Abstract
Microsatellite Instability (MSI) is displayed approximately by 15% of colorectal cancers (CRC). Defective DNA mismatch repair (MMR) generates mutations at repetitive DNA sequences such as those located in the double strand break (DSB) repair gene MRE11. We assessed the mutational status of MRE11 in a panel of 17 CRC cell lines and 46 primary tumors finding a strong correlation with MSI status in both cell lines and tumors. Therefore, we hypothesized that deficiency in MRE11 may sensitize CRC cells to PARP-1 inhibition based on the concept of synthetic lethality. We further assessed the activity of the PARP-1 inhibitor, ABT-888, in CRC cell lines and observed preferential cytotoxicity in those MSI cell lines harboring mutations in MRE11 compared to both wild-type cell lines and microsatellite stable (MSS) cell lines. A significant correlation between MRE11 expression levels and cytotoxicity to ABT-888 at 10 μM was observed (R2=0.915, P<0.001). Using two experimental approaches, including shRNA knocking down MRE11 in the wild-type and MSS cell line SW-480 and a second cell line model transfected with mutant MRE11, we experimentally tried to confirm the role of MRE11 in conferring sensitivity to PARP-1 inhibition. Both models led to changes in proliferation in response to ABT-888 at different concentrations, and a drug-response effect was not observed, suggesting a possible contribution of additional genes. We conclude that MSI colorectal tumors deficient in DSB repair secondary to mutation in MRE11 show a higher sensitivity to PARP-1 inhibition. Further clinical investigation of PARP-1 inhibitors is warranted in MSI CRCs.
Keywords: microsatellite instability, colorectal cancer, MRE11, poly(ADP-ribose) polymerase inhibitors, connectivity map, gene expression
Introduction
Tumors displaying microsatellite instability (MSI) either due to germline or epigenetic inactivation of one the mismatch repair (MMR) genes account for approximately 10–15% of colorectal cancers (CRC)(1, 2). Sporadic MSI tumors have recognizable clinicopathological features such as right-sided location, older age of diagnosis, lower pathological stage and better prognosis(3–5). Genetic instability in this subgroup primarily reflects variation in microsatellite tracts due to a defective functioning of the surveillance mechanism performed by the MMR system. This instability is further reflected by more than 30 genes known to have mutations in microsatellite tract repeats, including DNA repair proteins involved in double strand break (DSB) repair through the homologous recombination pathway, such as MRE11 and RAD50(6). In particular, a microsatellite tract of 11(T) located at intron 4 of MRE11 is mutated in approximately 80% of MSI tumors and leads to aberrant splicing and a truncated protein(7). The multiprotein complex integrated by MRE11-RAD50-NBS (MRN complex) is the primary sensor of DSB and recruits other signaling proteins at DSB sites(8). These microsatellite mutations constitute a specific genetic background that can be exploited as potential drug target and predictor of sensitivity to specific therapies focused in DNA damage pathways. In fact, mutations in MRE11 has been shown to sensitize cells to agents causing replication fork stress as a result of a lack of 3′-5′ exonuclease activity, absence of formation of MRE11 foci and ATM autophosphorylation(9).
Poly(ADP-ribose) polymerase (PARP-1) cooperates with DNA ligases in order to repair single strand breaks whenever the ends need processing(10). Therefore, inhibitors of PARP-1 increase the levels of persisting single-strand breaks that will lead to DNA DSB upon replication(11). DSBs are one of the most important threats to genomic integrity and trigger repair proteins involved in the non-homologous end joining and homologous recombination pathways. Experimental evidence shows a direct interaction between PARP-1 and MRE11 reflected by the fact that PARP-1 is apparently required for rapid accumulation of MRE11 at DSB sites(12). Therefore, tumor cells harboring mutations of genes involved in homologous recombination such as BRCA1, BRCA2 and MRE11 are particularly vulnerable to DNA damage. Our aim in this study was to assess the activity of the PARP-1 inhibitor ABT-888 in CRC cell lines harboring a mutation in the homologous recombination gene MRE11 that frequently accompany MMR deficiency.
Materials and Methods
Cell lines and primary tumor samples
A total of 17 CRC cell lines were selected for experiments based on microsatellite status obtained from the Wellcome Trust Sanger Institute Cancer Genome Project web site(13) as detailed in Supplementary Table S1. Cell lines were purchased from the American Type Culture Collection (Manassas, VA). MSI status was also confirmed independently in cell lines, as well as the presence of mutations in the principal oncogenes for CRC. Cells were grown in DMEM/F12 medium supplemented with 10% of fetal bovine serum and 1% penicillin/streptomycin. SW480/SN3 and its derivative SM1.3 expressing a construct for Δ5-7MRE11 were generously provided by Dr. Mark Meuth (Institute for Cancer Studies, The University of Sheffield, Sheffield, UK) and has been reported previously(9). These cell lines were grown in DMEM medium. In addition 46 CRC samples from the Molecular Epidemiology of Colorectal Cancer (MECC)(14) study were assessed for the frequency mutation of MRE11 poly(T)11 in primary colon tumors (Supplementary Table S2).
Analysis of MRE11 poly(T)11 and RAD50 poly(A)9 in cell lines and primary tumors
Analyses for the length of the MRE11 poly(T)11 and RAD50 poly(A)9 were performed on genomic DNA extracted from cell lines and CRC samples microdissected from paraffin embedded tissue blocks. Forward and reverse primers for MRE11 and RAD50 were labeled with FAM. PCRs were performed using GeneAmp Fast PCR Master Mix (Applied Biosystems, Foster City, CA) in separate reactions. The PCR fragments were detected by capillary electrophoresis on ABI370 at the University of Michigan Sequencing Core. The electropherograms were analyzed using GeneMarker v1.51 software (SoftGenetics, LLC., State Collage, PA). Primer sequences and PCR conditions are available upon request.
Expression of MRE11 wild type and mutant transcripts and PARP-1
Total RNA from 10 cell lines was extracted using the TRIzol protocol according to manufacturer’s instructions (Invitrogen, Carlsbad, CA). Adequate quantities of high-quality total RNA were determined by Agilent 2100 BioAnalyzer (Agilent Technologies, Palo Alto, CA). cDNA was synthesized using High Capacity cDNA Reverse Transcription Kit from 200 ng of RNA. All samples were tested in triplicate. Real-time qPCR was carried out using SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA) on Applied Biosystems Prism 7900 HT Sequence Detection System. Two sets of primers were designed to assess the independent expression levels of the MRE11 wild type and the mutant transcript, one set for PARP-1 and one for GAPDH that was used as an endogenous control. Primer sequences and PCR conditions are available upon request. The relative expression of the wild type and mutant transcript of MRE11 and PARP1 was calculated by ΔCt normalization to the expression of GAPDH.
Immunofluorescence
Cells were cultured on coverslips, fixed, and processed as previously described(15). For each cell line, Rad51 and γH2AX foci were quantified following fixation of cells 24 hours after being seeded (t=0) and 18 hours after irradiation (7.5 Gy) (t=18). Samples were imaged with an Olympus FV500 confocal microscope (Olympus America) with a 60× objective. For quantification of Rad51 and γH2AX foci, at least 100 cells were visually scored for each condition by two independent observers. Cells with ≥5 foci were scored as positive and percent positivity was compared at baseline and 18 hours following irradiation for three cell lines (SW620, HCT15, and SW48).
Irradiation
Cells were irradiated using a Philips RT250 (Kimtron Medical) at a dose rate of ~2 Gy/min in the University of Michigan Comprehensive Cancer Center Experimental Irradiation Core.
Statistical analysis of association between expression and mutational status of MRE11
Comparisons of expression levels between cell lines grouped by MRE11 mutational status across three categories were performed using the rank-based, Kruskal-Wallis ANOVA test followed by posthoc Dunn’s test for pairwise differences among groups (MRE11 homozygous versus heterozygous mutants, MRE11 homozygous mutants versus wild-type, MRE11 heterozygous mutants versus wild-type) using SAS version 9.1 (The SAS Institute, Cary, NC). Nonparametric methods were used to protect against violation of normality assumption given the limited sample size.
Analysis of gene expression data of tumor samples
RNA isolation and Microarray procedures. Primary tumors were obtained at the time of surgical resection, then they were snap frozen in liquid nitrogen at −80°C, embedded in optimum cutting temperature freezing media (MilesScientific, Naperville, IL), cryotome sectioned, stained with H&E, and evaluated by a surgical pathologist. Areas with >70% tumor cellularity were identified for RNA isolation. Selected sections of tumor samples were homogenized in Trizol (Invitrogen, Carlsbad, CA), and total cellular RNA was purified according the instructions of the manufacturer, with additional purification using RNeasy spin columns (Qiagen, Valencia, CA). RNA quality was assessed by 1% agarose gel electrophoresis, and samples were included only if the 18S and 28S bands were discrete and approximately equal. Expression levels were measured in two batches using Affymetrix U133A and U133A Plus 2.0 arrays (Santa Clara, CA). Preparation of target cRNA, hybridization, and scanning were performed according to the protocols of the manufacturer.
Statistical Analysis of Microarray Data
Expression analyses were carried out in the R-software using the package Bioconductor(16). Expression data in both batches were first subjected for quality assessment by creating the density plot of the log-intensity and RNA degradation plot corresponding to each sample. For all 331 samples, MAS 5.0-calculated signal intensities were normalized using the quantile normalization procedure implemented in robust multiarray analysis(17, 18) and the normalized data were log2 transformed. Sample specific median centering and scaling by the standard deviation were additionally applied. Filtering was done to exclude probe sets that were not expressed or probe sets that exhibited low variability across samples. Expression values were required to be above the lower quartile of all expression measurements in at least 25% of samples, and the interquartile range across the samples on log2 scale was required to be at least 0.5. After preprocessing and quality assessment the two batches were aligned. Probes of the U133A array present in the U133A Plus 2.0 were selected and quantile normalized to mimic the distribution of the U133A array. Five samples had been hybridize in both arrays and served to verify that the correlation of the expression was adequate for the majority of the probes. A total of 331 samples and 419,473 probe sets were subjected to further analysis. Two class comparison of MSI versus MSS tumors were carried out by two-sample t-test to yield a list of differentially expressed probe sets, discriminating between the two biologic states of interest under consideration. We had information on MSI status available from 300 patients whose characteristics are detailed in Supplementary Table S3. The issue of multiple testing was addressed by using adjusted P-values after controlling for the overall false discovery rate by Benjamini and Hochberg method (BH-adjusted P-values)(19). We also evaluated the local false discovery rate associated with our probe selection procedure by using the locfdr package in R(20).
Gene expression data sources, selection of probes from gene expression data sets submitted to the Connectivity Map Build 02 and generation of compound lists
Bioinformatic approaches to identify the level of enrichment between gene expression profiles characterizing MSI tumors and gene changes induced in vitro by the PARP-1 inhibitor Phenanthridinone and others were assessed using the Connectivity Map(21). We have used five different data sets characterizing MSI-H tumors. Four of them were previously published and retrieved directly from their original publications(22–25). Criteria followed for selection of probe sets and detailed lists have been published previously(26). The fifth dataset was generated from a total of 300 colorectal fresh frozen tumors collected from the MECC study and analyzed in two batches. The first batch was hybridized to the Affymetrix U133A chip and the second one to U133A Plus 2.0 (Supplementary Table S3). The final list of probe sets defining gene expression of MSI-H compared to MSS tumors was selected based on the strength of multiplicity adjusted P-values (cut-off P-value of < 0.001) and ratio of mean expression values across the two groups (cut-off Fold-change >1.3 and <0.7). It contained 442 upregulated and 480 downregulated probe sets as detailed in Supplementary Table S4 and S5.
Cytotoxicity experiments, dose-response data and IC50 calculations of ABT-888 in cell line models
ABT-888 (A-861695, Abbott Laboratories, IL, USA) was obtained from Axxora (San Diego, CA). Stock concentrations of 1 mM were maintained in DMSO and were dissolved in supplemented medium to obtain 6 serial dilutions (1 nM to 50 μM). Cytotoxicity experiments were performed in four homozygous MRE11 mutant, one heterozygous and three wild-type cell lines. 1,000 cells were seeded in triplicate per well and after 24 hours medium was replaced with medium containing 6 different concentrations of ABT-888. In control wells cells were fed with standard medium. After 6 days on treatment cell viability was estimated using WST-1. Cell viability was estimated on the basis of their ability to metabolize the tretazolium salt WST-1 to formazan by mitochondrial dehydrogenases. Quantification of absorbances was analyzed using a Spectramax 190 (Molecular Devices Corporation, Sunnyvale, CA). The percentage of surviving cells at each concentration relative to the non-treated group was calculated. The drug concentrations resulting in 50% of growth inhibition (IC50) were then estimated by fitting a sigmoid shaped dose-response curve. Differences in drug sensitivity comparing the mean IC50 of three independent experiments for every cell line and cell proliferation data at a certain drug concentration (10 μM) between cell lines classified by MRE11 polyT(11) mutational status determined using Kruskal-Wallis ANOVA as described previously. In addition, correlation between the levels of expression of the mutant transcript of MRE11 poly(T)11 and the percentage of growth inhibition induced by ABT-888 at 10 uM were examined by Spearman’s coefficient of rank correlation test. P-values <0.05 were considered to indicate statistical significance. These calculations were performed using SAS version 9.1.
Cell cycle analysis
For cell cycle analysis cells were washed in cold PBS, fixed in 70% ethanol, washed, resuspendedin 25 μg/ml Propidium Iodine (PI) with 100 μg/ml RNase A (BD Pharmigen, San Jose, CA), andincubated for 30 min at 37°C. Fluorescence was measured on a BD Biosciences FACSCaliburs flow cytometer within 1 h. Data were analyzed using the MODFIT 2.0 program (Verity Software House, Topsham, ME).
Stable knockdown of MRE11 in SW-480 cells
Several lentiviral-based plasmid containing a short hairpin RNA (shRNA) to human MRE11 was purchased from Open Biosystems (Huntsville, AL). Nonsilencing(mock) shRNAs were used as negative controls. The sequence of the MRE11-specific 22-mer shRNA with highest efficacy and reported here was AGGCCATGAACATGAGTGTAAA. Experimental procedures for shRNA transfection were done according to the Open Biosystems technical manual. Stable clones were generated by transfecting SW480 cells in 6-well dishes with 2 μg of each of the shRNA plasmids using FugeneHD (Roche) according to the manufacturer recommendations. Forty-eight hours after transfection, the cells were placed under selection with 2.0 μg/mL puromycin (Sigma-Aldrich), splitting them when the cells reached confluency. Multiple clones from the same transfection were pooled and grown under puromycin selection. Successful knockdown of specific gene products was confirmed by qRT-PCR as described above.
Results
Microsatellite instable colorectal cancer cell lines are associated with mutations in MRE11 poly(T)11
Mutations in coding microsatellite tracts of MRE11 and RAD50 were assessed in a panel of 17 CRC cell lines. None of the MSS cell lines were mutated in MRE11 or RAD50. All MSI-high (MSI-H) cell lines harbored a mutation in the polyT(11) tract located in the intron 4 of MRE11. Three had monoallelic and five had biallelic mutations. Only three cell lines harbored mutations in an exonic polyA(9) repeat of RAD50 (Table 1 and Supplementary Table S1), and all of them were MSI. Therefore we decided to focus our attention on MRE11. To confirm the frequency of the MRE11-intron 4 mutation we genotyped 46 tumors from the MECC study (Supplementary Table S2). 18/22 (82%) of MSI tumors harbored a mutation in MRE11 compared to 0% of Microsatellite Stable (MSS) (P-value<0.001). Among the 22 MSI tumors with MRE11 mutations, 36% of tumors had biallelic mutations.
Table 1.
Frequency of the MRE11 polyT(11) mutation in cell lines and primary CRC.
| MSI | MSS | P-value | |
|---|---|---|---|
| Cell lines – N | 8 | 9 | |
| MRE11 poly(T)11 mutation – N (%) | <0.001 | ||
| Yes | 8 (100) | 0 (0) | |
| No | 0 (0) | 9 (100) | |
| Primary tumors – N | 22 | 24 | |
| MRE11 poly(T)11 mutation – N (%) | <0.001 | ||
| Yes | 18 (82) | 0 (0) | |
| No | 4 (18) | 24 (100) | |
Mutations of MRE11 are not associated with an increase of PARP-1 expression
Due to the fact that the Intron 4-MRE11 mutation generates a change in splicing that leads to skipping of exon 5, we decided to assess the expression of MRE11 in 10 colorectal cancer cell lines. Two sets of primers for the mutant and the wild-type transcripts of MRE11 were designed (Figure 1A). Those cell lines harboring biallelic mutations in MRE11 showed a significant decrease in the levels of the wild-type transcript of MRE11 and an increase in the mutant transcript compared to wild-type cells (P-value<0.05, Figure 1B and 1C). In contrast, cell lines with monoallelic mutations showed intermediate expression levels of the mutant transcript to wild-type cells (P-value<0.05, Figure 1C). In addition, we assessed the basal levels of PARP-1 expression and no differences were observed between MRE11 mutant and wild-type cell lines (Figure 1D).
Figure 1.
A) a polyT(11) tract located at the Intron4 is the target for mutations in MSI tumors. Shortening in one or more nucleotides causes changes in the splicing can induce complete skipping of exon 5 and protein truncation. Two sets of primers were designed to measure the levels of the wild type (wt) and the mutant transcript (mut) of MRE11; B), C) and D) levels of expression of the wt and mut transcript of MRE11, as well as PARP-1 assessed by qRT-PCR in 10 CRC cell lines.
MSI-H tumors present with changes in gene expression related to the homologous recombination pathway
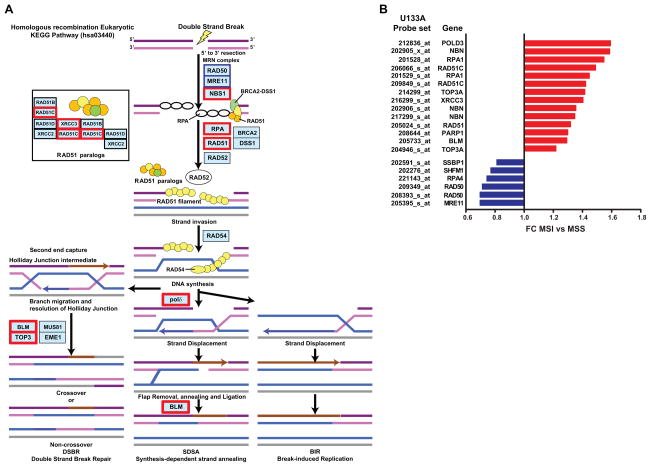
Once we observed that deficiency in MRE11 exists among MSI tumors, our interest was focused on assessing if the homologous recombination pathway showed evidence of deregulation in MSI tumors. Therefore we examined the expression levels of those genes integrated in the KEGG pathway hsa03440 using data from a total of 300 CRCs from the MECC study (Supplementary Table S3). As shown in Figure 2A and Supplementary Table S6 a total of 14 genes out of 30 were significantly differentially expressed in MSI-H compared to MSS tumors (Multiplicity adjusted Benjamini-Hochberg P-value<0.05) (19). MRE11 and RAD50 probe sets showed a lower expression in MSI-H tumors and simultaneously other probe sets such as PARP-1 were significantly upregulated, probably secondary to the deficiency in the MRN complex proteins (Figure 2B). This data provides evidence of significant differential expression of the homologous recombination pathway in MSI tumors.
Figure 2.
Schematic representation of the homologous recombination pathway hsa03440 retrieved from the KEGG database. In red are those genes upregulated and in blue those downregulated. Note that not all of the proteins acting in this pathway are shown in this figure; B) Probe sets from hsa03440 significantly deregulated in MSI compared to MSS tumors and their corresponding fold-changes;
Mutations of MRE11 induce higher levels of unrepaired DNA damage and attenuate the formation of Rad51 foci after irradiation
In order to determine the effect of MRE11 mutation on homologous recombination repair, we measured Rad51 focus formation at baseline and in response to radiation-induced DNA damage. We found that Rad51 focus formation following irradiation in the wild-type and monoallelic MRE11 mutant cell lines was significantly increased from baseline. However, the cell line harboring a biallelic mutation did not exhibit Rad51 mobilization upon irradiation. This observation highlights the impairment of double strand break repair by the homologous recombination pathway in cell lines deficient in MRE11 (Figure 3). Furthermore, we investigated the degree of unrepaired DNA damage in cell lines with different MRE11 mutational statuses by quantifying γH2AX with immunofluorescence (data not shown). In all three cell lines, irradiation induced an increase in the levels of unrepaired DNA damage, as expected. Under baseline conditions, the cell line SW-48 harboring biallelic mutations in MRE11 presented with more cells positive for γH2AX foci as compared to the wild-type cell line SW-620 and the monoallelic mutant HCT-15, suggesting that MRE11 mutation results in accumulation of DNA damage in otherwise unperturbed cells.
Figure 3.
Rad51 foci were analyzed under baseline conditions (B, t=0) and 18 hours after irradiation (IR, t=18). Percentages of foci-positive cells at each time point are presented. Error bars represent standard errors of the mean for two independent observations. Asterisks designate statistically significant differences in percent positivity at baseline vs. 18 hours post-irradiation (P-value<0.05). Representative images are presented for both markers and time points.
A gene expression profile of MSI-H tumors is anti-correlated with changes induced by the PARP-1 inhibitor Phenathridinone
Based on this deficiency in DSB repair exhibited by MSI tumors, we hypothesized that PARP-1 inhibitors may have a role in the treatment of this tumor subtype based on the concept of synthetic lethality. In addition we searched for in silico data supporting this biologically-driven hypothesis using a systems biology tool. We interrogated the Connectivity Map(21) database in order to determine if gene expression changes induced in cell line models after treatment with first generation PARP-1 inhibitor such as Phenanthridinone, NU-1025 and 1,5-Isoquinolinediol were anti-correlated with different gene expression profiles characterizing MSI-H tumors using two different measures: the enrichment score and the connectivity score. Strong evidence of anti-correlation between both Phenanthridinone and NU-1025 and MSI-H tumors was found in three out of five gene expression data sets. In the case of Phenanthridinone enrichment (~ −0.95) and connectivity scores (~ −0.60, Table 2) were very similar across signatures. 1,5-Isoquinolinediol was correlated in only one of the data sets. However, based on previous in vitro studies this compound is the least specific in terms of PARP inhibition of the three compounds studied.
Table 2.
Enrichment results obtained from the application to the Connectivity Map to five different data sets defining MSI CRCs. Enrichment* and Connectivity† scores for every data set are presented. These score are intended to reflect the correlation between gene expression changes induced in cell line models after treatment with first generation PARP-1 inhibitor and those gene expression profiles characterizing MSI-H tumors coming from different data sets. The enrichment score indicates the strength of this correlation and the connectivity score is relative to the rest of drugs tested in the Connectivity Map. The absolute value of both ranges from +1 to −1 and refers to the level of correlation or anti-correlation with the original signature of interest. Phenanthridinone, NU-1025 and 1,5-Isoquinolinediol are first generation PARP inhibitors, being Phenanthridinone the compound with the most consistent results.
| Data set | Compound name | Dose | Cell line | Connectivity score* | Enrichment score† |
|---|---|---|---|---|---|
| Vilar | Phenanthridinone | 51 μM | MCF7 | −0.68 | −0.98 |
| Banerjea | Phenanthridinone | 51 μM | MCF7 | −0.61 | −0.94 |
| Watanabe | Phenanthridinone | 51 μM | MCF7 | 0 | 0.62 |
| Koinuma | Phenanthridinone | 51 μM | MCF7 | −0.60 | −0.95 |
| Kruhoffer | Phenanthridinone | 51 μM | MCF7 | 0 | 0.54 |
| Vilar | NU-1025 | 100 μM | MCF7 | 0 | 0.523 |
| Banerjea | NU-1025 | 100 μM | MCF7 | −0.243 | −0.4 |
| Watanabe | NU-1025 | 100 μM | MCF7 | −0.56 | −0.93 |
| Koinuma | NU-1025 | 100 μM | MCF7 | 0 | 0.408 |
| Kruhoffer | NU-1025 | 100 μM | MCF7 | 0.46 | 0.81 |
| Vilar | 1,5-Isoquinolinediol | 100 μM | HL60 | 0 | 0.650 |
| Banerjea | 1,5-Isoquinolinediol | 100 μM | HL60 | −0.51 | −0.86 |
| Watanabe | 1,5-Isoquinolinediol | 100 μM | HL60 | 0 | −0.642 |
| Koinuma | 1,5-Isoquinolinediol | 100 μM | HL60 | 0 | −0.53 |
| Kruhoffer | 1,5-Isoquinolinediol | 100 μM | HL60 | 0 | 0.656 |
Low levels of MRE11 wild type transcripts increase the sensitivity to the PARP-1 inhibitor ABT-888 in MSI-H cell lines
We selected ABT-888 to assess the effects of a novel PARP-1 inhibitor in a colorectal cancer model deficient in MRE11. We used three wild-type, one monoallelic and four biallelic mutants of MRE11 cell lines for these experiments. Since the monoallelic mutant cell line has intermediate expression levels of the MRE11 mutant transcript but closer to wild-type cells we decided to group it along with them for IC50 and cytotoxicity at 10 μM comparisons. A significant difference in cytotoxicity was found at 10 μM concentration between biallelic mutants and wild-type plus monoallelic mutants (44.5% versus 80.65%, P-value=0.028, Figure 4A) and a 2.5-fold difference in terms of IC50 when compared between these two groups (8.9 versus 22.23 μM, P-value=0.028, Figure 4B). Then we studied the correlation between levels of expression of the mutant transcript of MRE11 and cytotoxicity to ABT-888 at a concentration of 10 μM showing a significant association between both (R=−0.9048, P-value=0.0046, Figure 4C). Cell cycle changes revealed an arrest in G1 and a decrease in S-phase after treatment with ABT-888 in MRE11 biallelic mutants (Figure 4D), consistent with expectation given the mechanism of PARP-inhibitors.
Figure 4.
A) and B) Comparison of cytotoxicity at 10 μM and IC50 in biallelic mutants and wild-type plus monoallelic mutants; C) Correlation between expression levels of the MRE11 mutant transcript and growth inhibition at 10 μM of ABT-888; D) Cell cycle changes after treatment with two different concentrations of ABT-888. Note that cell cycle changes were more pronounced in biallelic than monoallelic mutants and wild type cells.
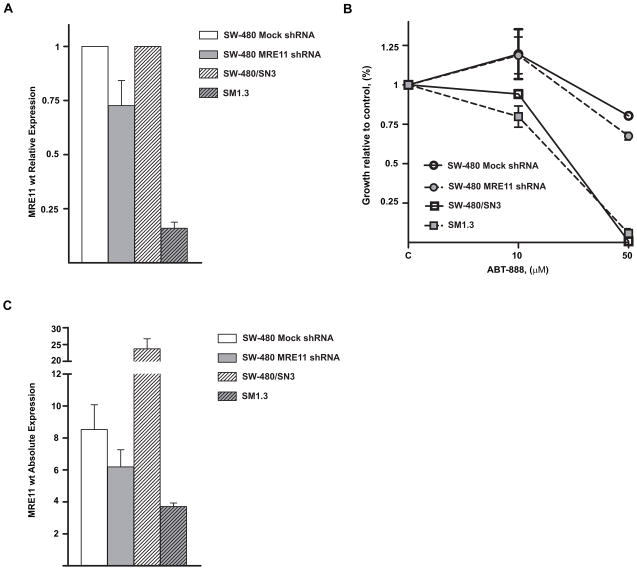
Finally we conducted experiments using two different approaches to diminish the expression of MRE11. First, shRNA was used to knock down the expression of MRE11 in a wild type MRE11 and MSS cell line to further assess its role as a mediator of the effect of PARP-1 inhibition. As depicted in Figure 5A we were able to achieve a significant, but incomplete, knockdown of MRE11 gene expression in the SW-480 cell line. We observed differences in proliferation between mock shRNA and MRE11 shRNA stable transfectants when 50 μM of ABT-888 was used, pointing towards a potential role of MRE11 as responsible for the drug effects (Fig 5B). Therefore, a second validation experiment using the cell line SW480/SN3 and its derivative SM1.3 was performed. SM1.3 was originally transfected with an expression construct for Δ5-7MRE11 lacking exons 5–7 that was originally cloned from the CRC cell line HCT-116. Experiments using this cell line were designed to confirm the role of MRE11 mutations in conferring sensitivity to PARP inhibitors. We assessed the expression of the wt transcript of MRE11 in the parental SW-480/SN3 and in SM1.3 observing that the derivative cell line SM1.3 expressed less than 20% of the parent line’s MRE11 transcript levels, supporting its utility as an appropriate model (Figure 5A). Cytotoxicity assays comparing ABT-888 in these two cell lines showed a higher response by SM1.3 to PARP inhibition at 10 μM, consistent with the trends of the results obtained with the shRNA approach. However, SM1.3 did not show a differential response at 50 μM, suggesting either a threshold effect or an imperfect dose-response relationship when compared with the previous shRNA knocking down approach (Figure 5C).
Figure 5.
A) Levels of MRE11 wild type transcript in transfected cells with a shRNA plasmid against MRE11 and in the derivative cell line SM1.3 that has been transfected with an expression construct for Δ5-7MRE11 lacking exons 5–7. Levels of expression were normalized to mock cells and to the parental cell line SW480/SN3, respectively. B) Proliferation after treatment with ABT-888. Note a difference in proliferation between SW-480 transfected with shRNA anti-MRE11 plasmid and SW-480 transfected with a mock plasmid at 50 μM and also between SM1.3 and its parental cell line at 10 μM. C) Expression levels of the wild type transcript of MRE11 compared to levels expressed in the original panel of cell lines.
Discussion
Genetic instability in MSI tumors is secondary to the presence of a deficiency in the MMR system that introduces a myriad of mutations in downstream genes(27). One of these gene targets is MRE11, a gene that is implicated in homologous recombination. MRE11 forms a multiprotein complex with RAD50 and NBS1 that signals double strand DNA breaks and then recruits other proteins that initiate DNA repairing. We have observed frequent mutations in an specific poly(T)11 tract located in the intron 4 of MRE11 in MSI colorectal tumors that is consistent with previous reports(28, 29), thus confirming that MSI is strongly associated with this MRE11 mutation.
At the present time deficiency in homologous recombination has been therapeutically exploited therapeutically in those tumors exhibiting mutations in BRCA1 and BRCA2(30). This novel therapeutic approach is based on the fact that simultaneous deficiency in two genes may introduce lethality in a biologic system that otherwise would be tolerant to the loss of one of them(31). Although the role of BRCA1 and BRCA2 is more predominant in homologous recombination than MRE11, our hypothesis is that other components of this pathway may also predict an increase in the sensitivity to PARP-1 inhibitors. In terms of the biology of double strand break repair, we have shown that cell lines harboring biallelic mutations in MRE11 did not effectively promote homologous recombination expressed by the mobilization of Rad51. Moreover, biallelic mutant cell lines presented with higher levels of unrepaired DNA damage in basal conditions. Consistent with our hypothesis, we then demonstrated that those colorectal cancer cell lines displaying MSI and harboring biallelic mutations in MRE11 have greater sensitivity to PARP-1 inhibition. However, the effect of PARP-1 inhibition is abrogated when one wild-type allele of MRE11 is retained. This point has been illustrated in our data by the fact that a heterozygous cell line had similar sensitivity to PARP-1 inhibition to those with both alleles intact. A similar effect has been reported in the preclinical testing of these compounds in BRCA-1 and 2 deficient models. Therefore we suggest that those patients with MSI tumors harboring biallelic mutations in MRE11 may represent a target population for the use of PARP-1 inhibitors in patients with MSI colorectal cancers. We have shown that tumors with bialleic mutations represent approximately 36% of the total of MRE11 mutants. Finally, we tested several shRNA constructs in two different microsatellite stable CRC cell lines, SW-480 and HT-29 in order to functionally validate the contribution of MRE11 to PARP-1 sensitivity. We initially observed that those transfections with highest efficacy knocking-down the expression of MRE11 induced lethality in the cells. The validation experiments were first performed in a SW-480 transfected with a shRNA that achieved a level of down-regulation only 25%. Despite this limited suppression, we observed a higher sensitivity to PARP-1 inhibitors at 50 μM of ABT-888 following this level of decreased MRE11 expression. In addition, we performed a second set of validation studies using a cell line model transfected with a transcript that contains a mutation in MRE11 lacking exons 5–7 and leads to dramatically lower levels of the wt MRE11 at 80% of the baseline compared to the parental cell line. Again we observed that PARP inhibition exerted a higher effect on the derivative cell line SM1.3 at a lower concentration of the drug of 10 μM but not at 50 μM. Therefore, these data are not entirely consistent with dose-response inhibition across all concentrations, and suggest that either inhibitory thresholds might not be perfectly modeled by our transfection system, or that other genetic variation may contribute to PARP sensitivity. In fact, we have been able to identify by using gene expression profiling other candidate genes that are involved in the homologous recombination pathway and significantly deregulated in MSI-H tumors.
To our knowledge, this is the first communication on the activity of PARP-1 inhibitor in a solid tumor harboring a deficiency in a DNA repair pathway other than BRCA1 and 2, thus suggesting broader applications of this therapeutic strategy. Therefore, our study also provides preclinical rationale for an ongoing phase II clinical trial exploring the activity of a different PARP-1 inhibitor in colorectal tumors stratified by MSI status (NCT00912743). In addition, we suggest that combinations of other therapies inducing DSB such as radiation or irinotecan may enhance the effects of PARP-1 inhibitors in MSI CRCs and are warranted in the future.
Supplementary Material
Acknowledgments
Grant support: This work was supported in part by Fundacion “la Caixa”, Barcelona, Spain (EV), NCI 1R01CA81488 (SBG), NIH R03 CA130045 (BM), University of Michigan Comprehensive Cancer Center Core Support grant (NIH 5P30CA46592) and Michigan Institute for Clinical & Research Health (UL1RR024986).
We want to thank Mark Meuth and Anil Ganesh from the Institute of Cancer Studies (Sheffield, UK) for providing us with cell lines for validation experiments.
References
- 1.Aaltonen LA, Salovaara R, Kristo P, Canzian F, Hemminki A, Peltomaki P, et al. Incidence of hereditary nonpolyposis colorectal cancer and the feasibility of molecular screening for the disease. N Engl J Med. 1998;338(21):1481–7. doi: 10.1056/NEJM199805213382101. [DOI] [PubMed] [Google Scholar]
- 2.Hampel H, Frankel WL, Martin E, Arnold M, Khanduja K, Kuebler P, et al. Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer) N Engl J Med. 2005;352(18):1851–60. doi: 10.1056/NEJMoa043146. [DOI] [PubMed] [Google Scholar]
- 3.Gryfe R, Kim H, Hsieh ET, Aronson MD, Holowaty EJ, Bull SB, et al. Tumor microsatellite instability and clinical outcome in young patients with colorectal cancer. N Engl J Med. 2000;342(2):69–77. doi: 10.1056/NEJM200001133420201. [DOI] [PubMed] [Google Scholar]
- 4.Greenson JK, Bonner JD, Ben-Yzhak O, Cohen HI, Miselevich I, Resnick MB, et al. Phenotype of microsatellite unstable colorectal carcinomas: Well-differentiated and focally mucinous tumors and the absence of dirty necrosis correlate with microsatellite instability. Am J Surg Pathol. 2003;27(5):563–70. doi: 10.1097/00000478-200305000-00001. [DOI] [PubMed] [Google Scholar]
- 5.Greenson JK, Huang SC, Herron C, Moreno V, Bonner JD, Tomsho LP, et al. Pathologic predictors of microsatellite instability in colorectal cancer. Am J Surg Pathol. 2009;33(1):126–33. doi: 10.1097/PAS.0b013e31817ec2b1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duval A, Hamelin R. Mutations at coding repeat sequences in mismatch repair-deficient human cancers: toward a new concept of target genes for instability. Cancer Res. 2002;62(9):2447–54. [PubMed] [Google Scholar]
- 7.Giannini G, Ristori E, Cerignoli F, Rinaldi C, Zani M, Viel A, et al. Human MRE11 is inactivated in mismatch repair-deficient cancers. EMBO Rep. 2002;3(3):248–54. doi: 10.1093/embo-reports/kvf044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bekker-Jensen S, Lukas C, Kitagawa R, Melander F, Kastan MB, Bartek J, et al. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J Cell Biol. 2006;173(2):195–206. doi: 10.1083/jcb.200510130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wen Q, Scorah J, Phear G, Rodgers G, Rodgers S, Meuth M. A Mutant Allele of MRE11 Found in Mismatch Repair-deficient Tumor Cells Suppresses the Cellular Response to DNA Replication Fork Stress in a Dominant Negative Manner. Mol Biol Cell. 2008;19(4):1693–705. doi: 10.1091/mbc.E07-09-0975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoeijmakers JH. DNA damage, aging, and cancer. N Engl J Med. 2009;361(15):1475–85. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- 11.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434(7035):917–21. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 12.Haince J-F, McDonald D, Rodrigue A, Dery U, Masson J-Y, Hendzel MJ, et al. PARP1-dependent Kinetics of Recruitment of MRE11 and NBS1 Proteins to Multiple DNA Damage Sites. Journal of Biological Chemistry. 2008;283(2):1197–208. doi: 10.1074/jbc.M706734200. [DOI] [PubMed] [Google Scholar]
- 13.Wellcome Trust Sanger Institute [Internet] Cambridge: Wellcome Trust Sanger Institute Genome Research Ltd; 2010. Nov 30, [cited 2008 July 1]; Available from: http://www.sanger.ac.uk/genetics/CGP/CellLines/ [Google Scholar]
- 14.Poynter JN, Gruber SB, Higgins PD, Almog R, Bonner JD, Rennert HS, et al. Statins and the risk of colorectal cancer. N Engl J Med. 2005;352(21):2184–92. doi: 10.1056/NEJMoa043792. [DOI] [PubMed] [Google Scholar]
- 15.Parsels LA, Morgan MA, Tanska DM, Parsels JD, Palmer BD, Booth RJ, et al. Gemcitabine sensitization by checkpoint kinase 1 inhibition correlates with inhibition of a Rad51 DNA damage response in pancreatic cancer cells. Mol Cancer Ther. 2009;8(1):45–54. doi: 10.1158/1535-7163.MCT-08-0662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–93. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 18.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 19.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Statist Soc Ser. 1995;B V 57:289–300. [Google Scholar]
- 20.Efron B. Large-scale simultaneous hypothesis testing: the choice of a null hypothesis. Jour Amer Stat Assoc. 2004;99:96–104. [Google Scholar]
- 21.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313(5795):1929–35. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 22.Watanabe T, Kobunai T, Toda E, Yamamoto Y, Kanazawa T, Kazama Y, et al. Distal colorectal cancers with microsatellite instability (MSI) display distinct gene expression profiles that are different from proximal MSI cancers. Cancer Res. 2006;66(20):9804–8. doi: 10.1158/0008-5472.CAN-06-1163. [DOI] [PubMed] [Google Scholar]
- 23.Banerjea A, Ahmed S, Hands RE, Huang F, Han X, Shaw PM, et al. Colorectal cancers with microsatellite instability display mRNA expression signatures characteristic of increased immunogenicity. Mol Cancer. 2004;3:21. doi: 10.1186/1476-4598-3-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kruhoffer M, Jensen JL, Laiho P, Dyrskjot L, Salovaara R, Arango D, et al. Gene expression signatures for colorectal cancer microsatellite status and HNPCC. Br J Cancer. 2005;92(12):2240–8. doi: 10.1038/sj.bjc.6602621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koinuma K, Yamashita Y, Liu W, Hatanaka H, Kurashina K, Wada T, et al. Epigenetic silencing of AXIN2 in colorectal carcinoma with microsatellite instability. Oncogene. 2006;25(1):139–46. doi: 10.1038/sj.onc.1209009. [DOI] [PubMed] [Google Scholar]
- 26.Vilar E, Mukherjee B, Kuick R, Raskin L, Misek DE, Taylor JM, et al. Gene Expression Patterns in Mismatch Repair-Deficient Colorectal Cancers Highlight the Potential Therapeutic Role of Inhibitors of the Phosphatidylinositol 3-Kinase-AKT-Mammalian Target of Rapamycin Pathway. Clin Cancer Res. 2009;15(8):2829–39. doi: 10.1158/1078-0432.CCR-08-2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vilar E, Gruber SB. Microsatellite instability in colorectal cancer-the stable evidence. Nat Rev Clin Oncol. 2010;7(3):153–62. doi: 10.1038/nrclinonc.2009.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giannini G, Rinaldi C, Ristori E, Ambrosini MI, Cerignoli F, Viel A, et al. Mutations of an intronic repeat induce impaired MRE11 expression in primary human cancer with microsatellite instability. Oncogene. 2004;23(15):2640–7. doi: 10.1038/sj.onc.1207409. [DOI] [PubMed] [Google Scholar]
- 29.Miquel C, Jacob S, Grandjouan S, Aime A, Viguier J, Sabourin JC, et al. Frequent alteration of DNA damage signalling and repair pathways in human colorectal cancers with microsatellite instability. Oncogene. 2007;26(40):5919–26. doi: 10.1038/sj.onc.1210419. [DOI] [PubMed] [Google Scholar]
- 30.Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361(2):123–34. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 31.O’Connor MJ, Martin NM, Smith GC. Targeted cancer therapies based on the inhibition of DNA strand break repair. Oncogene. 2007;26(56):7816–24. doi: 10.1038/sj.onc.1210879. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.