Abstract
In archaea and eukaryotes aminoacyl-tRNA synthetases (aaRSs) associate in multi-synthetase complexes (MSCs), however the role of such MSCs in translation is unknown. MSC function was investigated in vivo in the archaeon Thermococcus kodakarensis, wherein six aaRSs were affinity co-purified together with several other factors involved in protein synthesis, suggesting that MSCs may interact directly with translating ribosomes. In support of this hypothesis, the aminoacyl-tRNA synthetase (aaRS) activities of the MSC were enriched in isolated T. kodakarensis polysome fractions. These data indicate that components of the archaeal protein synthesis machinery associate into macromolecular assemblies in vivo and provide the potential to increase translation efficiency by limiting substrate diffusion away from the ribosome, thus facilitating rapid recycling of tRNAs.
Keywords: Aminoacyl-tRNA synthetase, archaea, ribosome, translation, tRNA
1. Introduction
Aminoacyl-tRNA synthetases (aaRSs) attach amino acids (aa) to their cognate tRNAs to form aa-tRNA, which is then delivered to the ribosome for protein synthesis as a ternary complex with translation elongation factor 1A (EF1A) and GTP [1]. AaRSs have been found in a variety of complexes with each other and with other factors, potentially expanding the functions of aaRSs both within and beyond translation [2]. In mammalian cells nine aaRSs (ArgRS, AspRS, GlnRS, GluRS, IleRS, LeuRS, LysRS, MetRS and ProRS) associate with three non-synthetase protein factors (p18, p38, and p43) to form a large multi-aminoacyl-tRNA synthetase complex (MSC). The accessory proteins are important for the formation and stability of the complex, promote binding of tRNAs to the complex, and also play other roles outside translation [3-6]. In addition to the MSC, another aaRS, ValRS forms a complex with the human multi-subunit translation elongation factor 1H (EF1H), which increases the catalytic efficiency of tRNAVal aminoacylation [7]. In lower eukaryotes, including Saccharomyces cerevisiae, complexes have been characterized between GluRS, MetRS and Arc1p, and between SerRS and the peroxisome biosynthesis factor Pex21p, both of which enhance tRNA binding to the respective aaRSs [8-10].
Although many of these associations were first described in eukaryotic cells, numerous multi-enzyme complexes containing aaRSs have also been identified in both Bacteria and Archaea. In Bacteria, complexes comprised of one aaRS and a second non-aaRS protein have been implicated in diverse cellular functions including editing of misacylated tRNAs, indirect synthesis of aa-tRNA and metabolite biosynthesis [11-13]. In Archaea, aaRS-containing complexes were first described in Haloarcula marismortui, with many aaRSs purified in one or possibly two large complexes, and in Methanocaldococcus jannaschi where ProRS was found to interact with components of the methanogenesis machinery [14-16]. In another archaeal methanogen, Methanothermobacter thermautotrophicus, one complex composed of LeuRS, LysRS, ProRS and EF1A was identified while another contained SerRS and ArgRS. In both cases the formation of MSCs was found to improve the catalytic efficiency of tRNA aminoacylation by the aaRSs present in the corresponding complexes [17-21].
Previous studies on aaRS subcellular localization led to the proposal that MSCs directly channel aa-tRNAs to EF1A without dissociation in the cytoplasm [18,22]. This channeling could potentially provide a sequestered pool of aa-tRNAs specifically for utilization in protein synthesis, although a direct interaction between the ribosome and aaRSs in the MSC has not been demonstrated. The mammalian MSC (MARS) has been shown to interact with polysomes, but whether this reflects substrate channelling during protein synthesis is unclear given the presence of three essential aaRS-interacting factors that also function outside the complex [6,23]. MSCs have been identified in Archaea that do not require aaRS-interacting factors for assembly and function, potentially providing suitable systems to investigate MSC interactions with other components of the translation machinery. Previously attempts to characterize, purify and reconstitute archaeal MSCs have met with some limited success, in part due to the comparative instability of the complex [19]. Here we describe a more systematic investigation of an archaeal aaRS interactome, and define a polysome-associated MSC in the archaeon Thermococcus kodakarensis, providing evidence for the interaction of aaRSs with the mRNA translation machinery and consistent with substrate channeling during protein synthesis.
2. Materials and methods
2.1. Strain construction and protein purification
Construction of shuttle vector pHis6-HA LeuRS and its use to transform T kodakarensis strain KW128 were performed as previously described [24,25]. N-terminally tagged intein fusion derivatives of LeuRS, ProRS and EF1A, and N-terminally His6 tagged TyrRS were produced in Escherichia coli using standard procedures (see Supplementary Materials for details of construction of the corresponding plasmids). Intein tagged LeuRS, ProRS and EF1A were produced by transforming E. coli BL21(DE3)pLysS (Stratagene) with pTYB11 vectors containing the respective genes. Protein was produced by first incubating a starter culture at 37 °C (240 rpm) until mid log phase was reached and then using this to inoculate a larger culture (1L). The larger culture was grown to an OD600 of 0.2 at 37 °C and then transferred to 18 °C (190 rpm) for 90 min. Protein expression was induced with 0.5 mM IPTG for 12 h. Cell-free extract was produced by sonication of cells in buffer A [20 mM Tris-HCl (pH 8.0), 500 mM NaCl and 10 % glycerol] containing a protease inhibitor mixture tablet (Complete Mini, EDTA-free; Roche Applied Science) followed by centrifugation at 150000 x g for 45 min. The resulting supernatant was loaded onto a chitin column, washed extensively with buffer A, and cleavage of the intein tag was induced by incubation of the protein on the chitin column with buffer A containing 50 mM dithiothreitol (DTT) for 24 h (all of the following procedures were performed at 4 °C unless otherwise stated). The protein was then eluted with buffer A containing 50 mM DTT. Fractions containing the respective proteins were pooled (judged by Coomassie Brilliant Blue staining after SDS-PAGE) and buffer exchanged into buffer B (25 mM Tris-HCl, pH 7, 50 mM KCl, 10 mM MgCl2, 5 mM 2-mercaptoethanol, 10 % glycerol) before loading on to a Resource Q column, or in the case of EF1A a Resource S column (GE Healthcare), to which a KCl gradient (0–500 mM) in buffer B was applied. Eluted fractions containing the desired protein were pooled and dialyzed against storage buffer [25 mM Tris-HCl (pH 7.5), 100 mM KCl, 10 mM MgCl2, 5 mM 2-mercaptoethanol and 50 % glycerol; for EF1A 10 μM GDP was also included] prior to storage at -80 °C. His6-TyrRS was purified as previously described and stored in the above mentioned storage buffer at -80 °C [18].
2.2. Isolation and identification of His6-HA-LeuRS associated proteins from T. kodakarensis
T. kodakarensis KW128/pHis6-HA-LeuRS ([25]; Supplementary Fig. 1) was grown to mid exponential phase (OD600 = 0.4) at 85 °C in ASW-YT medium supplemented with 5 g sodium pyruvate/L and 5 μM mevinolin. Cells were harvested by centrifugation, resuspended in 20 mL of buffer A with 10 mM imidazole and lysed by sonication. After centrifugation, the resulting clarified lysate was loaded onto a 1ml HiTrap chelating column (GE Healthcare) pre-equilibrated with NiSO4. The column was washed with buffer A and proteins were eluted using a linear imidazole gradient from buffer A to 100 % buffer C (25 mM Tris-HCl (pH 8), 100 mM NaCl, 500 mM imidazole and 10 % glycerol). Fractions containing His6-HA-LeuRS were then identified via western blot analyses using anti-HA antibodies, and 30 μg aliquots of the positive fractions subsequently precipitated by adding trichloroacetic acid (TCA; 15 % final concentration). TCA-precipitated proteins were identified by multidimensional protein identification technology (MuDPIT) at the Ohio State University mass spectrometry facility (http://www.ccic.ohio-state.edu/MS/proteomics.htm) using the MASCOT search engine. MASCOT scores >100, indicative of a minimum of two unique peptide fragments identified from the same protein, were considered significant. Protein isolation and mass spectrometry analyses were also performed with lysates of two independent cultures of wild-type T. kodakarensis KW128 to identify untagged proteins that non-specifically bound and eluted from the Ni2+-charged matrix. All proteins identified in the experimental samples that had MASCOT scores >100, but were absent from the control samples, are listed in Supplementary Table 1.
2.3. Aminoacylation Assays
T. kodakarensis total tRNA was prepared as previously described [26]. The gene encoding T. kodakarensis tRNALeu (GAG anticodon) cloned into pUC19 was used for synthesis and purification of the corresponding in vitro transcribed tRNA using standard procedures [27]. L-[U-14C] leucine (324 mCi/mmol), L-[U-14C] serine (163mCi/mmol), L-[U-14C] arginine (346 mCi/mmol), L-[U-14C] phenylalanine (487 mCi/mmol), L-[U-14C] glutamic acid (260mCi/mmol) and L-[U-14C] tyrosine (482mCi/mmol) were from PerkinElmer Life Sciences. L-[U-14C] proline (269 mCi/mmol), L-[U-14C] aspartic acid (207 mCi/mmol), L-[U-14C] alanine (164 mCi/mmol) and L-[U-14C] lysine (309 mCi/mmol) were from Amersham Biosciences. A reaction mixture containing 100 mM HEPES (pH 7.5), 250 mM KCl, 10 mM MgCl2, 10 mM DTT, bovine serum albumin (200 μg/mL), T. kodakarensis total tRNA or in vitro transcribed tRNA, and aaRSs at concentrations indicated for specific experiments was first prepared and preincubated for 15 min at room temperature. The appropriate radiolabeled amino acid was then added to the reaction mixture and the temperature raised to 65 °C. After 1 min of incubation, the reaction was initiated with the addition of 5 mM ATP. Aliquots of reaction mixture were spotted on 3MM filter paper presoaked in 5 % TCA (w/v) at required time intervals, washed in 5 % TCA acid, dried and level of radioactivity was determined by scintillation counting. Experiments to determine the effect of EF1A on aminoacylation were performed as described above, except that T. kodakarensis EF1A (15 μM) was first activated by incubation at 37 °C for 30 min in a buffer containing 20 mM Tris HCl (pH 7.5), 1 mM GTP, 7 mM MgCl2, 100 mM KCl, 5 mM DTT, 3 mM PEP and 30 mg/ml pyruvate kinase.
2.4. Polysome Preparation
T. kodakarensis cells (KW128) were grown to mid exponential phase (OD600 = 0.4) at 85 °C in ASW-YT medium supplemented with 5g sodium pyruvate/l, and mupirocin was then added to a final concentration of 12 μg/ml and incubated at 85 °C for 5 min. Cell cultures were then chilled on ice, centrifuged at 10000 g for 5 min at 4 °C, the resulting pellet resuspended in 0.5 ml chilled cell lysis buffer [10 mM Tris-acetate buffer (pH 7.5), 14 mM magnesium acetate, 60 mM potassium acetate and 1 mM DTT] and frozen in liquid nitrogen followed by three freeze thaw cycles. 1 μl of RNase inhibitor (Roche) was added to the lysed cells, which were then centrifuged at 16000 x g for 10 min at 4 °C and 0.25 ml of the resulting supernatant was loaded onto an 11 ml 10-40 % sucrose gradient containing 20 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 100 mM NH4Cl and 2 mM DTT. Sucrose gradients were spun in an ultracentrifuge using an SW-41 rotor at 150000 x g for 4 h at 4°C. Gradients were fractionated using an ISCO model 183 syringe-pump at 1.48 ml/min while monitoring absorbance at 254 nm.
3. Results
3.1. T. kodakarensis contains an MSC that enhances tRNA aminoacylation
Previous studies identified an MSC in M. thermautotrophicus that contained LeuRS as a core scaffolding protein. To determine if an MSC exists in T. kodakarensis, a plasmid expressing His6-HA-LeuRS was used to produce a bait for affinity co-purification experiments (supplementary Figure 1). Cell-free lysates of T. kodakarensis/pHis6-HA LeuRS were loaded onto a Ni2+ matrix, washed, and bound proteins were then eluted using an imidazole gradient. His6-HA-LeuRS-containing fractions were identified using anti-HA antibodies, and the corresponding eluates further analyzed by mass spectrometry. A substantial number of components of the translation machinery were found associated with LeuRS including five aaRSs (TyrRS, ProRS, GlyRS, MetRS and CysRS), EF1A, IF2, IF2B, EF2 and several ribosomal proteins (Table 1; Supplementary Table 1). Consistent with previous reports describing interactions between the MSC and other pathways outside translation in archaea and eukaryotes [15,28], proteins involved in metabolism, transcription and protein modification were also found associated with LeuRS (Supplementary Table 1). As this analysis was performed with cells grown to mid-log phase, and given that ribosomal proteins were abundant in the interactome, some of the detected proteins may have co-purified as nascent polypeptides and may not reflect direct interactions with LeuRS.
Table 1.
Identification of components of the protein synthesis machinery that interact with LeuRS
| ORF a | Description b |
|---|---|
| TK0568 | tyrosyl-tRNA synthetase |
| TK0978 | glycyl-tRNA synthetase |
| TK0550 | prolyl-tRNA synthetase |
| TK1461 | leucyl-tRNA synthetase |
| TK0444 | cysteinyl-tRNA synthetase |
| TK1049 | methionyl-tRNA synthetase |
| TK0308 | elongation factor 1A |
| TK0556 | translation initiation factor 2B subunit beta |
| TK1305 | translation initiation factor 2 |
| TK0309 | elongation factor 2 |
| TK0506 | translation-associated GTPase |
| TK1254 | 30S ribosomal protein S3Ae |
| TK1529 | 30S ribosomal protein S4e |
| TK1538 | 30S ribosomal protein S19P |
| TK1276 | 30S ribosomal protein S19e |
| TK1504 | 30S ribosomal protein S11P |
| TK1505 | 30S ribosomal protein S4 |
| TK1496 | 30S ribosomal protein S2 |
| TK0307 | 30S ribosomal protein S10P |
| TK1521 | 30S ribosomal protein S5P |
| TK1526 | 30S ribosomal protein S8P |
| TK1099 | 30S ribosomal protein S27e |
| TK1500 | 30S ribosomal protein S9P |
| TK1078 | 30S ribosomal protein S12P |
| TK1251 | 30S ribosomal protein S15P |
| TK1532 | 30S ribosomal protein S17P |
| TK1951 | 30S ribosomal protein S6e |
| TK0187 | glutamine amidotransferase, class I |
| TK1239 | peptide chain release factor 1 |
| TK1671 | tRNA-modifying enzyme |
| TK0981 | N2, N2-dimethylguanosine tRNA methyltransferase |
| TK0970 | N(2),N(2)-dimethylguanosine tRNA methyltransferase |
| TK0704 | SAM-dependent methyltransferase |
Open reading frames (ORFs) are numbered according to the annotated genome sequence of T. kodakarensis [35].
Description of the corresponding proteins are taken from the TIGR Comprehensive Microbial Resource (cmr.tigr.org/tigr-scripts/CMR/shared/AnnotationSearch.cgi).
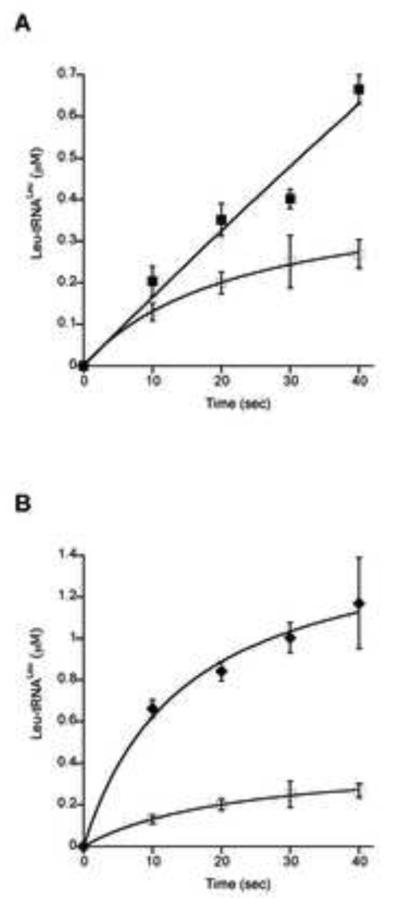
The effect of MSC formation on the aminoacylation activities of several of the corresponding aaRSs was further investigated in vitro. Functional effects of association between LeuRS, TyrRS, ProRS and EF1A were investigated by monitoring the aminoacylation activities of the aaRSs in the presence of other enzymes. An excess of each partner protein was used in order to minimize the free fraction of the monitored enzyme. Addition of a 10-fold excess of ProRS increased the aminoacylation activity of LeuRS approximately 2-fold; a 40-fold excess was necessary to produce a comparable change in the presence of TyrRS, suggesting this aaRS forms a weaker interaction with LeuRS than does ProRS (Fig. 1). The possible enhancement of aminoacylation activities of ProRS and TyrRS in the presence of excess partner aaRSs was also investigated, but no significant differences were observed compared with the activities of the respective enzymes alone. Previously, it has been shown in archaea and eukaryotes that addition of EF1A increases the activity of LeuRS and ValRS, respectively [7,18], prompting us to investigate if this was also the case for T. kodakarensis LeuRS. The GDP-bound form, as well as the GTP activated state of EF1A were tested for their possible effect on the aminoacylation activity of LeuRS. The largest increase in the rate of Leu-tRNALeu formation was observed in the presence of EF1A•GTP, indicating that enhancement of LeuRS activity depends on the presence of activated elongation factor (Fig. 2). The inclusion of more than two partners, for example the co-incubation of LeuRS, ProRS, and EF1A together, did not further enhance aminoacylation rates above those observed when only two partners were present (data not shown).
Figure 1. Effects of ProRS and TyrRS on aminoacylation by LeuRS.
T. kodakarensis total tRNA (2 mg/ml) was aminoacylated under standard conditions (7 μl samples) in the presence of LeuRS alone (80 nM) (◻) or with addition of A, ProRS (800 nM) (∎) or B, TyrRS (3.2 μM) (◆). Non-specific increase in the activity of aaRSs were excluded by adding BSA in the reaction mixtures. Further addition of BSA did not change the activities of the aaRSs being monitored (data not shown).
Figure 2. Effect of EF1A on aminoacylation by LeuRS.
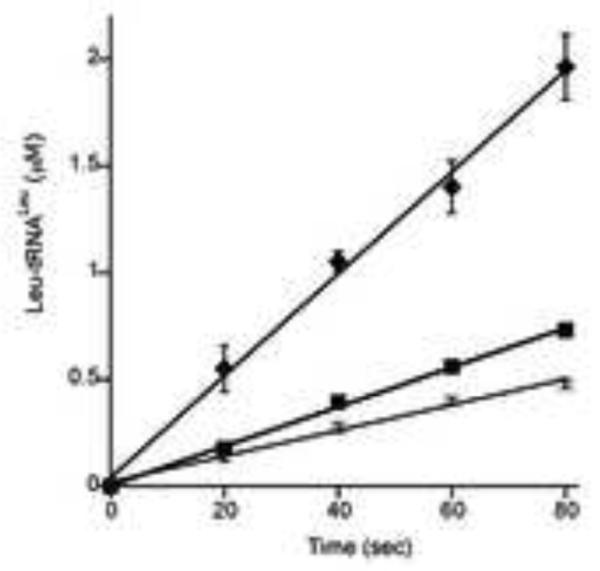
T. kodakarensis total tRNA (2 mg/ml) was aminoacylated under standard conditions (7 μl samples) in the presence of LeuRS alone (15 nM) (◻), or with addition of EF1A-GDP (3.5 μM) (∎) or EF1A-GTP (3.5 μM) (◆).
MetRS and CysRS were not included in these experiments as they did not co-associate with other putative MSC components during polysome analyses (see 3.2 below), and GlyRS was also excluded as the recombinant protein was not active when tested for in vitro aminoacylation activity (data not shown).
3.2 Association of the MSC with the protein synthesis machinery
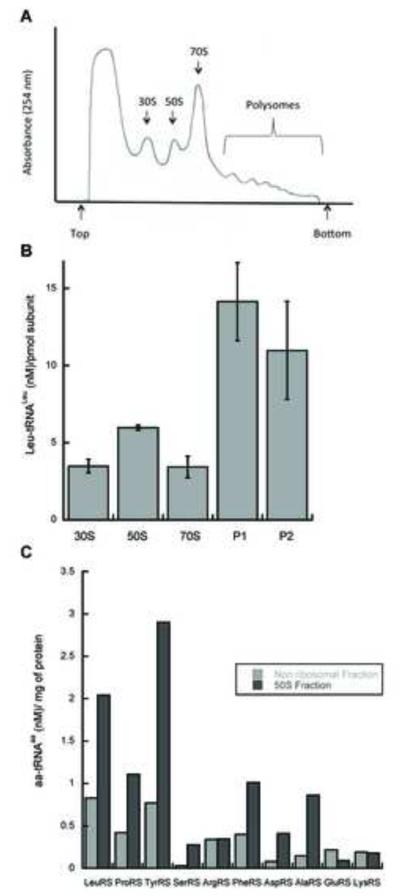
Affinity co-purification of proteins interacting with LeuRS identified several ribosomal proteins, suggesting that the archaeal MSC might interact with actively translating ribosomes. To investigate whether aaRSs are associated with actively translating ribosomes in T. kodakarensis, we attempted to isolate polysomes from exponentially growing cells and subsequently separate them from other ribosomal fractions by sucrose gradient sedimentation. Several antibiotics were tested for their ability to stall translation in T. kodakarensis cells (a requirement for stable polysome isolation) and of these mupirocin was found to be the most effective (Fig. 3A). Mupirocin is a polyketide antibiotic produced by Pseudomonas fluorescens that inhibits archaeal IleRS in vivo, and it was used here to facilitate polysome isolation from T. kodakarensis [29,30]. Polysomes, monosomes and individual small and large ribosomal subunits were fractionated over sucrose density gradients, and the distribution of tRNA aminoacylation activities associated with each fraction were investigated. The distribution of LeuRS was examined in detail and found to be enriched in polysomes and, to a lesser extent in other ribosomal fractions as compared to non-ribosomal fractions (Fig. 3B). The presence of other aaRSs in ribosomal fractions was investigated by monitoring activities associated with the 50S ribosomal fraction, which was chosen due to its relatively high abundance, middle position among the ribosomal fractions, and good separation from non-ribosomal fractions. Seven aaRSs out of the ten tested were found enriched in the ribosomal fraction, as compared to the non-ribosomal fraction S1 (Fig. 3C). Among the 7 aaRS activities identified were ProRS, LeuRS and TyrRS, which were also shown to associate in T. kodakarensis by affinity co-purification (see above), providing further support for the direct association of the MSC with actively translating ribosomes. The other aaRSs found to be associated with ribosomes might also be present in the MSC, but may have escaped detection by mass spectrometry due to the inherent limitations of pulling down large complexes using an epitope-tagged bait protein.
Figure 3. Association of aminoacylation activities with the protein synthesis machinery.
T. kodakarensis cells were grown under anaerobic conditions to mid log phase and then subjected to polysome analysis. Polysome lysates were fractionated by sucrose gradient centrifugation and the ribosomal subunits, monosomes and polysome fractions collected. A, absorbance profile (A254) of fractions eluted from the gradient. The top of the gradient is on the left and peaks representing the 30S and 50S ribosomal subunits, monosomes and polysomes are denoted. B, Leu-tRNA aminoacylation activity in ribosomal subunits, monosomes and polysomal fractions (P1 and P2). C, Aminoacylation activities associated with 50S ribosomal subunits.
4. Discussion
In the present study the LeuRS interactome of T. kodakarensis was determined using a combination of co-affinity purification and proteomics. The interactome contained, among other proteins, five additional aaRSs (ProRS, TyrRS, GlyRS, MetRS and CysRS), EF1A, EF2, IF2B and IF2, and a number of ribosomal proteins. LeuRS, ProRS and EF1A have previously been shown to be part of a complex in M. thermautrophicus [18,19], while ProRS, MetRS and LeuRS are components of the mammalian MSC [6]. Interactions between T. kodakarensis LeuRS with TyrRS or ProRS enhanced tRNALeu aminoacylation, similar to previous reports of improved aminoacylation upon aaRS complex formation in archaea and yeast [10,19,21]. The catalytic enhancement observed here was less extensive than previously described in M. thermautrophicus, being limited to LeuRS with no comparable improvements observed in the aminoacylation activities of TyrRS or ProRS. LeuRS activity was also enhanced by the GTP-activated form of archaeal EF1A but not by GDP-bound EF1A. EF1A-GTP preferentially binds aa-tRNA [31], and the observed enhancement is consistent with previous reports of eukaryotic aaRSs forming specific interactions with elongation factors [7,32]. The ability of activated elongation factors to increase aaRS product release rates was previously shown to accelerate aa-tRNA synthesis [33], and our findings now suggest this is also the case for Leu-tRNALeu in the T. kodakarensis MSC.
Enhanced aa-tRNA synthesis within MSCs provides a possible mechanism to increase the rate of protein synthesis, as does the association of such complexes directly with ribosomes [34]. The presence of sixteen 30S ribosomal proteins, IF2, and IF2B and EF2, in the LeuRS interactome , and the detection of several aaRS activities enriched in polysomes, both support the association of the identified MSC with the translation machinery. The detection of IF2, IF2B, MetRS and EF2 in the interactome along with 30S, but not 50S, ribosomal proteins suggests the MSC might first interact with the 30S preinitiation complex; alternatively, these findings may instead indicate that any interactions between the MSC and the 50S subunit occur primarily via rRNA, or that protein-protein interactions are not sufficiently persistent to survive our purification procedures.
While the precise details of some interactions remain to be resolved, as does the possible structural role of tRNA [21], our data indicate the presence in T. kodakarensis of a larger macromolecular assembly including the MSC, the translational machinery, and perhaps other major complexes such as the proteasome, as also recently observed in Schizosaccharomyces pombe [28]. The channeling of aa-tRNAs directly to ribosomes without diffusion into the cytoplasm has been proposed to improve translation efficiency, and the observation in T. kodakarensis of MSCs associated with active ribosomes is consistent with this model. Direct investigation of the importance of MSCs in vivo may now be possible in T. kodakarensis by targeted disruption of protein-protein interactions between specific aaRSs, which would also help to address long-standing questions as to the impact on the cell of tRNA channeling during translation.
Supplementary Material
Highlights.
Six aaRSs copurified with protein synthesis factors from T. kodakarensis
AaRS activities were enriched in archeael polysome fractions
Archaeal translation machinery components associate into macromolecular assemblies
Structured summary of protein interactions.
LeuRS physically interacts with DNA methylase 115, flagellin 118, acetylpolyamine aminohydrolase 140, Pyruvoyl-dependent arginine decarboxylase 154, RecJ-like exonuclease 476, acetyl-CoA acetyltransferase (mevanolate Pathway) 110, glutamine amidotransferase, class I 437, GMP synthase subunit B 184, pyridoxine biosynthesis protein 1183, quinolinate synthetase 206, L-aspartate oxidase 322, uridylate kinase 164, putative molybdenum cofactor biosynthesis protein C 127, bifunctional carboxypeptidase/aminoacylase 214, aspartate racemase 166, serine/threonine protein kinase 111, SAM-dependent methyltransferase 144, GTP cyclohydrolase 398, DNA topoisomerase VI subunit A 209, DNA topoisomerase VI subunit B 192, Type A Flavoprotein 911, NAD(P)H:rubredoxin oxidoreductase (Fatty acid metabolism) 120, NAD(P)H:rubredoxin oxidoreductase 120, cofactor-independent phosphoglycerate mutase 909, bis(5′-adenosyl)-triphosphatase 205, thiamine monophosphate kinase 179, pyruvate formate lyase family activating protein 298, 3-hydroxy-3-methylglutaryl-CoA reductase (mevanolate, N(2),N(2)-dimethylguanosine tRNA methyltransferase 145, N2, N2-dimethylguanosine tRNA methyltransferase 170, putative 5-methylcytosine restriction system, GTPase subunit 947, D-aminopeptidase 540, calcineurin superfamily metallophosphoesterase 118, rubrerythrin-related protein 317, 30S ribosomal protein S12P 161, DNA-directed RNA polymerase subunit beta 373, protein disulfide oxidoreductase 139, 30S ribosomal protein S27e 178, ribonuclease Z 122, 2-oxoglutarate ferredoxin oxidoreductase subunit gamma 352, 2-oxoglutarate ferredoxin oxidoreductase subunit alpha 407, methylmalonyl-CoA mutase, N-terminus of large subunit 172, AP endonuclease (base excision repair pathway) 365, CTP synthetase 105, PBP family phospholipid-binding protein 272, lipoate-protein ligase A, C-terminal section 234, peptide chain release factor 1 331, 30S ribosomal protein S15P 143, NADH oxidase 432, Putative oxidoreductase 538, NAD(P)H-flavin oxidoreductase 471, ferredoxin--NADP(+) reductase subunit alpha 471, Lrp/AsnC family transcriptional regulator 378, glycine dehydrogenase subunit 2 255, glycerol kinase 257, phosphomannomutase-related protein 321, ribose-5-phosphate isomerase A 107, phosphate transport regulator 193, isopentenyl pyrophosphate isomerase (mevanolate Pathway) 500, amino acid kinase 203, NADH:polysulfide oxidoreductase 203, 5′-methylthioadenosine phosphorylase 158, 30S ribosomal protein S9P 171, DNA-directed RNA polymerase subunit D 302, cytidylate kinase 305, adenylate kinase 109, 30S ribosomal protein S8P 180, 30S ribosomal protein S17P 131, serine--glyoxylate aminotransferase, class V (transferasetransaminase), predicetd ATPase 204, metallo-beta-lactamase superfamily hydrolase 134, metallo-beta-lactamase superfamily hydrolase 134, metal-dependent hydrolase 253, putative RNA-associated protein 167, proteasome subunit alpha 174, tRNA-modifying enzyme 172, sugar-phosphate nucleotydyltransferase 108, cytidylyltransferase 128, N-acetylchitobiose deacetylase 124, transcriptional regulator 364, glutamine synthetase 120, N6-adenine-specific DNA methylase 194, ArsR family transcriptional regulator 113, 5′-methylthioadenosine phosphorylase II 280, DNA repair and recombination protein RadA 323, 30S ribosomal protein S6e 106, pyruvate ferredoxin oxidoreductase subunit beta 282, cysteine desulfurase 521, hydrogenase maturation protein HypF 235, iron-molybdenum cofactor-binding protein 192, ATPase 260, 4Fe-4S cluster-binding protein 254, phosphopyruvate hydratase 650, fructose-1,6-bisphosphatase 140, aspartate carbamoyltransferase catalytic subunit 158, Bipolar DNA helicase 448, bipolar DNA helicase 448, molybdenum cofactor biosynthesis protein A 182, proteasome-activating nucleotidase 474, deoxycytidylate deaminase 163, cell division protein FtsZ 821, ribulose bisophosphate carboxylase 1767, chaperonin beta subunit 460, DEAD/DEAH box RNA helicase 175, 30S ribosomal protein S10P, elongation factor 1A, elongation factor 2, cysteinyl-tRNA synthetase, translation-associated GTPase, prolyl-tRNA synthetase, translation initiation factor 2B subunit beta, tyrosyl-tRNA synthetase, glycyl-tRNA synthetase, methionyl-tRNA synthetase, 30S ribosomal protein S3Ae, 30S ribosomal protein S19e, translation initiation factor 2, 30S ribosomal protein S2, 30S ribosomal protein S11P, 30S ribosomal protein S4, 30S ribosomal protein S5P, 30S ribosomal protein S4e and 30S ribosomal protein S19P by pull down (View interaction)
Acknowledgments
This work was supported by the National Institutes of Health (GM65183, M.I.; GM100329-01, T.J.S). We thank J. Shepherd for critical reading of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Ibba M, Soll D. Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- [2].Ray PS, Arif A, Fox PL. Macromolecular complexes as depots for releasable regulatory proteins. Trends Biochem. Sci. 2007;34:158–164. doi: 10.1016/j.tibs.2007.02.003. [DOI] [PubMed] [Google Scholar]
- [3].Quevillon S, Mirande M. The p18 component of the multisynthetase complex shares a protein motif with the beta and gamma subunits of eukaryotic elongation factor 1. FEBS Lett. 1996;395:63–67. doi: 10.1016/0014-5793(96)01005-8. [DOI] [PubMed] [Google Scholar]
- [4].Quevillon S, Agou F, Robinson JC, Mirande M. The p43 component of the mammalian multi-synthetase complex is likely to be the precursor of the endothelial monocyte-activating polypeptide II cytokine. J. Biol. Chem. 1997;272:32573–32579. doi: 10.1074/jbc.272.51.32573. [DOI] [PubMed] [Google Scholar]
- [5].Quevillon S, Robinson J-C, Berthonneau E, Siatecka M, Mirande M. Macromolecular assemblage of aminoacyl-tRNA synthetases: identification of protein-protein interactions and characterization of a core protein. J. Mol. Biol. 1999;285:183–195. doi: 10.1006/jmbi.1998.2316. [DOI] [PubMed] [Google Scholar]
- [6].Kaminska M, Havrylenko S, Decottignies P, Le Marechal P, Negrutskii B, Mirande M. Dynamic organization of aminoacyl-tRNA synthetase complexes in the cytoplasm of human cells. J. Biol. Chem. 2009;284:13746–13754. doi: 10.1074/jbc.M900480200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Negrutskii BS, Shalak VF, Kerjan P, El’skaya AV, Mirande M. Functional interaction of mammalian valyl-tRNA synthetase with elongation factor EF-1alpha in the complex with EF-1H. J. Biol. Chem. 1999;274:4545–4550. doi: 10.1074/jbc.274.8.4545. [DOI] [PubMed] [Google Scholar]
- [8].Simos G, Segref A, Fasiolo F, Hellmuth K, Shevchenko A, Mann M, Hurt EC. The yeast protein Arc1p binds to tRNA and functions as a cofactor for the methionyl- and glutamyl-tRNA synthetases. EMBO J. 1996;15:5437–5448. [PMC free article] [PubMed] [Google Scholar]
- [9].Godinic V, Mocibob M, Rocak S, Ibba M, Weygand-Durasevic I. Peroxin Pex21p interacts with the C-terminal noncatalytic domain of yeast seryl-tRNA synthetase and forms a specific ternary complex with tRNA(Ser) FEBS J. 2007;274:2788–2799. doi: 10.1111/j.1742-4658.2007.05812.x. [DOI] [PubMed] [Google Scholar]
- [10].Simos G, Sauer A, Fasiolo F, Hurt EC. A conserved domain within Arc1p delivers tRNA to aminoacyl-tRNA synthetases. Mol. Cell. 1998;1:235–242. doi: 10.1016/s1097-2765(00)80024-6. [DOI] [PubMed] [Google Scholar]
- [11].An S, Musier-Forsyth K. Trans-editing of Cys-tRNA Pro by Haemophilus influenzae YbaK protein. J. Biol. Chem. 2004;279:42359–42362. doi: 10.1074/jbc.C400304200. [DOI] [PubMed] [Google Scholar]
- [12].Buddha MR, Crane BR. Structures of tryptophanyl-tRNA synthetase II from Deinococcus radiodurans bound to ATP and tryptophan. Insight into subunit cooperativity and domain motions linked to catalysis. J. Biol. Chem. 2005;280:31965–31973. doi: 10.1074/jbc.M501568200. [DOI] [PubMed] [Google Scholar]
- [13].Bailly M, Blaise M, Lorber B, Becker HD, Kern D. The transamidosome: a dynamic ribonucleoprotein particle dedicated to prokaryotic tRNA-dependent asparagine biosynthesis. Mol. Cell. 2007;28:228–239. doi: 10.1016/j.molcel.2007.08.017. [DOI] [PubMed] [Google Scholar]
- [14].Goldgur Y, Safro M. Aminoacyl-tRNA synthetases from Haloarcula marismortui: an evidence for a multienzyme complex in a procaryotic system. Biochem. Mol. Biol. Int. 1994;32:1075–1083. [PubMed] [Google Scholar]
- [15].Lipman RS, Chen J, Evilia C, Vitseva O, Hou YM. Association of an aminoacyl-tRNA synthetase with a putative metabolic protein in archaea. Biochemistry. 2003;42:7487–7496. doi: 10.1021/bi0344533. [DOI] [PubMed] [Google Scholar]
- [16].Oza J, Sowers KR, Perona JJ. Linking energy production and protein synthesis in hydrogenotrophic methanogens. Biochemistry. 2012 doi: 10.1021/bi300106p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Praetorius-Ibba M, Rogers TE, Samson R, Kelman Z, Ibba M. Association between archaeal prolyl-and leucyl-tRNA synthetases enhances tRNA Pro aminoacylation. J. Biol. Chem. 2005;280:26099–26104. doi: 10.1074/jbc.M503539200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hausmann CD, Praetorius-Ibba M, Ibba M. An aminoacyl-tRNA synthetase:elongation factor complex for substrate channeling in archaeal translation. Nucleic Acids Res. 2007;35:6094–6102. doi: 10.1093/nar/gkm534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Praetorius-Ibba M, Hausmann CD, Paras M, Rogers TE, Ibba M. Functional association between three archaeal aminoacyl-tRNA synthetases. J. Biol. Chem. 2007;282:3680–3687. doi: 10.1074/jbc.M609988200. [DOI] [PubMed] [Google Scholar]
- [20].Hausmann CD, Ibba M. Structural and functional mapping of the archaeal multi-aminoacyl-tRNA synthetase complex. FEBS Lett. 2008;582:2178–2182. doi: 10.1016/j.febslet.2008.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Godinic-Mikulcic V, Jaric J, Hausmann CD, Ibba M, Weygand-Durasevic I. An archaeal tRNA-synthetase complex that enhances aminoacylation under extreme conditions. J. Biol. Chem. 2011;286:3396–3404. doi: 10.1074/jbc.M110.168526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Mirande M, Le Corre D, Louvard D, Reggio H, Pailliez JP, Waller JP. Association of an aminoacyl-tRNA synthetase complex and of phenylalanyl-tRNA synthetase with the cytoskeletal framework fraction from mammalian cells. Exp. Cell Res. 1985;156:91–102. doi: 10.1016/0014-4827(85)90264-2. [DOI] [PubMed] [Google Scholar]
- [23].Kaminska M, Havrylenko S, Decottignies P, Gillet S, Le Marechal P, Negrutskii B, Mirande M. Dissection of the structural organization of the aminoacyl-tRNA synthetase complex. J. Biol. Chem. 2009;284:6053–6060. doi: 10.1074/jbc.M809636200. [DOI] [PubMed] [Google Scholar]
- [24].Li Z, Santangelo TJ, Cubonova L, Reeve JN, Kelman Z. Affinity purification of an archaeal DNA replication protein network. MBio. 2010;1 doi: 10.1128/mBio.00221-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Santangelo TJ, Cubonova L, Reeve JN. Shuttle vector expression in Thermococcus kodakaraensis: contributions of cis elements to protein synthesis in a hyperthermophilic archaeon. Appl. Environ. Microbiol. 2008;74:3099–3104. doi: 10.1128/AEM.00305-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Polycarpo C, et al. Activation of the pyrrolysine suppressor tRNA requires formation of a ternary complex with class I and class II lysyl-tRNA synthetases. Mol. Cell. 2003;12:287–294. doi: 10.1016/s1097-2765(03)00280-6. [DOI] [PubMed] [Google Scholar]
- [27].Sampson JR, Uhlenbeck OC. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc. Natl. Acad. Sci. U.S.A. 1988;85:1033–1037. doi: 10.1073/pnas.85.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Sha Z, Brill LM, Cabrera R, Kleifeld O, Scheliga JS, Glickman MH, Chang EC, Wolf DA. The eIF3 interactome reveals the translasome, a supercomplex linking protein synthesis and degradation machineries. Mol. Cell. 2009;36:141–152. doi: 10.1016/j.molcel.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Jenal U, Rechsteiner T, Tan PY, Buhlmann E, Meile L, Leisinger T. Isoleucyl-tRNA synthetase of Methanobacterium thermoautotrophicum Marburg. Cloning of the gene, nucleotide sequence, and localization of a base change conferring resistance to pseudomonic acid. J. Biol. Chem. 1991;266:10570–10577. [PubMed] [Google Scholar]
- [30].Rechsteiner T, Leisinger T. Purification of isoleucyl-tRNA synthetase from Methanobacterium thermoautotrophicum by pseudomonic acid affinity chromatography. Eur. J. Biochem. 1989;181:41–46. doi: 10.1111/j.1432-1033.1989.tb14691.x. [DOI] [PubMed] [Google Scholar]
- [31].Vitagliano L, Masullo M, Sica F, Zagari A, Bocchini V. The crystal structure of Sulfolobus solfataricus elongation factor 1alpha in complex with GDP reveals novel features in nucleotide binding and exchange. EMBO J. 2001;20:5305–5311. doi: 10.1093/emboj/20.19.5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Sang LJ, Gyu PS, Park H, Seol W, Lee S, Kim S. Interaction network of human aminoacyl-tRNA synthetases and subunits of elongation factor 1 complex. Biochem. Biophys. Res. Commun. 2002;291:158–164. doi: 10.1006/bbrc.2002.6398. [DOI] [PubMed] [Google Scholar]
- [33].Zhang CM, Perona JJ, Ryu K, Francklyn C, Hou YM. Distinct kinetic mechanisms of the two classes of aminoacyl-tRNA synthetases. J. Mol. Biol. 2006;361:300–311. doi: 10.1016/j.jmb.2006.06.015. [DOI] [PubMed] [Google Scholar]
- [34].Fredrick K, Ibba M. How the sequence of a gene can tune its translation. Cell. 2010;141:227–229. doi: 10.1016/j.cell.2010.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Fukui T, Atomi H, Kanai T, Matsumi R, Fujiwara S, Imanaka T. Complete genome sequence of the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1 and comparison with Pyrococcus genomes. Genome Res. 2005;15:352–363. doi: 10.1101/gr.3003105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.