Abstract
Grape seed proanthocyanidins (GSPs) have been shown to have anti-skin carcinogenic effects in in vitro and in vivo models. However, the precise epigenetic molecular mechanisms remain unexplored. This study was designed to investigate whether GSPs reactivate silenced tumor suppressor genes following epigenetic modifications in skin cancer cells. For this purpose, A431 and SCC13 human squamous cell carcinoma cell lines were used as in vitro models. The effects of GSPs on DNA methylation, histone modifications and tumor suppressor gene expressions were studied in these cell lines using enzyme activity assays, western blotting, dot-blot analysis and real-time polymerase chain reaction (RT-PCR). We found that treatment of A431 and SCC13 cells with GSPs decreased the levels of: (i) global DNA methylation, (ii) 5-methylcytosine, (iii) DNA methyltransferase (DNMT) activity and (iv) messenger RNA (mRNA) and protein levels of DNMT1, DNMT3a and DNMT3b in these cells. Similar effects were noted when these cancer cells were treated identically with 5-aza-2′-deoxycytidine, an inhibitor of DNA methylation. GSPs decreased histone deacetylase activity, increased levels of acetylated lysine 9 and 14 on histone H3 (H3-Lys 9 and 14) and acetylated lysine 5, 12 and 16 on histone H4, and reduced the levels of methylated H3-Lys 9. Further, GSPs treatment resulted in re-expression of the mRNA and proteins of silenced tumor suppressor genes, RASSF1A, p16INK4a and Cip1/p21. Together, this study provides new insight into the epigenetic mechanisms of GSPs and may have significant implications for epigenetic therapy in the treatment/prevention of skin cancers in humans.
Keywords: Grape seed proanthocyanidins, DNA methylation, skin cancer cells, histone acetylation, tumor suppressor genes and proteins
Introduction
Skin cancer is the manifestation of a series of genetic and epigenetic events. Epigenetic processes result in heritable phenotypical changes in gene expression that do not involve alterations in the actual DNA sequence. Instead, epigenetic changes involve variations in DNA methylation, chromatin structure or microRNA profiles that then modify gene expression (Jones and Baylin, 2002). The hallmarks of epigenetic gene regulation are DNA methylation and histone modifications. Because epigenetic changes are reversible and can be manipulated pharmacologically, this area has become a focus of particular interest for the development of therapeutic agents to treat cancer, including cutaneous carcinogenesis. Epigenetic alterations in various genes play crucial roles in the development of cancers (Jones and Baylin, 2002). Epigenetic inactivation of genes by promoter hypermethylation has been recognized as an important mechanism by which tumor suppressor genes are shut down during development of cancers. Hypermethylation of CpG islands in the promoter region leads to silencing either by direct inhibition of transcription factor binding, or by attracting methylated-DNA binding proteins, recruiting other transcriptional repressors such as histone deacetylases (HDACs) and histone methyl transferases (Jones and Baylin, 2002; Hegi et al., 2009). DNA methylation at the 5′ position of cytosine is the most well characterized epigenetic mechanism which can be inherited without changing the DNA sequence (Jones and Baylin, 2002; Hegi et al., 2009). The mammalian DNA methylation machinery is made up of two components: DNA methyltransferases (DNMTs), which establish and maintain genome-wide DNA methylation patterns, and the methyl-CpG-binding proteins, which are involved in “reading” and interpreting the methylation patterns (Jones, 2002; Antequera and Bird, 1993; Howell Jr et al., 2009). DNA hypermethylation is one of the major epigenetic mechanisms in the silencing of expression of tumor suppressor genes (Jones, 2002; Antequera and Bird, 1993; Herman and Baylin, 2003). DNA methylation is commonly associated with increased levels of functionally aberrant DNMTs, which then initiate the methylation of cytosine at the 5′ position in CpG dinucleotides (Jones and Baylin, 2002; Counts and Goodman, 1995). In addition to DNA methylation, histone modifications, particularly methylation and acetylation, are involved in transcriptional silencing of a number of genes in cancers. As the epigenetic mechanisms are reversible in nature, some dietary phytochemicals, which possess anti-carcinogenic properties, have been assessed for their effect of epigenetic processes (Davis and Milner, 2004; Davis and Uthus, 2004; Huang, 2002).
Bioactive phytochemicals, particularly dietary, offer promising options for the development of more effective strategies for the prevention or treatment of cancers and they can be utilized as complementary and alternative medicine. While phytochemicals have been used for thousands of years in various cultures/civilizations for the treatment of many diseases, wound healing or to preserve skin beauty, their active ingredients and mechanisms of action are not well characterized. Grape seed proanthocyanidins (GSPs) are promising bioactive molecules that have demonstrated anti-carcinogenic effects in some animal tumor models and exhibit no apparent toxicity in vivo (Nandakumat et al., 2008; Mittal et al., 2003; Meeran et al., 2009). As previously described, this product contains primarily proanthocyanidins (89%), which constitute dimers, trimers, tetramers, and oligomers of monomeric catechins and/or (−)-epicatechins (Mittal et al., 2003). It is likely that at least some of the components present in the GSPs act synergistically and thus this product may be more effective than any single component. GSPs have been shown to inhibit ultraviolet radiation-induced skin tumors in SKH-1 hairless mice (Mittal et al., 2003) as well as in 2,4-dimethylbenz(a)anthracene-initiated and 12-O-tetradecanoylphorbol-13-acetate-promoted skin tumors in C3H/HeN mouse model (Meeran et al., 2009). However, an epigenetic basis for their chemopreventive effects remains unexplored.
In this study, we investigated whether GSPs would reactivate silenced tumor suppressor genes and determined the molecular mechanism underlying these effects using two well known human skin cancer cell lines, A431 and SCC13, as an in vitro model. Our study demonstrates that treatment of skin cancer cells with GSPs results in decreased levels of DNA methylation, inhibited histone deacetylase (HDAC) activity and increased levels of acetylated histones in cancer cells. The ultimate result of these effects was re-expression of tumor suppressor genes (RASSF1A, p16INK4a and Cip1/p21). Interestingly, the epigenetic effects of GSPs were non-significant in normal human epidermal keratinocytes (NHEK).
Material and methods
Chemicals and antibodies
GSPs were obtained commercially from Kikkoman Corporation (Noda, Japan). The quality of this product is maintained by the vendor on lot-to-lot basis. GSPs contain about 89% proanthocyanidins with dimers (6.6%), trimers (5.0%), tetramers (2.9%) and oligomers (74.8%), as reported previously (Mittal et al., 2003). The Methylamp™ Global DNA Methylation Quantification Kit and the EpiQuik DNA Methyltransferase Activity Assay Kit were purchased from Epigentek, Inc. (New York, NY). Standardized real-time PCR primers for DNMT1, DNMT3a, DNMT3b, RASSF1A, p16INK4a and Cip1/p21 were obtained from SuperArray Biosciences (Fredrick, MD). Antibodies were procured as follows: 5-methylcytosine (5-mC) from Calbiochem (New Jersey, NJ), DNMT1, DNMT3a and DNMT3b from Imgenex Corporation (San Diego, CA), HDAC3 and HDAC4 from Cell Signaling Technology Inc. (Danvers, MA), RASSF1A, acetyl histone H4 and H3 related antibodies were from Upstate Biotechnology and Abcam (Cambridge, MA), and p16INK4a and Cip1/p21 from Santa Cruz Biotechnology (Santa Cruz, CA).
Skin cancer cell lines and cell culture
A431 and SCC 13 cells were obtained from the American Type Culture Collection (Manassas, VA). The cells were cultured as monolayers in 100 mm tissue culture dishes in DMEM supplemented with 10% heat-inactivated fetal bovine serum (Hyclone, Logan, UT) and 100 μg/ml penicillin-streptomycin (Invitrogen). They were maintained in an incubator with a humidified atmosphere of 95% air and 5% CO2 at 37°C. The NHEK were obtained from Cell Culture Core Facility of Skin Diseases Research Centre at the University of Alabama at Birmingham, AL. The NHEK were cultured in keratinocyte growth medium supplemented with 5ng/ml human recombinant epidermal growth factor and 0.05 mg/ml bovine pituitary extract (Gibco/Invitrogen, Carlsbad, CA) and maintained in an incubator under the conditions as described above. Cells were seeded at a density of 1 × 106 cells per petri dish and allowed to attach for 24 h before treatment with testing agents for either 3 or 5 days. Media and treatment agents were refreshed every 3 days. The sub-confluent cells (60–70% confluent) were treated with either varying concentrations of GSPs (0, 5, 10, 15 and 20 μg/ml) or 5-aza-2′-deoxycytidine (5-aza-dc) or trichostatin (TSA) after dissolving in DMSO. The cells treated only with vehicle (DMSO) served as a control [maximum concentration of DMSO, 0.1% (v/v) in media].
Assay for Global DNA methylation
For the analysis of global DNA methylation levels, the total genomic DNA was extracted from the cells which were treated with GSPs or 5-aza-dc using the DNeasy Kit (Qiagen Sciences, MD) following the manufacturer’s protocol. The Global DNA methylation levels were determined using the Methylamp™ Global DNA Methylation Quantification Kit following the manufacturer’s instructions. The methylated fraction of DNA is recognized by a 5-mC antibody. With this colorimetric assay kit, the amount of methylated DNA, which is proportional to the optical density, is quantified through an enzyme-linked immunosorbent assay (ELISA)-like reaction. This analysis provides the levels of global DNA methylation and is not specific to any particular gene.
Assay for DNMT activity
Cells were treated with various concentrations of GSPs or 5-aza-dc for 3 or 5 days. Thereafter, cells were harvested and nuclear extracts were prepared using Epiquik Nuclear Extraction Kit (Epigentek Inc., New York, NY) following the manufacturer’s instructions. DNMT activity was determined in nuclear extracts using Epiquik DNA Methyltransferase Activity Assay Kit (Epigentek Inc., New York, NY) following manufacturer’s protocol.
Analysis of 5-mC in DNA following dot-blot assay
Cells were treated with GSPs (0, 5, 10 and 20 μg/ml) or 5-aza-dc for 5 days. Genomic DNA was isolated using the DNA Isolation Kit (Qiagen Sciences, MD), and dot-blot analysis was performed as detailed previously (Katiyar et al., 2010). Briefly, genomic DNA (1 μg) was denatured and then blotted onto Hybond-ECL nitrocellulose membranes using Bio-Dot Microfiltration Apparatus (Bio-Rad Laboratories, Inc. Hercules, CA). This was fixed by baking the membrane for 30 min at 80°C. The membrane was incubated with an antibody specific to 5-mC (1:500, v/v) followed by incubation with a horseradish peroxidase-conjugated secondary antibody. The membrane was then treated with enhanced chemiluminescence detection reagents and exposed to Kodak autoradiograph films. Equal DNA loading was verified by staining the membranes with 0.2% methylene blue. The intensity of each dot was measured by densitometry and normalized to total DNA.
Assay for HDAC activity
The effect of GSPs on HDAC activity in skin cancer cells was measured using the HDAC Colorimetric Activity Assay Kit (Active Motif Inc., Carlsbad, CA) following the manufacturer’s protocol. Briefly, the cells were treated with GSPs or TSA for 3 or 5 days. Cells were harvested and nuclear protein fractions were isolated using Epiquik Nuclear Extraction Kit (Epigentek Inc., New York, NY). Forty micrograms of nuclear proteins from each treatment group were incubated with a colorimetric HDAC substrate for 3 h after which the reaction was developed with assay development buffer supplied with the kit. Nuclear protein extracts treated with TSA (100 nM), an inhibitor of HDAC activity, served as a positive control.
Assay for histone acetyltransferase (HAT) activity
The effect of GSPs on HAT activity in cancer cells was measured using EpiQuik™ HAT Activity Assay Kit from Epigentek Group Inc. (Brooklyn NY, www.Epigenetek.com). This kit is designed to measure total HAT activity. In this assay, the unique histone substrate is stably captured in the strip wells. Active HATs present in nuclear lysates bind and acetylate the substrate. The acetylated substrate is then recognized by a high affinity anti-acetylated histone antibody provided with the kit. The amount of acetylated histone was then colorimetrically quantified through an ELISA-like reaction following the manufacturer’s protocol. The color absorbance was read on a microplate reader at 450 nm.
mRNA analysis of DNMTs and tumor suppressor genes using real-time PCR
For the mRNA analysis, the total RNA was first extracted from the cells of different treatment groups using Trizol Reagents (Invitrogen, Carlsband, CA). Then, cDNA was synthesized using the reverse transcription reaction (iScript cDNA Synthesis Kit, Bio-Rad, Hercules, CA). The cDNA was amplified using real-time PCR with a Bio-Rad MyiQ thermocycler and SYBR Green detection system (Bio-Rad, Hercules, CA). Manufacturer-supplied standardized primer pairs were then used to measure the following: DNMT1, DNMT3a, DNMT3b, RASSF1A, p16INK4a and Cip1/p21. The standard PCR conditions used were: 95°C for 15 min and then 40 cycles at 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s, as recommended by the supplier (SuperArray Bioscience Corporation, Frederick, MD). The mRNA expression levels of genes were normalized to the sample expression level of the GAPDH, and relative to the average of all delta Ct-values in each using the cycle threshold (Ct) method. All samples were run in duplicate to ensure amplification integrity.
Analysis of protein levels using western blotting
For this purpose, cell lysates from different treatment groups were prepared as previously described (Sharma et al., 2010; Singh et al., 2011). Proteins (30–50 μg) were electrophoresed on 10% Tris-glycine gels (Invitrogen, Carlsband, CA) and then transferred onto nitrocellulose membranes. After blocking the membrane in freshly prepared phosphate-buffered saline (PBS)-containing 3% non-fat dry milk, the membranes were incubated with desired antibodies at 4°C overnight followed by incubation with an appropriate peroxidase-conjugated secondary antibody. Specific proteins were visualized on X-ray film using an enhanced chemiluminescence reagent system (Amersham Life Science, Inc.). The equal loading of proteins of DNMT1, DNMT3a, DNMT3b, Cip1/p21, RASSF1A and p16INK4a was verified using anti-β actin antibody, while in case of histones, anti-histone H3 antibody was used. Representative blots are shown from 3 independent experiments.
Statistical analysis
The statistical analysis of the data on global DNA methylation levels, DNMT activity and HDAC activity are expressed as percentages with the basal levels in skin cancer cells taken as 100%. The results of the real-time PCR are expressed as the means ± standard deviation. Student’s t-test was used to determine the statistical difference between treatment groups. The data were considered significant if P<0.05.
Results
Analysis of global DNA methylation level in human cutaneous squamous cell carcinoma cells and comparison with normal human epidermal keratinocytes
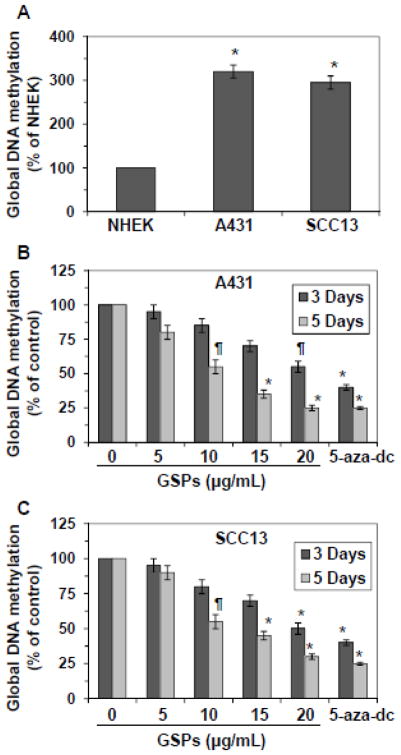
To compare if there is any difference in global DNA methylation level between skin cancer cells and normal skin cells (NHEK), A431 and SCC13 cells were subjected to the analysis of global DNA methylation level and data were compared with the global DNA methylation level in NHEK. As shown in Figure 1A, the levels of global DNA methylation were nearly 3-fold higher in A431 and SCC13 cells (P<0.001) as compared to the NHEK.
Figure 1.
Effect of GSPs and 5-aza-dc on global DNA methylation levels in human squamous cell carcinoma A431 and SCC13 cells. (A) Comparative levels of global DNA methylation in NHEK, A431 and SCC13 cells. Data are presented in terms of percent of NHEK, which were used as normal control cells. Significantly higher level of global DNA methylation level versus NHEK, *P<0.001. (B & C) Effect of GSPs on the global DNA methylation levels in A431 and SCC13 cells. A431 and SCC13 cells were treated with various concentrations of GSPs (0, 5, 10, 15 and 20 μg/ml) for 3 and 5 days and the levels of global DNA methylation were determined using Global DNA Methylation kit. 5-aza-dc is a well known inhibitor of DNA methylation and was used as a positive control. Data are presented in terms of percent of control (non-GSPs-treated) group, which was assigned a value of 100%, and as means ± SD, n=3. Significant difference versus non-GSPs-treated controls, *P<0.001, **P<0.01.
GSPs and 5-aza-dc (a demethylating agent) reduce the levels of global DNA methylation in skin cancer cells
To examine whether GSPs have epigenetic effects on DNA methylation levels in skin cancer cells, A431 and SCC13 cells were treated with various concentrations of GSPs (0, 5, 10, 15 and 20 μg/ml) for 3 days and 5 days. At the termination of the experiments, cells were harvested and DNA was isolated and subjected to the analysis of global DNA methylation levels using Global DNA Methylation Kit. Data were presented in terms of percent of control (non-GSPs-treated). As shown in Figure 1B, treatment of A431 cells with GSPs for 3 days resulted in reduction (5–45%) in the levels of DNA methylation in a concentration-dependent manner. However, the levels of global DNA methylation in A431 cells showed greater decrease (20–75%, P<0.01 and P<0.001) after 5 days of treatment with GSPs than the treatment of cells with GSPs for 3 days. Similar to A431 cells, treatment of SCC13 cells with GSPs for 3 days resulted in 5–50% reduction in a dose-dependent manner. However, the inhibitory effect of GSPs on global DNA methylation was greater (10–70%) when the cells were treated for 5 days, as shown in Figure 1C. Treatment of A431 cells or SCC13 cells with 5-aza-dc for 3 and 5 days also significantly reduced the levels of global DNA methylation in these cells. The inhibitory effect of 5-aza-dc on global DNA methylation in A431 cells after the treatment of cells for 3 and 5 days varied from 60–75% (P<0.001). Similar inhibitory effect of 5-aza-dc on global DNA methylation was observed in SCC13 cells under identical conditions. The inhibitory effect of GSPs and 5-aza-dc on DNA methylation after the treatment of cells for 24 h was not significantly lower than non-GSPs- or non-5-aza-dc-treated controls.
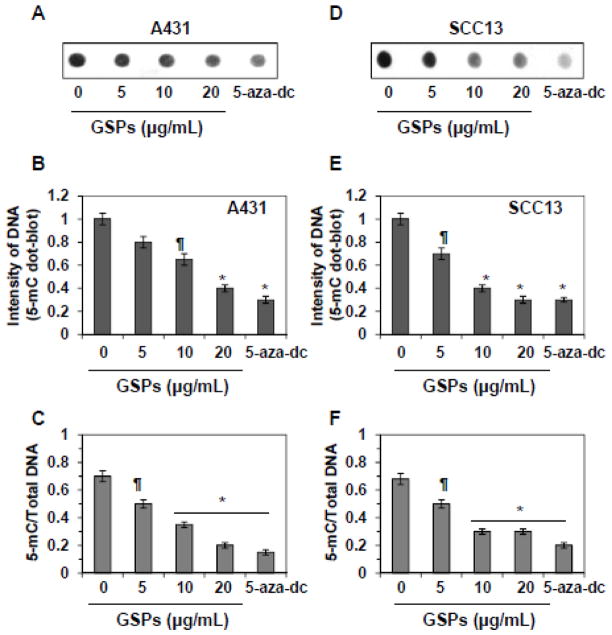
GSPs and 5-aza-dc reduce the levels of 5-mC in DNA of skin cancer cells
To examine the effect of GSPs on DNA methylation, A431 and SCC13 cells were treated with GSPs for 5 days, and the cells were then harvested. Cells were also treated with 5-aza-dc for 5 days, which served as a positive control. Total genomic DNA was isolated from cancer cells and analyzed by dot-blot assay using an antibody specific to 5-mC. As shown in Figure 2A, treatment of A431 cells with GSPs or 5-aza-dc decreased methylation of total DNA. This is evident from the reduced intensities of the dot-blots. The intensity of individual dot-blot was measured by densitometry, as shown in Figure 2B. These results indicate that treatment of A431 cells with varying concentrations of GSPs for 5 days decreased the intensities of dot-blots of 5-mC by 20–60% compared with non-GSPs-treated cells. Identical effect of GSPs was also found when SCC13 cells were treated with GSPs (30–70% decrease), as shown in Figure 2E. The levels of 5-mC were normalized by total DNA levels (Figure 2C). The analysis of these data indicates a significant decrease (28–71%, P<0.05 and P<0.001) in DNA methylation in terms of total amount of DNA by the treatment of GSPs in A431 cells. Identical experiments were conducted with SCC13 cells. As shown in Figures 2D, 2E and 2F, treatment of SCC13 cells with GSPs or 5-aza-dc significantly decreased the levels of 5-mC compared with non-GSPs-treated control group.
Figure 2.
Treatment of A431 and SCC13 cells with GSPs for 5 days decreases the levels of 5-mC dose-dependently. 5-aza-dc, an inhibitor of 5-mC, was used as a positive control. (A & D) Dot-blot analysis of 5-mC in DNA extracted from various groups treated with or without GSPs. (B & E) Dot-blot data are presented in terms of relative density of dot-blots as means ± SD in different treatment groups, n=3. (C & F) Effect of GSPs and 5-aza-dc on genomic DNA methylation levels in A431 and SCC13 cells. DNA methylation at 5-cytosine was detected using an anti-5-mC antibody and samples loading was determined by staining total DNA with methylene blue. The intensity of individual dot was measured by densitometry and levels of 5-mC were normalized by total DNA. Data are mean ± SD from three separate experiments. Significant difference versus control (non-GSPs-treated group), *P<0.001, **P<0.05.
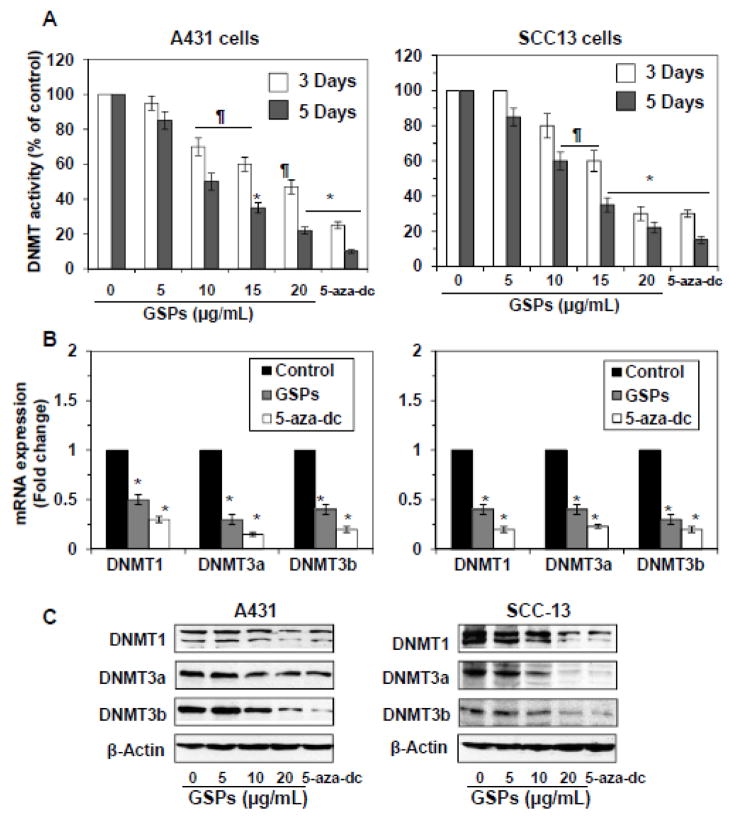
Both GSPs and 5-aza-dc decrease activity, the mRNA and protein expressions of DNMTs in skin cancer cells
DNMTs have been shown to play important roles in DNA methylation (Jones, 2002). Therefore, we determined the activity of DNMT in skin cancer cells after treatment of cells with GSPs for 3 days and 5 days. GSPs decreased DNMT activity greater in A431 (15–78%, P<0.05-P<0.001) and SCC13 (15–80%, P<0.05–P<0.001) cells after treatment of cells for 5 days compared to the treatment of cells for 3 days, and this effect of GSPs was dose-dependent, as shown in Figure 3A. Similar inhibitory effect on DNMT activity was also seen in the treatment of cells with 5-aza-dc (75–90%, P<0.001). These results were consistent with the quantitative analysis of the mRNA expression of the DNMTs using real-time PCR (Figure 3B). The mRNA levels of DNMT1, DNMT3a and DNMT3b were significantly reduced (P<0.05 to P<0.001) in both A431 (50–70%) and SCC13 (60–70%) cells after the treatment of cells with GSPs for 5 days. The decrease in mRNA expression of DNMTs was greater after treatment with GSPs for 5 days than treatment with GSPs for 3 days (data not shown). We also determined the effect of GSPs on the levels of DNMTs in skin cancer cells. Western blot analysis revealed that there was a decrease in the protein expression levels of DNMT1, DNMT3a and DNMT3b after the treatment of A431 and SCC13 cells with GSPs for 5 days in comparison to non-GSPs-treated controls. Treatment of A431 and SCC13 cells with 5-aza-dc for 5 days also decreased the protein expression levels of DNMT1, DNMT3a and DNMT3b, as shown in Figure 3C.
Figure 3.
Treatment of A431 or SCC13 cells with GSPs or 5-aza-dc decreases DNMT activity and reduces the levels of mRNA and protein expressions of DNMT1, DNMT3a and DNMT3b. (A) Total DNMT activity in nuclear extracts was determined using the DNA Methyltransferase Activity Assay Kit. Data are presented in terms of percentage versus non-GSPs-treated controls, which was assigned a value of 100% and as means ± SD from three independent experiments. (B) Quantitative real-time PCR analyses of mRNA expressions of DNMT1, DNMT3a and DNMT3b in A431 and SCC13 cells after treatment of cells with GSPs or 5-aza-dc for 5 days. The results are presented as the expression of the individual mRNA with normalization to β-actin, and as mean values ± SD from three independent experiments. Significant difference versus non-GSPs-treated controls, *P<0.001. (C) The levels of DNMT1, DNMT3a and DNMT3b in cell lysates of A431 and SCC13 cells were determined using western blot analysis after treating the cells with GSPs or 5-aza-dc for 5 days. Representative blots are shown from three independent experiments.
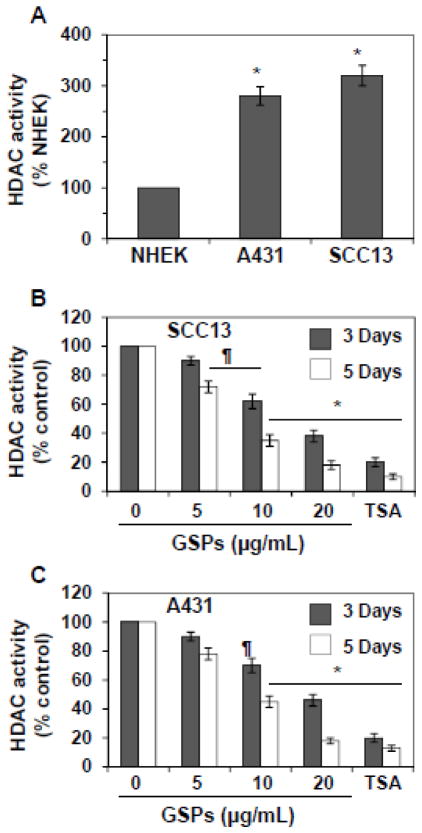
Analysis of HDAC activity in skin cancer cells and comparison with NHEK
Nuclear extracts from A431, SCC13 and NHEK were prepared and subjected to the analysis of HDAC activity, as detailed in Materials and methods. As shown in Figure 4A, the levels of HDAC activity were approximately three-fold higher in A431 and SCC13 cells compared to the NHEK. This data also indicates that the two skin cancer cell lines have identical levels of HDAC activity.
Figure 4.
Effect of GSPs and TSA on HDAC activity in A431 and SCC13 cells. (A) Comparative levels of HDAC activity in NHEK, A431 and SCC13 cells. Data are presented in terms of percent of NHEK, which were used as a normal control cells. Significant greater HDAC activity versus NHEK, *P<0.001. (B & C) SCC13 and A431 cells were treated with various concentrations of GSPs (0, 5, 10 and 20 μg/ml) for 3 or 5 days, and HDAC activity was assessed with proteins of nuclear extracts prepared from various treatment groups using HDAC Activity Assay Kit following the manufacturer’s instructions. Cells were also treated with TSA (0.2 μM) for 3 and 5 days and was used as a positive control. Data are presented as mean ± SD (n=3) and presented in terms of percentage versus non-GSPs-treated controls. Significant difference versus controls; **P<0.05, *P<0.001.
Effect of GSPs and TSA on HDAC activity in skin cancer cells
To examine the effect of GSPs and TSA on HDAC activity in skin cancer cells, SCC13 cells were treated with GSPs (5, 10 and 20 μg/ml) for 3 and 5 days. At the termination of the experiments, cells were harvested and HDAC activity was determined as detailed under Materials and methods. As shown in Figure 4B, treatment of SCC13 cells with GSPs for 3 days resulted in reduction of HDAC activity (10–62%). However, this reduction on HDAC activity by GSPs was comparatively greater (28–82%, P<0.01 to P<0.001) when cells were incubated with GSPs for 5 days. Similar to GSPs, TSA also decreased HDAC activity significantly (80–87%, P<0.001) in SCC13 cells, which served as a positive control for this experiment. Identical experiments were also conducted with A431 cell line and the effect of GSPs and TSA was determined on HDAC activity. As shown in Figure 4C, treatment of A431 cells with GSPs or TSA for 3 and 5 days significantly decreased (P<0.05, P<0.001) the HDAC activity. This effect of GSPs was dose-dependent.
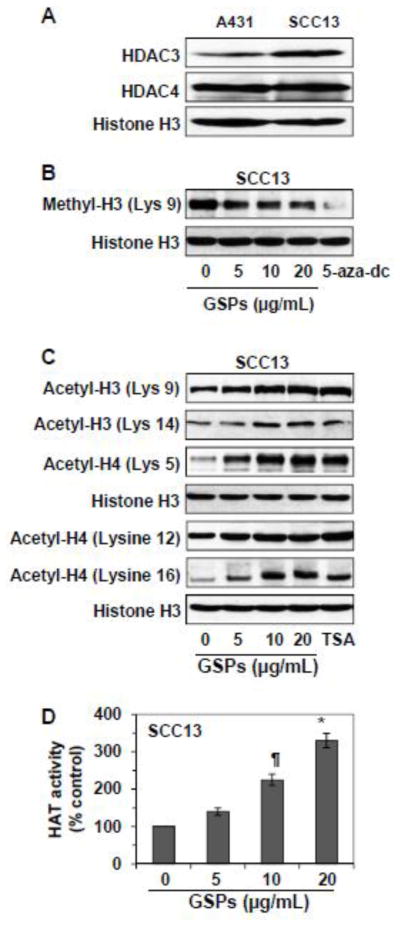
Effect of GSPs, 5-aza-dc and TSA on histone methylation and histone acetylation
Like DNA methylation, acetylation and/or methylation of histone proteins also play a crucial role in modification of chromatin structure, which can result in silencing of tumor suppressor genes. To determine whether GSPs also affect the histones, we determined the effect of GSPs on acetylation of histone H3 and H4. Before examining the effect of GSPs on histone modifications, the levels of HDAC3 and HDAC4 were analyzed and compared between A431 and SCC13 cells. The basal levels of HDAC3 and HDAC4 in both cell lines are shown with some difference in HDAC3 levels (Figure 5A). As the effects of GSPs on HDAC activity in both cell lines were identical, the effect of GSPs on histone modifications were only determined in SCC13 cells. Also, as the effect of GSPs on HDAC activity was greater after 5 days of treatment than for 3 days of treatment, we selected the time point of 5 days for further studies. For this purpose SCC13 cells were treated with GSPs, 5-aza-dc or TSA for 5 days. Cells were harvested and cell lysates were subjected to the analysis of histone methylation and histone acetylation using western blot analysis. It has been reported that deacetylation and methylation of H3-Lys 9 are associated with silencing of gene expression, whereas acetylation of H3-Lys 9 is involved in activation of gene expression (Katiyar, 2008). Therefore, we selected to analyze H3-Lys 9 to determine whether GSPs would alter its acetylation and methylation states. Western blot analysis revealed that treatment of SCC13 cells with GSPs decreased the levels of methylated H3-Lys 9 (Figure 5B), while increased the levels of acetylated H3-Lys 9 (Figure 5C) in a dose-dependent manner compared to non-GSPs-treated control. Treatment of 5-aza-dc also decreased levels of methylated H3-Lys 9 (Figure 5B). The effect of GSPs was also determined on acetylation status of H3 at Lys 14, and acetylation status of H4 on different lysine residues. We found that treatment with GSPs increased the levels of acetylated H3 at Lys 14, and acetylated H4 at Lys 5, Lys 12 and Lys 16. The effect of TSA on H3 and H4 acetylation on different Lys residues was similar to that of GSPs.
Figure 5.
Effect of GSPs or TSA on acetylation of histones in human skin cancer cells. (A) The basal levels of HDAC3 and HDAC4 were determined in A431 and SCC13 cells using western blot analysis. (B) The effect of GSPs and 5-aza-dc was determined on the levels of methyl H3 (Lys 9) in SCC13 cells. Cells were treated for 5 days with GSPs or 5-aza-dc, and cell lysates were prepared for western blot analysis. Cells treated with 5-aza-dc served as a positive control. (C) Cells were treated with GSPs or TSA for 5 days, and cell lysates were analyzed for modifications in histone acetylation levels using western blotting. Representative blots are shown from 3 independent experiments. (D) Effect of GSPs on HAT activity in SCC13 cells after treating the cells for 5 days, as detailed in Materials and methods. Significant increase versus non-GSPs-treated control, **P<0.05, *P<0.001.
GSPs increased HAT activity in SCC13 cells
Inhibition of HAT is a vital step in several diseases, including cancer. We have shown that treatment of skin cancer cells with GSPs enhanced acetylation of histone proteins. We further evaluated the effect of GSPs on HAT activity in cancer cells using EpiQuik™ HAT Assay Kit following the manufacturer’s protocol (Epigentek Group Inc., Brooklyn, NY). For this purpose, SCC13 cells were treated with GSPs for 5 days. Nuclear extracts were prepared from different treatment groups and subjected to the analysis of HAT activity. As shown in Figure 5D, GSPs increased the HAT activity (40–230%) in SCC13 cells in a concentration-dependent manner.
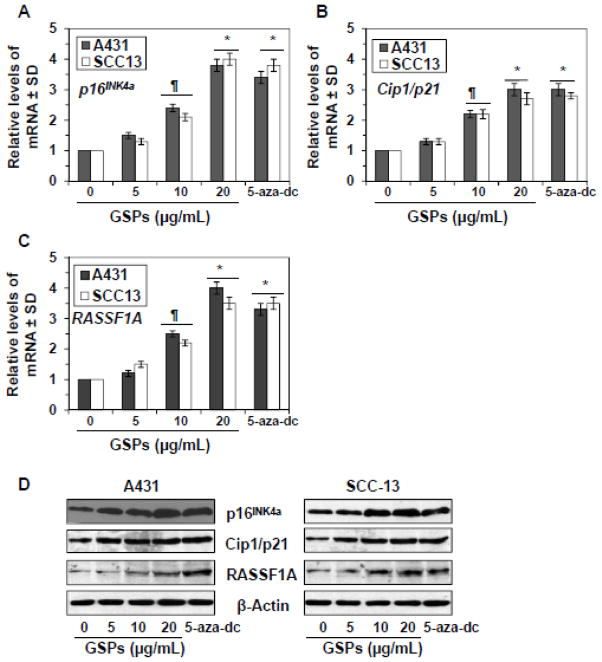
GSPs and 5-aza-dc reactivate tumor suppressor genes and proteins in human skin cancer cells
Next, we determined whether inhibitory effect of GSPs and 5-aza-dc on DNA methylation and HDAC activity in SCC13 and A431 cells leads to the reactivation of tumor suppressor genes in these cells. For this purpose SCC13 and A431 cells were treated with GSPs (0, 5, 10 and 20 μg/ml) and 5-aza-dc for 5 days. Cells were harvested and cellular RNA from different treatment groups was isolated and subjected to RT-PCR analysis as detailed in Materials and methods. RT-PCR analysis revealed that treatment of SCC13 and A431 cells with GSPs resulted in a significant re-expression of silenced mRNA of p16INK4a (1.3 to 4.0 fold), Cip1/p21 (1.2–4.0 fold) and RASSF1A (1.3–3.0 fold) (P<0.05 and P<0.001) in a dose-dependent manner compared to the control. Treatment of cells with 5-aza-dc also significantly reactivated (2.8 to 3.8 fold, P<0.001) the mRNA expression of these tumor suppressor genes, as shown in Figure 6A, 6B and 6C. We also determined the levels of tumor suppressor proteins using western blot analysis. As shown in Figure 6D, GSPs treatment reactivated the protein expression levels of silenced tumor suppressor proteins, p16INK4a, Cip1/p21 and RASSF1A in a dose-dependent manner. Under identical experimental conditions, similar reactivation effect on tumor suppressor proteins was also observed when SCC13 and A431 cells were treated with 5-aza-dc (Figure 6D).
Figure 6.
Treatment of human skin cancer cells, A431 and SCC13, with GSPs or 5-aza-dc for 5 days reactivates silenced tumor suppressor genes, p16INK4a, Cip1/p21 and RASSF1A. (A, B and C) RNA was isolated from the cells of different treatment groups and subjected to the quantification of mRNA expression levels of tumor suppressor genes using real-time PCR using the procedure detailed in Materials and methods. The mRNA levels were normalized to housekeeping gene (β-actin) and are presented as relative change compared to non-GSPs-treated controls, which was assigned an arbitrary unit 1 in each case. The mRNA levels are expressed in terms of mean values ± S.D. from three independent experiments. Significant difference versus non-GSPs-treated controls, *P<0.001, **P<0.01. (D) The levels of tumor suppressor proteins, RASSF1A, Cip1/p21 and p16INK4a, were determined in cell lysates using western blotting under identical experimental conditions. Representative blots are shown from three experiments.
Discussion
Dietary bioactive phytochemicals have low potentials for toxicity and should be considered as potential alternatives for the prevention and treatment of various cancers. Intake of GSPs prevents skin cancer through their anti-inflammatory, anti-oxidative and DNA repair mechanisms (Nandakumar et al., 2008; Katiyar, 2008; Sharma et al., 2007; Vaid et al., 2010). It is well recognized that cancer is a manifestation of both genetic and epigenetic modifications. The epigenetic effects of GSPs should be explored to define their chemopreventive role in prevention of skin cancer risk. Epigenetic changes are heritable changes in gene expression that do not affect the actual DNA sequence but have been recognized to play a crucial role in the etiology of cancer. DNA methylation and histone modifications are important epigenetic events in regulation of gene expression and maintenance of cellular function, which contribute to the development of cancer (Jones and Baylin, 2002; Jones, 2002; Counts and Goodman, 1995). These epigenetic changes in DNA often lead to silencing of tumor suppressor genes, which play a crucial role in tumor development and progression.
Emerging evidence suggests that dietary components/phytochemicals are involved in epigenetic modifications, regulate cellular function and modify the risk of cancer (Davis and Uthus, 2004; Huang, 2002). Here, we examined epigenetic effects induced by GSPs and elucidated the underlying mechanisms using the human skin cancer cell lines, SCC13 and A431, as an in vitro model. We also used 5-aza-dc, a well known DNA demethylating agent, and TSA, a well characterized HDAC inhibitor, as positive controls. Our study reveals that the levels of global DNA methylation were higher in skin cancer cell lines compared to that of NHEK and also that GSPs significantly decreased the levels of global DNA methylation as well as the levels of 5-mC in DNA in both skin cancer cell lines in a dose-dependent manner. The most profound alterations in levels of DNA methylation were observed after 5 days of treatment with GSPs. Treatment of skin cancer cells with 5-aza-dc also significantly decreased the levels of global DNA methylation. 5-aza-dc also decreased enzymatic activity as well as mRNA and protein expressions of DNMT1, DNMT3a and DNMT3b in both cancer cell lines. The inhibitory effects of GSPs on the DNMT activity, mRNA and protein expressions of DNMT1, DNMT3a and DNTM3b were also dose and time dependent. These results indicate that the epigenetic action of GSPs is similar to that of 5-aza-dc. Our preliminary screening experiments also suggest that treatment of cells at higher concentrations of GSPs induced cell death, which masked the epigenetic effects of GSPs in skin cancer cells (data not shown). This data suggests that the DNA methylation process in skin cancer cells can be reversed in cancer cells by bioactive phytochemicals such as GSPs.
Histone deacetylases are linked to transcriptional activation and repression of genes and therefore play an important role in development of human cancers (Rice and Allis, 2001; Tollefsbol, 2009). Histone deacetylation leads to a closed chromatin structure with transcriptional repression while histone acetylation results in an open chromatin structure associated with transcriptional activation (Tollefsbol, 2009; Margueron et al., 2005). Post-transcriptional modifications of histone methylation and acetylation may contribute to cancer development by modulation of the expression of tumor suppressor genes and oncogenes. HDACs play a crucial role in regulation of histone deacetylation. Deacetylation and methylation of H3-Lys 9 are the most common histone modifications involved in epigenetic repression of genes (Nakayama et al., 2001). In our in vitro model, GSPs significantly decrease HDAC activity of skin cancer cells. GSPs decreased levels of methylated H3-Lys 9, but increased levels of acetylated H3-Lys 9 and H3-Lys 14. Similar results were obtained when cells were treated with TSA. These data indicate that GSPs decrease HDAC activity to maintain H3-Lys 9 at a high level of acetylation and a low level of methylation, which may result in transcriptional activation of genes. The effects of 5-aza-dc were similar to GSPs in methylation of H3-Lys 9 in cancer cells. These observations suggest that GSPs and 5-aza-dc inhibit methyltransferase to prevent histone methylation. Treatment of SCC13 cells with GSPs also increases acetylation of histone H4 at Lys 5, Lys 12 and Lys 16, which may also favor transcriptional activation of tumor suppressor genes. These observations also suggest that GSPs are involved in both DNA demethylation and modifications of histone acetylation and methylation. Some other dietary phytochemicals, such as diallyl disulfide and genistein, have also been shown to affect DNA methylation and histone modifications in colon and prostate cancer cells (Druesne et al., 2004; Majid et al., 2008). (−)-Epigallocatechin-3-gallate from green tea has been shown to induce epigenetic modifications including the reactivation of tumor suppressor genes in human prostate, colon, esophageal and skin cancer cell lines (Fang et al., 2003; Nandakumar et al., 2011). Lycopene and genistein demethylated the GSTP1 promoter and reactivated GSTP1 expression in human breast cancer cells (King-Batoon et al., 2008).
It has been reported that hypermethylation of the p16INK4a gene is frequently observed in some cancers (Gazzeri et al., 1998; Merlo et al., 1995). Studies also suggest that RASSF1A inactivation is closely related to Ras activation in human cancers and thus contributes to malignant transformation by inhibiting Ras-mediated apoptosis (Dammann et al., 2000). The inactivation of RASSF1A by CpG island hypermethylation induced cyclin D1 accumulation and direct cell cycle progression (Shivakumar et al., 2002) and thus increase the proliferation potential of cancer cells. Chronic exposure of the skin to ultraviolet radiation enhances DNA hypermethylation patterns that leads to transcriptional silencing of tumor suppressor genes (RASSF1A and p16INK4A) in UVB-exposed skin and UVB-induced skin tumors of mice (Nandakumar et al., 2011). In our system, GSPs reactivate or re-express mRNA levels as well as proteins of tumor suppressor genes, such as p16INK4a, Cip1/p21 and RASSF1A, in both A431 and SCC13 human skin cancer cells. Reactivation of these genes by GSPs may block deregulated proliferation of cancer cells and may inhibit cancer cell progression. Identical observations were also found when these skin cancer cells were treated with 5-aza-dc, which is a well characterized DNA demethylating agent.
Together, our study demonstrates that proanthocyanidins from grape seeds have the ability to reactivate or restore the expression of the DNA hypermethylation-silenced tumor suppressor genes, p16INK4a, Cip1/p21 and RASSF1A, in A431 and SCC13 human skin cancer cells by reducing the activity of DNMTs and HDAC. This new information suggests that epigenetic regulation of tumor suppressor genes by GSPs may have a role in skin cancer chemoprevention. These findings are of importance for clinical applications of GSPs. In vivo animal studies are underway to understand the epigenetic mechanism of cancer chemoprevention by dietary GSPs.
Highlights.
Epigenetic modulations have been shown to have a role in cancer risk.
Proanthocyanidins decrease the levels of DNA methylation and histone deacetylation
Proanthocyanidins inhibit histone deacetylase activity in skin cancer cells.
Proanthocyanidins reactivate tumor suppressor genes in skin cancer cells.
Grape seed proanthocyanidins can prevent skin cancer through epigenetic modulation.
Acknowledgments
This work was supported by funds from the Veterans Administration Merit Review Award (ONCA-029-11S, S.K.K.) and National Institutes of Health (CA140832, S.K.K.). The human keratinocytes were obtained from the UAB Skin Diseases Research Center (AR050948). Editorial assistance from Dr. Fiona Hunter is gratefully acknowledged. The content of this manuscript does not necessarily reflect the views or policies of the funding sources.
Abbreviations used
- GSPs
grape seed proanthocyanidins
- DNMT
DNA methyltransferase
- HDAC
histone deacetylase
- H3-Lys 9
lysine 9 on histone H3
- PCR
polymerase chain reaction
- TSA
trichostatin A
- 5-aza-dc
5-aza-2′-deoxycytidine
Footnotes
Conflict of Interest Statement
The authors declare that there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Antequera F, Bird A. CpG islands. EXS. 1993;64:169–185. doi: 10.1007/978-3-0348-9118-9_8. [DOI] [PubMed] [Google Scholar]
- Counts JL, Goodman JI. Alterations in DNA methylation may play a variety of roles in carcinogenesis. Cell. 1995;83:13–15. doi: 10.1016/0092-8674(95)90228-7. [DOI] [PubMed] [Google Scholar]
- Dammann R, Li C, Yoon JH, Chin PL, Bates S, Pfeifer GP. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat Genet. 2000;25:315–319. doi: 10.1038/77083. [DOI] [PubMed] [Google Scholar]
- Davis CD, Milner J. Frontiers in nutrigenomics, proteomics, metabolomics and cancer prevention. Mutat Res. 2004;551:51–64. doi: 10.1016/j.mrfmmm.2004.01.012. [DOI] [PubMed] [Google Scholar]
- Davis CD, Uthus EO. DNA methylation, cancer susceptibility, and nutrient interactions. Exp Biol Med (Maywood) 2004;229:988–995. doi: 10.1177/153537020422901002. [DOI] [PubMed] [Google Scholar]
- Druesne N, Pagniez A, Mayeur C, Thomas M, Cherbuy C, Duée PH, Martel P, Chaumontet C. Diallyl disulfide (DADS) increases histone acetylation and p21(waf1/cip1) expression in human colon tumor cell lines. Carcinogenesis. 2004;25:1227–1236. doi: 10.1093/carcin/bgh123. [DOI] [PubMed] [Google Scholar]
- Fang MZ, Wang Y, Ai N, Hou Z, Sun Y, Lu H, Welsh W, Yang CS. Tea polyphenol (−)-epigallocatechin-3-gallate inhibits DNA methyltransferase and reactivates methylation-silenced genes in cancer cell lines. Cancer Res. 2003;63:7563–7570. [PubMed] [Google Scholar]
- Gazzeri S, Gouyer V, Vour’ch C, Brambilla C, Brambilla E. Mechanisms of p16INK4A inactivation in non small-cell lung cancers. Oncogene. 1998;16:497–504. doi: 10.1038/sj.onc.1201559. [DOI] [PubMed] [Google Scholar]
- Hegi ME, Sciuscio D, Murat A, Levivier M, Stupp R. Epigenetic deregulation of DNA repair and its potential for therapy. Clin Cancer Res. 2009;15:5026–5031. doi: 10.1158/1078-0432.CCR-08-1169. [DOI] [PubMed] [Google Scholar]
- Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- Howell PM, Jr, Liu S, Ren S, Behlen C, Fodstad O, Riker AI. Epigenetics in human melanoma. Cancer Control. 2009;16:200–218. doi: 10.1177/107327480901600302. [DOI] [PubMed] [Google Scholar]
- Huang S. Histone methyltransferases, diet nutrients and tumor suppressors. Nat Rev Cancer. 2002;2:469–476. doi: 10.1038/nrc819. [DOI] [PubMed] [Google Scholar]
- Jones PA. DNA methylation and cancer. Oncogene. 2002;21:5358–5360. doi: 10.1038/sj.onc.1205597. [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- Katiyar SK. Grape seed proanthocyanidines and skin cancer prevention: Inhibition of oxidative stress and protection of immune system. Mol Nutr Food Res. 2008;52:S71–S76. doi: 10.1002/mnfr.200700198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar SK, Vaid M, van Steeg H, Meeran SM. Green tea polyphenols prevent UV-induced immunosuppression by rapid repair of DNA damage and enhancement of nucleotide excision repair genes. Cancer Prev Res. 2010;3:179–189. doi: 10.1158/1940-6207.CAPR-09-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King-Batoon A, Leszczynska JM, Klein CB. Modulation of gene methylation by genistein or lycopene in breast cancer cells. Environ Mol Mutagen. 2008;49:36–45. doi: 10.1002/em.20363. [DOI] [PubMed] [Google Scholar]
- Majid S, Kikuno N, Nelles J, Noonan E, Tanaka Y, Kawamoto K, Hirata H, Li LC, Zhao H, Okino ST, Place RF, Pookot D, Dahiya R. Genistein induces the p21WAF1/CIP1 and p16INK4a tumor suppressor genes in prostate cancer cells by epigenetic mechanisms involving active chromatin modification. Cancer Res. 2008;68:2736–2744. doi: 10.1158/0008-5472.CAN-07-2290. [DOI] [PubMed] [Google Scholar]
- Margueron R, Trojer P, Reinberg D. The key to development: interpreting the histone code? Cur Opin Gen Dev. 2005;15:163–176. doi: 10.1016/j.gde.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Meeran SM, Vaid M, Punathil T, Katiyar SK. Dietary grape seed proanthocyanidins inhibit 12-O-tetradecanoyl phorbol-13-acetate-caused skin tumor promotion in 7, 12-dimethylbenz(a)anthracene-initiated mouse skin, which is associated with the inhibition of inflammatory responses. Carcinogenesis. 2009;30:520–528. doi: 10.1093/carcin/bgp019. [DOI] [PubMed] [Google Scholar]
- Merlo A, Herman JG, Mao L, Lee DJ, Gabrielson E, Burger PC, Baylin SB, Sidransky D. 5′ CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat Med. 1995;1:686–692. doi: 10.1038/nm0795-686. [DOI] [PubMed] [Google Scholar]
- Mittal A, Elmets CA, Katiyar SK. Dietary feeding of proanthocyanidins from grape seeds prevents photocarcinogenesis in SKH-1 hairless mice: relationship to decreased fat and lipid peroxidation. Carcinogenesis. 2003;24:1379–1388. doi: 10.1093/carcin/bgg095. [DOI] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- Nandakumar V, Singh T, Katiyar SK. Multi-targeted prevention and therapy of cancer by proanthocyanidins. Cancer Lett. 2008;269:378–387. doi: 10.1016/j.canlet.2008.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandakumar V, Vaid M, Katiyar SK. (−)-Epigallocatechin-3-gallate reactivates silenced tumor suppressor genes by reducing DNA methylation and increasing histones acetylation in human skin cancer cells. Carcinogenesis. 2011;32:537–544. doi: 10.1093/carcin/bgq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandakumar V, Vaid M, Tollefsbol TO, Katiyar SK. Aberrant DNA hypermethylation patterns lead to transcriptional silencing of tumor suppressor genes in UVB-exposed skin and UVB-induced skin tumors of mice. Carcinogenesis. 2011;32:597–604. doi: 10.1093/carcin/bgq282. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Rice JC, Allis CD. Histone methylation versus histone acetylation: new insights into epigenetic regulation. Curr Opin Cell Biol. 2001;13:263–273. doi: 10.1016/s0955-0674(00)00208-8. [DOI] [PubMed] [Google Scholar]
- Sharma SD, Meeran SM, Katiyar SK. Dietary grape seed proanthocyanidins inhibit UVB-induced oxidative stress and activation of mitogen-activated protein kinases and nuclear factor-κB signaling in in vivo SKH-1 hairless mice. Mol Cancer Ther. 2007;6:995–1005. doi: 10.1158/1535-7163.MCT-06-0661. [DOI] [PubMed] [Google Scholar]
- Sharma SD, Meeran SM, Katiyar SK. Proanthocyanidins inhibit in vitro and in vivo growth of human non-small cell lung cancer cells by inhibiting the prostaglandin E2 and prostaglandin E2 receptors. Mol Cancer Ther. 2010;9:569–580. doi: 10.1158/1535-7163.MCT-09-0638. [DOI] [PubMed] [Google Scholar]
- Shivakumar L, Minna J, Sakamaki T, Pestell R, White MA. The RASSF1A tumor suppressor blocks cell cycle progression and inhibits cyclin D1 accumulation. Mol Cell Biol. 2002;22:4309–4318. doi: 10.1128/MCB.22.12.4309-4318.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh T, Vaid M, Katiyar N, Sharma S, Katiyar SK. Berberine, an isoquinoline alkaloid, inhibits melanoma cancer cell migration by reducing the expressions of cyclooxygenase-2, prostaglandin E2 and prostaglandin E2 receptors. Carcinogenesis. 2011;32:86–92. doi: 10.1093/carcin/bgq215. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Tollefsbol TO. Cancer epigenetics. CRC Press/Taylor & Francis Group; Boca Raton, USA: 2009. [Google Scholar]
- Vaid M, Sharma SD, Katiyar SK. Proanthocyanidins inhibit photocarcinogenesis through enhancement of DNA repair and xeroderma pigmentosum Group A-dependent mechanism. Cancer Prev Res. 2010;3:1621–1629. doi: 10.1158/1940-6207.CAPR-10-0137. [DOI] [PubMed] [Google Scholar]