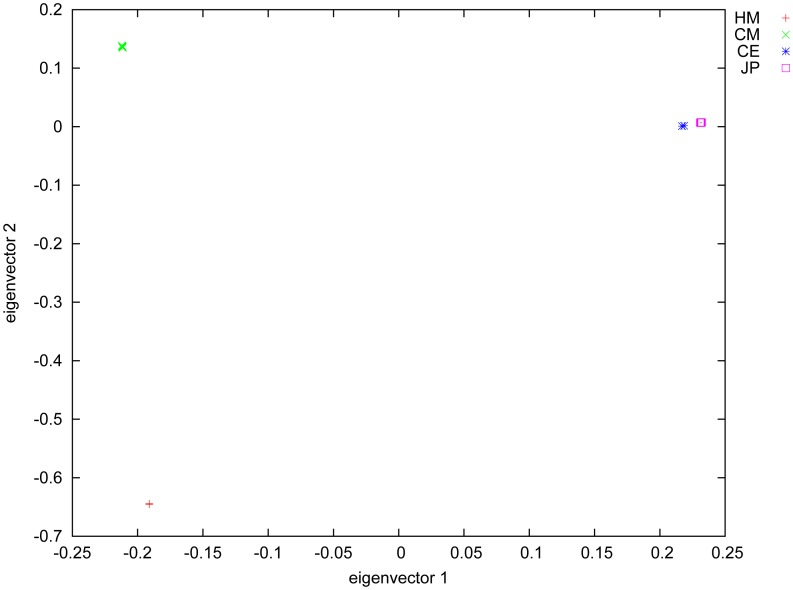
Since the nineteen non-human primate exomes included three different species, we used the polymorphism data to evaluate the evolutionary relationships among individuals. We constructed a phylogenetic tree (Figure 4) using the neighbor-joining (NJ) method. The phylogenetic tree is consistent with the ancestry analysis shown in previous genome studies [17], [18], as well as with results based upon microsatellite markers [1]. Principal component analysis (PCA) also showed that the individuals in each species were genetically most similar (Figure 5).
Figure 4. Phylogenetic tree.
Evolutionary distance in human, chimpanzee and macaque lineages, showing the inferred evolutionary relationships among 21 samples based upon similarities and differences in their genetic characteristics. The taxa joined together in the tree are implied to have descended from a common ancestor.
Figure 5. Principle Component Analysis.
Principal components analysis plot of 9 chimpanzees, 2 crab-eating macaques, 8 Japanese macaques and 2 human samples. Chimpanzees and humans cluster well within their own group, while crab-eating and Japanese macaques spread away are in relatively close proximity to each other, indicating recent genetic divergence between them.
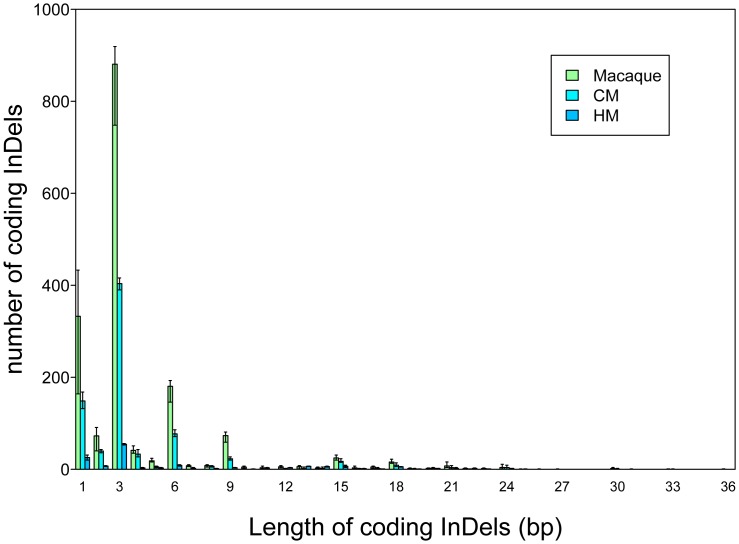
We also identified short insertions and deletions (indels) in the exons, which are likely to be functionally important and may contribute to species divergence (Table S3). Here we mapped the reads from each individual to the human reference sequence using BWA and Samtools [28] to identify indels (see methods for details). In total, 140∼1,883 (range for all individuals) human-primate coding indels were discovered in each individual (Table 6), and 54.61%∼77.32% of them were 3 base pairs in length. The distribution of indel lengths in the coding regions is consistent with a previous study [29] across all of the 21 exomes (Figure 6), reflecting the evolutionary pressure to preserve intact reading frames.
Table 6. Indel summary.
| CE1 | CE2 | JP (mean ± s.d.) | CM (mean ± s.d.) | HM1 | HM2 | |
| Total number of indels | 13,890 | 17,417 | 17,482.8±475.4 | 6,693.7±334.3 | 773 | 697 |
| Ins-coding | 698 | 910 | 1,072.4±40.3 | 427.6±17.6 | 93 | 84 |
| Del-coding | 607 | 698 | 696.5±17.6 | 360.3±8.4 | 59 | 56 |
| Splice site | 885 | 1,078 | 833.1±17.4 | 303.7±49.5 | 45 | 33 |
| Intron | 10,813 | 13,615 | 13,771.6±398.6 | 5,079.8±267.4 | 518 | 465 |
| 5′ UTRs | 256 | 341 | 344.5±8.2 | 190.3±6.3 | 22 | 29 |
| 3′ UTRs | 579 | 709 | 701.4±8.3 | 280.9±14.8 | 30 | 25 |
| Intergenic | 52 | 66 | 63.3±2.3 | 51.1±2.3 | 6 | 5 |
| Total insertion | 7,460 | 9,589 | 11,127.4±321.8 | 3,673.7±177.5 | 437 | 395 |
| Total deletion | 6,430 | 7,828 | 6355.4±155.8 | 3,020±177.6 | 336 | 302 |
| Heterozygous indels | 378 | 536 | 295.0±19.3 | 452.4±60.2 | 280 | 240 |
| Homozygous indels | 13,512 | 16,881 | 17,187.8±457.9 | 6,241.2±277.5 | 493 | 457 |
Each exome from 21 individuals was aligned to the human reference genome for indel identification.
Figure 6. The length distribution of coding indels for each individual.
140∼1,883 coding indels were called in each individual, and 54.61%∼77.32% of them were 3 bp in length. All indels were relative to the human reference genome.
We carried out Sanger sequencing for validation on a randomly selected 41 SNPs and 18 indels in two exome sequenced CE individuals. The validation rate was 85.4% for SNPs and 84.2% for indels (Table S4), indicating a high accuracy rate.
Discussion
Here we evaluated the efficiency of exome capture and sequencing in non-human primates using a human capture array. We determined that although capture specificity decreased as sequence divergence increased, it is still a viable option for crab-eating macaque, Japanese macaque and Chimpanzee exome enrichment. We also identified inter-species and intra-species variation by comparing recovered sequences to the rhesus macaque reference genome, and then validated the call accuracy using Sanger resequencing.
One limitation of this method is the inability to detecting large structural variations, such as large indels, inversions and copy number variations (CNV). In addition, the particular human exon capture design may miss some important 5′ and 3′ noncoding as well as small RNAs. Further, a subset of the genomic target regions were not recovered.
Despite the limitations, use of human exome-enrichment has some distinct advantages. We were able to efficiently capture gene coding region for both species with reference genomes, as well as those without a species-specific reference genome. Applications of this method could enable the efficient, large scale exon-sequencing of non-human primates, potentially identifying DNA variants of relevance to human disease, and guiding animal model development and translational research. Finally, exome-capture approaches could expedite the comparative genomic analysis of non-human primate species, for example between gorilla, orangutan, gibbons, baboon, and even New World monkeys, providing insight to the evolution of the human genome.
The nineteen non-human primate exomes presented here also highlight the degree of variation existing between these widely used non-human primate animal models. The abundant genetic diversity evident in individual primates from distinct geographic populations is of direct interest to primatology, medical research, population genetics and phylogeographic studies.
Materials and Methods
Ethics Statement
Blood samples of 2 Han Chinese individuals were collected from Anhui Medical University. We have obtained ethics approval for our study from the Ethical Committee of Anhui Medical University. We have also obtained informed written consent from all participants involved in this study. The authors of this paper did not collect the blood samples themselves; all non-human primate blood samples used in this study were initially collected for routine health check purposes.
Samples from nine wild born, unrelated chimpanzees (Pan troglodytes schweinfurthii) were collected at the Ngamba Island Chimpanzee Sanctuary in Uganda. The samples were taken during routine health checks by the sanctuary veterinarians and were exported under CITES export permit Sn.UG 002249. The Ugandan Wildlife Authorities and Uganda National Council for Science and Technology approved the research that resulted in the CITES export permission.
Two adult cynomolgus macaque (Macaca fascicularis) were individually housed in cages with the size of 70 cm×70 cm×80 cm by the South-China Primate Research & Development Center (Guangdong Landau Biotechnology Co., China) under the temperature of, between 16°C and 29°C and 50∼80% relative air humidity, with four air changes per hour and a 12 h light:12 h dark cycle. Animals were fed apples (100 g/day each) and monkey Chow (Feed Research Institute, Guangzhou, Guangdong) twice daily and allowed free access to water. The blood samples were collected during routine veterinary health checks. Guangdong Landau Biotechnology Co. is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC). The macaques were originally exported from Kampuchea under CITES export permit Sn. IC 0381. All macaque experiments were subject to approval and surveillance by the Institutional Animal Care and Use Committee of Guangdong Landau Biotechnology Co., Ltd. Genomic DNA was extracted from blood samples using the Nucleon kit (TaKaRa, Japan). Blood derived DNA was used to minimize somatic and cell-line derived false positives.
Eight Japanese macaques (Macaca fuscata) were born and raised at the Oregon National Primate Research Center (ONPRC); all animal procedures were approved by the ONPRC Institutional Animal Care and Use. Committee and conformed to the NIH guidelines on the ethical care and use of animals in research. The selected animals were members of captive breeding group that lived within in a 2 acre, outdoor corral, and consumed Primate Diet no. 5000, (Lab Diet, Richmond, IN). Blood samples were collected during routine veterinary care, by venipuncture and collection into an EDTA vacutainer. Genomic DNA was isolated using the Wizard DNA Extraction Kit (Promega, Inc.).
Exon Capture and Sequencing
The qualified genomic DNA samples were randomly fragmented by Covaris and the sizes of the library fragments were distributed between 150 to 200 bp. Adapters were then ligated to both ends of the resulting fragments. The adapter-ligated templates were purified by the Agencourt AMPure SPRI beads and fragments with insert size of about 250 bp were excised. Extracted DNA was amplified by ligation-mediated PCR (LM-PCR), purified, and hybridized to the SureSelect Biotinylated RNA Library (BAITS) for enrichment. Hybridized fragments were bound to the strepavidin beads whereas non-hybridized fragments were washed out after 24 h. Captured LM-PCR products were subjected to Agilent 2100 Bioanalyzer to estimate the magnitude of enrichment.
Each captured library was loaded on the Illumina Hiseq2000 platform for high-throughput sequencing to the desired average sequencing depth (The exomes of all 21 individuals were sequenced with a mean depth ≥28 fold on the array design target region). Raw image files were processed by Illumina basecalling Software 1.7 for base-calling with default parameters and the sequences of each individual were generated as 90 bp pair-end reads.
Read Mapping and Genotype Calling
SOAPaligner (soap 2.21) was used to align reads to the human reference genome (hg19) allowing a maximum of 3 mismatches per 90 bp read. Full SOAP parameters were: -a -b -D -o -2 -t -v 3 -l 35 -s 40 -m 0 -x 500 -p 4 -r 1 (-v 5 for macaques). Reads that aligned to the designed target region (TR) were collected for genotype calling and subsequent analysis.
Based on SOAP alignment results, the software SOAPsnp was used to call genotypes. The following parameters were set: -r 0.0005 -e 0.001 -t -u -2 -i -d -o -M -L 90 -s -T(http://soap.genomics.org.cn/ for details).
Raw data has NCBI Short Read Archive accession no. SRA038809.
SNP Identification
Single base differences between human and non-human primates references (ref-ref variants) were identified from pairwise blastz alignments result. *.axt files for Human/Chimp (panTro2) and Human/Rhesus (rheMac2) were downloaded from UCSC Genome Browser (http://hgdownload.cse.ucsc.edu/downloads.html#human).
Raw point variants from exome sequencing data of each individual were extract from SOAPsnp generated genotype with filter criteria Q20 and depth ≥10. Homozygous variants consistent with ref-ref variants were defined as interspecies variants. Remaining raw point variants were considered to be potential intraspecies polymorphisms.
Targeted Orthologous Region (TOR)
Orthologous score (OS) for every site in TR was calculated with pairwise blastz alignments result. The *.axt files for Human/Chimp, Chimp/Human (panTro2) and Human/Rhesus, Rhesus/Human (rheMac2) were downloaded from UCSC Genome Browser (http://hgdownload.cse.ucsc.edu/downloads.html#human).
Alignment of fragments from the TR between human and non-human primates were used to calculate OS. Alignment hits count = 0, OS = 0; alignment hits count = 1, OS = 1; alignment hits count≥2, OS = 2. Regions in TR where OS = 1 were defined as TOR.
Phylogenetic Tree
We use high quality genotypes on chromosome 10 (TOR, depth≥10 and consensus quality≥20 in every individual) generated by SOAPsnp to build a phylogenetic tree with the program TreeBeST (treebest-1.9.2, http://treesoft.sourceforge.net/treebest.shtml; L.Heng, A.J. Vilella, E.Birney, and R.Durbin, in prep) with the model of neighbour-joining tree, SDI, rooting (threshold: nj -b 500).
Principal Component Analysis (PCA)
We use high quality genotypes on chromosome 10 (TOR, depth≥10 and individuals whose quality score under 20 were discarded) generated by SOAPsnp to perform principal component analysis (PCA) using EIGENSOFT3.0 (parameter: -i all.evec -c 1∶2 -p HM:CM:CE:JP -x).
Identification of Insertions and Deletions
Gap tolerant alignments to human reference (hg19) were used to call indels with program BWA using parameters: aln -o 1 -e 63 -i 15 -L -l 31 -k 2 -t 4. Insertions and deletions (indels) were identified using samtools, with the command lines as following:
samtools mpileup -ugf ref.fa -b bam.list | bcftools view -bvcg - > var.raw.bcf
bcftools view var.raw.bcf | vcfutils.pl varFilter -D10000> var.flt.vcf.
Filter criteria were ≥3 reads support and number of indel supported reads ≥30% of all reads mapped to the genomic position. Indels were called as heterozygous if the indel supported reads were 30–70% of all reads at that position, and homozygous if they were greater than 70%.
Validation of SNP and Indel Variants
Forty one SNPs and 18 indels were selected randomly for validation of SNPs and indels. The selected SNP and indels were genotyped by PCR and Sanger sequencing. Primers for each selected SNP and indel were designed based on the IR genome; the detail sequences of each primer pairs were supplied in Table S4. The polymerase chain reaction (PCR) was performed in a final volume of 50 ul with 30 cycles at 94°C for 30 s, annealing temperature (Table S4) for 30 s, and 72°C for 30 s. The PCR product was purified and sequenced by BGI (BGI-Shenzhen, Shenzhen 518083, China).
Supporting Information
Coverage of targeted orthologous genes. Coverage of 18,594 gene coding regions of human CCDS by theoretical target region, target orthologous region and sequencing data from 3 species.
(RAR)
Data production. Summary of captured target sequence coverage for each non-human primate exome and two human exomes.
(XLSX)
Indels. Indels of 21 individuals identified through aligning the exomes to the human reference genome.
(XLSX)
Sanger sequencing validation result of SNP and Indels.
(XLS)
Acknowledgments
We thank Prof. Rasmus Nielsen from UC-Berkley for his suggestions and comments. We are also indebted to many additional faculty and staff of BGI-Shenzhen who contributed to this teamwork. For generously providing chimpanzee samples, we thank Ngamba Island Chimpanzee Sanctuary.
Funding Statement
This study was funded by the National Science and Technology Major Project of Key Drug Innovation and Development (2011ZX09307-303-03), the Science and Technology Planning Project of Guangdong Province, China (2010B060200007) and the Fundamental Research Funds for the Central Universities (2012zz0091, 2012ZZ0093 and 2011ZM0111). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Kumar S, Hedges SB (1998) A molecular timescale for vertebrate evolution. Nature 392: 917–920. [DOI] [PubMed] [Google Scholar]
- 2. Francis PJ, Appukuttan B, Simmons E, Landauer N, Stoddard J, et al. (2008) Rhesus monkeys and humans share common susceptibility genes for age-related macular disease. Hum Mol Genet 17: 2673–2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schwandt ML, Lindell SG, Higley JD, Suomi SJ, Heilig M, et al. (2011) OPRM1 gene variation influences hypothalamic-pituitary-adrenal axis function in response to a variety of stressors in rhesus macaques. Psychoneuroendocrinology 36: 1303–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen GL, Novak MA, Meyer JS, Kelly BJ, Vallender EJ, et al. (2010) TPH2 5′- and 3′-regulatory polymorphisms are differentially associated with HPA axis function and self-injurious behavior in rhesus monkeys. Genes Brain Behav 9: 335–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dissen GA, Lomniczi A, Heger S, Neff TL, Ojeda SR. (2012 ) Hypothalamic EAP1 (Enhanced at Puberty 1) Is Required for Menstrual Cyclicity in Nonhuman Primates. Neuroendocrinology 153: 350–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Axthelm MK, Bourdette DN, Marracci GH, Su W, Mullaney ET, et al. (2011) Japanese macaque encephalomyelitis: A spontaneous multiple sclerosis–like disease in a nonhuman primate. Annals of neurology 70: 362–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yukie M, Yamaguchi K, Yamashima T (2006) Impairments in recognition memory for object and for location after transient brain ischemia in monkeys. Reviews in the neurosciences 17: 201–214. [DOI] [PubMed] [Google Scholar]
- 8. Matsuura S, Egi Y, Yuki S, Horikawa T, Satoh H, et al. (2011) MP-124, a novel poly (ADP-ribose) polymerase-1 (PARP-1) inhibitor, ameliorates ischemic brain damage in a non-human primate model. Brain research 1410: 122–131. [DOI] [PubMed] [Google Scholar]
- 9. Uchida A, Sasaguri H, Kimura N, Tajiri M, Ohkubo T, et al. (2012) Non-human primate model of amyotrophic lateral sclerosis with cytoplasmic mislocalization of TDP-43. Brain 135: 833–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Willard SL, Shively CA (2012) Modeling depression in adult female cynomolgus monkeys (Macaca fascicularis). American Journal of Primatology 74: 528–542. [DOI] [PubMed] [Google Scholar]
- 11. Langergraber K, Schubert G, Rowney C, Wrangham R, Zommers Z, et al. (2011) Genetic differentiation and the evolution of cooperation in chimpanzees and humans. Proceedings of the Royal Society B: Biological Sciences 278: 2546–2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hvilsom C, Qian Y, Bataillon T, Li Y, Mailund T, et al. (2012) Extensive X-linked adaptive evolution in central chimpanzees. Proceedings of the National Academy of Sciences 109: 2054–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kosiol C, V T, Da Fonseca RR, Hubisz MJ, Bustamante CD, et al. (2008) Patterns of positive selection in six Mammalian genomes. PLoS genetics 4: e1000144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dean L, Kendal R, Schapiro S, Thierry B, Laland K (2012) Identification of the Social and Cognitive Processes Underlying Human Cumulative Culture. Science 335: 1114–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ludwig VU, Adachi I, Matsuzawa T (2011) Visuoauditory mappings between high luminance and high pitch are shared by chimpanzees (Pan troglodytes) and humans. Proceedings of the National Academy of Sciences 108: 20661–20665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Koops McGrew, W.C, de Vries (2012) Matsuzawa (2012) Nest-Building by Chimpanzees (Pan troglodytes verus) at Seringbara, Nimba Mountains: Antipredation, Thermoregulation, and Antivector Hypotheses. International Journal of Primatology 33: 356–380. [Google Scholar]
- 17. Gibbs RA, Rogers J, Katze MG, Bumgarner R, Weinstock GM, et al. (2007) Evolutionary and biomedical insights from the rhesus macaque genome. Science 316: 222–234. [DOI] [PubMed] [Google Scholar]
- 18. Chimpanzee Sequencing, Analysis Consortium (2005) Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437: 69–87. [DOI] [PubMed] [Google Scholar]
- 19. Metzker ML (2010) Sequencing technologies - the next generation. Nature reviews Genetics 11: 31–46. [DOI] [PubMed] [Google Scholar]
- 20. Ng SB, Buckingham KJ, Lee C, Bigham AW, Tabor HK, et al. (2010) Exome sequencing identifies the cause of a mendelian disorder. Nat Genet 42: 30–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang JL, Yang X, Xia K, Hu ZM, Weng L, et al. (2010) TGM6 identified as a novel causative gene of spinocerebellar ataxias using exome sequencing. Brain 133: 3510–3518. [DOI] [PubMed] [Google Scholar]
- 22. Ozgül RK, Siemiatkowska AM, Yücel D, Myers CA, Collin RW, et al. (2011) Exome sequencing and cis-regulatory mapping identify mutations in MAK, a gene encoding a regulator of ciliary length, as a cause of retinitis pigmentosa. The American Journal of Human Genetics 89: 253–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yi X, Liang Y, Huerta-Sanchez E, Jin X, Cuo ZX, et al. (2010) Sequencing of 50 human exomes reveals adaptation to high altitude. Science 329: 75–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Burbano HA, Hodges E, Green RE, Briggs AW, Krause J, et al. (2010) Targeted investigation of the Neandertal genome by array-based sequence capture. Science 328: 723–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li R, Yu C, Li Y, Lam TW, Yiu SM, et al. (2009) SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25: 1966–1967. [DOI] [PubMed] [Google Scholar]
- 26. Li H, Durbin R (2010) Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26: 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li R, Li Y, Fang X, Yang H, Wang J, et al. (2009) SNP detection for massively parallel whole-genome resequencing. Genome Res 19: 1124–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, et al. (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang J, Wang W, Li R, Li Y, Tian G, et al. (2008) The diploid genome sequence of an Asian individual. Nature 456: 60–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Coverage of targeted orthologous genes. Coverage of 18,594 gene coding regions of human CCDS by theoretical target region, target orthologous region and sequencing data from 3 species.
(RAR)
Data production. Summary of captured target sequence coverage for each non-human primate exome and two human exomes.
(XLSX)
Indels. Indels of 21 individuals identified through aligning the exomes to the human reference genome.
(XLSX)
Sanger sequencing validation result of SNP and Indels.
(XLS)