Abstract
Although the number of phylotypes present in a microbial community may number in the hundreds or more, until recently, fluorescence in situ hybridization has been used to label, at most, only a handful of different phylotypes in a single sample. We recently developed a technique, CLASI-FISH for Combinatorial Labeling and Spectral Imaging – Fluorescence in situ Hybridization, to greatly expand the number of distinguishable taxa in a single FISH experiment. The CLASI technique involves labeling microbes of interest with combinations of probes coupled with spectral imaging to allow the use of fluorophores with highly overlapping excitation and emission spectra. Here, we present the basic principles and theory of CLASI-FISH along with some guidelines for performing CLASI-FISH experiments. We further include a protocol for creating fluorescence spectral reference standards, a vital component of successful CLASI-FISH.
Keywords: Spectral Imaging, Combinatorial Labeling, Fluorescence in situ hybridization, FISH, Microbial Diversity
Introduction
Theory of CLASI-FISH
Molecular sequence-based surveys suggest that microbial communities may be extremely species-rich, with hundreds of phylotypes present in each sample habitat [3,4,14]. These studies have generated “parts lists” for many microbial communities—lists of organisms and genes present in a microbial ecosystem [13]. Because the function of any biological system is implicit in its structure, a full understanding of microbial community physiology requires an understanding of the physical structure of these communities. To that end, fluorescence in situ hybridization (FISH) using oligonucleotide probes targeted to ribosomal RNA has proven to be an effective tool for the identification, quantification, and spatial analysis of microbes in situ [2,6].
Statement of the Problem
Although in principle, FISH probes specific for nearly any microbial phylotype or taxon could be designed, in practice, the use of bandpass filters in fluorescence image acquisition limits the number of fluorophores that can be simultaneously differentiated. This is due to the photo-physical properties of available fluorophore reporters, namely that many organic fluorophores have highly overlapping excitation and emission spectra. As a result, FISH has been used routinely to distinguish only a few types of microbes in any single experiment.
The CLASI Solution
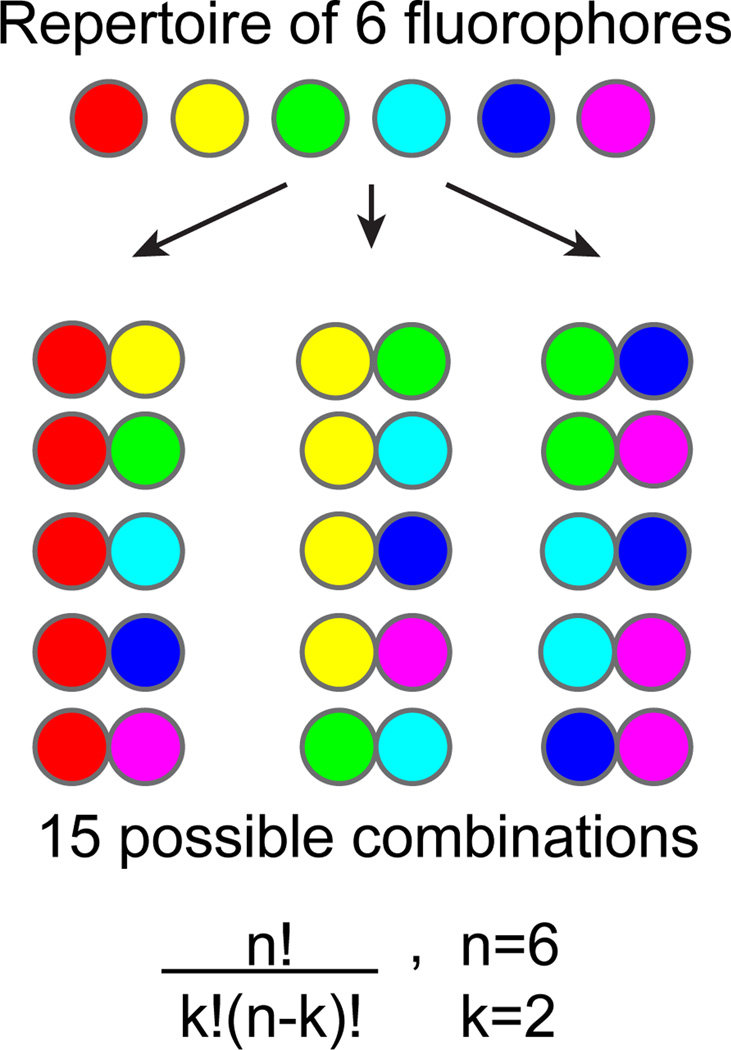
We recently reported the development of a biological labeling and fluorescence image analysis method, which we call Combinatorial Labeling and Spectral Imaging (CLASI), and its application to the simultaneous identification of tens to potentially hundreds of microbial taxa in a single microscope image by FISH [16]. Our combinatorial labeling strategy entails the labeling of a given type of microorganism with two or more fluorophores selected from a repertoire of chosen fluorophores. With this combinatorial labeling approach we greatly expand the number of different kinds of microbes distinguishable in a field of view (Fig. 1).
Figure 1. The power of combinations.
With a relatively small number of fluorophores, it is possible to create many different specific combinations. When the number of dyes, n, is 6, and the combination size, k, is set to 2 there exist 15 different combinations. The binary label restriction gives a more robust assay because the a priori knowledge that all cells in an image are labeled with exactly two fluorophores is used when assigning objects their label-type.
Recent advances in fluorescence spectral image acquisition and the application of linear unmixing to spectrally-recorded image data allow the unambiguous identification of fluorophores with overlapping spectra [18]. The simplest definition of spectral imaging is that it is the combination of spectroscopy with microscopy, such that at every pixel in a digital fluorescence image, a full spectrum is recorded [8]. Many different commercially available and custom-built spectral microscope systems exist. The protocols included in this article are written for use with commercially available Laser Scanning Confocal Microscopes (LSCMs), which are equipped with multi-anode spectral detectors, such as those available from Nikon or Zeiss. The general protocols of CLASI-FISH should be compatible with spectral microscope systems from any commercial vendor.
Once a spectral image of combinatorially-labeled microbes has been acquired, every pixel in the recorded image must be assigned its fluorophore composition. For this purpose, a computational analysis called linear unmixing is often used [9]. If the spectra of all fluorophores used in an experiment are known, then the unknown recorded image and the known reference spectra may be treated as a system of linear equations and a least squares approach is used to assign an abundance for all fluorophores used in the experiment, at each pixel in the image (Fig. 2). Because the instrument may impart artifacts in the recorded spectra of the unknown sample, known reference spectra should be acquired directly on the instrument, using labeled standard specimens. Appropriate standards are non-autofluorescent bacteria, such as E. coli, which have been labeled individually with all the fluorophores that will be used in an experiment, and imaged using identical settings on the instrument that are used to image the unknown sample. For this purpose, we include with this article a protocol for creating spectral fluorescence reference standards.
Figure 2. Spectral Imaging and Linear Unmixing of microbes.
(A). 24-channel spectral image of a mixture of E. coli cells labeled either singly or in binary combination with BODIPY-FL and Oregon Green-514. (B). Emission spectra of the two fluorophores as recorded on the instrument used to acquire the image in (A). (C). Pseudocolor image after application of linear unmixing. BODIPY-FL intensity is colored green and Oregon Green 514 is red. Cells labeled with a combination of both fluorophores appear yellow in this merged image.
Application
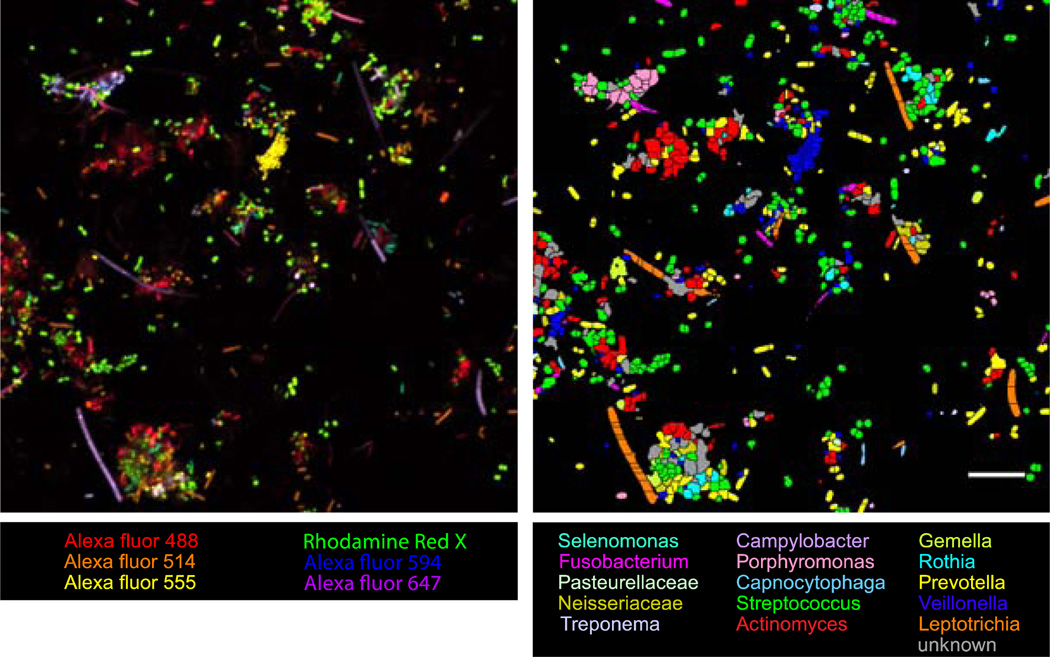
We have applied CLASI-FISH to study the systems-level structure of human oral microbial communities [16]. Fifteen of the most abundant taxa in extracted human dental plaque were labeled in a single experiment and a proximity analysis was performed to characterize the spatial distribution of the fifteen different taxa in the biofilm. All fifteen taxa were identified in the natural community (Fig. 3).
Figure 3. Example CLASI-FISH image.
Images of CLASI-FISH-labeled semi-dispersed human dental plaque. Colors in the raw spectral image (Left) represent one of six different fluorophores, used in all the possible binary combinations to label 15 different taxa in the community. Color in the segmented image (Right) of the same field of view represents one of each of the 15 probed taxa. Cells, which are ambiguous in their label type because of errors in image segmentation, are declared unknown and colored grey. Bar = 10 µm.
Methods
Specific Considerations for CLASI-FISH
FISH protocols may be broken down into 4 steps: Sample collection and fixation, hybridization, image acquisition, and image analysis. Established protocols such as previously reported by Pernthaler, et al., [11], are generally appropriate for CLASI-FISH hybridization. What follows are some considerations specific for the CLASI-FISH protocol.
Fixation
The simultaneous labeling of different types of microbes in a complex community poses the challenge of adequately fixing microbes differing significantly in cell wall composition, e.g. mixtures of both Gram (+) and Gram (−) organisms. Typically Gram (−) organisms are fixed in 2–4 % paraformaldehyde (PFA) and Gram (+) organisms in ethyl or methyl alcohol. Gram positive organisms may then be subsequently treated to enzymatically degrade the cell wall, e.g., with lysozyme or proteinase K [12]. Several strategies have been proposed for simultaneous or multiplexed FISH of mixed microbial communities, starting with modifications of fixation protocols.
Fixation of microbial samples in 2% PFA, followed by extended hybridization time and stringent washing [16].
Fixation of microbial samples in 2–4% PFA, then light enzymatic treatment, e.g. with low concentration or for short incubation time [15].
Fixation with ethyl alcohol [7].
As described below, we typically use treatment with PFA followed by ethyl alcohol.
Hybridization
Modifications of hybridization protocols for CLASI-FISH are minor. Each organism is labeled with two or more probes, rather than a single probe. At least two strategies are available.
Synthesize two (or more) versions of the same oligonucleotide probe, each version conjugated to a different fluorophore. The advantage of this approach is simplicity—only one oligonucleotide sequence need be validated for FISH. It may be that there exists only one oligo sequence suitable for identification of an organism in the context of its natural community. A disadvantage of this single oligo approach is competition between the two labeled versions of the oligo, thus reducing the fluorescence signal in each of the labeling channels.
Synthesize two, or more, different oligonucleotide sequences specific for labeling a particular organism in the context of its community. Each probe sequence targets a different part of the 16S rRNA. The advantage of this approach is that no reduction in probe fluorescence intensity is expected compared to a singly–labeled organism. Another advantage is confirmation of taxon identity by independent probes. The disadvantage is that multiple probes need to be designed and validated and there may be difficulties in designing two or more probes that label an organism with approximately equal efficiency under the same hybridization conditions.
Our practice to date is to begin the design of a probe set by synthesizing a single oligo in two fluorescent versions for each taxon and to subsequently improve the probe set by designing second or third oligos for taxa where possible.
Spectral Image Acquisition
Spectral Image Acquisition
Singly-labeled pure samples of E. coli or other reference microbes, labeled with the universal bacterial probe, EUB-338 make excellent test specimens and are useful for checking spectral image acquisition set-up. If the known samples can be unmixed correctly, that is, within an acceptable uncertainty range, using a particular set of spectral image acquisition parameters, then those conditions should be appropriate for imaging unknown environmental specimens.
It may be necessary to use multiple wavelength excitation to efficiently excite all the fluorophores used in an experiment. This is because fluorophores have both characteristic emission and excitation spectra.
At least two general strategies are available if more than one excitation wavelength is needed to excite all the fluorophores used to label a particular sample:
Illuminate the specimen with all excitation wavelengths simultaneously. This is possible with the use of lasers in laser scanning confocal microscopy or with LEDs in widefield spectral imaging. In addition, it will be necessary to use a multi-notched dichroic mirror if the specimen is illuminated via the epi path. Two, three, or more wavelength notch dichroics are available. Simultaneous illumination with multiple wavelengths may not be possible with a white light source, such as a mercury arc or metal halide lamp, because multiple different filters would be required to generate different excitation wavelength bands. The advantage of simultaneous multi-wavelength excitation is simplicity—all fluorophores are excited in one step. The main disadvantage is that the wavelength notches in a multi-wavelength dichroic introduce artifacts in the shape of recorded emission spectra, and can make different fluorophores appear to have similar spectral characteristics.
Illuminate the specimen sequentially with each wavelength excitation. The advantage is that no spectral shape artifacts will be introduced in the recorded spectra due to notch filters. The main disadvantage is that because of excitation cross-talk, fluorophores may be excited more than once—some fluorophores will absorb light at more than one wavelength used to excite the sample. This may result in an increase in fluorophore bleaching; it also leaves the user with the dilemma of what to do with emission spectra recorded from more than one excitation wavelength. To minimize bleaching, sequential excitation should start at the longest wavelength employed, then, move progressively toward the shortest wavelength. See the section on Image Analysis for suggestions on how to deal with emitted light from a particular fluorophore recorded from more than one excitation wavelength. In general, analysis is carried out with the strongest emission spectra.
Reference Spectra
In order to assign to each pixel in the image its fluorophore composition, the linear unmixing algorithm requires, as input, the known reference spectra for all of the fluorophores used in an experiment. See Appendix for a protocol for creating microbial spectral reference standards.
E. coli make excellent standard specimens for CLASI-FISH. Separate aliquots of E. coli are labeled in standard FISH reactions with a different version of the EUB-338 probe, each version conjugated to a different fluorophore from the pool of all fluorophores used in an experiment. Aliquots of these pure, singly-labeled E. coli cells are then spotted on separate slides for imaging.
Reference spectrum specimens should be treated exactly as unknown microbial specimens. They should be mounted in the same mounting medium used for mixed environmental communities. A mounting medium with anti-fade properties and appropriate refractive index are crucial and many options are available from different vendors. Mounting media with DAPI is not appropriate for reference standards, as the DAPI fluorescence will add to, and be recorded as part of, the reference fluorophore spectrum. In addition, reference spectra should be acquired under identical imaging conditions as unknown labeled microbial specimens. For example, for any image of mixed microbes, reference spectra should have been acquired on the same instrument, using the same objective and the same detector settings. In addition, microscope specimens should always be mounted with coverslips of 0.17 mm thickness. These #1.5 coverslips are matched for modern microscope objectives
Autofluorescence
Cells from natural environments and other non-cellular debris may exhibit autofluorescence. In this case, an unlabeled specimen may be imaged using the same acquisition parameters as labeled images. The spectra from autofluorescent cells is recorded and added to the fluorophore library along with the E. coli reference spectra. Autofluorescence is thus treated as another spectral character in the labeled sample and may be easily identified after unmixing.
Image Analysis
Linear unmixing
Once a spectral image is recorded, we wish to identify at each pixel in the image the fluorophore composition of the recorded signal. To achieve this task, many microscope and spectral imaging device manufacturers include linear unmixing algorithms with their instrument software. In addition there exists a free linear unmixing plugin for ImageJ [1]. Linear unmixing is applied to recorded images using reference spectra generated from E. coli reference cells.
Some pre-processing of spectrally recorded images may help with linear unmixing. For example, a low pass filter may be applied to raw spectral images before unmixing. This will reduce noise in the image, but may cause spectral mixing of neighboring pixels which may consist of separate spectral characters, for example, background may get mixed into pixels of cells, especially around the edges of the cells. A second recommended processing step is background correction for non-uniform illumination of the sample, either in software or with the use of an image of a uniform fluorescent specimen. Both spatial filtering and background correction may be performed on raw spectral images or on the images after unmixing, depending upon the capabilities of the linear unmixing software used.
If multiple excitations were used sequentially to excite all the fluorophores, then each excitation is unmixed separately. All of the reference spectra for fluorophores maximally excited by a particular wavelength excitation are used for unmixing that spectral data set. In addition, it may be necessary to include additional reference spectra which are more efficiently excited at other wavelengths, but which nonetheless are somewhat excited by the wavelength used in the imaging regime. One may then discard the unmixing results for these off-peak fluorophores because they are redundant. The remaining image channels are then collated as an image stack, with one channel for every fluorophore used in the experiment.
Combinatorial-label assignment
After applying linear unmixing the final image analysis step unique to CLASI-FISH is the conversion of the unmixed image data, in which each pixel is assigned an abundance for every fluorophore used in the experiment, to a particle-based data set in which each particle in the image is assigned its combinatorial label, or taxon identity. The combinatorial label assignment is a multi-step process beginning with image segmentation.
Image Segmentation
First, cells need to be segmented from background and from each other. For this task, it is helpful to have an image of all the cells in the sample, for example labeled with the EUB-338 probe conjugated to a fluorophore that does not overlap spectrally with any of the fluorophores used with taxon-specific probes. The EUB image should then be collected with a standard detector using a high quality bandpass filter, rather than with a spectral detector. This is because spectral images are in general, more noisy than standard fluorescence images because the same amount of fluorescence signal that is recorded in a single channel with a standard detector is spread out over multiple channels with a spectral detector.
The best image segmentation will come from an image with the highest possible signal to noise ratio. By simultaneously labeling one’s sample of interest with both taxon-specific probes and a general probe for bacteria, one can identify the near total number of cells in the image and determine the fraction of total cells in the image that are labeled with taxon-specific probes. If such an image is not possible for a specimen, then a spectral image after linear unmixing may be used.
Many procedures for image segmentation are available in both commercial and non-commercial image processing software. We have found the following protocol suitable for CLASI-FISH image segmentation. Unmixed images are imported into ImageJ and a global intensity threshold is applied. The threshold is set suitably low such that all pixels that represent cells are brought to foreground. This process generally results in multiple touching cells identified as one contiguous object. To separate touching cells, a watershed algorithm is applied to the images. Local maxima are identified in the unmixed image, then, used to skeletonize the image using the “Find maxima” command in ImageJ. The skeleton lines are superimposed onto the thresholded image to create a segmented image in which touching cells are separated.
Once image segmentation is complete, it becomes possible to transform the pixel-based fluorophore data from the linear unmixing operation(s) into a cell or particle-based data set, in which each cell is identified in its taxon identity. For this, one need analyze the average computed fluorophore intensity of all the fluorophores used in the experiment over all of the pixels that make up each segmented cell. The fluorophores with the highest abundance in the cell are considered that cell’s combinatorial label, and that specific fluorophore combination may be mapped back to a specific taxon identity. All other fluorophore intensities are discarded as artifacts from the linear unmixing operation due to noise in the recorded spectral image and unavoidable shortcomings of the unmixing algorithm. Although the absolute intensities of each fluorophore in every cell may vary, the a priori knowledge that every identified cell in the image is labeled with exactly two fluorophores allows the use of the simple logic above to confidently assign taxonomic identity. After cells have been identified in their taxon identity, stereological and spatial analyses may be performed using other available software [5].
Discussion
Recent improvements in organic fluorescent reporter brightness and photostability; improvements in microscope optics and detector sensitivity; as well as the increasing performance to cost ratios of computer technology have fostered the development of fluorescence spectral imaging, in which multiple fluorophores with highly overlapping emission spectra may be simultaneously distinguished in a single image. CLASI-FISH is a combinatorial labeling and spectral imaging technique that greatly increases the number of different taxa of microbes that can be distinguished in a single sample.
One requirement for successful CLASI-FISH is that of reference spectra. The linear unmixing algorithm used to computationally assign fluorophore identities to every pixel in a spectral image requires prior knowledge of the spectra for every fluorophore used to label an ecological sample. To that end, we have provided a protocol for reference spectra acquisition in the appendix to this manuscript.
A second requirement for CLASI-FISH is that the fluorescent signal from labeled microbes must be suitably bright. With spectral imaging, the emitted light from any sample is spread over multiple channels, resulting in lower signal to noise data compared to images collected in one or a few channels through bandpass filters. In any single environmental microbial sample, some cells may be bright after FISH labeling while others may show low signal intensity, presumably due to low density of ribosomes in cells from oligotrophic environments or to a low accessibility of the cells or probe target sites to oligonucleotide probes [10,17]. This problem of high dynamic range in signal intensities is particularly problematic for CLASI-FISH because images that are acquired such that bright cells are overexposed in the recorded image in order to visualize dim cells are incompatible with linear unmixing. Overexposed pixels impart artifacts on the recorded spectral shape and are therefore ambiguous in their fluorophore identity. Improvements in signal amplification, microscope detectors with improved sensitivity, and the use of adaptive optics to record high dynamic range images will be important to make the CLASI technique more practical for the study of microbial communities from oligotrophic environments.
Nonetheless, with suitable oligonucleotide probes and the CLASI-FISH protocols included above, researchers in diverse fields of microbial ecology should be well-poised to begin answering the crucial questions of microbial community composition and dynamics that require a full ecosystems-level approach to investigation.
Acknowledgements
We are grateful to Rudolf Oldenbourg for help revising this manuscript. This work was supported in part by a grant from the Alfred P. Sloan Foundation to G.G.B., and NIH Grants F31DE019576 to A.M.V. and RC1DE20630 to G.G.B. from the National Institute for Dental and Craniofacial Research.
Appendix
Protocol for creating E. coli spectral reference standards
Fixation
E. coli are grown to an approximate OD600 of 0.5 in 5 mL of Luria Bertani (LB) broth.
An equal volume (5 mL) of fresh 4% PFA is added to the culture broth.
Cells are incubated at room temperature for 1.5 hours.
Cells are washed 3× in phosphate buffered saline (PBS).
After the first wash, cells are transferred to a 1.6 mL microcentrifuge tube and washed 2× in 1.5 mL PBS.
After the third wash, cell pellet is re-suspended in 750 µL PBS. Pellet is pipetted up and down vigorously and tube is vortex mixed to thoroughly re-suspend cells and break up the pellet.
750 µL of 100% ethyl alcohol is added to the tube to give a final concentration of 50% PBS / 50% ethyl alcohol. Cells may be stored at −20°C for up to 6 months before FISH.
Hybridization
Protocol modified from Pernthaler, et al [11].
For each reference fluorophore standard, 10 µL of fixed E. coli cells are pipetted into a separate 0.6 mL microcentrifuge tube.
90 µL of Hybridization buffer is added to each tube [0.9 M NaCl, 0.02 M Tris (pH 7.5), 0.01% SDS, 20% formamide].
1 µL of EUB-338-fluorophore probe [0.2 mM] is added to the tube to give a final probe concentration of 2 µM. The tube is immediately vortex mixed.
Steps 2–4 are repeated for every fluorophore needed.
Tubes are placed in a suitable plastic rack and covered with aluminum foil to protect them from exposure to room light.
Rack is placed in a hybridization oven at 46°C and cells are incubated for 3 hours.
Each tube of cells is washed once in 100 µL of Wash 1 buffer [0.9 M NaCl, 0.02 M Tris (pH 7.5), 0.01% SDS, 20% formamide] and incubated at 48°C for 15 minutes.
Each tube of cells is washed once in 200 µL of Wash 2 buffer [0.9 M NaCl, 0.02 M Tris (pH 7.5), 0.01% SDS] and incubated at 48°C for 15 minutes.
Each tube of cells is resuspended in 100–200 µL of Resuspension buffer [0.025 M NaCl + 0.02 M Tris (pH 7.5)].
A hydrophobic ring of ~1cm diameter is drawn on one Ultrastick microscope slide (GoldSeal) for each E. coli labeltype using a Pap pen (Thermofisher).
40 µL of washed cells are pipetted onto the slides within the ring.
Slides are placed in a humid chamber and cells are allowed to settle onto the slides for 45 minutes in the dark.
Specimens are dehydrated in an ethyl alcohol series. Slides are first placed in a 50 mL centrifuge tube containing 35 mL of 50% ethyl alcohol and left in the tube for 10 seconds. Each slide is then removed and immediately placed in a 50 mL tube with 35 mL of 75% ethyl alcohol for 10 s. Slides are then sequentially incubated for 10 s in three tubes of 100% ethyl alcohol.
Slides are air dried in the dark for 10 minutes.
-
Slides are mounted in either Vectashield (Vector Laboratories) or Prolong Gold (Invitrogen) mounting medium.
If specimens are mounted in Vectashield they may be imaged immediately
If slides are mounted in Prolong Gold they are incubated at room temperature for 3 days in the dark to allow the mounting medium to cure.
Reference Spectra Collection
The following protocol is for E. coli reference samples labeled with the following fluorophores: Pacific Blue, Pacific Orange, BODIPY-FL, Oregon Green 514, Alexa fluor 532, Alexa fluor 546, Rhodamine Red-X and Texas Red-X on a laser scanning confocal microscope equipped with 32-anode spectral detector as in Valm, et al 2011.
-
Use prior knowledge about reference spectra absorption to determine for each fluorophore its maximum absorption wavelength available on the instrument. For example we recommend the “Spectra Viewer” web application from Invitrogen http://www.invitrogen.com.
- Fluorophores and illumination wavelengths for maximum excitation are as follows:
Pacific Blue 405 nm Pacific Orange 405 nm BODIPY-FL 488 nm Oregon Green 514 488 nm Alexa fluor 532 488 nm Alexa fluor 546 561 nm Rhodamone Red-X 561 nm Texas Red-X 561 nm
Acquire the best possible image of Pacific Blue labeled E. coli using 405 nm excitation. Aim for highest possible signal to noise image, using all the imaging parameters that will be used for acquiring images of experimental samples, i.e., same objective, main beam dichroic, and detector bandwidth settings.
Acquire the best possible image of Pacific Orange with 405 nm excitation.
Acquire the best possible image of BODIPY-FL with 405 nm excitation. Even though this fluorophore is better excited by 488 nm light than 405 nm, we still acquire a reference spectrum for this fluorophore using 405 nm excitation.
Acquire BODIPY-FL, Oregon Green 514, Alexa 532, Alexa fluor 546, and Rhodamine Red-X images with 488 nm excitation.
Acquire Alexa fluor 532, Alexa fluor 546, Rhodamine Red-X, and Texas Red-X with 561 nm excitation.
Use the microscope instrument software to define a region of interest (ROI) in each cell in each image. The software calculates the average intensity over all the pixels in the ROI to generate a reference spectrum.
Validation of CLASI-FISH Imaging and Analysis Settings
In addition to a means for acquiring reference spectra, labeled E. coli make excellent test specimens for CLASI-FISH. If labeled test specimens can be imaged and correctly unmixed using a particular set of image acquisition and analysis parameters, then the user is well-positioned to image and unmix unknown environmental samples labeled with the same fluorophores as the test specimens.
Start with separate aliquots of E. coli suspended in resuspension buffer, each aliquot labeled with a different fluorophore version of the EUB-338 probe.
Prepare a mixture of all the different label-types. The mixture may be an equal mixture, in which all label-types are added in an equal amount, or it may be a stepped mixture, in which a different volume of each label-type is added to the mixture to give different relative proportions in the final mixture. It is important to know what the expected proportion of each label-type should be in the final mixture.
Spot the mixture onto a slide for imaging.
Acquire spectral images and perform linear unmixing on the raw data as would be done for an unknown environmental sample.
Analyze the image and count the frequency of each label-type in the unmixed images. The output from the CLASI-FISH analysis should correlate with the known input into the mixture within an acceptable uncertainty range.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Abramoff M, Magalhaes P, Ram S. Image processing with ImageJ. Biophotonics. 2004;11:36–42. [Google Scholar]
- 2.Amann RI, Krumholz L, Stahl DA. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashby MN, Rine J, Mongodin EF, Nelson KE, Dimster-Denk D. Serial analysis of rRNA genes and the unexpected dominance of rare members of microbial communities. Appl. Environ. Microbiol. 2007;73:4532–4542. doi: 10.1128/AEM.02956-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 5.Daims H, Lücker S, Wagner M. daime, a novel image analysis program for microbial ecology and biofilm research. Environ Microbiol. 2006;8:200–213. doi: 10.1111/j.1462-2920.2005.00880.x. [DOI] [PubMed] [Google Scholar]
- 6.DeLong EF, Wickham GS, Pace NR. Phylogenetic stains: ribosomal RNA- based probes for the identification of single cells. Science. 1989;243:1360–1363. doi: 10.1126/science.2466341. [DOI] [PubMed] [Google Scholar]
- 7.Fazi S, Amalfitano S, Pizzetti I, Pernthaler J. Efficiency of fluorescence in situ hybridization for bacterial cell identification in temporary river sediments with contrasting water content. Syst. Appl. Microbiol. 2007;30:463–470. doi: 10.1016/j.syapm.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 8.Garini Y, Young IT, McNamara G. Spectral imaging: Principles and applications. Cytometry. 2006;69A:735–747. doi: 10.1002/cyto.a.20311. [DOI] [PubMed] [Google Scholar]
- 9.Neher R, Neher E. Optimizing imaging parameters for the separation of multip.e labels in a fluorescence image. J Microsc. 2004;213:46–62. doi: 10.1111/j.1365-2818.2004.01262.x. [DOI] [PubMed] [Google Scholar]
- 10.Pernthaler A, Pernthaler J, Amann R. Fluorescence in situ hybridization and calalyzed reporter deposition for the identification of marine bacteria. Appl. Environ. Microbiol. 2002;86:3094–3101. doi: 10.1128/AEM.68.6.3094-3101.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pernthaler J, Glöckner F-O, Schönhuber W, Amann R. In: Methods in Microbiology. Paul J, editor. London: Academic Press, Ltd; 2001. pp. 207–226. [Google Scholar]
- 12.Quevedo B, et al. Phylogenetic group- and species-specific oligonucleotide probes for single-cell detection of lactic acid bacteria in oral biofilms. BMC Microbiol. 2011;11:14. doi: 10.1186/1471-2180-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raes J, Bork P. Molecular eco-systems biology: towards an understanding of community function. Nat. Rev. Microbiol. 2008;6:693–699. doi: 10.1038/nrmicro1935. [DOI] [PubMed] [Google Scholar]
- 14.Sogin ML, et al. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc. Natl. Acad. Sci. U.S.A. 2006;103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thurnheer T, Gmür R, Guggenheim B. Multiplex FISH analysis of a six-species bacterial biofilm. J. Microbiol. Meth. 2004;56:37–47. doi: 10.1016/j.mimet.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 16.Valm AM, et al. From the Cover: Systems-level analysis of microbial community organization through combinatorial labeling and spectral imaging. Proc. Natl. Acad. Sci. U.S. A. 2011;108:4152–4157. doi: 10.1073/pnas.1101134108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yilmaz LS, Okten HE, Noguera DR. Making all parts of the 16S rRNA of Escherichia coli accessible in situ to single DNA oligonucleotides. Appl. Environ. Microbiol. 2006;72:733–744. doi: 10.1128/AEM.72.1.733-744.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zimmermann T. Spectral imaging and its applications in live cell microscopy. FEBS Letters. 2003;546:87–92. doi: 10.1016/s0014-5793(03)00521-0. [DOI] [PubMed] [Google Scholar]