Abstract
Background
Prenatal alcohol exposure can result in Fetal Alcohol Spectrum Disorder (FASD). Not all women who consume alcohol during pregnancy have children with FASD and studies have shown that genetic factors can play a role in ethanol teratogenesis. We examined gene expression in embryos and placentae from C57BL/6J (B6) and DBA/2J (D2) mice following prenatal alcohol exposure. B6 fetuses are susceptible to morphological malformations following prenatal alcohol exposure while D2 are relatively resistant.
Methods
Male and female B6 and D2 mice were mated for two hours in the morning, producing four embryonic genotypes: true-bred B6B6 and D2D2, and reciprocal B6D2 and D2B6. On gestational day 9dams were intubated with either 5.8 g/kg ethanol, an is caloric amount of maltose-dextrin, or nothing Four hours later dams were sacrificed and embryos and placentae were harvested. RNA was extracted, labeled and hybridized to Affymetrix Mouse Genome 430 v2 microarray chips. Data were normalized, subjected to analysis of variance and tested for enrichment of gene ontology (GO) molecular function and biological process using the Database for Annotation, Visualization and Integrated Discovery (DAVID).
Results
Several gene classes were differentially expressed in B6 and D2 regardless of treatment, including genes involved in polysaccharide binding and mitosis. Prenatal alcohol exposure altered expression of a subset of genes, including genes involved in methylation, chromatin remodeling, protein synthesis and mRNA splicing. Very few genes were differentially expressed between maltose-exposed tissues and tissues that received nothing, so we combined these groups for comparisons with ethanol. While we observed many expression changes specific to B6 following prenatal alcohol exposure, none were specific for D2. Gene classes up-or down regulated in B6 following prenatal alcohol exposure included genes involved in mRNA splicing, transcription and translation.
Conclusions
Our study identified several classes of genes with altered expression following prenatal alcohol exposure, including many specific for B6, a strain susceptible to ethanol teratogenesis. Lack of strain specific effects in D2 suggests there are few gene expression changes that confer resistance. Future studies will begin to analyze functional significance of the expression changes.
Keywords: fetal alcohol spectrum disorders, development, gene expression, inbred strains
Introduction
Prenatal alcohol exposure can have detrimental effects on the developing fetus. The most severe cases are diagnosed with Fetal Alcohol Syndrome (FAS), a disorder characterized by prenatal and/or postnatal growth retardation, craniofacial abnormalities, and CNS dysfunction (Jones and Smith 1973, 1975). Because some children with prenatal alcohol damage do not display the full spectrum of FAS symptoms, particularly facial dysmorphology, the term Fetal Alcohol Spectrum Disorders (FASD) has been coined to describe varying degrees of ethanol teratogenesis, including FAS (Koren et al., 2003; Sokol et al., 2003). The estimated incidence of FASD in the United States is generally acknowledged to be around 1% of live births (May and Gossage, 2001; Sampson et al., 1997), although more recent studies examining FASD in specific populations, and in other parts of the world, suggest that the incidence maybe considerably higher (Garcia-Algar et al., 2008; Hutson et al., 2010; Landgren et al., 2010; Miller et al. 2006).
Many variables play a role in susceptibility to FASD, including genetics. Human studies have shown that monozygotic twins are more similarly affected than dizygotic twins following prenatal alcohol exposure (Chasnoff, 1985; Christoffel and Salafsky, 1975; Palmer et al., 1974 Riikonen, 1994; Streissguth and Dehaene, 1993). Other studies have shown that different alleles of the alcohol dehydrogenase gene (ADH), an enzyme involved in ethanol metabolism, can influence the severity of teratogenesis in different ethnic populations (Warren & Li, 2005). While these studies have shown a role for genetics in the development of FASD, they are few in number, and the range of genetic variation is unknown.
Mice are an excellent model organism for investigating genetic effects on many genotypes, and are well suited for studying the effects of prenatal ethanol exposure (Driscoll et al., 1990). Inbred strains and selectively bred mice have been shown to differ in teratogenic outcomes following in utero ethanol exposure. These differences affect embryolethality; development of various brain structures; fetal weight gain; and digit, skeletal, ocular, renal and heart anomalies (Boehm et al., 1997; Downing et al., 2009; Giknis et al., 1980; Gilliam et al., 1989, 1997; Webster et al., 1980). Results from these mouse models provide additional evidence for the importance of genetics in susceptibility to FASD.
Identification of susceptible and resistant strains also allows elucidation of the genetic architecture underlying ethanol teratogenesis. Work from our laboratory and others has shown that C57BL/6J (B6) mice are susceptible to growth retardation and a number of morphological malformations following in utero ethanol exposure, while DBA/2J (D2) mice are relatively resistant (Boehm et al., 1997; Downing and Gilliam, 1999; Downing et al., 2009; Gilliamet al., 1997; Webster et al., 1980). In a reciprocal cross between B6 and D2, we identified a maternal effect on skeletal malformations following in utero ethanol exposure (Downing and Gilliam, 1999); genetically identical F1 fetuses carried in a B6 dam had a significantly higher percentage of skeletal malformations (digit and vertebral) than F1 fetuses carried in a D2 dam. This showed that maternal genotype can play an important role in susceptibility to the teratogenic effects of ethanol.
We have recently completed quantitative trait locus (QTL) mapping in recombinant inbred mice derived from a cross between B6 and D2 and identified regions on chromosomes harboring genes mediating susceptibility to several measures of ethanol teratogenesis (Downing et al. 2008; Downing et al., under review). In this study we further elucidate the genetic pathways mediating susceptibility to ethanol teratogenesis by examining global changes in gene expression in embryos and placenta from B6, D2, and reciprocal F1s. Results from this study will allow us to correlate changes in gene expression with susceptibility and resistance to the teratogenic effects of ethanol.
Methods
Mating, Ethanol Administration and Tissue Harvesting
Male and female C57BL/6J (B6) and DBA/2J (D2) mice were purchased from the Jackson Laboratory, Bar Harbor, Maine. Two females were placed with a singly-housed male from 7:00–9:00 each morning. Females were checked for seminal plugs as evidence of pregnancy, and the day of plug detection was designated gestational day 0 (GD 0). Plugged females were weighed and singly housed. At 12 pm on GD 9, dams were weighed to verify pregnancy and then intragastrically intubated with either 5.8 g/kg ethanol (20% w:v) or an is caloric amount of maltose dextrin (MD); a nonintubated (no-gavage) control group was also included. Our mating scheme generated four embryonic genotypes: true-bred B6xB6 (maternal genotype first) and D2xD2 and reciprocal B6xD2 and D2xB6 F1 genotypes; B6xD2 F1s will be referred to as BxD, while D2xB6 F1s will be referred to as DxB. Five litters per genotype and treatment were generated. Four hours after intubation, dams were sacrificed and c-sectioned. The four-hour time point was chosen because a previous study from our laboratory showed a decrease in expression of the Igf2 gene in B6 embryo and placenta following prenatal alcohol exposure (Downing et al., 2011). Seminal plugs were detected at approximately 9 am on GD 0 and tissues were harvested 9 days + 7 hours later, which is approximately GD 9.3. Embryo and placenta were extracted under a dissecting microscope and immediately placed in a −80 freezer for subsequent RNA extraction. At this stage of development, chorioallantoic attachment of the placenta to the decidua has taken place and villi have begun to form; placental tissue included mostly chorioallantois with a bit of decidua. Within a litter, all embryos were pooled for RNA extraction and all placentae were pooled for RNA extraction. We did not stage embryos by number of somites, so it is probable that our embryos (both within strain and between strains) contained different numbers of somites.
RNA Preparation
Approximately 30 mg of tissue was homogenized for ~30s in Qiagen RLT buffer (RNeasy kit) using a Kinematica Polytron rotor-type homogenizer (Lucerne, Switzerland). Total RNA longer than 200 nucleotides was extracted using a Qiagen RNeasy kit (Duesseldorf, Germany ) per manufacturer’s instructions. The polyadenylated RNA was converted to double-stranded cDNA using the Affymetrix Gene Chip One-Cycle Target Labeling and Control Reagent skit (Santa Clara, CA). cDNA was converted to biotin-labeled cRNA using the Affymetrix IVT Labeling kit. Isolated cRNA was fragmented, quality checked, and hybridized to the Affymetrix Mouse Genome 430 v2 Arrays per the Affymetrix EukGE-WS2v4_450 Hybridization protocol; arrays were scanned using an Affymetrix Gene Array scanner. Data are available publicly at PhenoGen (http://phenogen.ucdenver.edu).
Data Analysis
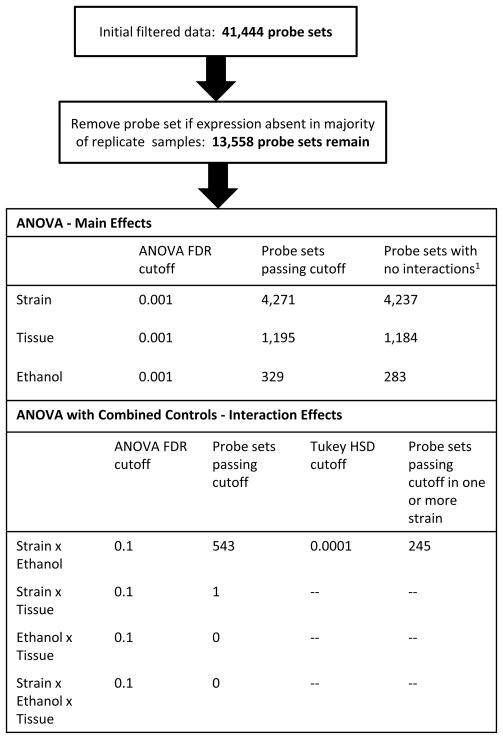
The 120 microarrays (4 strains × 2 tissues × 3 treatments × 5 mice) were quality checked using the simpleaffy R software package of Bioconductor (Wilson & Miller, 2005). Quality control measures for all arrays used in this study were with in acceptable limits. Probes on the arrays were excluded from the analysis if their DNA sequences did not match the National Center for Biotechnology Information (NCBI) build m37, matched the genome at multiple sites or harbored a SNP among any of the 19 strains (including B6 and D2) used in PhenoGen, whose genotype data is available at the Imputed Genotype Resource from the Jackson Laboratory (http://cgd.jax.org/datasets/popgen/imputed.shtml). Entire probe sets were excluded if more than 7 of the 11 associated probes were excluded. As a result, 111,297 probes and 3,657 probe sets were eliminated from the study, leaving 41,444 probe sets (92%) representing 19,806 genes in the NCBI database.
The signal intensities from the micro arrays were normalized using the affy and gcrma R software packages of Bioconductor (Gentleman et al., 2004) and adjusted for batch effects using the ComBat package (Johnson, et al., 2007). The mas5 Bioconductor package was used to generate “Present” and “Absent” calls. Following the suggestions of McClintick and Edenberg (2006), we retained a probe set in our analysis only if it was called “Present” in at least 3 of the 5 replicates for strain, treatment, and tissue (Fig. 1). The remaining 13,558 probe sets deemed present represented 8,520 genes in the NCBI database.
Figure 1. Analysis procedure.
Probe sets were excluded from the analysis if expression was present in less than three of the five replicates per strain and treatment. The 13,558 probe sets considered present represented 8521 distinct known genes. The table shows the statistical cutoffs used to select informative probe set lists for the analysis of each variable.
We used analysis of variance (ANOVA) to test for expression effects due to strain, treatment, tissue, and their interactions. Because of the large number of probe sets, we used a false discovery rate (FDR) cutoff rather than an ANOVA p-value to assess statistical significance (Benjamini & Hochberg, 1995). For the main effects of strain, ethanol, and tissue, we used an FDR threshold of 0.001 (Fig. 1). The probe sets passing this criterion were filtered further by removing those suggestive (FDR < 0.1) for an interaction among the three variables (Fig. 1). We made repeated use of Tukey’s honest significant difference test (Tukey’s HSD) to evaluate statistical differences between factor combinations (Kutner et al., 2005). The difference between MD and no-gavage controls was assessed by making pair-wise comparisons between each of the treatment factors, using a Tukey HSD p-value threshold of p < .0001 on the probe sets previously selected for response to treatment; there was no significant difference. Therefore, to analyze the interaction between strain and ethanol, we reran the ANOVA with MD and no-gavage controls combined. We found suggestive evidence (FDR < 0.1) of a strain × ethanol interaction for 543 probe sets. We then used Tukey’s HSD test with a p-value cutoff of .0001 as our criterion for defining probe sets significantly affected by a strain × treatment interaction (Fig. 1). Specifically, a probe set was selected if the HSD p-value for at least one within-strain comparison between ethanol and control was < .0001. This particular test will not select those probe sets that have similar patterns of differential expression in response to ethanol across all strains. Thus, our list of probe sets with significant strain × ethanol interaction effect is a separate list from probe sets with significant ethanol effects.
Many probe sets had differential expression for ethanol only in B6, but none only in D2. It is possible that differential expression in D2 was masked by extreme variability. To compare the variability in expression between the two strains, for each probe set in the filtered list, we tested for significant differences in variance between the strains and calculated the p-value for the F-statistic on the ratio Var (D2E)/Var (B6 E), where D2E and B6 E are the vectors of expression values for the DBA and B6 strains respectively, using the approach in Ho et al., 2008.
We tested for enrichment of gene ontology (GO) molecular function and biological process terms in our gene lists using the Database for Annotation, Visualization and Integrated Discovery (DAVID ) v6.7 Gene Functional Classification Tool at http://david.abcc.ncifcrf.gov/tools.jsp (Huang et al. 2009, Dennis et al. 2003). Enrichment was assessed against the background of 13,558 probe sets considered present on the chips in our analysis of the main effects of strain, tissue, ethanol, and the 243 probe sets specific to the B6 ethanol by strain interaction response. For the 23 probe sets that overlapped between B6 and DBA in the ethanol by strain interaction response, we used the 243 probe sets affected in B6 as the background. For group classification stringency, we set the Kappa similarity term overlap to 5 and Kappa similarity threshold setting to 0.70 when the number of probe sets being analyzed was <1,000 and 0.80 when the number was> 1,000. We used an enrichment score >1.3 (equivalent to p < 0.05, uncorrected for multiple hypotheses) as our criterion for notable enrichment as suggested by Huang et al (2009). We also conducted a visual inspection of the input gene lists to identify genes belonging to enriched molecular functions and biological processes in the outputs that were missed by DAVID, a known problem caused by variability in the depth of annotation (http://david.abcc.ncifcrf.gov/forum/cgi-bin/ikonboard.cgi?act=ST;f=3;t=1389). Consequently, we have added Clns1a to Table 1 (Chloride ion binding); Mmp14 to Table 1 (Collagen metabolic process); Kif2a, Kif13a to Table 1 (Mitotic motor activity); Sfrs6 and Sfwsap to Table 1 (main effect ethanol, mRNA splicing); Sfrs6, Sfrs18, Sfswap, and Srsf11 to Table 2; Rpl30 to Table 3, Ccnt2 to Table 4, Mbd5, Mettl2, Mthfd2, Mthfr and Rg9mtd2 to supplementary table 4 (methylation) and Brd2, Brwd1, Hdac2, Hist2h3c2, Iws1, Mbtd1, Naa15, Naa25 and Uba5 to supplementary table 4 (chromatin remodeling). Therefore, even though DAVID correctly identifies enriched biological processes (under the assumption that missing annotations are infrequent and randomly distributed), we cannot guarantee that all of the input genes belonging to enriched categories have been listed in our tables.
Table 1.
Enriched molecular functions and biological processes
| Enrichment score | Process Term | Term Enrichment | Number of genes | Mean fold difference |
|---|---|---|---|---|
| S Strain Main effect (most significant 3000 probe sets) | DBA/B6 | |||
| 4.3 | Polysaccharide binding | 1.9 | 34 | 1.32 |
| 4.2 | Mitosis | 1.6 | 59 | 0.90 |
| 2.0 | Calcium-dependent phospholipid binding (Annexins) | 2.5 | 7 | 1.12 |
| 1.7 | Chloride ion binding | 2.1 | 11 | 0.89 |
| Tissue Main Effect (1195 probe sets): | emb/plac1 | |||
| 7.0 | Cell adhesion | 2.0 | 61 | 1.25 |
| 3.8 | Polysaccharide binding | 2.5 | 19 | 1.32 |
| 3.5 | Ion homeostasis | 2.1 | 30 | 1.17 |
| 2.8 | Muscle organ development | 2.1 | 22 | 1.27 |
| 2.6 | Peptidase inhibitor | 2.5 | 15 | 1.31 |
| 2.6 | Regulation of phosphorylation | 1.7 | 33 | 1.14 |
| 2.5 | Regulation of cell migration | 2.2 | 14 | 1.12 |
| 2.1 | Response to hormone stimulus | 2.1 | 21 | 1.16 |
| 2.0 | Neuron development | 1.7 | 22 | 1.24 |
| 1.9 | Vasculature development | 1.7 | 33 | 1.20 |
| 1.9 | Negative regulation of transcription | 1.5 | 37 | 1.10 |
| 1.7 | IGF binding | 3.4 | 6 | 1.19 |
| 1.7 | Lung development | 2.0 | 15 | 1.19 |
| 1.4 | Negative regulation of apoptosis | 1.5 | 25 | 1.08 |
| Ethanol main effect (283 probe sets) | EtOH/ctrls2 | |||
| 2.1 | Methylation | 4.2 | 7 | 0.94 |
| 1.9 | Chromatin organization | 2.5 | 13 | 0.91 |
| 1.6 | Ribosome biogenesis | 3.4 | 7 | 0.87 |
| 1.5 | mRNA splicing | 2.0 | 12 | 0.88 |
| Strain-specific effects of ethanol | B6/DBA | |||
| 8.4 | mRNA splicing | 5.3 | 23 | 0.68 |
| 2.6 | Translation | 2.5 | 10 | 0.71 |
| 2.1 | Transcription | 1.5 | 36 | 0.76 |
emb/plac = embryo/placenta
EtOH/ctrls = ethanol exposure/mean of maltose dextrin and no-gavage controls
Table 2.
mRNA splicing genes altered in expression in B6 in response to ethanol
| Gene Symbol1 | Gene Name | Fold Change (Etoh/Cont) |
|---|---|---|
| Ddx5 | DEAD-box polypeptide 5 [helicase] | 0.74 |
| Hnrnpa2b1 | Heterogeneous nuclear ribonucleoprotein A2/B1 | 0.64 |
| Ncbp2 | Nuclear cap binding protein subunit 2 | 0.75 |
| Pabpc1 | Poly (A) binding protein, cytoplasmic 1 | 0.71 |
| Pnn | Pinin [splicing regulator] | 0.65 |
| Ppp4r2 | Protein phosphatase 4, regulatory subunit 2 | 0.76 |
| Prpf38a | PRP38 pre-mRNA processing factor 38 (yeast) domain containing A | 0.72 |
| Prpf38b | PRP38 pre-mRNA processing factor 38 (yeast) domain containing B | 0.63 |
| Prpf4b | PRP4 pre-mRNA processing factor 4 homolog B (yeast) | 0.69 |
| Rbm5 | RNA binding motif protein 5 | 0.78 |
| Rbm39 | RNA binding motif protein 39 | 0.76 |
| Sfpq | Splicing factor proline/glutamine rich (polypyrimidine tract binding protein associated) | 0.77 |
| Sfrs18 | Serine/arginine-rich splicing factor 18 | 0.66 |
| Sfswap | splicing factor, suppressor of white-apricot homolog (Drosophila) | 0.81 |
| Srek1 | Splicing regulatory glutamine/lysine-rich protein 1 | 0.64 |
| Srsf1 | Serine/arginine-rich splicing factor 1 | 0.72 |
| Srsf6 | Serine/arginine-rich splicing factor 6 | 0.64 |
| Srsf7 | Serine/arginine-rich splicing factor 7 | 0.67 |
| Srsf10 | Serine/arginine-rich splicing factor 10 | 0.62 |
| Srsf11 | Serine/arginine-rich splicing factor 11 | 0.78 |
| Syncrip | Synaptotagmin binding, cytoplasmic RNA interacting protein | 0.76 |
| Tardbp | TAR DNA binding protein | 0.66 |
| Thoc6 | THO complex 6 homolog (Drosophila) [mRNA transport to cytoplasm] | 0.77 |
Mouse Genome Informatics database (http://www.informatics.jax.org).
Fold change < 1 indicates reduced expression in response to ethanol relative to a value of 1. Mean = 0.68, SD = 0.16
Table 3.
Translation genes altered in expression in B6 in response to ethanol
| Gene Symbol1 | Gene Name | Fold Change (Etoh/Cont)2 |
|---|---|---|
| Aimp2 | aminoacyl tRNA synthetase complex-interacting multifunctional protein 2 | 0.79 |
| Eef1e1 | eukaryotic translation elongation factor 1 epsilon 1 | 0.68 |
| Rpl5 | ribosomal protein L5 | 0.72 |
| Rpl7l1 | ribosomal protein L7-like 1 | 0.80 |
| Rpl12 | ribosomal protein L12 | 0.53 |
| Rpl15 | ribosomal protein L15 | 0.77 |
| Rpl30 | ribosomal protein L30 | 0.79 |
| Rpl31 | ribosomal protein L31 | 0.84 |
| Rps24 | ribosomal protein S24 | 0.49 |
| Rpl41 | ribosomal protein L41 | 0.66 |
Mouse Genome Informatics database (http://www.informatics.jax.org).
Fold change < 1 indicates reduced expression in response to ethanol relative to a value of 1. Mean = 0.71, SD = 012.
Table 4.
Transcription genes altered in expression in B6 in response to ethanol
| Gene Symbol1 | Gene Name | Fold Change (Etoh/Cont)2 |
|---|---|---|
| 2210018M11Rik | RIKEN cDNA 2210018M11 gene | 0.82 |
| Aebp2 | AE binding protein 2 | 0.63 |
| Bcl3 | B-cell leukemia/lymphoma 3 | 0.57 |
| Bclaf1 | BCL2-associated transcription factor 1 | 0.75 |
| Brms1l | breast cancer metastasis-suppressor 1-like | 0.80 |
| Ccnh | cyclin H | 0.62 |
| Ccnt1 | cyclin T1 | 0.79 |
| Ccnt2 | cyclin T2 | 0.63 |
| Cnot4 | CCR4-NOT transcription complex, subunit 4 | 0.60 |
| Creb1 | cAMP responsive element binding protein 1 | 0.68 |
| Crebzf | CREB/ATF bZIP transcription factor | 0.68 |
| Crem | cAMP responsive element modulator | 0.66 |
| Foxc1 | forkhead box C1 | 1.35 |
| Foxk2 | forkhead box K2 | 0.75 |
| Gtf2h3 | general transcription factor IIH, polypeptide 3 | 0.81 |
| Hdac9 | histone deacetylase 9 | 0.60 |
| Kdm4c | lysine (K)-specific demethylase 4C | 0.77 |
| Nfya | nuclear transcription factor-Y alpha | 0.75 |
| Pias2 | protein inhibitor of activated STAT 2 | 0.73 |
| Pnn | Pinin | 0.65 |
| Pura | purine rich element binding protein A | 0.76 |
| Rbm39 | RNA binding motif protein 39 | 0.76 |
| Sfpq | splicing factor proline/glutamine rich (polypyrimidine tract binding protein associated) | 0.77 |
| Suv420h1 | suppressor of variegation 4–20 homolog 1 (Drosophila) | 0.75 |
| Tardbp | TAR DNA binding protein | 0.66 |
| Trip4 | thyroid hormone receptor interactor 4 | 0.81 |
| Zbtb6 | zinc finger and BTB domain containing 6 | 0.81 |
| Zfp26 | zinc finger protein 26 | 0.79 |
| Zfp275 | zinc finger protein 275 | 0.78 |
| Zfp322a | zinc finger protein 322A | 0.73 |
| Zfp654 | zinc finger protein 654 | 0.72 |
| Zfp672 | zinc finger protein 672 | 0.74 |
| Zfp715 | zinc finger protein 715 | 0.80 |
| Zfp719 | zinc finger protein 719 | 0.72 |
| Zkscan3 | zinc finger with KRAB and SCAN domains 3 | 0.76 |
| Zxdc | ZXD family zinc finger C | 1.27 |
Mouse Genome Informatics database (http://www.informatics.jax.org).
Fold change < 1 indicates reduced expression in response to ethanol relative to a value of 1. Mean = 0.76, SD = 0.15
Quantitative PCR
A subset of differentially expressed genes was verified using real-time, quantitative PCR, including Adamts1, Bcl3, Bfar, Cd44, Crebzf, Ddx5, Ppp4r2, Rbm5, Suv420h1, Stab2, Thbs2 and Vcan. Mice were mated and treated, and tissue was excised, as described above. Three litters were generated per treatment; within a litter all embryonic tissue was pooled for RNA extraction. Tissue was homogenized using a Power Gen 125 Homogenizer (Fisher Scientific) and RNA extracted using a Qiagen RNeasy Tissue kit, following manufacturer’s instructions. Total RNA was then reverse transcribed to yield single-strand cDNA using a Promega ImPromII Reverse Transcription System, following manufacturer’s instructions. Primers were designed using Primer Express software (ABI). Quantitative RT-PCR was performed on cDNAs using SYBR green chemistry and a Realplex2 MasterCycler (Eppendorf). Relative quantification of mRNA levels was determined by normalizing against a control gene, Gapdh, using the comparative CT method (Livak and Schmittgen, 2001; Schmittgen and Livak, 2008). Each time a sample was assayed, it was run in triplicate.
Results
Analysis of Variance: Main Effects
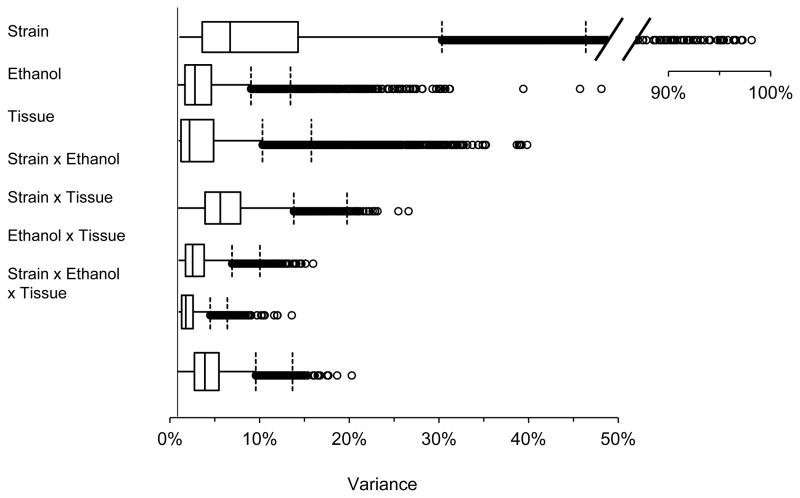
Across all probe sets, the effect of strain, independent of ethanol or tissue, explained the most variation (Fig. 2): > 25% of the variation in 1,709 probe sets (13%), > 50% in 570 probe sets (4%), and > 90% in 33 probe sets. The 3,000 probes sets (input limit of DAVID) most affected by strain on the basis of statistical significance (FDR < 3×10*−5) indicated enrichment for polysaccharide binding genes (enrichment score = 4.3, 1.9-fold enrichment, Bonferroni-corrected p = 0.047, Table 1). These 34 polysaccharide binding genes were expressed 32% higher on average in DBA than B6 (SE = 13%; 19 genes expressed higher in DBA, 15 lower; Supplementary Table 1). There was also suggestive enrichment for an effect on mitosis genes (enrichment score 4.2, 1.6-fold enrichment; Table 1). On average, the 59 mitosis genes were expressed 10% lower in DBA than B6 (SE= 4%, 50 genes lower in DBA, 9 higher; Supplementary Table 2). As shown in Table 1, calcium-dependent phospholipid binding and chloride ion binding also had enrichment scores greater than the suggestive criterion of 1.3 (uncorrected p < 0.05; Huang et al. 2009).
Figure 2. Box-and-whiskers plots showing the variance explained by each variable across all probe sets.
The data set consists of 13,558 probe sets. The line segment to the left of the box is the first quartile, the box segment to the right of this line is the second quartile, the vertical line within each box represents the median, the box segment to the right of the median is the third quartile, and the line segment to the right of the box is the fourth quartile. Points to the right of the first vertical dashed line are outliers (> 1 box distance from third quartile). Points to the right of the second vertical dashed line (> 2 box distances from third quartile) are extreme outliers. Outliers are genes for which the variable explains an exceptionally large amount of the variation in expression level.
Tissue, independent of strain or treatment, explained > 10% of the variation in 1,100 probe sets (8%) and > 25% in 104 probe sets (Fig. 2). The 1,184 probe sets having an FDR < 0.001 and no interaction effects (Fig. 1) were enriched for a large variety of molecular functions and biological processes (Table 1). All of these processes were expressed at a higher level in embryo than placenta; across all 1,184 probe sets, expression was 14% higher on average in embryo. The most significant difference was a 2-fold enrichment for genes affecting cell adhesion (enrichment score 7.0, Bonferroni-corrected p = 0.0004). The 61 cell adhesion genes were expressed 25% higher on average in embryo than placenta (SE = 3%; 52 genes expressed higher, 9 lower; Supplementary Table 3). There was also suggestive enrichment and higher expression in embryo for polysaccharide binding genes (enrichment score = 3.8, Table 1), a category also suggested above as one of the main effects of strain. The gene expression levels were also higher in embryo for processes such as muscle, neuron, and lung development (Table 1).
Ethanol exposure, independent of strain or tissue, explained > 10% of the expression variation in 500 probe sets (4%) and > 25% in 28 probes sets (Fig. 2). The 283 probe sets having an FDR < 0.001 and no interaction effects were suggestively enriched for genes affecting methylation (RNA, DNA and protein), chromatin organization, ribosome biogenesis, and mRNA splicing (Table 1; Supplementary Table 4). Ethanol had an inhibitory effect on all of these processes (Table 1; across all 283 probes, ethanol inhibited expression by 7% on average).
ANOVA: Interactions
The only substantial interaction effect among the three variables (strain, ethanol exposure, and tissue) was between strain and ethanol exposure (Fig. 2). This interaction explained > 10% of the variation in 1,062 genes and > 15% in 126 genes. The strain × tissue interaction explained > 10% of the variation in only 45 genes (Fig. 2), with only one gene having an FDR < 0.1 (Cryab: crystallin alpha B). The ethanol x tissue interaction explained > 10% of the variation in only 4 probe sets (Fig. 2); no gene had an FDR < 0.1. The 3-way interaction of strain × ethanol × tissue explained > 10% of the variation in 203 probes (Fig. 2), but no genes had an FDR < 0.1.
Ethanol controls
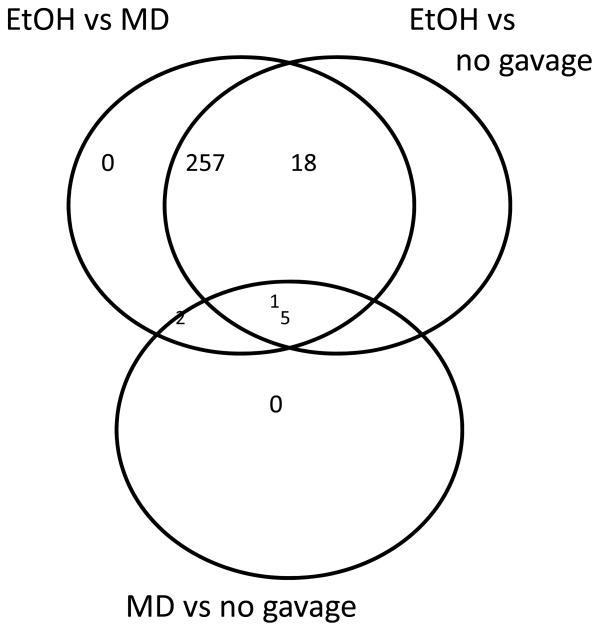
Maltose dextrin is commonly used in fetal alcohol studies to control for the calorie content of the ethanol dose. However, MD could have its own effects on gene expression that could confound the comparison to ethanol. Therefore, we included controls that were not gavaged (no-gavage controls). In response to ethanol relative to MD, there were 260 probe sets that were significantly altered in their expression across all strains (Fig. 3). Of these, 258 were also significantly altered relative to the no-gavage control (Fig. 3). Therefore, the choice of control had a negligible effect on the identification of ethanol responsive genes. The direct comparison of MD to no gavage also suggested only 8 expression differences (Fig. 3). Consequently, we combined the MD and no-gavage controls and reran the ANOVA to analyze the strain-specific effects of ethanol.
Figure 3. Venn diagram showing the large overlap in ethanol response genes defined by comparison to either the maltose dextrin or no-gavage controls.
EtOH = ethanol, MD = maltose dextrin. ANOVA indicated 283 probe sets with an FDR < 0.001. Tukey’s HSD test was then used to subdivide the list according to treatment (p < 0.001) Results indicate that very few genes were differentially altered in their response to ethanol when comparing the response to maltose dextrin or no gavage.
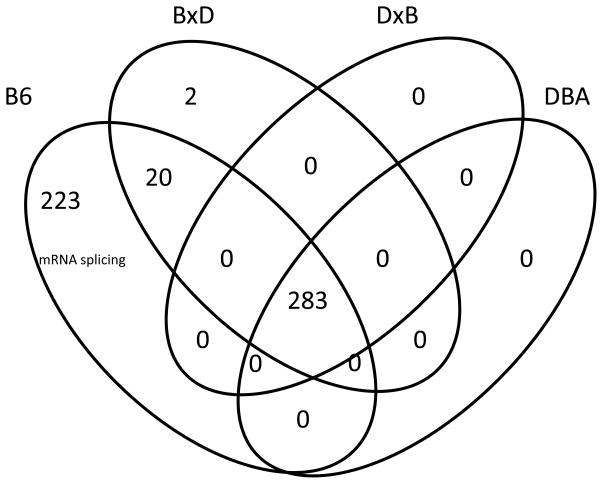
Strain-specific effects of ethanol
In addition to the 283 probe sets whose expression was altered by ethanol across all four strains, there were 245 probe sets whose response was specific to just one or two strains. The vast majority of these responses (223) were specific to B6, two were specific to the BxDF1s, and twenty were shared by B6 and the BxDF1s (Fig. 4). In contrast, there were no ethanol expression effects specific to DBA or the DxBF1s (Fig. 4). The possibility exists that greater variability among the DBA samples masked differential expression between ethanol and control. We compared the variances for our filtered probe sets between the two strains using an F-test (see Methods) and the distribution of p-values indicates that there are at least as many probe sets with greater variance in the B6 samples as there are in the D2 samples. After correction for multiple testing, there are 16 probe sets with FDR < 0.1. We concluded that differential variability is not a factor in the selection of probe sets with differential expression between ethanol and controls in B6 and not in DBA
Figure 4. Venn diagram showing the number of ethanol response genes unique to each strain or strain combination.
Only B6 and the BxD F1s had genes affected by ethanol that were not affected across all strains. Tukey HSD test was used to determine differential expression between ethanol and control within each strain. The genes specific to B6 were significantly enriched for mRNA splicing genes (all showing decreased expression, Table 2). The number of genes affected across all strains (283) comes from the ANOVA for the main effect of ethanol (Fig. 2).
Ninety-three percent of the 243 probe sets affected by ethanol in B6 (Fig. 4) were decreased in expression. They were also highly enriched for genes affecting mRNA splicing (enrichment score 8.4, 5-fold enrichment, Bonferroni-corrected p < 10−5); all 23 splicing genes were inhibited 20 to 40% by ethanol (mean 32%; Table 2). An inhibitory effect on mRNA splicing was also suggested as one of the main effects of ethanol as indicated above. There was suggestive enrichment for genes affecting translation as well (enrichment score 2.6, 2.5-fold enrichment, Table 1); all 10 genes were inhibited 20 to 50% (mean 29%; Table 3). There was also suggestive enrichment for genes affecting transcription (enrichment score 2.1, 1.5-fold enrichment, Table 1). The 20 probe sets affected by ethanol in both B6 and the BXD F1s were not enriched for a molecular function or biological process.
Quantitative PCR
We used q-PCR to verify expression changes of several genes. Fold changes were determined using the method of Livak and Schmittgen (2001; Schmittgen and Livak, 2008). Adamts1, Cd44, Vcan, Stab2 and Thbs2 are all genes involved in polysaccharide binding and are differentially expressed between B6 and D2 (Supplementary Table 1). For q-PCR analysis, after normalizing against Gapdh, fold changes were calculated as 2−(CTD2-CTB6), where CT is the cycle threshold. Expression changes observed in these 4 genes in the microarray analyses were verified with q-PCR (Table 5). Bcl3, Crebzf, Ddx5, Ppp4r2, Suv420h1 and Rbm5 were genes down regulated in B6, but not D2, following prenatal alcohol exposure. For q-PCR analysis, after normalizing against Gapdh, fold changes (B6) were calculated as 2−(CTetoh-CTcontrol). As can be seen in Table 5, all expression changes were verified except Ppp4r2. Microarray analyses showed that in B6, Ppp4r2 was down regulated following prenatal alcohol exposure (0.76 Ethanol/Control), while q-PCR showed a 0.84-fold increase in Ppp4r2 (B6) following prenatal alcohol exposure. Bfar and Crebzf are genes that were down regulated following prenatal alcohol exposure. For q-PCR analysis, after normalizing against Gapdh, fold changes were calculated as 2−(CTetoh-CTcontrol). Q-PCR verified down regulation of Bfar but not Crebzf (Table 5). As discussed above, Crebzf was down regulated in B6, but not D2, following prenatal alcohol exposure in both the microarray and q-PCR analyses (Table 5). Microarray analysis also showed a small but statistically significant decrease in Crebzf following ethanol exposure regardless of genotype (0.88 Etoh/Cont; Table 5); this small decrease was not verified by q-PCR, where we observed a small (non-significant) increase in Crebzf expression.
Table 5.
Changes in gene expression verified by quantitative, real-time PCR.
| Gene | FC MA | FC qPCR |
|---|---|---|
| Adamts1a | 1.2 | 0.45 |
| Cd44a | 0.75 | −0.62 |
| Vcana | 0.62 | −0.93 |
| Stab2a | 4.10 | 2.8 |
| Thbs2b | 1.90 | 1.15 |
| Bcl3b | 0.57 | −1.03 |
| Crebzfb | 0.68 | −1.21 |
| Ddx5b | 0.74 | −0.97 |
| Ppp4r2b | 0.76 | 0.86 |
| Suv420h1b | 0.75 | −1.42 |
| Rbm5b | 0.78 | −1.33 |
| Bfarc | 0.86 | −0.77 |
| Crebzfc | 0.88 | 0.11 |
FC MA = fold change from the microarray analysis; FC Q-PCR = fold change from the q-PCR analysis.
These genes were differentially expressed between B6 and D2, regardless of treatment. Fold change is expressed as D2/B6 for the MA data, while for the q-PCR data fold change is expressed as 2−(CTD2-CTB6), where CT = cycle threshold. For example, Adamts1 showed a 0.45-fold increase in expression in D2 compared to B6, while Cd44 showed a 0.62 decrease in expression in D2 compared to B6.
These genes showed decreased expression in B6 following prenatal alcohol exposure but not D2. Fold change is expressed as Ethanol/Control in B6 for the microarray data, while for q-PCR fold change (B6) is expressed as 2−(CTetoh-CTcontrol).
These genes showed decreased expression following prenatal alcohol exposure. Fold change is expressed as Ethanol/Control for the microarray data, while for q-PCR fold change is expressed as 2−(CTetoh-CTcontrol).
Discussion
Genetic variation in susceptibility to the teratogenic effects of alcohol has been reported in both humans and mice. Studies are few, however, and the range of genetic variation is relatively unknown. We have shown that B6 fetuses are susceptible to a number of morphological malformations following ethanol exposure on GD 9, including digit and axial skeletal malformations, hydronephrosis and dilated brain ventricles, while D2 fetuses are relatively resistant (Boehm et al., 1997; Downing and Gilliam, 1999; Downing et al., 2009). Recently we have finished quantitative trait locus (QTL) mapping for ethanol teratogenesis in recombinant inbred mice derived from crossing B6 and D2 (BXD RIs) and have identified several candidate genes mediating teratogenesis (Downing et al., 2008; Downing et al., in revision). Results from this study will identify changes in gene expression in susceptible and resistant strains and further elucidate the genetic architecture underlying ethanol teratogenesis.
B6 and D2 have been shown to differ in several aspects of development, including somitogenesis and brain development (Thiel at al., 1993; Luzzatto et al., 1988). Many genes and transcripts were differentially expressed between B6 and D2 (regardless of treatment), including 34 polysaccharide binding genes;19 were expressed higher in D2 and 15 expressed higher in B6. Polysaccharides are a large class of carbohydrate structures, including starches, glycogen, proteoglycans and glycosaminoglycans such as hyaluronan, and play a role in embryonic development. Of particular interest are the genes Adamts1, Vcan and Cd44. Adamts1 lies in a QTL region mediating fused digits and is expressed in the interdigital web during digit formation (Downing et al., in revision); most of our fused digits appear to result from failure of the interdigital web to regress properly. We found that Adamts1 is expressed at a significantly higher level in D2 embryos compared to B6 embryos. B6 and D2 are polymorphic for many single nucleotide polymorphisms (SNPs) in Adamts1, including several in 5′ and 3′ UTR. Adamts1 cleaves the protein versican (Vcan), a chondroitin sulfate proteoglycan and major component of the extracellular matrix (ECM); versican is also involved in interdigital web regression during development. Vcan is expressed at a significantly higher level in B6 embryos compared to D2 and is polymorphic for several SNPs in B6 and D2, including 3 in the 5′ UTR. Cd44 is located in a QTL region for vertebral malformations and is expressed at a higher level in D2 embryos compared to B6 embryos; it is highly polymorphic in B6 and D2. Cd44 codes for a cell-surface glycoprotein that functions as a receptor for hyaluronan (a polysaccharide) and other ligands. Both Cd44 and hyaluronan are highly expressed in the developing sclerotome and axial skeleton, including vertebral arches, bodies and intervertebral discs (Fenderson et al., 1993; Wheatley et al., 1993). While differential expression of these 3 genes was observed across treatments (not in response to ethanol), these baseline expression differences could play a role in differential sensitivity to ethanol observed in B6 and D2 fetuses. Future studies need to examine expression of these genes in tissues where we observe malformations, specifically in developing digits and axial skeleton. Interestingly, Hashimoto-Torri et al. (2011) reported that Vcan was downregulated in cerebral cortex of CD-1 mice following prenatal alcohol exposure. In addition to polysaccharide binding genes, mitosis genes (higher expression in B6), calcium-dependent phospholipid binding genes (higher expression in D2) and genes involved in chloride ion binding (higher expression in B6) showed differential at the suggestive level. Thus, many genes and several gene classes are differentially expressed between B6 and D2, independent of treatment.
Following prenatal alcohol exposure several classes of genes showed differential expression, including genes involved in methylation (RNA, DNA and protein), chromatin organization and remodeling, ribosome biogenesis/protein synthesis and mRNA splicing. Other studies have supported similar findings. Gutala et al. (2004) exposed cultured cortical neurons from GD 14 embryos (B6) to ethanol and reported that expression of genes related to the ubiquitin-proteasome pathway and genes involved in protein synthesis were altered. We also observed downregulation of several genes in the ubiquitin pathway, including Usp34, Ube2n, Uba5 and Ubxn2b; Otud7b was upregulated. Liu et al. (2009) exposed cultured B6 embryos to ethanol and found altered expression of Ueb3c; they also reported altered expression of several genes involved in chromatin remodeling, including Brwd1, a gene we found was downregulated following prenatal alcohol exposure. While they did not report altered gene expression, Liu et al. reported altered DNA methylation at the Ilf3 promoter, another gene involved in chromatin remodeling; we observed downregulation of Ilf3 following prenatal alcohol exposure. In a subsequent study from the Zhou laboratory (Zhou et al., 2011), GD 8.25 embryos (B6) were exposed to ethanol in culture. Using the same Affymetrix Mouse 430 2.0 microarrays, they reported altered expression of several epigenetic genes, including downregulation of Hist3h2a, a gene we also observed was downregulated following in utero ethanol exposure. Zhou et al. (2011) also reported altered expression of several ubiquitin genes and genes involved in protein synthesis. It appears as if genes involved in the ubiquitin pathway, genes involved in protein synthesis and epigenetic genes involved in chromatin remodeling and methylation are important targets of prenatal alcohol exposure.
One of the advantages of the present study is the use of strains of mice that are susceptible and resistant to the ethanol teratogenesis. Observation of expression changes in susceptible, but not resistant strains (and vice versa) greatly narrows the list of candidate genes mediating teratogenic susceptibility. Surprisingly, all strain-specific expression changes occurred in B6 or BXD; none were specific for D2 or DXB. This lack of strain-specific effects in D2 and DXB was not due to greater variability among these samples; variability in expression was actually greater among B6 samples. This suggests that there are no (or very few) gene expression changes that confer resistance to the teratogenic effects of alcohol, at least in the D2 strain. However, one caveat that must be kept in mind in comparing gene expression between B6 and D2 is that they may be at slightly different developmental stages. The Zhou laboratory reported that on GD 8.25, no embryos from four D2 dams had any somites, while embryos from B6 dams had 3–6 somites; they concluded that D2 embryos were developmentally about 6 hours behind B6 embryos (Ogawa et al. 2005). Thiel et al. (1993) showed that on GD 9, B6 embryos had a mean of 13.3 somites, while D2 embryos had an average of 10.3 somites. There was a great deal of within strain variation; B6 embryos had a range of 6–20 somites while D2 embryos had 5–18 somites. These results raise the possibility that when intubated at approximately GD 9.3, B6 and D2 embryos are at slightly different developmental time points, and results from the present study must be interpreted in this light. While they may be slightly behind B6 developmentally, the D2 genotype is relatively resistant to ethanol teratogenesis. As noted above, we observe a number of morphological malformations in B6 fetuses exposed to 5.8 g/kg alcohol at 12:00 pm on GD 9. When dosed at 12:00, 4:00, 6:00 or 8:00 pm on GD 9, D2 fetuses show very few malformations (Downing et al., unpublished data). A recent study from the Zhou laboratory, using cultured embryos, showed that D2 embryos exposed to ethanol at the same developmental time point as B6 embryos are less susceptible to developmental delay of midbrain, hindbrain, heart, caudal neural tube and hind limbs, and are less susceptible to apoptosis (Chen et al., 2011). Thus, while B6 and D2 may be at slightly different developmental stages and this may confound comparison of gene expression changes in the present study, B6 are susceptible to teratogenesis when exposed to ethanol on GD 9 while D2 are relatively resistant.
In B6, 23 genes involved in mRNA splicing were downregulated following prenatal alcohol exposure, 10 genes involved in translation were downregulated and 34 genes involved in transcription were downregulated, while 2 were upregulated. Expression of several of these genes has previously been shown to be altered following prenatal alcohol exposure. We observed a large decrease in expression of Rps24, a ribosomal protein, in B6 tissue following prenatal alcohol exposure. Gutala et al. (2004) also found this gene to be downregulated following prenatal alcohol exposure, while Green et al. (2007) showed altered expression of Rps24 in head fold tissue from two C57BL/6 sub strains following prenatal alcohol exposure. We also observed a large decrease in expression of Sfrs18 in B6 following in utero ethanol exposure and a decrease in expression of Sfrs8 in both B6 and D2; these genes are involved in mRNA splicing. Interestingly, in cortex from CD-1 mice exposed to ethanol prenatally, Hashimoto-Torii et al. (2011) reported a significant increase in expression of these two genes. Several genes involved in transcription identified in the present study have also been shown previously to be affected by prenatal alcohol exposure. In B6 we found a decrease in expression of Creb1 and Zfp322a, while we found a significant increase in expression of Zbtb44 in B6 and D2; Hashimoto-Torii et al. (2011) found a significant decrease in expression in all three of these genes. In addition, we observed a decrease in expression of Zfp143 in both strains following prenatal alcohol exposure; Green et al. also reported altered expression of this gene. Interestingly, we also observed a large decrease in B6 in the transcription factor Crebzf; this gene lies in a QTL region mediating rib malformations following in utero ethanol exposure (Downing et al., under review). The finding that genes involved in transcription, translation and mRNA splicing are affected by prenatal alcohol exposure in B6 but not D2 suggests that they play a key role in susceptibility to ethanol teratogenesis.
Of note are genes we did not find differentially expressed following prenatal alcohol exposure. We found no expression changes in genes coding for alcohol or aldehyde dehydrogenase, or Cyp2e1, a cytochrome P450 enzyme involved in ethanol metabolism. This is not surprising given that B6 and D2 show few differences in ethyl alcohol metabolism. We previously observed a maternal effect on ethanol teratogenesis (reciprocal F1 differences) in B6 and D2 fetuses (Downing and Gilliam, 1999). One source of variation that can account for a maternal effect is genomic imprinting, a type of parent-of-origin gene expression. Recently we found small decreases in DNA methylation at the imprinted Igf2 locus in B6 embryos following prenatal alcohol exposure and observed 1.0–1.5-fold decreases in four Igf2 transcripts (Downing et al., 2011). This decrease in Igf2 expression was not verified in the present study, although there was only one probe set for Igf2 that was “present”. Most imprinted genes were present on the chip but we observed no significant changes in expression following prenatal alcohol exposure. This does not support a role for genomic imprinting mediating differential susceptibility to the teratogenic effects of alcohol in B6 and D2, but we examined expression at only one time point and two tissues. Additional studies are needed to fully characterize the role of genomic imprinting in ethanol teratogenesis. Interestingly, there were 22 genes that were differentially expressed in BxD embryos, but not DxB embryos, following prenatal alcohol exposure. These included several genes involved in apoptosis: Bcl3, Chac1, Cirbp, Fem1b, Kctd11 and Thap2. Bcl3 and Cirbp are particularly intriguing because they are maternal effect genes (Jugessur et al. 2010; Park et al. 2009; Peng et al. 2000). It is possible that apoptosis may act through the maternal uterine environment, or through other maternally-mediated mechanisms, to affect teratogenesis differently in fetuses carried in B6 and D2 moms.
Several of the significant expression changes observed in our study were rather small. This is perhaps not too surprising because development is a finely tuned process. Small expression changes in any one of a number of developmental pathways could have significant effects on subsequent developmental processes. Our fold changes are in line with fold changes reported by others following in utero ethanol exposure (Green et al. 2007; Gutala et al. 2004). The direction of change in expression was remarkably consistent. Prenatal alcohol exposure decreased expression of many genes, while it increased expression of relatively few.
In summary, we found many gene expression differences between developing B6 and D2, including a number of genes involved in polysaccharide binding. Several classes of genes showed altered expression following prenatal alcohol exposure, including genes involved in methylation, chromatin remodeling, ribosome biogenesis and mRNA splicing, which supports findings from several previous studies. Interestingly, we found no expression changes that were specific for the resistant D2 strain. Gene classes that were preferentially downregulated in B6 following prenatal alcohol exposure included genes involved in transcription, translation and mRNA splicing. Future studies will begin to analyze the function of these differentially expressed genes in order to determine their roles in susceptibility to the teratogenic effects of alcohol.
Supplementary Material
Acknowledgments
This research was supported by grants R01AA016676, R24AA01466 and K01AA16922
This research was supported by NIAAA grants R01AA016676 (TEJ), R24AA01466 (BT) and K01AA16922 (KJK). We thank Dr. Brad Rikke for Gene Ontology analysis. We also thank Jami Biers and Alexi for their assistance in intubations and tissue harvesting.
References
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. JR Statistical Soc B. 1995;57:289–300. [Google Scholar]
- Boehm SL, Lundahl KR, Caldwell J, Gilliam DM. Ethanol teratogenesis in the C57BL/6J, DBA/2J and A/J inbred mouse strains. Alcohol. 1997;14:389–395. doi: 10.1016/s0741-8329(97)87950-5. [DOI] [PubMed] [Google Scholar]
- Chasnoff IJ. Fetal Alcohol Syndrome in twin pregnancy. Acta Genet Med Gemellol (Roma) 1985;34:229–232. doi: 10.1017/s0001566000004797. [DOI] [PubMed] [Google Scholar]
- Chen Y, Ozturk NC, Ni L, Goodlett C, Zhou FC. Strain differences in developmental vulnerability to alcohol exposure via embryo culture in mice. Alcohol Clin Exp Res. 2011;35:1293–1304. doi: 10.1111/j.1530-0277.2011.01465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoffel KK, Salafsky I. Fetal alcohol syndrome in dizygotic twins. J Pediatr. 1975;87:963–967. doi: 10.1016/s0022-3476(75)80919-x. [DOI] [PubMed] [Google Scholar]
- Da Lee R, Rhee GS, An SM, Kim SS, Kwack SJ, Seok JH, Chae SY, Park CH, Yoon HJ, Cho DH, Kim HS, Park KL. Differential gene profiles in developing embryo and fetus after in utero exposure to ethanol. J Toxicol Environ Health (A) 2004;67:2073–2084. doi: 10.1080/15287390490515001. [DOI] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- Downing C, Gilliam DM. Cytoplasmic factors do not contribute to a maternal effect on ethanol teratogenesis. Behav Genet. 1999;29:31–39. doi: 10.1023/a:1021485821842. [DOI] [PubMed] [Google Scholar]
- Downing C, Carosone-Link P, Gaudreau C, Kimball A, Broncucia H, Biers J, Johnson TE, Gilliam DM. Quantitative trait locus mapping for ethanol teratogenesis in BXD recombinant inbred mice. Alcohol Clin Exp Res. 2008;32 (6 Supplement):11A. doi: 10.1111/j.1530-0277.2012.01754.x. [DOI] [PubMed] [Google Scholar]
- Downing C, Balderrama-Durbin C, Broncucia H, Gilliam D, Johnson TE. Ethanol teratogenesis in five inbred strains of mice. Alcohol Clin Exp Res. 2009;33:1238–1245. doi: 10.1111/j.1530-0277.2009.00949.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downing C, Johnson TE, Larson C, Leakey TI, Siegfried RN, Rafferty TM, Cooney CA. Subtle decreases in DNA methylation and gene expression at the mouse Igf2 locus following prenatal alcohol exposure: Effects of a methyl-supplemented diet. Alcohol. 2011;45:65–71. doi: 10.1016/j.alcohol.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downing C, Balderrama-Durbin C, Kimball A, Wright H, Biers J, Gilliam DM, Johnson TE. Quantitative trait locus mapping for ethanol teratogenesis in BXD recombinant inbred mice. Alcohol Clin Exp Res. doi: 10.1111/j.1530-0277.2012.01754.x. (Under Review) [DOI] [PubMed] [Google Scholar]
- Driscoll CD, Streissguth AP, Riley EP. Prenatal alcohol exposure: Comparability of effects in humans and animal models. Neurotoxicol Teratol. 1990;12:231–237. doi: 10.1016/0892-0362(90)90094-s. [DOI] [PubMed] [Google Scholar]
- Fenderson BA, Stamenkovic I, Aruffo A. Localization of hyaluronan in mouse embryos during implantation, gastrulation and organogenesis. Differentiation. 1993;54:85–98. doi: 10.1111/j.1432-0436.1993.tb00711.x. [DOI] [PubMed] [Google Scholar]
- Garcia-Algar O, Kulaga V, Gareri J, Koren G, Vall O, Zuccaro P, Pacifici R, Pichini S. Alarming prevalence of fetal alcohol exposure in a Mediterranean city. Ther Drug Monit. 2008;30:249–254. doi: 10.1097/FTD.0b013e31816a8657. [DOI] [PubMed] [Google Scholar]
- Giknis MLA, Damjanov I, Rubin E. The differential transplacental effects of ethanol in four mouse strains. Neurobehav Toxicol. 1980;2:235–237. [Google Scholar]
- Gilliam DM, Kotch LE, Dudek BC, Riley EP. Ethanol teratogenesis in selectively bred Long-Sleep and Short-Sleep mice: A comparison to inbred C57BL/6J mice. Alcohol Clin Exp Res. 1989;13:667–672. doi: 10.1111/j.1530-0277.1989.tb00402.x. [DOI] [PubMed] [Google Scholar]
- Gilliam DM, Mantle MA, Barkhausen DA, Tweden DR. Effects of acute prenatal ethanol administration in a reciprocal cross of C57BL/6J and Short-Sleep mice: Maternal effects and nonmaternal factors. Alcohol Clin Exp Res. 1997;21:28–34. [PubMed] [Google Scholar]
- Green ML, Singh AV, Zhang Y, Nemeth KA, Sulik KK, Knudsen TB. Reprogramming of genetic networks during initiation of the Fetal Alcohol Syndrome. Devel Dyn. 2007;236:613–631. doi: 10.1002/dvdy.21048. [DOI] [PubMed] [Google Scholar]
- Gutala R, Wang J, Kadapakkam S, Hwang Y, Ticku M, Li MD. Microarray analysis of ethanol-treated cortical neurons reveals disruption of genes related to the ubiquitin-proteasome pathway and protein synthesis. 2004. [DOI] [PubMed] [Google Scholar]
- Hashimoto-Torii K, Kawasawa YI, Kuhn A, Rakic P. Combined transcriptome analysis of fetal human and mouse cerebral cortex exposed to alcohol. PNAS. 2011;108:4212–4217. doi: 10.1073/pnas.1100903108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho JW, Stefani M, dos Remedios CG, Charleston MA. Differential variability analysis of gene expression and its application to human diseases. Bioinformatics. 2008;24:i390–8. doi: 10.1093/bioinformatics/btn142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nature Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hutson JR, Magri R, Gareri JN, Koren G. The incidence of prenatal alcohol exposure in Montevideo Uruguay as determined by meconium analysis. Ther Drug Monit. 2010;32:311–317. doi: 10.1097/FTD.0b013e3181dda52a. [DOI] [PubMed] [Google Scholar]
- Jones KL, Smith DW. Recognition of the fetal alcohol syndrome in early infancy. Lancet. 1973;2 (7836):999–1001. doi: 10.1016/s0140-6736(73)91092-1. [DOI] [PubMed] [Google Scholar]
- Jones KL, Smith DW. The fetal alcohol syndrome. Teratology. 1975;12:1–10. doi: 10.1002/tera.1420120102. [DOI] [PubMed] [Google Scholar]
- Jugessur A, Shi M, Gjessing HK, Lie RT, Wilcox AJ, Weinberg CR, Christensen K, Boyles AL, Daack-Hirsch S, Nguyen TT, Christiansen L, Lidral AC, Murray JC. Maternal genes and facial clefts in offspring: A comprehensive search for genetic associations in two population-based cleft studies from Scandinavia. PLoS One. 2010;5:e11493. doi: 10.1371/journal.pone.0011493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren G, Nulman I, Chudley AE, Loocke C. Fetal Alcohol Spectrum Disorder. CMAJ. 2003;169:1181–1185. [PMC free article] [PubMed] [Google Scholar]
- Kutner MH, Nachtsheim CJ, Neter J, Li W. Applied Linear Statistical Models. 5. New York, NY: McGraw-Hill/Irwin; 2005. series Operational and decision sciences. [Google Scholar]
- Landgren M, Svensson L, Stromland K, Andersson Gronlund M. Prenatal alcohol exposure and neurodevelopmental disorders in children adopted from eastern Europe. Pediatrics. 2010;125:e1178–e1185. doi: 10.1542/peds.2009-0712. [DOI] [PubMed] [Google Scholar]
- Liu Y, Balaraman Y, Wang G, Nephew KP, Zhou FC. Alcohol exposure alters DNA methylation profiles in mouse embryos at early neurulation. Epigenetics. 2009;4:500–511. doi: 10.4161/epi.4.7.9925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KL, Schmittgen TS. Analysis of relative gene expression using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Luzzatto AC, Mangano G, Vonesch N. Prenatal development of the hippocampus in two strains of inbred mice. Int J Dev Neurosci. 1988;6:211–216. doi: 10.1016/0736-5748(88)90001-9. [DOI] [PubMed] [Google Scholar]
- May PA, Gossage JP. Estimating the prevalence of fetal alcohol syndrome: A summary. Alcohol Res Health. 2001;25:159–167. [PMC free article] [PubMed] [Google Scholar]
- McClintick JN, Edenberg HJ. Effects of filtering by Present call on analysis of microarray experiments. BMC Bioinformatics. 2006;7:49. doi: 10.1186/1471-2105-7-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller LC, Chan W, Litvinova A, Rubin A, Comfort K, Tirella L, Cermak S, Morse B, Kovalev I Boston-Murmansk Orphanage Research Team. Fetal Alcohol Spectrum Disorders in children residing in Russian orphanages: A phenotypic survey. Alcohol Clin Exp Res. 2006;30:531–538. doi: 10.1111/j.1530-0277.2006.00059.x. [DOI] [PubMed] [Google Scholar]
- Ogawa T, Kuwagata M, Ruiz J, Zhou FC. Differential teratogenic effect of alcohol on embryonic development between C57BL/6 and DBA/2 mice: A new view. Alcohol Clin Exp Res. 2005;29:855–863. doi: 10.1097/01.alc.0000163495.71181.10. [DOI] [PubMed] [Google Scholar]
- Palmer RH, Ouellette EM, Warner L, Leichtman SR. Congenital malformations in the offspring of a chronic alcoholic mother. Pediatrics. 1974;53:490–494. [PubMed] [Google Scholar]
- Park BY, Sull JW, Park JY, Jee SH, Beaty TH. Differential parental transmission of markers in BCL3 among Korean cleft case-parent trios. J Prev Med Pub Health. 2009;42:1–4. doi: 10.3961/jpmph.2009.42.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Kok KH, Xu R-E, Kwok KHH, Tay D, Fung PCW, King H-f, Lin MCM. Maternal cold inducible RNA binding protein is required for embryonic kidney formation in Xenopus laevis. FEBS Letters. 2000;482:37–43. doi: 10.1016/s0014-5793(00)02019-6. [DOI] [PubMed] [Google Scholar]
- Riikonen RS. Difference in susceptibility to teratogenic effects of alcohol in discordant twins exposed to alcohol during the second half of gestation. Pediatr Neurol. 1994;11:332–336. doi: 10.1016/0887-8994(94)90012-4. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KD. Analyzing real-time PCR data by the comparative CT method. Nature Prot. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Sokol RJ, Delaney-Black V, Nordstrom B. Fetal alcohol spectrum disorder. JAMA. 2003;290:2996–2999. doi: 10.1001/jama.290.22.2996. [DOI] [PubMed] [Google Scholar]
- Streissguth AP, Dehaene P. Fetal alcohol syndrome in twins of alcoholic mothers: concordance of diagnosis and IQ. Am J Med Genet. 1993;47:857–861. doi: 10.1002/ajmg.1320470612. [DOI] [PubMed] [Google Scholar]
- Thiel R, Chahoud I, Jurgens M, Neubert D. Time-dependent differences in the development of somites of four different mouse strains. Teratog Carcinog Mutagen. 1993;13:247–257. doi: 10.1002/tcm.1770130602. [DOI] [PubMed] [Google Scholar]
- Warren K, Li TK. Genetic polymorphisms: impact on the risk of fetal alcohol spectrum disorders. Birth Defects Res A Clin Mol Teratol. 2005;73:195–203. doi: 10.1002/bdra.20125. [DOI] [PubMed] [Google Scholar]
- Webster WS, Walsh DA, Lipson AH, McEwen SE. Teratogenesis after acute alcohol exposure in inbred and outbred mice. Neurobehav Toxicol. 1980;2:227–234. [Google Scholar]
- Wheatley SC, Isacke CM, Crossley PH. Restricted expression of the hyaluronan receptor, CD44, during postimplantation mouse embryogenesis suggests key roles in tissue formation and patterning. Develop. 1993;119:295–306. doi: 10.1242/dev.119.2.295. [DOI] [PubMed] [Google Scholar]
- Wilson CL, Miller CJ. Simpleaffy: a BioConductor package for Affymetrix Quality Control and data analysis. Bioinformatics. 2005;21:3683–3685. doi: 10.1093/bioinformatics/bti605. [DOI] [PubMed] [Google Scholar]
- Zhou FC, Zhao Q, Liu Y, Goodlett CR, Liang T, McClintick JN, Edenberg HJ, Li L. Alteration of gene expression by alcohol exposure at early neurulation. BMC Genomics. 2011;12:124. doi: 10.1186/1471-2164-12-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.