Abstract
Cytogenetic analysis of a primary bone neoplasm with pericytic features in a 67 year-old-male revealed a t(7;12)(p22;q13) among other karyotypic abnormalities. Subsequent molecular studies confirmed the presence of an associated ACTB-GLI1 fusion transcript. An identical 7;12 translocation is known to characterize a discrete group of soft tissue tumors belonging to the myopericytic category termed “pericytoma with t(7;12).” To the best of our knowledge, this is the first case of pericytoma with t(7;12) arising in bone. Cytogenetic and molecular analyses were useful, if not essential, in classifying this rare diagnostic entity.
Keywords: perictyoma, ACTB, GLI1
Introduction
Bone tumors constitute a heterogeneous group of neoplasms that are frequently diagnostically challenging. Careful consideration and correlation of clinical and radiographic findings are important in the diagnostic workup. Chromosomal aberrations, particularly translocations and corresponding fusion genes, also play a central role in the classification of bone tumors [1].
“Pericytoma with t(7;12)” is a rare soft tissue neoplasm that displays a perivascular pattern of spindle-to-ovoid cell proliferation [2]. The underlying molecular event of this translocation is a fusion of the ACTB and GLI1 genes that offers a definitive means for identifying this tumor [3]. Only five cases of “pericytoma with t(7;12)” have been fully characterized in the literature [2,3] although lesions of this type may have gone previously unrecognized as a distinct entity. To the best of our knowledge, this is the first report of this neoplasm arising in bone.
Case History
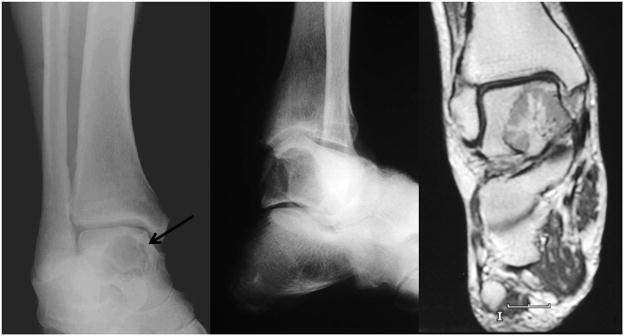
The patient was a 67-year-old man with a 7 month history of increasing pain in the right foot and ankle. Initial radiographic studies revealed a 2 cm lytic lesion of the right talus with focal overlying cortical erosion (Fig. 1). No extension into the soft tissue was identified. Subsequent physical examination and imaging studies failed to demonstrate evidence of other osseous or non-osseous lesions.
Figure 1.
A. Preoperative anteroposterior mortise and B. Broden’s view radiographs of the affected ankle showing a well-defined lytic lesion involving the body of the talus (arrow). No matrix calcification is identified. C. Coronal T1-weighted image after administration of gadolinium based contrast. Talus lesion demonstrates differential enhancement, most marked centrally with radial pattern. Several small foci of non-enhancement also present within lesion and persisted on other pulse sequences.
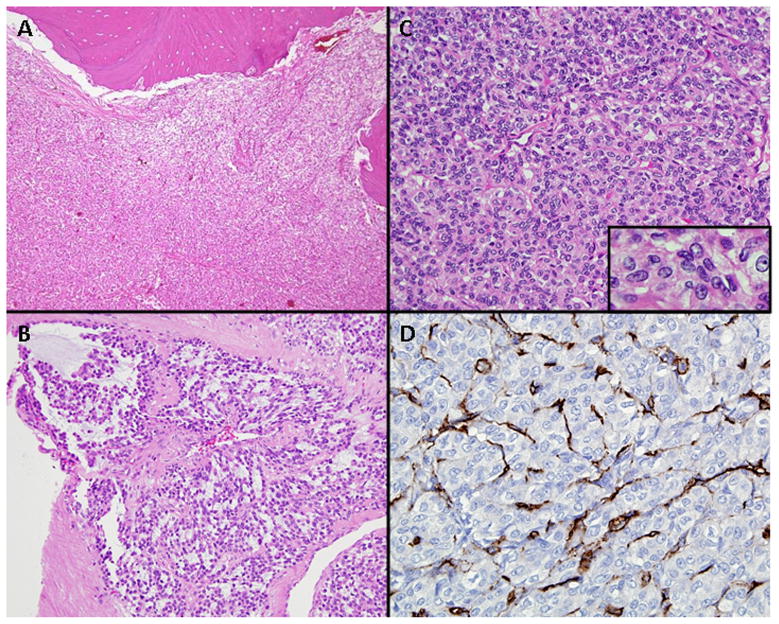
An open biopsy was performed. Histopathologic evaluation of this specimen demonstrated a cellular neoplasm with prominent lesional vasculature (Fig. 2A). The tumor was predominantly solid, although foci suggestive of a lobular growth pattern were noted as well as focal myxoid stroma (Fig. 2B). Individual cells were spindled to ovoid-shaped and exhibited round to oval nuclei with fine chromatin and occasional small nucleoli (Fig. 2C). Mitoses were infrequent. No pleomorphic tumor cells or tumoral vascular invasion was identified.
Figure 2.
A. Relatively dense cellular neoplasm occupies the intertrabecular bone marrow space. B. Tumor cells arranged around numerous thin-walled capillary vessels were spindle-to-ovoid shaped. Individual nuclei exhibited vesicular chromatin and frequently a small single nucleolus (insert). C. Focal myxoid stroma. D. CD34 immunostaining is negative in the tumor cells but highlights the vasculature.
The neoplastic cells were diffusely immunoreactive for vimentin and neuron-specific enolase. Focal immunoreactivity could be appreciated for SMA (smooth muscle actin) and EMA (epithelial membrane antigen). In contrast, the neoplastic cells uniformly failed to stain with antibodies to MAK-6 and AE1/AE3 (two pan-keratin cocktails), CAM 5.2 and AE1 (low molecular weight keratin antibodies), AE3 (high molecular weight keratin antibody), desmin, S-100 protein, synaptophysin, chromogranin, neurofilament protein, and CD34. CD34 immunostaining however, highlighted the numerous thin-walled capillary vessels (Fig. 2D). Ultrastructurally, the neoplastic cells featured abundant mitochondria, prominent rough endoplasmic reticulum, and Golgi zones. Also noted were scant cytoplasmic microfilaments but pinocytotic vesicles and basement membrane material were lacking.
At the time of diagnosis in 1996, following additional expert consultation with Dr. Juan Rosai (Memorial Sloan Kettering Cancer Center, New York City) and Drs. David Dahlin and K Krishnan Unni (Mayo Clinic, Rochester) this lesion was interpreted as a low-grade malignant neoplasm of uncertain histogenesis. The patient subsequently underwent a Syme amputation revealing a focally hemorrhagic neoplasm of the talus measuring 2.5 x 2.5 x 2.7 cm. Gross and microscopic examination of this specimen revealed local invasiveness as evidenced by extension of the tumor into the interosseous calcanean ligament but not into the adjacent calcaneous. There was no involvement of soft tissue and resection margins were negative.
Materials and Methods
Conventional Cytogenetic Analysis
Sterile, representative samples of both biopsy and resection specimens were submitted for cytogenetic analysis. Standard culture and harvest procedures were performed as described previously [4]. The karyotypes were expressed according to the International System for Human Cytogenetic Nomenclature 2009 [5].
Molecular Cytogenetic Analysis
In an effort to more fully characterize the karyotypic findings, fluorescence in situ hybridization (FISH) studies using whole chromosome paint probes for chromosomes 1, 2, 12, 13, 14, and 22, and chromosome enumeration probes for chromosomes 1, 7, 13/21 and 15 [Abbott Molecular, Des Plaines, IL and Oncor, Gaithersburg, MD] were performed on unstained metaphase cell preparations. Slide pretreatment included a 1 hr incubation in a 1X RNase solution (37ºC), followed by four consecutive rinses in 2X SSC at room temperature. Subsequently, slides were dehydrated in 70%, 85%, and 95% ethanol at −20º C for 2 min each and allowed to air dry. Denaturation was achieved by immersing slides for 2 min in a 70% formamide solution at 70ºC. Then, the slides were rinsed in 70%, 80%, 95%, and 100% ethanol washes at −20º C for 2 min each and air dried. Hybridization and counterstaining were performed according to the manufacturers’ instructions. Three to five metaphase cell images were acquired per probe using the Cytovision Image Analysis System (Applied Imaging, Santa Clara, CA).
RT-PCR (reverse transcription-polymerase chain reaction) Analysis
Total RNA was extracted from 100 mg of frozen tumor tissue using TRIzol® Reagent (Invitrogen, Carlsbad, CA). RNA integrity was assessed using the Agilent 2100 Bioanalyser (Agilent Technologies, Santa Clara, CA). Subsequently, cDNA synthesis was performed using the Advantage® RT-for-PCR kit (Clontech, Mountain View CA); 1 ug of total RNA was used as the template in 20 ul reaction with random hexamer primer according to the manufacturer’s instructions. The reaction was incubated at 42°C for 1 hr followed by 94°C for 5 min to arrest cDNA synthesis.
To detect the ACTB-GLI fusion transcript, PCR was performed using the Expand High Fidelity PCR System (Roche, Mannheim, Germany) with gene specific primer pairs ACTB61F-GLI868R and ACTB18F-GLI1246R (ACTB61F: 5’-CCGCCAGCTCAC CATGGATGATG, GLI 868R: 5’-GTGGCACACGAACTCCTTCCGCTC; ACTB 18 F: 5’-CACAGAGCCTC GCCTTTGCCGA, GLI 1246R: 5’-GCCGTTTGGTCACATGG GCGTC) [3]. 5 ul of diluted cDNA was used as template for PCR in 50 μ of assay containing 20 mM Tris-HCl, 50 mM KCL, 1.25 mM MgCl2, 0.4 mM dNTPs and 0.2 uM of each primer and 1 ul of High Fidelity Enzyme Mix. The PCR running program was optimized with an initial denaturation step of 94°C for 2 min, followed by the first 10 cycles with 94°C for 15 sec, 60°C for 30 sec, and 72°C for 45 sec, then the next 20 cycles with 94°C for 15 sec, 60°C for 30 sec, and 72°C for 45 sec with 5 sec increasing elongation time for each successive cycle, and a final elongation of 72°C for 10 min. The PCR products were analyzed on 1.5% agarose gels stained with ethidium bromide.
PCR products were cloned into pCR®4-TOPO® vector and transformed into E. coli One Shot® TOP 10 competent cells using TOPO TA Cloning® Kit for Sequencing (Invitrogen) according to the manufacturer’s instructions. Single colonies were picked and cultured in 5 ml of LB medium with 50ug/ml kanamycin at 37°C overnight. Plasmid DNA was isolated using QIAprep Spin Miniprep Kit (Qiagen, Valencia, CA) and digested with EcoR I (Invitrogen). The digestion reaction was then analyzed on a 1.5% agarose gel for confirmation of the presence and size of inserts. Plasmid DNA with different sizes of inserts were sequenced by the UNMC Sequencing Core Facility. When it was required, PCR products were separated on a 1.5% agarose gel and purifed with the QIAquick Gel Extraction Kit (Qiagen, Valencia, CA) prior to subcloning.
Results
Conventional and Molecular Cytogenetic Findings
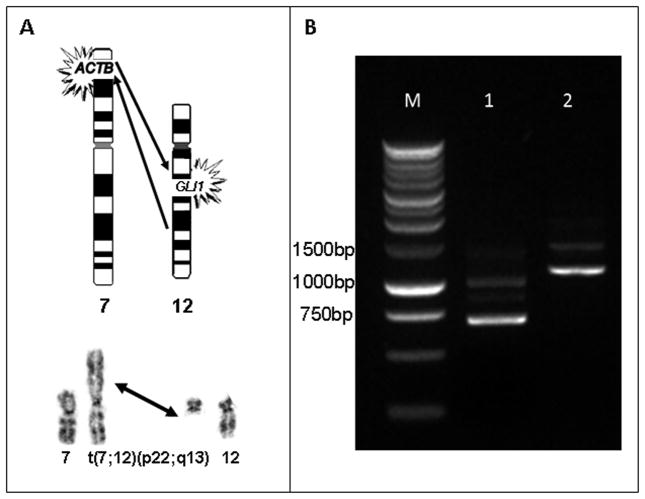
Conventional cytogenetic analysis of the biopsy specimen revealed a 7;12 translocation (Fig. 3A) among other clonal abnormalities in sixteen metaphase cells: 46,XY,+X,der(1)t(1;14) (p13;q11.2),der(2)t(2;13)(q31;q31),der(3)t(2;13;3) (q31;q11q31;p13),t(7;12)(p22;q13),−13,der(? 14)t(13;?14;?)(q11q12;?q11;?), der(15)t(3;15)(p13;q11). FISH analysis using the specified probes supported these findings: der(1)t(1;14)(wcp14+,D1Z1+,wcp1+),der(2)t(2;13) (wcp2+,wcp13+), der(3)t(2;13;3)(wcp2+,wcp13+),t(7;12) (wcp12+,D7Z1+;wcp12+),der(? 14)t(13;?14;?) (wcp13+),der(15)t(3;15)(D15Z1+). Similar karyotypic anomalies were identified in the subsequent resection specimen in addition to a single karyotypically normal cell: 46,XY, +X,der(1)t(1;14)(p13;q11.2),t(7;12)(p22;q13),−14[2]/46,idem,der(2)t(2;13)(q31;q31), der(3)t(2;13;3)(q31;q11q13;p13),−13,der(15)t(3;15)(p13;q11),+mar[10]/46,XY[1].
Figure 3.
A. Schematic illustration (top) and partial G-banded karyotype (bottom) illustrating the 7;12 translocation. B. RT-PCR detected ACTB-GLI1 fusion transcripts using ACTB 61F-868R and ACTB 18F-1246R primers, which amplified DNA products of 700 bp (lane 1) and 1119 bp (lane 2) respectively. M, 1 kb DNA ladder.
Molecular Genetic Findings
Primer pair ACTB61F-GLI868R amplified a 700 bp and a 950 bp product (Fig. 3B) and primer pair ACTB18F-GLI1246R yielded a 1119 bp and a 1400 bp product. The amplified fragments were cloned and analyzed by nucleotide sequencing (Fig. 4). The nucleotide sequence analysis of the amplified sequences of 700 bp and 1119 bp revealed a fusion gene composed of exons 1–3 of the ACTB gene and a portion of exon 5 as well as exons 6–12 of the GLI gene. Specifically, ACTB exon 3 at nucleotide position 436 was fused to exon 5 (nucleotide position 472) of the GLI1 gene, the latter containing an 18 bp (GCC CGG GGT GGG ATG) in-frame deletion. The fusion resulted in a transcript of 3318 nucleotides encoding a chimeric protein of 1106 amino acid residues. The sequencing results of fragments 950 bp and 1400 bp showed an identical molecular structure at the breakpoint and an intronic readthrough between exon 6 and 7 of GLI1 generating a transcript of 3783 nucleotides. The retention of the intronic sequences in the transcript introduced an early translational stop codon. The mRNA of 3783 nucleotides does not code for a functional protein.
Figure 4.
A. Schematic of ACTB, GLI1, and ACTB-GLI1 fusion gene. Individual exons are represented by boxes with non-translated regions in gray. Introns are shown as horizontal lines and drawn in scale. B. Partial nucleotide sequences around the breakpoint region of the ACTB- GLI1 fusion. ACTB exon 3 at nucleotide position 436 (solid horizontal bar) is fused in-frame with exon 5 of GLI1 at nucleotide position 547 (grey bar). Sequencing analysis demonstrated that exon 3 of ACTB is correctly spliced at the splice donor site (gt) and that exon 5 of GLI1 also has a twenty-one nucleotide long in-frame deletion (open bar) six nucleotides distal to the breakpoint in exon 5.
Discussion
Myopericytoma, a perivascular myoid neoplasm with a broad morphologic spectrum, typically arises in subcutaneous tissue of the extremities of adults [6,7]. Although myopericytomas distant from superficial soft tissue have been described, including locations such as the spine [8], oral cavity [9], and lung/pleura [10], among others, primary myopericytoma of bone has not been reported. Most myopericytomas behave in a benign fashion, although rare cases of malignant myopericytoma exist [7,11]. Metastasizing malignant myoepericytomas have presented as deep-seated rather than superficial lesions [7,11].
Pericytoma with t(7;12) is a distinct and exceedingly rare entity thought to fall within the category of myopericytic neoplasms [2]. Only five cases have been reported previously, describing primary lesions located in the tongue (3), stomach (1) and calf (1) [2]. The current case is quite exceptional as it represents the first pericytoma with t(7;12) to arise in bone.
The clinical behavior of the five previously described cases of pericytoma with t(7;12) was nonaggressive with no evidence of recurrence or metastasis (follow-up ranged from 22 to 120 months) [2]. Although the present tumor of bone exhibited relatively high cellularity and local invasiveness as evidenced by extension of the tumor into the interosseous calcanean ligament, there were no other features suggesting aggressive behavior such as pleomorphism, frequent mitotic figures or vascular invasion. The patient has remained free of disease 14 years following his resection.
The t(7;12)(p22;q13) chromosomal translocation produces a fusion of the GLI1 oncogene with the β-actin gene (ACTB), resulting in replacement of the promoter region of GLI1 with that of the ubiquitously expressed ACTB gene [2,3]. The (hedgehog) HH-GLI signaling pathway has been implicated in a large number of human cancers [12]. GLI1 has been identified as an amplified gene in glioblastoma and rhabdomyosarcoma [13,14]. The pathogenetic consequence of 7;12 translocation event is that the strong ACTB promoter leads to overexpression of GLI1 sequences important for transcriptional activation and deregulation of downstream target genes. At the level of molecular resolution, the 7;12 chromosomal translocation observed in the neoplasm described here is unbalanced as it is associated with a small deletion (a twenty-one nucleotide long in-frame deletion) in exon 5 of the GLI1 gene. The GLI1 gene included in the current ACTB-GLI1 fusion transcript starts with exon 5, thus integrating exons 7–10 encoding the five zinc finger domains. Four of the five zinc finger domains are believed to have DNA-binding activity that enable GLI1 to bind to DNA sequence to activate gene expression [15].
In summary, this primary bone tumor presented a significant diagnostic challenge in 1996. The terminology proposed by Dahlén et al. [2] to designate the soft tissue neoplasm they identified in 2004 as “pericytoma with t(7;12)” is useful in defining both morphological and cytogenetic characteristics. The current case demonstrates histopathologic features similar to those lesions arising in the soft tissue and recognition of identical cytogenetic and molecular findings further supports a common histopathogenesis between pericytoma with t(7;12) of soft tissue and bone origin.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Unni KK, Inward CY, Bridge JA, Kindblom L-G, Wold LE. Genetics of Bone Tumors. In: Silverberg SG, Sobin LH, editors. AFIP Fascicle on Tumors of Bone. 4. Washington, DC: American Registry of Pathology Press; 2005. pp. 11–26. [Google Scholar]
- 2.Dahlén A, Fletcher CDM, Mertens F, Fletcher JA, Perez-Atayde AR, Hicks MJ, Debiec-Rychter M, Sciot F, Wejde J, Wedin R, Mandahl N, Panagopoulos I. Activation of the GLI Oncogene through Fusion with the β-Actin Gene (ACTB) in a Group of Distinctive Pericytic Neoplasms. Am J Pathol. 2004;164:1645–53. doi: 10.1016/s0002-9440(10)63723-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dahlén A, Mertens F, Mandahl N, Panagopoulos I. Molecular genetic characterization of the genomic ACTB-GLI fusion in pericytoma with t(7;12) Biochem Biophys Res Commun. 2004;325:1318–23. doi: 10.1016/j.bbrc.2004.10.172. [DOI] [PubMed] [Google Scholar]
- 4.Bridge JA, Neff JR, Mouron BJ. (1992): Giant cell tumor of bone: chromosomal analysis of 48 specimens and review of the literature. Cancer Genet Cytogenet. 1992;58:2–13. doi: 10.1016/0165-4608(92)90125-r. [DOI] [PubMed] [Google Scholar]
- 5.Shaffer LG, et al., editors. ISCN: An International System for Human Cytogenetic Nomenclature. Basel, Switzerland: S. Karger; 2009. [Google Scholar]
- 6.Granter SR, Badizadegan K, Fletcher CDM. Myofibromatosis in adults, glomangiopericytoma and myopericytoma: a spectrum of tumors showing perivascular myoid differentiation. Am J Surg Pathol. 1998;22:513–25. doi: 10.1097/00000478-199805000-00001. [DOI] [PubMed] [Google Scholar]
- 7.Mentzel T, Dei Tos AP, Sapi Z, Kutzner H. Myopericytoma of skin and soft tissues. Clinicopathologic and immunohistochemical study of 54 cases. Am J Surg Pathol. 2006;30:104–13. doi: 10.1097/01.pas.0000178091.54147.b1. [DOI] [PubMed] [Google Scholar]
- 8.Cox D, Giltman L. Myopericytoma of the thoracic spine: a case report. Spine. 2003;28:E30–2. doi: 10.1097/00007632-200301150-00020. [DOI] [PubMed] [Google Scholar]
- 9.Dray MS, McCarthy SW, Palmer AA, Bonar SF, Stalley PD, Marjoniemi V, Millar E, Scolyer RA. Myopericytoma: a unifying term for a spectrum of tumours that show overlapping features with myofibroma. A review of 14 cases. J Clin Pathol. 2006;59:67–73. doi: 10.1136/jcp.2005.028704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edgecombe A, Peterson RA, Shamji FM, Commons S, Sekhon H, Gomes MM. Myopericytoma: a pleural-based spindle cell neoplasm off the beaten path. Int J Surg Pathol. 2011;19:247–51. doi: 10.1177/1066896910381897. [DOI] [PubMed] [Google Scholar]
- 11.McMenamin ME, Fletcher CD. Malignant myopericytoma: expanding the spectrum of tumours with myopericytic differentiation. Histopathology. 2002;41:450–60. doi: 10.1046/j.1365-2559.2002.01537.x. [DOI] [PubMed] [Google Scholar]
- 12.Ruiz i Altaba A, Mas C, Stecca B. The Gli code: an information nexus regulatingcell fate, stemness and cancer. Trends Cell Biol. 2007;17:438–47. doi: 10.1016/j.tcb.2007.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rao SK, Edwards J, Joshi AD, Siu IM, Riggins GJ. A survey of glioblastoma genomic amplifications and deletions. J Neurooncol. 2010;96:169–79. doi: 10.1007/s11060-009-9959-4. [DOI] [PubMed] [Google Scholar]
- 14.Gordon AT, Brinkschmidt C, Anderson J, Coleman N, Dockhorn-Dworniczak B, Pritchard-Jones K, Shipley J. A novel and consistent amplicon at 13q31 associated with alveolar rhabdomyosarcoma. Genes Chrom Cancer. 2000;28:220–26. [PubMed] [Google Scholar]
- 15.Pavletich NP, Pabo CO. Crystal structure of a five-finger GLI-DNA complex: new perspectives on zinc fingers. Science. 1993;261:1701–7. doi: 10.1126/science.8378770. [DOI] [PubMed] [Google Scholar]