Abstract
Chromatin remodeling factors are becoming known as crucial facilitators of recruitment of repair proteins to sites of DNA damage. Multiple chromatin remodeling protein complexes are now known to be required for efficient double strand break repair. In a screen for microRNAs that modulate the DNA damage response, we discovered that expression of the miR-99 family of microRNAs correlates with radiation sensitivity. These microRNAs were also transiently induced following radiation. The microRNAs target the SWI/SNF chromatin remodeling factor SNF2H/SMARCA5, a component of the ACF1 complex. We found that by reducing levels of SNF2H, miR-99a reduced BRCA1 localization to sites of DNA damage. Introduction of the miR-99 family of microRNAs into cells reduced the rate and overall efficiency of repair by both homologous recombination and non-homologous end joining. Finally, induction of the miR-99 family following radiation prevents an increase in SNF2H expression and reduces the recruitment of BRCA1 to sites of DNA damage following a second dose of radiation, reducing the efficiency of repair after multiple rounds of radiation as used in fractionated radiotherapy.
Keywords: microRNA, DNA repair, cancer, SNF2H, miR-99
Introduction
Mammalian cells have developed multiple complex mechanisms for identifying and repairing damage to their genomes caused by a wide variety of sources. Damage caused by UV, ionizing radiation, cross-linking and intercalating agents, as well as DNA replication errors are identified and repaired through multiple pathways dependent on the type of damage incurred. The ability of normal cells to detect and repair DNA damage, as well as initiate apoptosis when excess damage has occurred is crucial for maintaining genomic stability and preventing cancer. Conversely, the ability of cancer cells to sustain and repair large quantities of DNA damage contributes to their frequently observed resistance to DNA damaging chemotherapeutic and radiation treatment. Several histone modifications specifically designate sites of DNA damage, and are required for the recruitment of repair proteins(1). In particular, phosphorylation of H2AX (γH2AX) is required for DNA damage signaling and repair (2). There is a steadily growing body of evidence that indicates several other histone modifications including ubiquitination, methylation and acetylation occur that play a role in conjunction with, as well as independent of, γH2AX signaling to facilitate effective DNA repair (1, 3).
One of the functions of these histone marks is to recruit chromatin remodeling complexes to give repair proteins access to sites of DNA damage. ATP-dependent chromatin remodeling complexes were initially implicated in DSB repair when it was discovered that the INO80 complex is recruited to phosphorylated H2A in budding yeast and required for efficient conversion of double strand breaks into single stranded DNA prior to repair (4–7). The SWI/SNF complex is also recruited to γH2AX in conjunction with acetylation of histone H3, and required for maximal phosphorylation of H2AX (8). Conversely, the Tip60 complex is recruited to DNA damage sites to dephosphorylate γH2AX after the damage is repaired (9).
Double strand break(DSB) repair in mammalian cells typically occurs through two distinct pathways, homologous recombination(HR) or non-homologous end joining(NHEJ). DSB repair through HR is dependent on the recruitment of BRCA1 to DSB sites through a complex signaling cascade downstream of γH2AX (1). The protein MDC1 binds to γH2AX (10) and recruits the ubiquitin ligase RNF8, which subsequently ubiquitinates H2A and H2AX leading to eventual BRCA1 recruitment (11–14). NHEJ involves the binding of Ku proteins to double stranded ends of DNA at break sites(15) and the recruitment of DNA-PKcs and DNA ligase IV, which then facilitate the ligation of the free ends (16). Both processes require the action of chromatin remodeling proteins for proper recruitment of repair proteins to double strand break sites(17, 18).
Recently, the chromatin remodeling complex SNF2H (also known as SMARCA5) has been implicated in both the HR and NHEJ DNA repair pathways. Following DNA damage, the ACF1 chromatin remodeling complex containing SNF2H is required for the efficient recruitment of Ku70/80 proteins to laser stripe induced DNA damage sites, and depletion of components of this complex result in inefficient double strand break(DSB) repair by NHEJ (18). Interestingly, SNF2H is also required for BRCA1 recruitment to break sites in a pH2AX independent manner downstream of H2B ubiquitination by RNF20 (19).
We have previously characterized SNF2H as a target of microRNA regulation (20). microRNAs are small 17–25 nucleotide RNA molecules involved in post-transcriptional regulation of gene expression in metazoans in a sequence-specific manner (21). microRNAs in the same family (e.g. miR-99a, −99b and −100) have nearly identical sequence and target the same sets of mRNAs. microRNAs are crucial for development of higher organisms and each microRNA can regulate the expression of many proteins to enact complex changes in cellular phenotype. MicroRNAs have been found to be intimately involved in the differentiation of many tissue types during development (22–26). Additionally, in cancer, microRNAs are often misregulated, with certain miRNAs able to function as tumor suppressive and oncogenic factors (27–30).
In a screen for microRNAs that modulate a cell’s response to DNA damage, we found that expression of members of the miR-99 family were upregulated following DNA damage and that miR-99 expression correlated with radiation sensitivity. The downregulation of SNF2H, a miR-99 family target, mediated radiation sensitivity through its role in facilitating DNA repair. The upregulation of the miR-99 family following radiation decreased the efficiency of repair factor recruitment and the rate of DNA repair after a second exposure to ionizing radiation. Thus, the induction of miR-99 represents a switch by which cells subjected to multiple rounds of radiation are directed away from continuing to repair their DNA. Interestingly, fractionation of radiation in radiotherapy is based on the principle that multiple smaller doses of radiation are more effective than a single large dose of radiation. Although fractionated therapy is widely used in clinical practice, molecular mechanisms underlying the differential radiosensitivity of some tumors to fractionated therapy are not entirely understood (31, 32). We suggest that induction of anti-DNA repair microRNAs such as the miR-99 family is one such molecular mechanism.
Results
miR99 family members downregulated in radioresistant cancer sensitize cells to DNA damage by ionizing radiation
We were interested in examining microRNAs whose expression were misregulated in cancer and could alter the radiation sensitivity of cancer cells. We initially began by measuring differential microRNA expression in two breast cancer lines with differing radiation sensitivity. RNA was isolated from p53 positive MCF-7 and p53 negative SK-BR-3 cells before and 24 hours following treatment with 5 gy ionizing radiation(IR) and hybridized to Exiqon microarrays containing locked nucleic acid probes complementary to currently known microRNAs. We found 6 microRNAs to be more than 2 fold overexpressed in MCF7 cells and upregulated following irradiation (Fig. 1a). Among those upregulated were miR-21 and miR-26a, which are thought to be oncogenic microRNAs, targeting the PTEN tumor suppressor gene(33–35). miR-21* is also upregulated, but likely as an artifact of the increased expression of miR-21. Little is known about miR-105, though some evidence suggests it may be antiproliferative as it was found to reduce cyclin B expression in ovarian granulosa cells(36). The last two microRNAs among this group upregulated following irradiation and overexpressed in MCF7 were both members of the miR-99 family, miR-99a and miR-100. We had previously identified this family of microRNAs as being repressed during progression of prostate cancer and having a tumor suppressive role (20)(Fig. 1a). Their appearance in this screen suggested they may also play a role in altered radiation resistance during prostate and breast cancer progression.
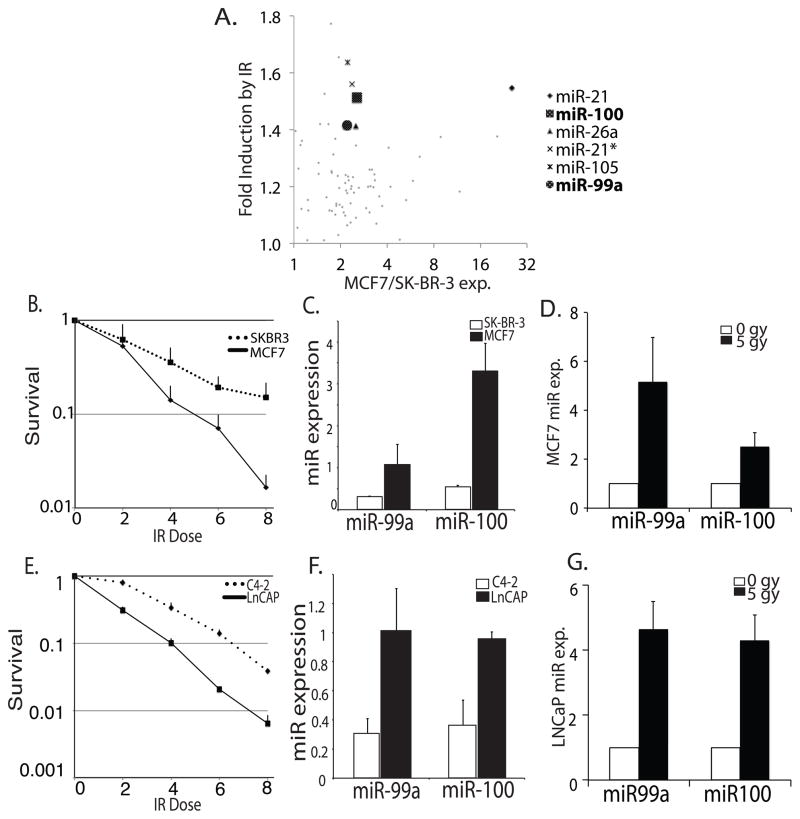
Figure 1.
miR99 family members are downregulated in more radioresistant breast and prostate cancer cell lines and are upregulated following DNA damage. (A) Microarray screen of microRNAs induced by radiation. Y-axis: fold upregulated following irradiation in MCF7. X-axis: fold expression in MCF7 (p53+) vs. SK-BR-3 (p53−). Highlighted microRNAs were at least 2 fold overexpressed in MCF7, and upregulated at least 1.4 fold 24 hours following treatment with IR. miR-99a and 100 were both found to meet these criteria. (B) Clonogenic assay of MCF7 and SK-BR-3 cells following IR. The colonies/well were counted to get fractional survival. Mean + SD (n=3). (C) miR-99a and miR-100 expression measured by qPCR in MCF7 and SK-BR-3 cells, normalized to u6snRNA. Mean + SD (n=6). (D) miR-99a and miR-100 expression measured by qPCR, normalized to β-Actin, 24 hours following IR treatment in MCF7 cells. Mean + SD (n=6). (E) Clonogenic assay of LNCap and C4-2 cells following IR. Fractional survival as in Fig. 1b (n=3). (F) miR-99a and miR-100 expression measured by qPCR in LNCaP and C4-2 cells, normalized to u6snRNA. Mean + SD (n=6). (G) miR-99a and miR-100 expression measured by qPCR, normalized to β-Actin, 24 hours following IR treatment in LNCaP cells. Mean + SD (n=6).
To confirm the radiation sensitivities of the cancer cell lines differentially expressing miR-99 family members, we performed clonogenic assays following IR comparing the survival of MCF7 and SK-BR-3 cells, as well as LNCaP cells and their more advanced daughter cell line C4-2. We found SK-BR-3 cells to be significantly more resistant to radiation treatment than MCF7 cells (Fig. 1b), and C4-2 cells more resistant than LNCaP cells (Fig. 1e), correlating with lower basal expression of miR-99a and miR-100 as measured by qPCR (Fig. 1c, 1f). qPCR was used to confirm that miR-99 family members were upregulated following IR treatment. We found miR-99a and miR-100 to be 3–4 fold upregulated in MCF7 and LNCaP cells 24 hours following IR, when normalized to β-actin, confirming the results of the microarray analysis, and suggesting their upregulation occurs following irradiation of prostate cancer cell lines as well (Fig. 1d, 1g), similar results were observed when normalized to u6snRNA or GAPDH (Fig. S1a, S1b).
Having observed a correlation between decreased expression of the miR-99 family and radiation resistance, we wanted to determine whether reintroduction of miR-99 family members into cells could increase DNA damage sensitivity. We introduced exogenous miR-99a or miR-100 into C4-2 and SK-BR-3 cells, treated with ionizing radiation and examined clonogenic survival. We found a significant reduction in survival following radiation in the presence of exogenous miR-99 family members (Fig. 2a, 2b). These data suggest that the miR-99 family may increase the sensitivity of cells to DNA damage and that a decrease in miR-99 family member expression may be a mechanism by which cancer cells can acquire resistance to DNA damaging agents.
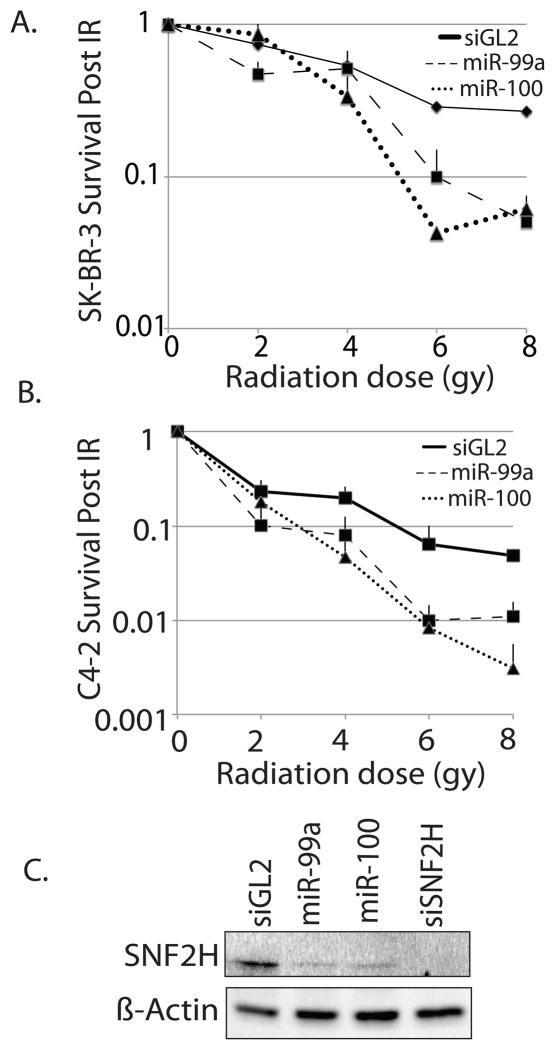
Figure 2.
Introduction of exogenous miR-99a and miR-100 sensitize cells to ionizing radiation. (A) Clonogenic assay of SK-BR-3 cells treated with IR following transfection with double stranded mimics of miR-99a or miR-100. siGL2: negative control RNA-duplex targeting luciferase. Fractional survival as in Fig. 1b. (B) Clonogenic assay of C4-2 cells following transfection with double stranded mimics of miR-99a or miR-100. Fractional survival as in Fig. 1b. (C) Western blot for level of SNF2H in C4-2 cells following transfection with double stranded mimics of miR-99a or miR-100, or siRNA targeting SNF2H.
miR-99 and siRNA of its target SNF2H reduce BRCA1 localization to sites of DNA damage
We previously identified the chromatin remodeling factor SNF2H as a direct target of miR-99a in LnCAP and C4-2 cells (20). SNF2H has recently been implicated in the repair of DNA damage by HR and NHEJ, and is an important member of complexes required for the recruitment and function of DNA repair proteins to sites of DSBs (18, 19). We were curious to see whether introduction of miR-99 family members would have an effect on DNA repair through SNF2H. We began by confirming that miR-99a and miR-100 could reduce the level of SNF2h in cells by performing western blots following transfection of double stranded mimic oligonucleotides. As expected, miR-99a and miR-100 significantly reduced the levels of SNF2H, though not to the same extent as siRNA targeting the 3′ UTR of SNF2H (Fig. 2c). To determine if the miR-99 family could affect the recruitment of DNA proteins to sites of damage, we introduced miR-99a or siRNA targeting the 3′ UTR of SNF2H into C4-2 cells and subjected them to gamma irradiation. We then examined BRCA1 and pH2AX foci formation by immunofluorescence. miR-99a, miR-100 and siSNF2H greatly reduced the number of cells expressing intense BRCA1 ionizing radiation induced foci (Fig. 3a, S1C, S2). Neither treatment reduced pH2AX foci following IR. We also examined MDC1 and conjugated ubiquitin foci formation following IR, and saw no significant reduction in the number of these foci (Fig. S3). This is consistent with the recent finding that SNF2H facilitates BRCA1 recruitment downstream of the ubiquitination of H2B by RNF20 in a pH2AX independent manner(19).
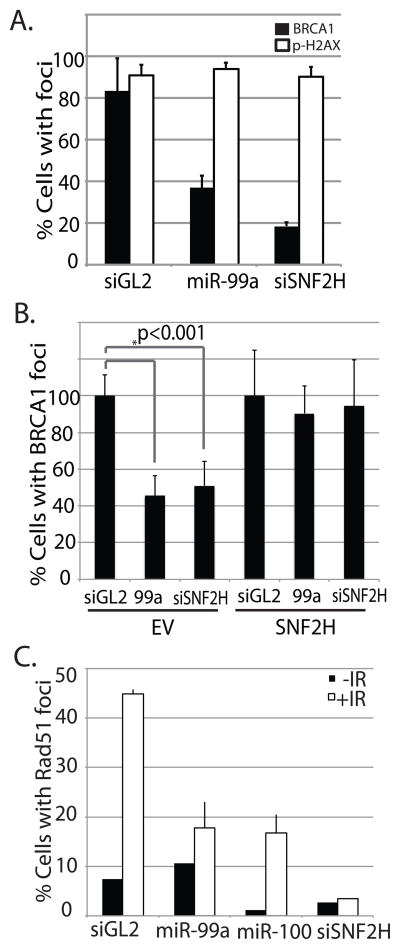
Figure 3.
miR-99a reduces foci formation of BRCA1 and Rad51, but not γH2AX following IR. (A) Quantitation of BRCA1 and γH2AX immunofluorescence in C4-2 cells transfected with miR-99a or with siRNA targeting SNF2H 3′ UTR 1hr following treatment with 5 gy IR. siGL2: negative control RNA duplex targeting luciferase. (B) Repression of BRCA1 foci is rescued by SNF2H. Quantitation of BRCA1 immunofluorescence in C4-2 cells transfected with miR-99a or siRNA targeting SNF2H 3′ UTR 1hr following treatment with 5 gy IR. EV: Empty vector. SNF2H: Cells expressing non-targetable SNF2H ORF. (C) Quantitation of Rad51 immunofluorescence in cells transfected with miR-99a, miR-100 or siRNA targeting SNF2H. Foci formation was examined 90min following treatment with 5 gy IR. Graphs show mean + SD, over 150 cells/sample.
Recruitment of DNA repair proteins is unaffected by miR99 in cells expressing non-targetable SNF2H
To determine if repression of SNF2H by miR-99a was responsible for the repression of BRCA1 recruitment by miR-99a, we stably expressed the SNF2H ORF in C4-2 cells. The absence of the 3′ UTR makes this form of SNF2H resistant to miR-99a and we examined BRCA1 foci in these cells following irradiation. The non-targetable SNF2H rescued BRCA1 foci formation both after treatment with miR-99a as well as siRNA targeted to the SNF2 3′UTR (Fig. 3B, S1D). This evidence shows that miR-99a inhibits BRCA1 recruitment through the downregulation of SNF2H.
miR-99 and siSNF2H reduce Rad51 recruitment to sites of DNA damage
Since BRCA1 is contained within multiple protein complexes that can either promote or repress DNA repair by homologous recombination, we examined whether miR-99 could repress recruitment of proteins directly involved in HR. miR-99a or 100, and siSNF2h were introduced into C4-2 cells, and Rad51 foci formation was examined following irradiation. Both the miR-99 family as well as siSNF2H greatly reduced Rad51 foci formation (Fig. 3c, S4), suggesting the recruitment of this essential component of the HR pathway requires SNF2H, and can be blocked by overexpression of the miR-99 family.
miR-99 reduces the rate of DNA repair through downregulation of SNF2H
To assess the effect of miR-99a on the efficiency of DNA repair, we transfected C4-2 cells with miR-99a or siSNF2H and performed neutral agarose comet assays, which detect DSBs, following treatment with IR. With control siRNA oligo, the majority of double-strand DNA breaks was repaired within 60 minutes of treatment with IR. In the presence of miR-99a or siSNF2H, the DNA damage persisted, resulting in a 3 fold greater tail moment than the control at 60 minutes following irradiation (Fig. 4a, S5a). A similar effect was seen with introduction of miR-100 (Fig. S6) However when we stably expressed the SNF2H ORF in these cells, the repression of DNA repair by miR-99a or siSNF2H was reversed (Fig. 4b, S5b). Therefore, miR-99a reduces double-strand break repair following IR through repression of SNF2H.
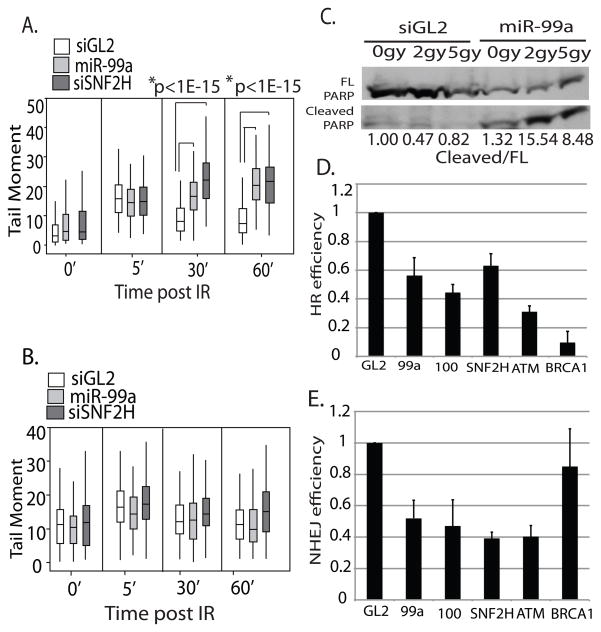
Figure 4.
miR-99 slows the kinetics of DNA repair following IR through repression of SNF2H, increases apoptosis and reduces the efficiency of DNA repair by HR and NHEJ. (A) DNA damage was measured by neutral agarose gel comet assay at indicated times following 10 gy IR treatment of C4-2 cells infected with control PMSCV retrovirus. The cells were irradiated 72 hours after transfection with miR-99a or siSNF2H 3′UTR. Box plots representing quantitation of tail moment. Horizontal line: median. Rectangle: interquartile range. Vertical line: 5th percentile to 95th percentile. At least 100 comets per sample were analysed. (B) DNA damage was measured by neutral comet assay in cells stably expressing SNF2H without its 3′UTR, 72 hours after transfection with miR-99a or siSNF2H 3′UTR. Rest as in A. (C) Western blot showing full length and cleaved PARP 48 hours following radiation in cells transfected with control siRNA or miR99a. Normalized to full length PARP, Quantitation shown below. Full length PARP shown at a lighter exposure than cleaved PARP. (D) Percentage of GFP positive cells representing successful HR 48 hours following I-Sce1 transfection. DR13-9 HeLa cells were transfected with I-Sce1 after a 24 hour treatment with miR-99a, miR-100, siSNF2H, siATM, or siBRCA1. GL2: negative control duplex against luciferase. Mean + SE (n=2). (E) Percentage of DS-Red positive cells representing successful NHEJ 48 hours following I-Sce1 induction. I-Sce1 was induced by doxycyline in 293B cells containing DS-Red-NHEJ reporter after a 24 hour treatment with miR-99a, miR-100, siSNF2H, siATM, or siBRCA1. Mean + SD, over 150 cells/sample.
Exogenous miR-99a increases the extent of apoptosis following IR
To determine if the deficit in DNA repair experienced by cells with exogenous miR-99 leads to an increase in apoptosis in these cells, we examined PARP cleavage in cells transfected with miR-99a following treatment with IR. While C4-2 cells treated with control siRNA displayed no increase in the ratio of cleaved PARP 48 hours following IR, miR-99a transfected cells displayed a marked increase in PARP cleavage after treatment with 2 and 5gy of IR (Fig. 4c). This suggests that miR-99a can modulate cell survival following DNA damage, likely through its ability to decrease DSB repair.
miR-99a, miR-100 and siSNF2H block homologous recombination
Having observed the effect of miR-99a and miR-100 on BRCA1 foci formation, we set forth to determine if the microRNAs reduced the rate of successful HR following DNA damage. miR-99a, miR-100 and siSNF2H, were transfected into DR13-9 HeLa cells, which contain two partial GFP cassettes containing I-Sce1 sites (37, 38). Successful HR after an I-Sce1 induced double-strand DNA break produces a functional GFP, whose expression can be measured by FACS. In the presence of miR-99a, miR-100 or siSNF2H, we observed a 40 percent reduction in HR activity as reported by the percentage of cells expressing GFP (Fig. 4d). By comparison, transfection with siRNA targeting ATM or BRCA1 resulted in 70 or 90 percent reductions in HR respectively. These data show that miR-99a, miR-100 and siSNF2H can reduce the efficiency of HR following double strand breaks, which correlates with the decreased recruitment of BRCA1 to sites of DNA damage.
miR-99a, miR-100 and siSNF2H block efficient repair of double strand breaks by non-homologous end joining
SNF2H is also important for the function of Ku70/80 at sites of DNA damage, facilitating efficient repair of double strand breaks by NHEJ(18). To determine if miR-99a and miR-100 could also reduce the efficiency of NHEJ we utilized a reporter assay that generates expression of DS-Red following successful NHEJ after I-Sce1-generated double strand breaks (39). siSNF2H caused a 60 percent reduction in NHEJ, represented by the percentage of DS-Red expressing cells. miR-99a or miR-100 produced a 50 percent reduction in NHEJ (Fig. 4e). siRNA targeting ATM also displays a significant reduction of NHEJ whereas siBRCA1 showed no significant reduction of NHEJ. This suggests that miR-99a/100 can also reduce the efficiency of repair of double strand breaks by NHEJ, as seen after repression of SNF2H.
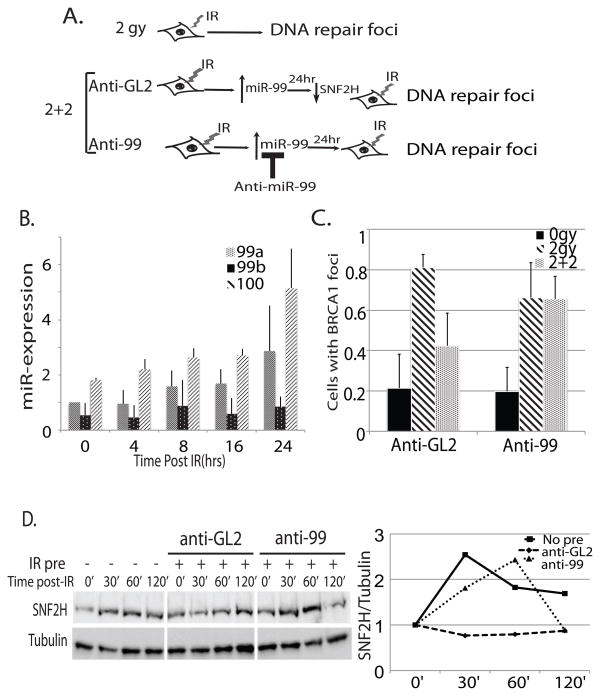
Induction of miR-99a and miR-100 following ionizing radiation reduces the efficiency of DNA repair when confronted with a second round of radiation by blocking SNF2H induction
Having observed that miR-99a and miR-100 are both induced following DNA damage and reduce the efficiency of DNA repair, we hypothesized that when confronted with multiple rounds of DNA damage, cells may become less efficient at DNA repair due to upregulation of the miR-99 family (Fig. 5a). We examined expression of the miR-99 family after radiation treatment over 24 hours and observed an increase in miR-99a and 100 expression of 3 fold over untreated cells (Fig. 5b). To determine if this induction resulted in an effect upon SNF2H expression, we examined SNF2H levels following irradiation in untreated cells, or in cells pretreated with IR 24 hours earlier. We found that in untreated cells, SNF2H is rapidly induced following radiation treatment, while in IR pretreated cells transfected with anti-GL2, this induction was no longer observed. However, when cells were treated with anti-99 (preventing the induction of miR99a or 100 (Fig. S6b), SNF2H was induced normally after the second radiation (Fig. 5d). Therefore the miR-99 induction after the first round of radiation prevents the upregulation of SNF2H protein following the second round of radiation. We also observed that SNF2H mRNA was transiently induced in all three treatment groups, indicating SNF2H is transcriptionally upregulated following IR (Fig. S6c). However, the increased miR-99 in IR-pretreated cells prevents the expected increase in SNF2H protein, as would be expected from the mechanism of action of a microRNA.
Figure 5.
Induction of miR-99 family members following radiation in LnCAP cells decreases the efficiency of repair after subsequent rounds of radiation by preventing induction of SNF2H. (A) Schematic representation of the experiment in Fig. 5c. and 5d Cells were treated with 2 gy, incubated for 24 hours and then treated with another round of 2 gy IR. miR-99a/miR-100 induction was blocked by introduction of 2′-O-methyl antisense oligonucleotides targeting mir-99a and miR-100 (anti-99) at the time of the first treatment (B) Expression of miR-99 family members by qRT-PCR over 24 hours following 2 gy IR. (C) Quantitation of BRCA foci formation in LNCaP with no radiation (0 gy) or conditions described above. At least 100 cells were counted in the irradiated samples and at least 45 in the unirradiated samples. 1:100% of cells have >10 foci. Mean + SD, over 150 cells counted/sample. (D) Western blot for SNF2H expression following 2 gy IR. Cells were left untreated or pretreated with 2 gy IR (IR pre) and incubated with 2′-O-methyl anti-GL2 or anti-miR-99 for 24 hours prior to second treatment of IR. Blots were quantitated and normalized to tubulin expression. Right: Expression relative to tubulin signal plotted relative to 0′ time point.
To test whether miR-99 induction, and prevention of SNF2H upregulation following IR had an effect on subsequent DNA repair, we performed a similar experiment and tested the efficiency of formation of DNA repair foci in untreated or IR pretreated cells. We found that after the first cycle of 2 gy IR, 80 percent of IR untreated cells formed strong BRCA1 foci. However, in IR pretreated cells transfected with anti-GL2, only 40 percent of cells formed strong BRCA1 foci after the second round of IR (Fig. 5d, S6a). In contrast, anti-99 treated cells had similar BRCA1 foci formation between IR untreated and pretreated cells, indicating that induction of the miR-99 family and subsequent inability to induce SNF2H is responsible for the reduction in BRCA1 recruitment to double strand break sites after a second round of DNA damage.
Upregulation of miR-99 following DNA damage decreases the rate of DSB repair after a second round of radiation
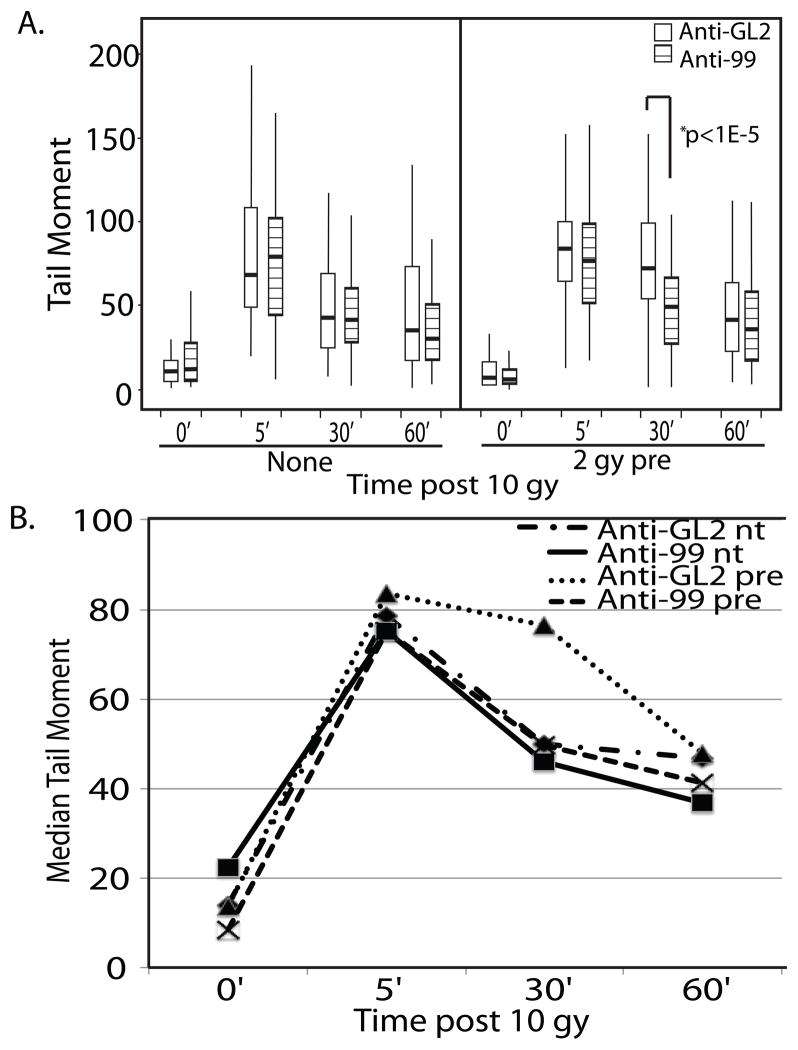
To examine whether upregulation of the miR-99 family following radiation could in fact slow the rate of DNA repair after a second round, we pretreated LNCaP cells with 2 gy of IR and performed neutral comet assays following a second 10 gy dose 24 hours later. During the time following pretreatment, cells were incubated with 2′-O-methyl antisense oligo inhibitors targeting either GL2(control) or the miR-99 family. Pretreatment with IR significantly increased DNA damage remaining 30 minutes following irradiation (Fig 6a, 6b, anti-GL2 pre). However, in cells containing anti-miR-99, there was no difference between the rate of repair of the pretreated and unpretreated cells, suggesting the decrease in repair following pre-irradiation of the control cells was due to miR-99 induction(Fig. 6a, 6b, anti-99 pre).
Figure 6.
Induction of miR-99 family members following IR delays repair if exposed to a subsequent round of radiation. (A) Boxplot of comet tail moments observed in LNCaP cells. Cells were either left untreated (nt) or exposed to a 2 gy pretreatment of IR (pre). After 24 hours treatment with 2′-O-methyl antisense oligonucleotides targeting either GL2 or miR-99a and miR-100 (anti-99), the cells were exposed to 10 gy. DNA damage was examined by comet assay at the given time points. (B) Plot of the median tail moment of each treatment group in the experiment shown in panel A. At least 100 comets per sample were analyzed.
Discussion
We have identified that the miR-99 family is upregulated in response to ionizing radiation and reduces the ability of cells to repair damaged DNA. In light of the recent findings demonstrating SNF2H facilitates HR and NHEJ DSB repair (18, 19), we determined that miR-99a could alter the efficiency of DNA repair by regulating the expression of SNF2H. Exposure to a single dose of DNA damage induces DNA repair pathways and also leads to increased miR-99 family expression. This subsequent induction of the miR-99 family prevents SNF2H from being rapidly induced following DNA damage, which decreases DNA repair efficiency if multiple rounds of DNA damage are experienced 24 hours apart. The expression of this microRNA is decreased in more advanced cancers, suggesting that the resultant greater DNA repair efficiency makes these cell lines more resistant to radiotherapy. Loss of this microRNA may represent a mechanism by which cancer cells acquire resistance to DNA damage by allowing efficient repair to continue after multiple mutagenic insults inflicted by radiotherapy. In addition, the observation that miR-99 expression correlates with p53 status suggests its induction also requires intact DNA damage checkpoint pathways, and the mechanism of this regulation warrants further study. The importance of the miR-99 family in regulating cellular response to DNA damage is further supported by recent findings that the miR-99 family targets pro-survival proteins IGF-1R and mTOR (20, 40), indicating that there are multiple pathways the miR-99 family targets that would presumably have an effect on cells survival following DNA damage.
microRNAs are regulated by the DNA damage response and alter the expression of many of the signaling and effector components of DNA repair (41). For example, the DNA damage signaling histone variant H2AX is regulated by miR-24 in post mitotic hematopoetic cells, rendering them hypersensitive to DNA damaging agents (42). The DNA damage checkpoint protein Cdc25a is regulated by miR-21, which is upregulated following DNA damage, resulting in slowing of cell cycle progression (43). The p53 tumor suppressor upregulates the transcription of the miR-34a-c family, which in turn alters gene expression, slowing cell cycle progression (44). Additionally, microRNA biogenesis is regulated at the processing step that converts primary miR to precursor miR by the association of p53 with the Drosha/DGCR8 complex, resulting in upregulation of several growth suppressive microRNAs (45). BRCA1 is directly downregulated by miR-182, resulting in impaired HR and sensitivity to ionizing radiation (46). The targeting of SNF2H, a protein required both for efficient HR as well as NHEJ repair by the miR-99 family is an additional level of control that microRNAs can exert over the DNA damage response, decreasing double strand break repair throughout the cell cycle. It will be of interest to examine the mechanism of regulation of the miR-99 family, how its upregulation is p53 dependent, and what epigenetic changes lead to a decrease of its expression in more advanced cancers. Additionally, while it is clear that chromatin remodeling factors such as SNF2H are required for efficient recruitment of DDR proteins, it will be of great interest to determine exactly which steps in the response require chromatin remodeling activity, and how expression and recruitment of these complexes to sites of DNA damage are regulated to facilitate repair.
A matter of great curiosity is how the regulation of DSB repair efficiency by the upregulation of miR-99 following DNA damage interacts with the timing and efficacy of fractionated radiation therapy in the treatment of cancers. Fractionation of radiotherapy has been in use since the early 20th century when it was found to be effective in maximizing damage to proliferating cells while minimizing damage to normal tissue. Fractionated therapy increases the therapeutic index of radiation by facilitating reoxygenation and cell cycle redistribution of rapidly dividing tumor cells while simultaneously allowing repair of sub-lethal DNA damage in non-dividing cells and repopulation of normal cells from the surrounding tissue. Recent attention has been focused on alternative regimens of fractionated therapy to increase their efficacy, as many protocols have been mostly unchanged since the 1930s (31). While altered regimens are being developed to minimize damage to surrounding tissues, consideration should be given to molecular mechanisms such as the induction of the miR-99 family to confer optimal tumor radiation sensitivity at given time points during treatment. Additionally, since decrease of miR-99 family expression in more advanced cancer cells correlates with their radioresistance and transfection of these microRNAs reduced cell survival following radiation treatment, in-vivo reintroduction of the miR-99 may be useful as a sensitizer to radiation therapy once microRNA delivery to tumor cells becomes feasible.
Materials and Methods
Cell Culture
Human prostate cancer cell lines LnCAP and C4-2, as well as human breast cancer cell lines MCF7 and SK-BR-3 were obtained from ATCC. HeLa DR13-9 cells (37, 38) were obtained from Dr. J.D. Parvin (Ohio State University). NHEJ-DSRed 293B cells were obtained from the Dr. J. Larner (UVA) and K. Valerie (VCU) (47). LnCAP and C4-2 were grown in RPMI 1640 medium supplemented with 10% FBS. MCF7, HeLa DR13-9 and NHEJ-DSRed 293B cells were cultured in DMEM with 10% FBS, and SK-BR-3 cells were cultured in McCoy’s 5a medium with 10% FBS. All cell lines were cultured under normal conditions.
Clonogenic assays
Cells were transfected in 10 cm dishes with 20 picomoles of miR-99a duplex or siRNA against firefly luciferase as a control. The oligonucleotides were incubated for 30 minutes with 8 uL Lipofectamine RNAimax per plate in 2 mL Optimem before mixing with trypsinized cells and plated at a 20% confluence. Plates were incubated for 48 hours before seeding to 6-well plates in the presence of 20nM duplex oligo at increasing seeding densities corresponding with the planned dose of radiation. 6-well plates were irradiated in a Shepherd Mark 1 Cs-137 irradiator. Cells were incubated until colonies arising from single cells reached 25 cells or more, with media changed every 72 hours. Colonies were counted, and clonogenic survival was calculated by dividing the number of colonies at a given dose by the number of colonies in the un-irradiated control samples, and multiplied by the seeding density coefficient.
miRNA expression analysis
RNA was isolated from cells using Trizol extraction (Invitrogen). For microarray analysis, RNA was further purified using RNAeasy RNA cleanup kit (Qiagen). For qPCR validation, Poly A tailing and cDNA preparation of mature microRNAs was performed using the NCODE miRNA amplification system (Invitrogen). qPCR amplification was performed using forward primers identical to the mature miRNA sequence, and NCODE universal reverse primers with Sybr Green ER (Invitrogen). Expression was normalized to u6snRNA, which used the primer sequence 5′-CTGCGCAAGGATGACACGCA-3′. The sequence of miR-99a is 5′-aacccguagauccgaucuugug-3′, and the sequence of miR-100 is 5′-aacccguagauccgaacuugug-3′.
Immunofluorescence
Cells were plated on glass cover slips in the presence of 20nM siRNA duplex. After 72 hours, the coverslips were irradiated with 5gy IR. After 60 minutes, the cover slips were fixed with 4% formaldehyde in PBS, and permeabilized in 0.5% Triton X-100 in PBS. Coverslips were blocked in 5% goat serum, or 5% donkey serum if primary antibody is of goat origin (MDC1). Coverslips were incubated at room temperature with primary antibody for 1 hour, and Alexa 488 or 549 conjugated secondary antibody for 1 hour, with three TBS washes following each antibody incubation. Coverslips were then mounted with Vectashield mounting solution (Vector Laboratories). Images were collected at equal exposures and foci were counted manually using ImageJ. Antibodies used were BRCA1 D-9(Santa Cruz), pH2AX (Cell signaling, Millipore), Rad51 H-92 (Santa Cruz) MDC1 C-20(Santa Cruz), Anti conjugated ubiquitin FK2 (Millipore). pH2AX primary antibodies was used at a 1:200 concentration in 5% goat serum, MDC1 antibody was used at a 1:100 concentration in 5% donkey serum, all other antibodies were used at a 1:100 concentration in 5% goat serum.
Western Blotting
Cells were lysed in IPH buffer, run on 10%-SDS-PAGE gels and transferred to nitrocellulose membranes. Blocking was performed with 3% BSA and membranes were incubated overnight with SNF2H antibody(Abcam) before washing and secondary antibody incubation and Incubation with Millipore Immobilon HRP substrate.
Microscopy
Fluorescence images were acquired on a Nikon Microphot SA upright microscope equipped with a Nikon NFX35 camera using SPOT imaging software(Diagnostic Instruments Inc.) and a Nikon PlanApo 60x oil objective lens. For each experiment, fluorescence images were acquired on the same day using the same exposure times, gamma, and gain between samples. Images were enhanced for brightness and contrast to the same extent within Photoshop.
HR/NHEJ assay
20% confluent HeLa DR13-9 cells and NHEJ-DS-Red 293B cells were transfected with 20nM siRNA using Lipofectamine RNAimax in 10 cm plates. After 24 hours, each plate was transfected with 20 ug I-Sce1 plasmid with Lipofectamine 2000. After another 48 hours, cells were trypsinized, resuspended in media and analyzed by FACS for GFP or DS-Red expression.
Comet Assay
Cells were transfected for 72 hours with 20nM siRNA oligo in 6cm plates, followed by 10 gy irradiation. Cells were then trypsinized following irradiation and resuspended in ice cold PBS, incubated on ice until all time points were collected. Neutral comet assays were then performed using the Trevigen Comet assay kit standard protocol.
Supplementary Material
Acknowledgments
AM was supported by DOD BCRP predoctoral traineeship BC073568. DS was supported by DOD PCRP predoctoral traineeship PC094499. This work was supported by P01CA104106 to AD. We thank members of the Dutta Laboratory for their help and discussion.
Footnotes
Conflict of interest.
The authors declare no conflict of interest.
References
- 1.van Attikum H, Gasser SM. Crosstalk between histone modifications during the DNA damage response. Trends in Cell Biology. 2009;19(5):207–17. doi: 10.1016/j.tcb.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 2.Celeste A, Petersen S, Romanienko PJ, Fernandez-Capetillo O, Chen HT, Sedelnikova OA, et al. Genomic Instability in Mice Lacking Histone H2AX. Science. 2002 May 3;296(5569):922–7. doi: 10.1126/science.1069398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Attikum H, Gasser SM. The histone code at DNA breaks: a guide to repair? Nat Rev Mol Cell Biol. 2005;6(10):757–65. doi: 10.1038/nrm1737. [DOI] [PubMed] [Google Scholar]
- 4.van Attikum H, Fritsch O, Hohn B, Gasser SM. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell. 2004 Dec 17;119(6):777–88. doi: 10.1016/j.cell.2004.11.033. [DOI] [PubMed] [Google Scholar]
- 5.van Attikum H, Fritsch O, Gasser SM. Distinct roles for SWR1 and INO80 chromatin remodeling complexes at chromosomal double-strand breaks. EMBO J. 2007;26(18):4113–25. doi: 10.1038/sj.emboj.7601835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morrison AJ, Highland J, Krogan NJ, Arbel-Eden A, Greenblatt JF, Haber JE, et al. INO80 and [gamma]-H2AX Interaction Links ATP-Dependent Chromatin Remodeling to DNA Damage Repair. Cell. 2004;119(6):767–75. doi: 10.1016/j.cell.2004.11.037. [DOI] [PubMed] [Google Scholar]
- 7.Tsukuda T, Fleming AB, Nickoloff JA, Osley MA. Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature. 2005;438(7066):379–83. doi: 10.1038/nature04148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee H-S, Park J-H, Kim S-J, Kwon S-J, Kwon J. A cooperative activation loop among SWI/SNF, [gamma]-H2AX and H3 acetylation for DNA double-strand break repair. EMBO J. 2010;29(8):1434–45. doi: 10.1038/emboj.2010.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jha S, Shibata E, Dutta A. Human Rvb1/Tip49 is required for the histone acetyltransferase activity of Tip60/NuA4 and for the downregulation of phosphorylation on H2AX after DNA damage. Mol Cell Biol. 2008 Apr;28(8):2690–700. doi: 10.1128/MCB.01983-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stewart GS, Wang B, Bignell CR, Taylor AMR, Elledge SJ. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature. 2003;421(6926):961–6. doi: 10.1038/nature01446. [DOI] [PubMed] [Google Scholar]
- 11.Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, et al. RNF8 Ubiquitylates Histones at DNA Double-Strand Breaks and Promotes Assembly of Repair Proteins. Cell. 2007;131(5):887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 12.Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FdrD, et al. Orchestration of the DNA-Damage Response by theRNF8 Ubiquitin Ligase. Science. 2007 Dec 7;318(5856):1637–40. doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huen MSY, Grant R, Manke I, Minn K, Yu X, Yaffe MB, et al. RNF8 Transduces the DNA-Damage Signal via Histone Ubiquitylation and Checkpoint Protein Assembly. Cell. 2007;131(5):901–14. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, et al. RNF168 Binds and Amplifies Ubiquitin Conjugates on Damaged Chromosomes to Allow Accumulation of Repair Proteins. Cell. 2009;136(3):435–46. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 15.Roberts SA, Ramsden DA. Loading of the Nonhomologous End Joining Factor, Ku, on Protein-occluded DNA Ends. Journal of Biological Chemistry. 2007 Apr 6;282(14):10605–13. doi: 10.1074/jbc.M611125200. [DOI] [PubMed] [Google Scholar]
- 16.Mladenov E, Iliakis G. Induction and repair of DNA double strand breaks: The increasing spectrum of non-homologous end joining pathways. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. 2011;711(1–2):61–72. doi: 10.1016/j.mrfmmm.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 17.Larsen DH, Poinsignon C, Gudjonsson T, Dinant C, Payne MR, Hari FJ, et al. The chromatin-remodeling factor CHD4 coordinates signaling and repair after DNA damage. The Journal of Cell Biology. 2010 Sep 6;190(5):731–40. doi: 10.1083/jcb.200912135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lan L, Ui A, Nakajima S, Hatakeyama K, Hoshi M, Watanabe R, et al. The ACF1 Complex Is Required for DNA Double-Strand Break Repair in Human Cells. Molecular Cell. 2010;40(6):976–87. doi: 10.1016/j.molcel.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 19.Nakamura K, Kato A, Kobayashi J, Yanagihara H, Sakamoto S, Oliveira DNP, et al. Regulation of Homologous Recombination by RNF20-Dependent H2B Ubiquitination. Molecular Cell. 2011;41(5):515–28. doi: 10.1016/j.molcel.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 20.Sun D, Lee YS, Malhotra A, Kim HK, Matecic M, Evans C, et al. miR-99 Family of MicroRNAs Suppresses the Expression of Prostate-Specific Antigen and Prostate Cancer Cell Proliferation. Cancer Research. 2011;71(4):1313–24. doi: 10.1158/0008-5472.CAN-10-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004 Jul;5(7):522–31. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 22.Dey BK, Gagan J, Dutta A. miR-206 and −486 Induce Myoblast Differentiation by Downregulating Pax7. Mol CellBiol. 2011 Jan 1;31(1):203–14. doi: 10.1128/MCB.01009-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gagan J, Dey BK, Layer R, Yan Z, Dutta A. MICRORNA-378 targets the myogenic repressor myor during myoblast differentiation. Journal of Biological Chemistry. 2011 Apr 6; doi: 10.1074/jbc.M111.219006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bruno IG, Karam R, Huang L, Bhardwaj A, Lou CH, Shum EY, et al. Identification of a MicroRNA that Activates Gene Expression by Repressing Nonsense-Mediated RNA Decay. Molecular Cell. 2011;42(4):500–10. doi: 10.1016/j.molcel.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen C-Z, Li L, Lodish HF, Bartel DP. MicroRNAs Modulate Hematopoietic Lineage Differentiation. Science. 2004 Jan 2;303(5654):83–6. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 26.Houbaviy HB, Murray MF, Sharp PA. Embryonic Stem Cell-Specific MicroRNAs. Developmental Cell. 2003;5(2):351–8. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- 27.Lee YS, Dutta A. MicroRNAs in Cancer. Annual Review of Pathology: Mechanisms of Disease. 2009 Feb 1;4(1):199–227. doi: 10.1146/annurev.pathol.4.110807.092222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007 May 1;21(9):1025–30. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435(7043):828–33. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang T-C, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, et al. Transactivation of miR-34a by p53 Broadly†Influences Gene Expression and†Promotes†Apoptosis. Molecular Cell. 2007;26(5):745–52. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marcu LG. Altered fractionation in radiotherapy: From radiobiological rationale to therapeutic gain. Cancer Treatment Reviews. 2010;36(8):606–14. doi: 10.1016/j.ctrv.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 32.Skvortsova I, Skvortsov S, Stasyk T, Raju U, Popper B-A, Schiestl B, et al. Intracellular signaling pathways regulating radioresistance of human prostate carcinoma cells. PROTEOMICS. 2008;8(21):4521–33. doi: 10.1002/pmic.200800113. [DOI] [PubMed] [Google Scholar]
- 33.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 Regulates Expression of the PTEN Tumor Suppressor Gene in Human Hepatocellular Cancer. Gastroenterology. 2007;133(2):647–58. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huse JT, Brennan C, Hambardzumyan D, Wee B, Pena J, Rouhanifard SH, et al. The PTEN-regulating microRNA miR-26a is amplified in high-grade glioma and facilitates gliomagenesis in vivo. Genes & Development. 2009 Jun 1;23(11):1327–37. doi: 10.1101/gad.1777409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 Is an Antiapoptotic Factor in Human Glioblastoma Cells. Cancer Research. 2005 Jul 15;65(14):6029–33. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 36.Sirotkin AV, Lauková M, Ovcharenko D, Brenaut P, Mlynček M. Identification of MicroRNAs controlling human ovarian cell proliferation and apoptosis. Journal of Cellular Physiology. 2010;223(1):49–56. doi: 10.1002/jcp.21999. [DOI] [PubMed] [Google Scholar]
- 37.Ransburgh DJR, Chiba N, Ishioka C, Toland AE, Parvin JD. Identification of Breast Tumor Mutations in BRCA1 That Abolish Its Function in Homologous DNA Recombination. Cancer Research. 2010 Feb 1;70(3):988–95. doi: 10.1158/0008-5472.CAN-09-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999 Oct 15;13(20):2633–8. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Golding SE, Morgan RN, Adams BR, Hawkins AJ, Povirk LF, Valerie K. Pro-survival AKT and ERK signaling from EGFR and mutant EGFRvIII enhances DNA double-strand break repair in human gliomacells. Cancer Biol Ther. 2009 Apr;8(8):730–8. doi: 10.4161/cbt.8.8.7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Doghman M, Wakil AE, Cardinaud B, Thomas E, Wang J, Zhao W, et al. Regulation of Insulin-like Growth Factor, ÄìMammalian Target of Rapamycin Signaling by MicroRNA in Childhood Adrenocortical Tumors. Cancer Research. 2010 Jun 1;70(11):4666–75. doi: 10.1158/0008-5472.CAN-09-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu H, Gatti RA. MicroRNAs: new players in the DNA damage response. Journal of Molecular Cell Biology. 2011 Jun 1;3(3):151–8. doi: 10.1093/jmcb/mjq042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lal A, Pan Y, Navarro F, Dykxhoorn DM, Moreau L, Meire E, et al. miR-24-mediated downregulation of H2AX suppresses DNA repair in terminally differentiated blood cells. Nat Struct Mol Biol. 2009;16(5):492–8. doi: 10.1038/nsmb.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang P, Zou F, Zhang X, Li H, Dulak A, Tomko RJ, et al. microRNA-21 Negatively Regulates Cdc25A and Cell Cycle Progression in Colon Cancer Cells. Cancer Research. 2009 Oct 15;69(20):8157–65. doi: 10.1158/0008-5472.CAN-09-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447(7148):1130–4. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460(7254):529–33. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- 46.Moskwa P, Buffa FM, Pan Y, Panchakshari R, Gottipati P, Muschel RJ, et al. miR-182-Mediated Downregulation of BRCA1 Impacts DNA Repair and Sensitivity to PARP Inhibitors. Molecular Cell. 2011;41(2):210–20. doi: 10.1016/j.molcel.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Golding SE, Rosenberg E, Valerie N, Hussaini I, Frigerio M, Cockcroft XF, et al. Improved ATM kinase inhibitor KU-60019 radiosensitizes glioma cells, compromises insulin, AKT and ERK prosurvival signaling, and inhibits migration and invasion. Mol Cancer Ther. 2009 Oct;8(10):2894–902. doi: 10.1158/1535-7163.MCT-09-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.