Naturalists have long known that copulation can be dangerous to females. For example, female waterfowl sometimes drown while being mated by multiple males (1). Beyond this type of anecdotal information, detailed quantification of the cost of mating in wild populations of waterstrides has demonstrated that: (i) males attempt to copulate with females at a rate that far exceeds what is needed to fertilize her eggs, (ii) males increase their fitness by mating at this high rate, and (iii) female fitness is reduced by supernumerary copulations (2, 3).
The first evidence for the cost of mating in a laboratory model system came in the mid-1970s during a study that was designed to measure at the relationship between temperature stress during development and the vigor of adults (4). This study of Drosophila melanogaster demonstrated that copulation harmed females by reducing their longevity, that females were harmed more when they had been reared under thermal stress, and that harm to females was reduced when their mates had been reared under thermal stress. A recent succession of experiments now extends this early research, especially those from the laboratory of Linda Partridge and her collaborators. The featured paper in this issue of PNAS by Civetta and Clarke (5) is an important advance in our understanding of the proximate and ultimate factors responsible for the phenomenon of male-induced harm to their mates. More specifically, Civetta and Clarke present data that explain what would otherwise be an evolutionary paradox: how a male's seminal fluid can both increase harm to the male's mate as well as increase that male's evolutionary fitness.
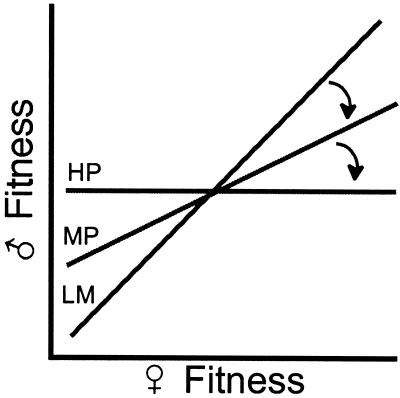
The idea that natural selection would favor a male that harmed his mate is counterintuitive. After all, reducing the survival or fecundity of a male's mate would seem to diminish rather than enhance his own reproductive fitness. To see why natural selection can sometimes favor males that harm their mates we need to consider the mating system of the species. If there is lifelong monogamy and random mating, then there is a perfect correlation between the fitness of a male and his mate and the potential for evolutionary conflict between the sexes is absent. But any deviation from this monogamous mating system reduces the correlation for lifetime fitness between a copulating pair, and this generates the potential for intersexual evolutionary conflict and the subsequent evolution of male-induced harm to their mates (Fig. 1).
Figure 1.
The progression from lifelong monogamy (LM) to moderate levels of promiscuity (MP) to high levels of promiscuity (HP) reduces the correlation for fitness between mated pairs.
Consider a promiscuous mating system where both males and females mate with many partners over their lifetime. Further assume, as is common in many species, that males do not provide material resources for their offspring, that mating pairs remain together only a short time during the act of copulation, and that females store sperm for a protracted period of time. In this case the lifetime fitness of a male is determined by the number of females that he mates and the average number of offspring that he sires per female. When a female mates with another male before the sperm from her previous mate is exhausted, then the first male's total offspring through that female will decline. So males are selected to suppress remating by their mates (to other males) and to maximize their mates' proximate fecundity, because their paternity rates at later times will be diluted each time one of their mates remates with another male. Females, on the other hand, will be selected to maximize their lifetime, as opposed to their short-term fecundity, and also will be selected to remate with other males at the rate that maximizes their fitness—as opposed to the zero remating rate favored by their previous mate(s).
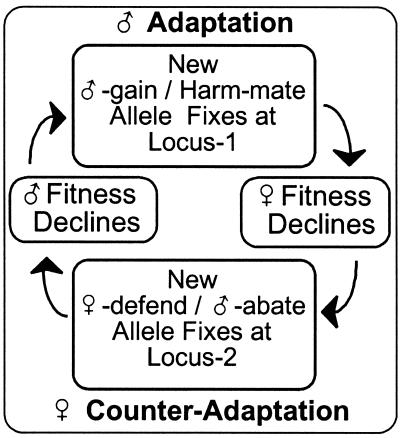
These intersexual differences in the optimal fecundity and remating rates set the stage for perpetual antagonistic coevolution between the sexes (Fig. 2). For example, suppose that at one locus (Locus-1) a mutation occurs that increases male fitness at the expense of his mate. When the mutation is male-limited in expression, or when selection on the mutation is not counterbalancing when expressed in females, then the mutation's advantage to males will cause the male-gain/mate-harm allele to fix in the population (male adaptation) despite the harm to his mates. Females are expected to evolutionarily respond, generally at another locus (Locus-2), when new mutations occur that reduce the male-induced harm to females. When the new mutation is female-limited in expression, or when selection on the new mutation is not counterbalancing when expressed in males, then the advantage to females will cause this female-defend/male-abate allele to fix in the population (female counteradaptation). Males can next recruit a new countercounter-adaptation and the cycle of intersexual antagonistic coevolution continues (6, 7). When females invest more in offspring compared with males, then males are expected to initiate most cycles of antagonistic coevolution. Although the cycle of antagonistic coevolution is framed in the context of males vs. females, the true genetic units that are antagonistically coevolving are different gene loci that mediate antagonistic interactions between different individuals, i.e., interlocus contest evolution (6, 7).
Figure 2.
Intersexual antagonistic coevolution between interacting alleles at different loci.
The simple evolutionary arguments described above suggest that males can evolve traits that harm their mates whenever the correlation for fitness between a copulating pair is less than one (6–8). Civetta and Clark (5) provide the first example of genetic polymorphism, probably at one or more loci coding for seminal fluid proteins, that produces a male-gain/mate-harm phenotype—genotypes that produce seminal fluid that increase harm to a male's mate also increase his fitness through that female. With lifelong monogamy a seminal fluid protein that reduces female survival would not be selectively favored because of the strong correlation between male and female fitness within a mated pair (Fig. 1). But in the Drosophila promiscuous mating system studied by Civetta and Clarke, most of the female's fitness loss will occur after she has remated with other males, and therefore most of the loss in the female's fitness will not reduce the target male's fitness. In this case, the lowered correlation for fitness between mates will cause the female-harm trait to be selectively favored whenever it caused the male to sire, on average, more offspring per mate—despite the cost to females. Such harm to females will be transient, however, whenever females can evolutionarily respond via counteradaptation (Fig. 2), in this case by changing the female receptor(s) that is targeted by the male's seminal fluid protein(s).
How strong is the evidence that males commonly harm their mates and thereby initiate cycles of intersexual antagonistic coevolution? Most of the quantitative information comes from studies of Drosophila melanogaster reared under laboratory conditions. In the late 1980s, a comprehensive study of the effect of mating on female fitness demonstrated that males harm females by reducing their survivorship when the two sexes are continuously housed together (9). This experiment and others (10, 11) showed that reduced survival of females was mediated by (i) a male behavioral component caused by persistent male courtship, including repeated attempts by males to mate with previously mated females, and (ii) an additional seminal fluid component that did not depend on the presence of sperm. In a particularly elegant experiment that used “genetic surgery,” the harm caused by seminal fluid was shown to be mediated by seminal fluid proteins that are produced by the mast cells within the male's reproductive tract (12). Some of the seminal fluid proteins remain within the female's reproductive tract while others enter her bloodstream and act as pheromones that suppress her propensity to remate and also boosts her fecundity (13, 14). Seminal fluid proteins potentially harm females, (i) as a byproduct of their toxic effect on resident sperm that are present in the female's sperm storage organs at the time of a second mating (e.g., one seminal fluid protein has high sequence homology with a gene coding for a spider toxin protein; ref. 13), and (ii) as a consequence of their pheromonal influence on her endocrine system.
Two microevolutionary experiments also support the conclusion that the sexes are locked in an evolutionary arms race. In the first experiment, cytogenetic constructs were used to create populations in which males could adapt to females but the females could not counterevolve (15). After only 30 generations the males rapidly adapted to the nonevolving females and increased their fitness by 25%. This evolutionary gain by the males was associated with a marked reduction in the survival of their mates. Female survival declined in response to the experimental males evolving to remate females at an elevated rate (so females received more seminal fluid, which is mildly toxic), and in one of the replicates, because of an increased toxicity of the males' seminal fluid.
In the second experiment, monogamy was experimentally enforced in populations of D. melanogaster that were normally promiscuous (16). In this context, intersexual antagonistic coevolution should be reversed and replaced by mutualistic coevolution between the sexes. As predicted, in the monogamy treatments male behavior and seminal fluid proteins evolved to enhance rather than reduce female survival, relative to promiscuous controls, and fitness of monogamous males declined in parallel, when they were placed back in their promiscuous ancestral population. Interestingly, females from the monogamy treatment evolved to be less resistant to male-induced harm when they were placed back in their promiscuous ancestral population.
Molecular studies of Drosophila seminal fluid proteins have demonstrated that they frequently diverge rapidly among closely related species and that they tend to be highly polymorphic (13, 17–19). It also has been shown that proteins of the male and female reproductive tracts evolve far more rapidly than proteins from other parts of the body (20, 21). Experimental studies have shown that seminal fluid proteins play a role in both the offense and defense phenotypes of a male (22–24). Offense is the capacity of a male to stimulate a female to mate with him (behavior component), and then to replace any resident sperm with his own (seminal fluid/sperm component). Defense appears to be mediated entirely by seminal fluid and sperm. It is the ability of a male to reduce his mate's propensity to remate with another male, and when she does, to prevent his sperm from being displaced by the second male.
In the mid-1990s, an experiment was done to look for associations between polymorphic seminal fluid proteins and a male's offense and defense phenotype (25). A survey of samples of seven polymorphic loci for seminal fluid proteins found no association with a male's offense phenotype, but four of the seven polymorphisms were associated with a male's defense phenotype (25). This does not mean that there is no polymorphic variation for male offense. But the fact that none of the sampled seven loci contributed to this phenotype, whereas more than half of the seven loci contributed to male defense, suggests that seminal fluid proteins may provide more heritable variation for male defense compared with offense. Correspondingly, Civetta and Clark (5) found no significant association (among lines isogenic for a major autosome) between a male's offense phenotype and the degree to which he harmed his mates, but a strong association was observed between harm to females and his defense phenotype.
Recent theory and experiments suggest that intersexual antagonistic coevolution may contribute importantly to speciation (7, 15, 26–28). Consider the traits that are expected to perpetually coevolve because of intersexual conflict: reproductive anatomy, physiology, and behavior. As these traits diverge among isolated populations, their inherent attributes will cause them to collaterally contribute to both pre- and post-zygotic reproductive isolation, and thereby accelerate the speciation process. This idea was recently tested (29) by comparing the rates of speciation among clades that are monandrous (females are monogamous, so intersexual conflict is low) vs. polyandrous (females have multiple mates, leading to higher intersexual conflict). Polyandrous clades were found to speciate four times faster, on average, supporting the conclusion that intersexual conflict is a major “engine of speciation.”
The experiments by Civetta and Clark (5) presented in this issue of PNAS make an important advance in our understanding of intersexual antagonistic coevolution by demonstrating that the same male genotypes that reduce female survival more also gain a greater fitness advantage (increased defense)—so a direct link between male-gain and harm to his mate is clearly established. Extending this experiment to the case of heterozygous chromosomes will determine how much of the genetic variation uncovered by Civetta and Clark is because of inbreeding depression vs. the additive genetic variation that can contribute to intersexual coevolution in an outbred population. Understanding the molecular mechanisms that underlie this male-gain/mate-harm intersexual interaction is the next challenge in understanding the perpetual “battle of the sexes.”
Footnotes
See companion article on page 13162.
References
- 1.McKinney F, Derrickson S R, Mineau P. Behaviour. 1983;86:250–294. [Google Scholar]
- 2.Arnqvist G, Danielsson I. Evolution. 1999;53:147–156. doi: 10.1111/j.1558-5646.1999.tb05340.x. [DOI] [PubMed] [Google Scholar]
- 3.Rowe L, Arnqvist G, Sih A, Krupa J. Trends Ecol Evol. 1994;9:289–293. doi: 10.1016/0169-5347(94)90032-9. [DOI] [PubMed] [Google Scholar]
- 4.Cohet Y A, David J R. Experientia. 1976;32:696–697. doi: 10.1007/BF01919838. [DOI] [PubMed] [Google Scholar]
- 5.Civetta A, Clark A G. Proc Natl Acad Sci USA. 2000;97:13162–13165. doi: 10.1073/pnas.230305397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rice W R. Proc Natl Acad Sci USA. 1998;95:6217–6221. doi: 10.1073/pnas.95.11.6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rice W R, Holland B. Behav Ecol Sociobiol. 1997;41:1–10. [Google Scholar]
- 8.Johnston R A, Laurent K. Am Nat. 2000;156:368–377. doi: 10.1086/303392. [DOI] [PubMed] [Google Scholar]
- 9.Fowler K, Partridge L. Nature (London) 1989;338:760–761. [Google Scholar]
- 10.Chapman T, Hutchings J, Partridge L. Proc R Soc London Ser B. 1993;253:211–217. doi: 10.1098/rspb.1993.0105. [DOI] [PubMed] [Google Scholar]
- 11.Chapman T. J Insect Physiol. 1992;38:223–227. [Google Scholar]
- 12.Chapman T, Liddle L F, Kalb J M, Wolfner M F, Partridge L. Nature (London) 1995;373:241–244. doi: 10.1038/373241a0. [DOI] [PubMed] [Google Scholar]
- 13.Wolfner M F. Insect Biochem Mol Biol. 1997;27:179–192. doi: 10.1016/s0965-1748(96)00084-7. [DOI] [PubMed] [Google Scholar]
- 14.Chen P S. Experientia. 1996;52:503–510. doi: 10.1007/BF01969718. [DOI] [PubMed] [Google Scholar]
- 15.Rice W R. Nature (London) 1996;381:232–234. doi: 10.1038/381232a0. [DOI] [PubMed] [Google Scholar]
- 16.Holland B, Rice W R. Proc Natl Acad Sci USA. 1999;96:5083–5088. doi: 10.1073/pnas.96.9.5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cirera S, Aguade M. Genetics. 1997;147:189–197. doi: 10.1093/genetics/147.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aguade M. Genetics. 1998;150:1079–1089. doi: 10.1093/genetics/150.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsaur S C, Ting C T, Wu C-I. Mol Biol Evol. 1998;15:1040–1046. doi: 10.1093/oxfordjournals.molbev.a026002. [DOI] [PubMed] [Google Scholar]
- 20.Civetta A, Singh R S. J Mol Evol. 1995;41:1085–1096. doi: 10.1007/BF00173190. [DOI] [PubMed] [Google Scholar]
- 21.Civetta A, Singh R S. Mol Biol Evol. 1998;15:901–909. doi: 10.1093/oxfordjournals.molbev.a025994. [DOI] [PubMed] [Google Scholar]
- 22.Harshman L G, Prout T. Evolution. 1994;48:758–766. doi: 10.1111/j.1558-5646.1994.tb01359.x. [DOI] [PubMed] [Google Scholar]
- 23.Price C S C, Dyer K A, Coyne J A. Nature (London) 1999;400:449–452. doi: 10.1038/22755. [DOI] [PubMed] [Google Scholar]
- 24.Chapman T, Neubaum D M, Wolfner M F, Partridge L. Proc R Soc London Ser B. 2000;267:1097–1105. doi: 10.1098/rspb.2000.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clark A G, Aguade M, Prout T, Harshman L G, Langley C H. Genetics. 1995;139:189–201. doi: 10.1093/genetics/139.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rice W R. In: Endless Forms: Species and Speciation. Howard D J, Berlocher S H, editors. New York: Oxford Univ. Press; 1998. pp. 261–270. [Google Scholar]
- 27.Parker G A, Partridge L. Philos Trans R Soc London B. 1998;353:261–274. doi: 10.1098/rstb.1998.0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Howard D J, Reece M, Gregory P G, Chu J, Cain M L. In: Endless Forms: Species and Speciation. Howard D J, Berlocher S H, editors. New York: Oxford Univ. Press; 1998. pp. 279–288. [Google Scholar]
- 29.Arnqvist G, Edvardsson M, Friberg U, Nilsson T. Proc Natl Acad Sci USA. 2000;98:10460–10464. doi: 10.1073/pnas.97.19.10460. [DOI] [PMC free article] [PubMed] [Google Scholar]