Abstract
We have investigated the ability of the 3′ exonuclease activity of S. cerevisiae DNA polymerase ε (Pol ε) to proofread newly inserted ribonucleotides (rNMPs). During DNA synthesis in vitro, Pol ε proofreads ribonucleotides with apparent efficiencies that vary from none at some locations to more than 90% at others, with rA and rU being more efficiently proofread than rC and rG. Previous studies show that failure to repair ribonucleotides in the genome of rnh201Δ strains that lack RNase H2 activity elevates the rate of short deletions in tandem repeat sequences. Here we show that this rate is increased by 2–4-fold in pol2–4 rnh201Δ strains that are also defective in Pol ε proofreading. In comparison, defective proofreading in these same strains increases the rate of base substitutions by more than 100-fold. Collectively, the results indicate that although proofreading of an ‘incorrect’ sugar is less efficient than is proofreading of an incorrect base, Pol ε does proofread newly inserted rNMPs to enhance genome stability.
Keywords: DNA replication, DNA polymerase ε, ribonucleotides, exonuclease, proofreading
1. Introduction
Replicative DNA polymerases almost always insert correct deoxynucleoside triphosphates (dNTPs) into correctly aligned primer-templates. When they occasionally do generate mismatches, these can be excised by the 3′ to 5′ exonuclease activities associated with many replicases, and rare mismatches that escape this proofreading can be corrected by mismatch repair (MMR). Operating in series, polymerase selectivity and the two error correction processes assure high fidelity DNA replication and stabilize the eukaryotic nuclear genome over many generations.
This understanding of high fidelity DNA replication has emerged from studies of base-base and insertion-deletion mismatches. DNA polymerases also discriminate well against inserting an ‘incorrect’ sugar, i.e., a ribonucleoside triphosphate (rNTP) (reviewed in [1, 2]). However they do so imperfectly, and the probability that a rNTP may be inserted is further increased by the fact that cellular rNTP concentrations are much higher than dNTP concentrations [3, 4]. In fact, during DNA synthesis in vitro in reactions containing the concentrations of dNTPs and rNTPs measured in extracts prepared from asynchronously growing, log phase S. cerevisiae cells, all three major yeast family B replicases, DNA polymerases α, δ, and ε (Pols α, δ, and ε), incorporate substantial numbers of rNTPs into DNA [5]. In these experiments, one ribonucleotide (rNMP) was stably incorporated for every 625, 5,000 or 1,250 deoxyribonucleotides (dNMPs) incorporated by Pols α, δ and ε, respectively. Interestingly, Pols δ and ε incorporate rNMPs despite the fact that they have intrinsic 3′ to 5′ exonuclease activities that are well known to efficiently proofread single base-base mismatches. This and the fact that urepaired ribonucleotides incorporated into DNA result in replicative stress and genome instability [6], motivated the current investigation of whether rNMPs inserted into DNA by Pol ε can be proofread by its intrinsic 3′ exonuclease.
The possibility that ribonucleotides might be proofread by Pol ε is suggested by previous studies of two family B homologs of Pol ε, T4 DNA polymerase [7] and ϕ29 DNA polymerase [8]. The intrinsic 3′ exonuclease activity of both polymerases can excise ribonucleotides from 3′-termini in primer-template DNA. Moreover, ϕ29 Pol extends a primer with a terminal rG less efficiently than it extends a primer with a terminal dG [8], thereby potentially increasing the probability of excision rather than extension. This may be important because studies of single base-base mismatches clearly show that the balance between excision and extension determines proofreading efficiency (reviewed in [9, 10]). However, neither the ϕ29 Pol nor the T4 Pol study measured actual proofreading, i.e., excision of a newly inserted ribonucleotide during an ongoing polymerization reaction. Thus, the efficiency with which a base pair containing an incorrect sugar is proofread during DNA synthesis, if at all, is largely unexplored. It is also currently unknown whether failure to proofread newly incorporated rNMPs has biological consequences. Interest in whether ribonucleotides can be proofread is increased by the demonstration that the other mechanism for correcting replication errors, DNA mismatch repair, does not prevent the genome instability associated with unrepaired ribonucleotides incorporated during DNA replication by Pol ε in yeast [11].
Here we investigate proofreading of ribonucleotides that are incorporated by S. cerevisiae Pol ε, which has been inferred to be the primary leading strand replicase [12]. This initial focus on Pol ε is based on the fact that Pol ε incorporates rNMPs during DNA synthesis in vitro [5] and in vivo [6], and failure to remove these rNMPs due to a defect in RNase H2-dependent repair increases the rate of 2–5 base pair deletions in tandem repeat DNA sequences [6]. Our biochemical and genetic results support the conclusion that during replication by Pol ε, exonucleolytic proofreading can remove newly inserted ribonucleotides and thereby enhance genome stability. We further show that editing an incorrect sugar in DNA is substantially less efficient than editing single base-base mismatches.
2. Material and Methods
2.1 Biochemistry
DNA modification and restriction enzymes were from New England Biolabs (Ipswich, MA), oligonucleotides were from Integrated DNA Technologies (Coralville, IA), ribonucleotide-containing oligonucleotides were from Dharmacon RNAi Technologies Thermo Scientific (Lafayette, CO), and dNTPs were from Amersham Biosciences (Piscataway, NJ).
2.2 Polymerases and DNA substrates
Wild type (WT) and exonuclease-deficient S. cerevisiae Pol ε were expressed and purified as previously described [13, 14]. Oligonucleotide primer-templates (Fig. 1A) were prepared as described [5].
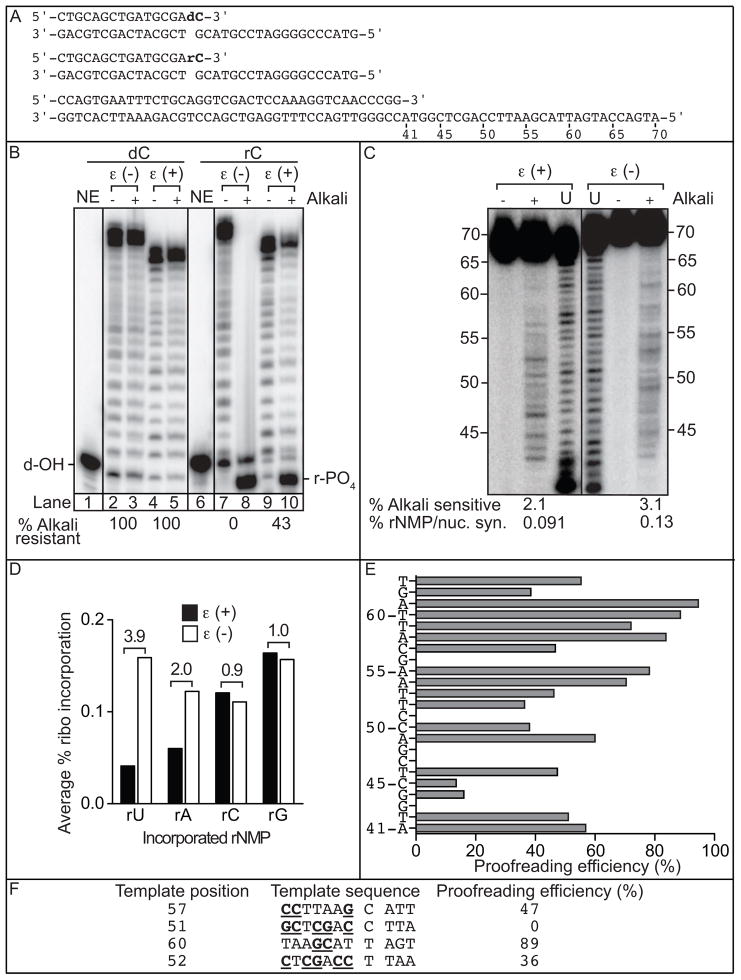
Fig. 1. Ribonucleotide extension, incorporation and proofreading by Pol ε.
(A) Sequences of primer-templates used for panel B (top two substrates) and panel C (lower substrate); (B) Alkali-cleavage of extension products. ε(+) and ε(−) refer to proofreading-proficient and proofreading-deficient Pol ε, respectively. NE indicates the no enzyme control. For the lanes under dC, the highest mobility band represents the unextended deoxy-terminated primer (d-OH). For lanes under rC, the highest mobility band (r-PO4) represents the 3′-terminal phosphate-containing product of extension followed by alkali cleavage. This molecule migrates faster due to the presence of the terminal-phosphate [19]. The percentage of alkali-resistant product is indicated below the image; (C) Stable rNMP incorporation. Lanes marked (U) depict products generated by Pol ε prior to gel purification, as described in [5]. The percentage of alkali sensitive products and the percentage of rNMP incorporation per nucleotide synthesized are shown below each lane. The mean and standard deviation for triplicate measurements was 2.1 ± 0.3 for wild type Pol ε and 3.1 ± 0.02 for exonuclease-deficient Pol ε; (D) Average frequency of ribonucleotide incorporation for rU, rA, rC and rG calculated from (C). The relative difference in ribonucleotide incorporation between proofreading-proficient and –deficient Pol ε is shown above each base; (E) Proofreading efficiency calculated as 1-(rNMP incorporation for proofreading proficient pol ε/rNMP incorporation for proofreading deficient pol ε) at 24 template positions; (F) Proofreading at two C and two T in four different sequence contexts: C57, C51, T60 and T52. The template base located at the site of proofreading is between the two spaces. G and C bases are underlined and in bold face to highlight the possibility that increased G+C content suppresses proofreading.
2.3 Extending a ribo-terminated primer terminus
Reaction mixtures contained 50 mM Tris (pH 7.5), 0.1 mg/ml BSA, 2 mM DTT, 8 mM MgCl2, 100 nM primer-template containing either a 3′-deoxy-terminated primer or a 3′-riboterminated primer (both labeled with 5′-γ-32P), and the concentrations of dNTPs (16 μM dATP, 30 μM dTTP, 12 μM dGTP, 14 μM dCTP) previously measured in extracts of asynchronous, logarithmically growing budding yeast cells [5]. Reactions were initiated by adding either exonuclease-proficient or exonuclease-deficient Pol ε(10 nM). For this assay, we used the catalytically active 152-kDa N-terminal fragments of the Pol ε catalytic subunit, enzymes that we previously demonstrated [14] are reasonable surrogates for the intact, four subunit Pol ε holoenzymes used for the proofreading assays described below. All components were mixed on ice and incubated for 30 minutes at 30°C. Aliquots of reaction products were incubated with either water or 0.3 M KOH for two hours at 55°C. An equal amount of formamide loading dye (95% deionized formamide, 25 mM EDTA, 0.1% bromophenol blue and 0.1% xylene cyanol) was added, the mixture was heated to 95°C for three minutes, and the products were separated by electrophoresis through an 8% denaturing polyacrylamide gel. A phosphoimager and Image quant software (GE Healthcare) were used to visualize and quantify the products.
2.4 Stable incorporation of rNMPs into DNA
Stable incorporation of rNMPs into DNA was assessed as previously described [5]. Reaction mixtures (20 μL) contained 2.0 pmol (100 nM) of a 70-mer template annealed to a 5′-[γ-32P]-labeled 40-mer DNA primer, and 10 nM Pol ε. The dNTP concentrations were as mentioned above, and the rNTP concentrations were rATP, 3000 μM; rCTP, 500 μM; rGTP, 700 μM; and rUTP, 1700 μM, as previously measured in extracts of asynchronous, logarithmically growing budding yeast cells [5].
2.5 Yeast strain construction
Saccharomyces cerevisiae strains used are isogenic derivatives of strain Δ|(−2)|-7BYUNI300 (MATa CAN1 his7-2 leu2-Δ::kanMX ura3-Δ trp1-289 ade2-1 lys2-ΔGG2899–2900 [15]). Relevant strain genotypes are listed in Supplemental Table S1. The pol2-M644G, rnh201Δ and pol2-M644G rnh201Δ strains were described previously [6, 12]. The pol2–4 strain was constructed using the integration-excision method. A wild type strain was transformed with BamHI-linearized pJB1 plasmid (containing a fragment of the POL2 gene with the D290A and E292A mutations [16]. Following verification of correct integration at the POL2 locus, URA3 was excised and pol2 mutations were confirmed by DNA sequencing. The pol2–4 rnh201Δ mutant was generated by deletion-replacement of RNH201 via transformation with a PCR product containing the hygromycin-resistance cassette (HYG-R) amplified from pAG32 [17] and flanked by 60 nucleotides of sequence homologous to the intergenic regions upstream and downstream of the RNH201 ORF. Transformants that arose from homologous recombination were verified by PCR analysis. The URA3 reporter gene was introduced into the pol2–4 and pol2–4 rnh201Δ strains in either orientation 1 or orientation 2 at position AGP1 [15] by transformation of a PCR product containing URA3 and its endogenous promoter flanked by sequence targeting the reporter to AGP1.
2.6 Strain growth and phenotypic analysis
Strains were grown in rich medium (YPDA: 1% yeast extract, 2% bacto-peptone, 250 mg/l adenine, 2% dextrose, 2% agar for plates). Spot assays were performed by plating 10-fold serial dilutions of exponentially growing cells onto YPDA in the absence or presence of the indicated concentrations of hydroxyurea (HU; Sigma H8627). Plates were incubated at 30°C and photographed after 3 days of growth.
2.7 Measurement of spontaneous mutation rates and sequence analysis
Spontaneous mutation rates were measured by fluctuation analysis as described previously [18]. For each 5-FOA-resistant mutant that was sequenced, an independent colony was patched to YPDA and then replica plated to medium containing 5-fluoro-orotic acid (5- FOA). Genomic DNA from a single 5-FOA resistant colony from each patched colony was isolated and the ura3 gene was PCR-amplified and sequenced. Rates of individual mutation classes were calculated by multiplying the fraction of that mutation type by the total mutation rate for each strain.
3. Results
3.1 rNTP insertion and extension in vitro by wild type and exonuclease-deficient Pol ε
In our initial study of ribonucleotide incorporation by yeast replicases [5], we reported that in reactions containing only one correctly paired dNTP or rNTP present at its cellular concentration, rNTP insertion by exonuclease-proficient Pol ε at a specific template position was lower than dNTP insertion by a factor of 500- to 6,700-fold, depending on the identity of the base. When the same method was used in a later study of rNTP insertion by exonuclease-deficient Pol ε[6], discrimination factors were at least 5-fold lower (see Fig. 1A in [6]), indicating that in these single nucleotide, single site assays, the majority of rNTPs inserted by Pol ε are removed by its intrinsic 3′ to 5′ exonuclease. On that basis, the present study began by monitoring the second step required for stable incorporation of a ribonucleotide into DNA, extension of a 3′-terminal ribonucleotide primer, versus a ‘control’ primer containing a 3′-terminal deoxynucleotide (Fig. 1A). In reactions containing the four dNTPs at their cellular concentrations, both wild type and exonuclease-deficient Pol ε extended the normal, i.e. deoxynucleotide-terminated primer to generate long DNA products (Fig. 1B, lanes 2 and 4). The products of these reactions were insensitive to alkaline hydrolysis (lanes 3 and 5), as expected because no ribonucleotide was present at the original terminus. The wild type and exonuclease-deficient polymerases also generated long DNA products with the substrate containing the 3′-terminal ribonucleotide (lanes 7 and 9). However in this case, all products of extension by exonuclease-deficient Pol ε were hydrolyzed by treatment with 0.3 M KOH, (lane 8), which generates ends with 3′-terminal phosphates that migrate as if they were about one nucleotide shorter than the original un-extended primer due to the negatively charged 3′-phosphate [19]. This complete hydrolysis is consistent with extension of the 3′-terminal ribonucleotide primer in all product molecules, without initial removal of the ribonucleotide by exonuclease-deficient Pol ε. In contrast, 43% of the products of extension by exonuclease-proficient Pol ε were resistant to alkaline hydrolysis (lane 10), indicating that in those products, the 3′-terminal ribonucleotide was excised prior to extension.
3.2 Proofreading reduces stable incorporation of rNMPs
The above results suggest that wild type Pol ε has the potential to proofread ribonucleotides that are incorporated when in competition with dNTPs during normal DNA synthesis. We therefore compared the ability of wild type and exonuclease-deficient four subunit Pol ε to stably incorporate rNMPs into DNA when copying a 70-mer template hybridized to a 40-mer primer (Fig. 1A), this time in reactions containing all four dNTPs and rNTPs at the cellular concentrations previously determined to be present in extracts of asynchronous, logarithmically growing yeast cells [5]. Full-length DNA products were isolated, subjected to alkaline hydrolysis and separated by denaturing polyacrylamide gel electrophoresis. The intensities of product bands measured by phosphoimagery were quantified in order to calculate the percentage of alkali sensitive, rNMP-containing products (values shown at the bottom of the lanes in Fig. 1C). The ribonucleotide incorporation value for wild type Pol ε was 2.1, which is similar to that reported in our initial study [5], and corresponds here to an average of about one rNMP incorporated per 1,000 dNMPs. The average ribonucleotide incorporation percentage for exonuclease-deficient Pol ε was 3.1% (Fig. 1C). This indicates that during ongoing DNA synthesis in vitro using cellular dNTP and rNTP concentrations, the exonuclease activity of Pol ε excises about one third of the rNTPs incorporated by the polymerase. Among the four bases, each of which is about equally (6 A, 5 C, 5 G, 7 T) represented in the substrate (Fig. 1A), the average rNMP proofreading efficiency varies in the order rU > rA > rC = rG (Fig. 1D). Examination of individual sites reveals that proofreading efficiency varies from no detectable proofreading at position G43, C47, G48, C51 and G56 to more than 90% at position T60 and A61 (Fig. 1E). Ranking of all template sites with respect to proofreading shows that even for the same base, proofreading efficiency varies with flanking sequence context. In general, the highest proofreading is observed when the flanking sequence contains low G+C content (Fig. 1F and Supplemental Table S2, discussed in section 4.1).
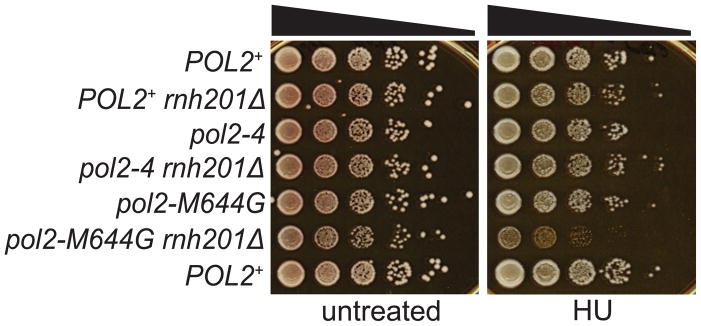
3.3 Growth analysis and sensitivity to replication stress
To determine if a defect in proofreading of ribonucleotides by Pol ε has phenotypic consequences, we compared growth and sensitivity to replication stress of the proofreading-proficient POL2+ (Pol ε) yeast strain to a pol2–4 mutant defective in Pol ε 3′ to 5′ exonuclease activity. The pol2–4 strain is proofreading-deficient due to alanine replacements of two conserved acidic amino acids in the exonuclease active site that cluster around the catalytic metal ions (Asp-290 and Glu-292; [16, 20, 21]). In parallel, we tested strains lacking RNase H2 activity due to deletion of the gene encoding the catalytic subunit of the enzyme, RNH201. RNase H2 is the enzyme that initiates repair of rNMPs incorporated into DNA by Pol ε during replication [6]. On rich medium, all mutants grew at a rate comparable to a wild type strain and colony size was normal, regardless of Pol ε proofreading and/or RNase H2 status (Fig. 2). Next, these strains were challenged with hydroxyurea (HU), a genotoxin that causes replication fork stalling and activation of the S-phase checkpoint due to depletion of dNTP pools [22, 23]. The pol2–4 and rnh201Δ mutants grew as well as the wild type control in the presence of HU (Fig. 2). A pol2-M644G mutant strain with increased capacity to incorporate rNMPs into genomic DNA was tested in parallel, and this mutant also grew normally in the presence of HU. However, deletion of RNH201 in the pol2-M644G strain conferred increased sensitivity to HU (Fig. 2, [24], indicating that failure to remove genomic rNMPs incorporated by Pol ε causes increased replication stress. In contrast, growth of the pol2–4 rnh201Δ double mutant was not impaired in the presence of HU (Fig. 2). The difference in HU sensitivity between the pol2-M644G rnh201Δ and pol2–4 rnh201Δ strains may be related to the fact that M644G Pol ε stably incorporates 10- fold more ribonucleotides into DNA in vitro and in vivo than does a wild type strain [6], whereas inactivation of the 3′ to 5′ exonuclease activity of Pol ε only increases stable ribonucleotide incorporation by 1.5-fold (Fig. 1C). Thus, the smaller increase in stable ribonucleotide incorporation correlates with the greater HU sensitivity of the pol2-M644Grnh201Δ strain as compared to the pol2–4 rnh201Δ strain.
Fig. 2. Phenotypic analysis of the pol2–4 rnh201Δ strain.
Ten-fold serial dilutions of exponentially growing cells from the indicated strains were grown on YPDA (untreated) or exposed to 150 mM hydroxyurea (HU). Plates were incubated at 30°C for 3 days and photographed. A pol2-M644G rnh201Δ mutant strain that shows moderate sensitivity to HU was included as a positive control.
3.4 Increased rate of short deletions in pol-4 rnh201Δ strains
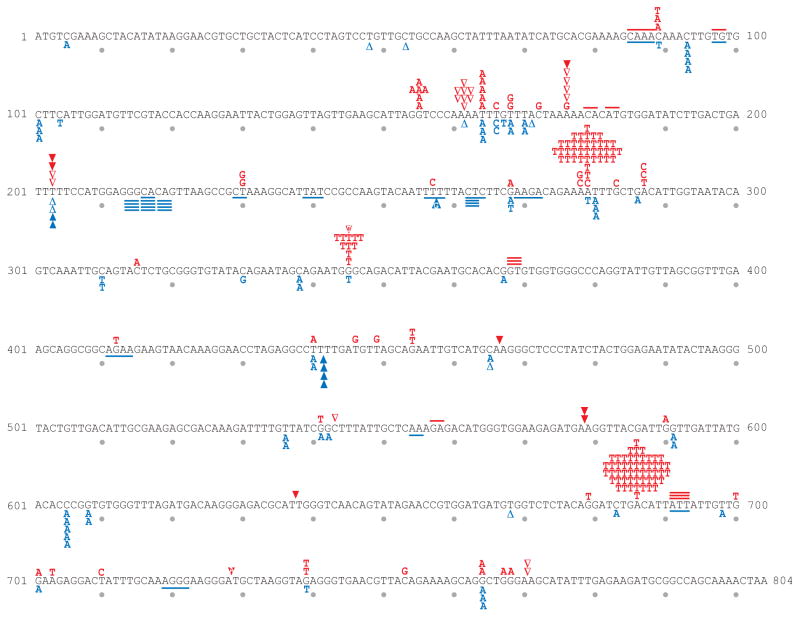
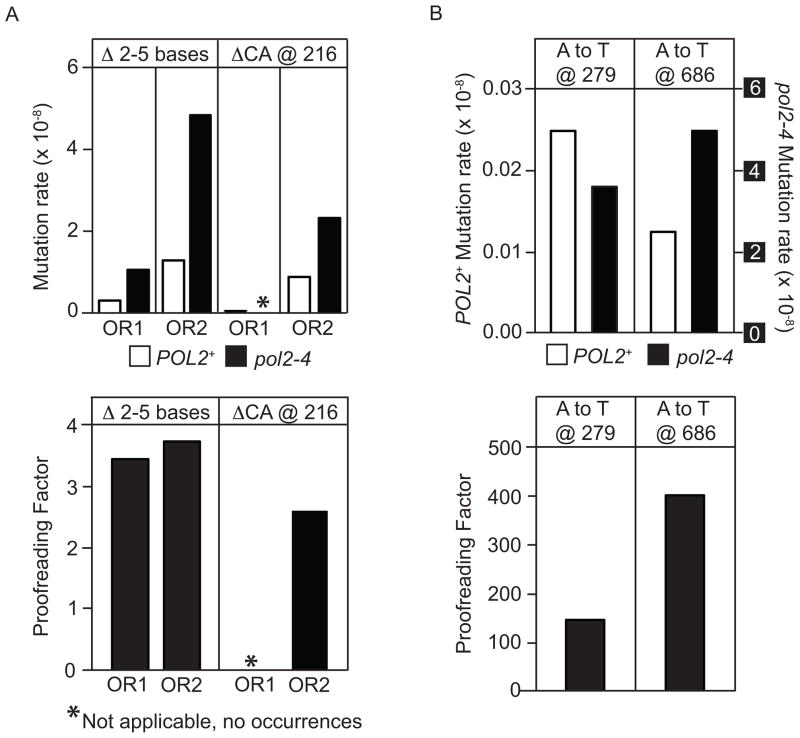
One cellular consequence of unrepaired rNMPs incorporated into DNA during replication by Pol ε is a 10-fold to 440-fold elevated rate of 2–5 base pair deletions in usually perfect but occasionally imperfect repetitive sequences [6, 11, 25]. These deletions dominate the mutational spectra of rnh201Δ strains encoding M644G Pol ε, or encoding wild type Pol ε, but not the spectra of isogenic RNH201+ strains, demonstrating that they result from unrepaired rNMPs incorporated by Pol ε during replication. To determine if defective proofreading of ribonucleotides by Pol ε increases the rate of these deletions, we compared spontaneous mutagenesis in POL2+ rnh201Δ strains that are proofreading-proficient to mutagenesis in pol2–4 rnh201Δ strains that lack Pol ε proofreading activity. Each strain was constructed with the URA3 mutational reporter gene placed near an efficient replication origin (ARS306), in one of two possible orientations (OR1 or OR2). Relative to the POL2+ rnh201Δ strains, total mutation rates were increased by 7-fold (OR1) and 10-fold (OR2) in the pol2–4 rnh201Δ strains (Table 1). Sequence analysis of the URA3 gene from 5-FOA resistant mutants in the pol2–4 rnh201Δ strain background revealed mostly single base substitutions, single base insertions/deletions and 2–5 base pair deletions (Table 1 and Fig. 3). The rates of specific mutations were then calculated and compared (Fig. 4A and Supplemental Table S3) to those for POL2+ rnh201Δ strains, using the spectra of mutations reported earlier (from [11] and listed in Table 1). The results demonstrate that loss of Pol ε proofreading increases the overall rate of 2–5 base pair deletions by 3.5-fold in OR1 (1.1 × 10−8 vs. 0.31 × 10−8) and by 3.7-fold in OR2 (4.9 × 10−8 vs. 1.3 × 10−8; Fig. 4A, lower panel and Supplemental Table S4). The rate increases by 2.6-fold for the OR2-specific CA deletion at base pairs 216–219 (2.3 × 0−8 vs. 0.9 × 10−8). Thus, depending on the comparison, the 3′-5′ exonuclease activity of Pol ε suppresses 62% to 73% of the short deletions resulting from rNTP incorporation by Pol ε. That the short deletions are due to unrepaired ribonucleotides in strains lacking RNase H2 activity is demonstrated by the fact that they are very rare in RNH201+ strains encoding either wild type Pol ε[26] or proofreading-deficient (pol2–4) Pol ε ([27, 28] and Williams and Kunkel, data not shown).
Table 1. Mutation rates and sequencing data for the yeast strains used in this study.
The 95% confidence intervals (CI) were calculated as described in reference [18]. The sequencing data for the POL2+ rnh201Δ strain are from reference [11]. The total number of FOAr mutants sequenced exceeds the number of mutations because some of the mutants had no sequence change in the URA3 open reading frame. These mutants may have arisen due to sequence changes in the URA3 promoter or another gene that affects resistance to 5-FOA. Because these mutants contribute to the overall mutation rate, they are included in the calculation of rates for individual mutation classes.
| URA3 orientation | OR1 | OR2 | OR1 | OR2 |
|---|---|---|---|---|
| Strain | POL2+ rnh201Δ | pol2–4 rnh201Δ | ||
| Mutation rate (× 10−8) | 3.1 | 3.7 | 21 | 36 |
| 95% CI | (2.7–3.5) | (3.3–4.5) | (18–26) | (29–45) |
| ura3 mutants sequenced | 251 | 273 | 216 | 200 |
| Single base substitutions | 86 | 89 | 154 | 57 |
| Single base indels | 27 | 14 | 26 | 15 |
| Total 2–5 base deletions | 25 | 96 | 11 | 27 |
| Other* | 19 | 13 | 3 | 0 |
Other mutations include mutations involving multiple bases (deletions of >5 bases, insertions of >1 base, duplications and complex mutations). Sequencing data for the POL2+ rnh201Δ strain is from [11].
Fig. 3. URA3 mutational spectrum for the pol2–4 rnh201Δ strain.
The coding strand of the 804 base pair URA3 open reading frame is shown with every tenth base indicated by a circle below the DNA sequence. The sequence changes observed upon sequence analysis of independent ura3 mutants are depicted above the coding sequence for URA3 orientation 1 in red, and below the coding sequence for URA3 orientation 2 in blue. Letters indicate single base substitutions, closed triangles indicate single base additions, open triangles indicate single base deletions, and short lines indicate deletions of 2–5 base pairs.
Fig. 4. Specific mutation rates and proofreading factor for URA3 mutation classes.
The mutation rate and proofreading values corresponding to these graphs are listed in Supplemental Tables S3 and S4; (A) Mutation rates and proofreading factors for the POL2+ rnh201Δ and pol2–4 rnh201Δ strains. Mutation rates for the indicated deletion mutations were calculated using the data in Table 1 and Fig. 3 as the fraction of each type of event among the total mutants sequenced, multiplied by the overall mutation rate for each strain. Rates for the POL2+ rnh201Δ strain were calculated from the URA3 spontaneous mutation rates in Table 1 and previously collected mutational spectra [11]. Proofreading factors were calculated by dividing the rate for the indicated mutation in the pol2–4 rnh201Δ strain by the rate for that mutation in the POL2+rnh201Δ mutant (Table 1 and Fig. 3; [11]). The orientation of the URA3 reporter (OR1 or OR2) is indicated. The asterisk (*) indicates that no events were observed for the pol2–4 rnh201Δ strain; (B) Mutation rates and proofreading factors for the indicated base substitution hotspots were calculated for the POL2+ rnh201Δ and pol2–4 rnh201Δ strains as described in part (A). Data correspond to strains in which URA3 is in orientation 1 (OR1).
Serving as a useful internal control for proofreading of single base-base mismatches are the many A to T substitutions seen at base pairs 279 and 686 in orientation 1 (Fig. 3). They dominate the OR1 spectrum for the pol2–4 rnh201Δ strain (Fig. 3), but they do not result from unrepaired ribonucleotides in DNA because they occur at a similar rate in an RNH201+ pol2–4 strain (data not shown). Importantly, the A to T substitutions at base pairs 279 and 686 occur in the pol2–4 rnh201Δ strain at rates that are increased by more than 100-fold compared to the POL2+ rnh201Δ (Fig. 4B), demonstrating that they are proofread much more efficiently than are the ribonucleotides responsible for the 2–5 base pair deletions (Fig. 4A).
4. Discussion
Given its critical role in stabilizing the genome by correcting replication errors, proofreading has been extensively studied (reviewed in [10, 29–32], and references therein). The vast majority of these studies have focused on proofreading of mispaired or misaligned DNA bases. By comparison, proofreading of a nucleotide containing a natural yet incorrect sugar is largely unexplored. Our interest in this topic is prompted by recent studies showing that yeast DNA polymerases α, δ and ε can incorporate large numbers of ribonucleotides into DNA in vitro [5], that Pol ε does this in vivo [6], and that failure to remove ribonucleotides from the yeast nuclear genome due to a defect in RNase H activity results in replicative stress and genome instability [6, 24]. The abundant and ultimately mutagenic incorporation of a non-canonical sugar during replication by Pol ε occurs despite the fact that Pol ε has an intrinsic 3′-5′ exonuclease that can efficiently proofread mismatched bases. This led us to wonder if Pol ε can proofread any of the ribonucleotides that it inserts into DNA, if so, with what efficiency, and if not, with what biological consequences. The present study provides insight into these issues.
4.1 Efficiency and specificity of Pol ε proofreading in vitro
The biochemical results demonstrate that the exonuclease activity of Pol ε can excise 3′-terminal ribonucleotides from primer-templates during polymerization. This includes excision of a pre-existing terminal ribonucleotide (Fig. 1A/B), i.e., one not inserted by the polymerase itself. This observation is consistent with earlier studies [7, 8] showing that the exonuclease activities of T4 DNA polymerase and ϕ29 DNA polymerase, B family homologs of yeast Pol ε, can excise a terminal ribonucleotide from DNA. Theoretically, the ability of Pol ε to excise a ribonucleotide it did not insert could be useful for removing a ribonucleotide incorporated by a separate, exonuclease-deficient DNA polymerase, (e.g., by Pol α at a replication origin [33]). Excision also occurs when Pol ε inserts a ribonucleotide during ongoing DNA synthesis in the presence of all eight nucleotide triphosphates at their estimated cellular concentrations (Fig. 1C/D). On average, this proofreading excises about one third of the inserted ribonucleotides that would otherwise be stably incorporated into full-length DNA chains (Fig. 1C). Having previously found that wild type Pol ε proofreads at least 92% of single base-base mismatches and at least 99% of single base insertion or deletion mismatches [28], we conclude that proofreading of ribonucleotides is less efficient than is proofreading of mismatched bases.
Pol ε proofreads rU with the highest efficiency, followed by rA, and then rC and rG (Fig. 1D). This order, and the sequence context dependence of proofreading (Fig. 1E), can be considered in light of previous studies of proofreading of single base mismatches (reviewed in [10, 29–32], and references therein). The polymerase and exonuclease active sites in proofreading-proficient DNA polymerases are physically separated, and the exonuclease active site binds single stranded DNA. Thus, proofreading requires a mismatched terminus to fray and form enough single stranded DNA to partition to the exonuclease active site for excision. It could be that rU-dA and rA-dT base pairs are less stable and fray more readily than rC-dG and rG-dC base pairs, increasing the likelihood of proofreading. Fraying may also partly underlie the sequence context dependence of ribonucleotide proofreading. For example, once rG is inserted opposite template C51 (Fig. 1F), it is not proofread (Fig. 1E/F). This observation correlates with the high G+C content in the upstream duplex (5 out of 7 bases; Fig. 1F) and strong stacking of rG with an upstream purine (an A). Both features would discourage fraying. In contrast, after rG is inserted opposite template C57, it is proofread with 47% efficiency (Fig. 1E/F). This correlates with a lower G+C content (3 out of 7 bases) in the upstream duplex and weaker stacking of rG with an upstream pyrimidine (a T; Fig. 1F), which may allow more frequent fraying. Similar logic holds for the difference in proofreading efficiency once rA is inserted opposite template T52 versus T60 (Fig. 1F). It is also worth noting that our reactions contain the naturally imbalanced dNTP concentrations measured in yeast cellular extracts [5]. These dNTP concentrations may also contribute to differences in ribonucleotide proofreading efficiency, because proofreading of single base mismatches is known to depend on the relative rates of excision of a primer-terminal error versus continued polymerization to bury the mismatch in duplex DNA and thus protect it from excision [34] (and reviewed in [9, 10]).
Because proofreading of ribonucleotides has not yet been extensively studied, additional parameters could also be relevant. Rules may emerge if proofreading is surveyed at a larger number of sites, which should be easier to do for ribonucleotide-containing base pairs than for base-base mismatches. This is because exonuclease-deficient Pol ε rarely generates single base mismatches (error rates of 10−4 to 10−5 [28]), whereas it incorporates ribonucleotides at 10 to 100- fold higher rates (Fig. 1D). Interestingly, exonuclease-deficient Pol ε incorporates the four different ribonucleotides at rates that differ by less than 2-fold (Fig. 1D, open bars). It therefore follows that the much greater site-to-site variations in ribonucleotide incorporation by wild-type Pol ε(variations of 10 to 70-fold, from Fig. 1C and [6]) are largely due to differences in proofreading rather than differences in initial ribonucleotide insertion.
4.2 Proofreading of ribonucleotides in vivo
Because ribonucleotides are very efficiently removed by RNase H2 and the mutational signature of 2–5 bp deletions in repetitive sequences is only apparent upon loss of RNase H2 activity [6, 11, 25], our current in vivo analysis has been conducted in an rnh201Δ strain background. The rate of 2–5 base pair deletions that depend on unrepaired ribonucleotides in DNA is 3.5 to 3.7-fold higher in pol2–4 rnh201Δ strains as compared to POL2+ rnh201Δ strains (Fig. 4A and Supplemental Table S3). These data support the interpretation that ribonucleotides incorporated into the nuclear genome by Pol ε are proofread by its intrinsic 3′ to 5′ exonuclease activity. These increases are consistent with the biochemical results showing that proofreading excises about one third of the ribonucleotides inserted by Pol ε. In comparison, proofreading of single base mismatches generated by yeast Pol ε is much more efficient (Fig. 4B and Supplemental Table S4). As a consequence, proofreading is relatively inefficient at preventing stable introduction of ribonucleotides into the yeast nuclear genome. Interestingly, this is not disastrous because a pol2–4 rnh201Δ strain lacking Pol ε proofreading and impaired for its ability to repair newly incorporated rNMPs grows relatively normally and is not sensitive when challenged with HU, an agent that causes replication fork stalling (Fig. 2). The ability to tolerate ribonucleotides in DNA is consistent with our previous speculation that the transient presence of ribonucleotides in the genome may have advantageous signaling roles [5].
Supplementary Material
Highlights.
DNA polymerase epsilon proofreads ribonucleotides inserted into DNA
Ribonucleotide proofreading by Pol epsilon stabilizes the genome
Proofreading of ribonucleotides is less efficient than proofreading of mismatched bases
Acknowledgments
Funding
This work was supported by Project Z01 ES065070 to T.A.K. from the Division of Intramural Research of the National Institutes of Health (NIH), National Institute of Environmental Health Sciences (NIEHS), and by the Swedish Research Council, the Swedish Cancer Society, Smärtafonden and the fund for Basic Science–oriented Biotechnology, Medical Faculty of Umeå University to E.J.
We thank Alan Clark and Danielle Watt for helpful comments on the manuscript, and Zachary Pursell and Youri Pavlov for strains and plasmids. We also acknowledge the Molecular Genetics Core Facility at the National Institute of Environmental Health Sciences for sequence analysis of 5-FOA-resistant mutants.
Footnotes
Conflict of Interest
None.
Supplementary data are available online: Supplemental Tables S1–S4.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Joyce CM. Choosing the right sugar: how polymerases select a nucleotide substrate. Proc Natl Acad Sci U S A. 1997;94:1619–1622. doi: 10.1073/pnas.94.5.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown JA, Suo Z. Unlocking the sugar “steric gate” of DNA polymerases. Biochemistry. 2011;50:1135–1142. doi: 10.1021/bi101915z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Traut TW. Physiological concentrations of purines and pyrimidines. Mol Cell Biochem. 1994;140:1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- 4.Chabes A, Georgieva B, Domkin V, Zhao X, Rothstein R, Thelander L. Survival of DNA damage in yeast directly depends on increased dNTP levels allowed by relaxed feedback inhibition of ribonucleotide reductase. Cell. 2003;112:391–401. doi: 10.1016/s0092-8674(03)00075-8. [DOI] [PubMed] [Google Scholar]
- 5.Nick McElhinny SA, Watts BE, Kumar D, Watt DL, Lundstrom EB, Burgers PM, Johansson E, Chabes A, Kunkel TA. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc Natl Acad Sci U S A. 2010;107:4949–4954. doi: 10.1073/pnas.0914857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nick McElhinny SA, Kumar D, Clark AB, Watt DL, Watts BE, Lundstrom EB, Johansson E, Chabes A, Kunkel TA. Genome instability due to ribonucleotide incorporation into DNA. Nat Chem Biol. 2010;6:774–781. doi: 10.1038/nchembio.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin TC, Wang CX, Joyce CM, Konigsberg WH. 3′-5′ Exonucleolytic activity of DNA polymerases: structural features that allow kinetic discrimination between ribo- and deoxyribonucleotide residues. Biochemistry. 2001;40:8749–8755. doi: 10.1021/bi0105936. [DOI] [PubMed] [Google Scholar]
- 8.Bonnin A, Lazaro JM, Blanco L, Salas M. A single tyrosine prevents insertion of ribonucleotides in the eukaryotic-type phi29 DNA polymerase. J Mol Biol. 1999;290:241–251. doi: 10.1006/jmbi.1999.2900. [DOI] [PubMed] [Google Scholar]
- 9.Johnson KA. Conformational coupling in DNA polymerase fidelity. Annu Rev Biochem. 1993;62:685–713. doi: 10.1146/annurev.bi.62.070193.003345. [DOI] [PubMed] [Google Scholar]
- 10.Kunkel TA. DNA replication fidelity. J Biol Chem. 2004;279:16895–16898. doi: 10.1074/jbc.R400006200. [DOI] [PubMed] [Google Scholar]
- 11.Clark AB, Lujan SA, Kissling GE, Kunkel TA. Mismatch repair-independent tandem repeat sequence instability resulting from ribonucleotide incorporation by DNA polymerase epsilon. DNA Repair (Amst) 2011;10:476–482. doi: 10.1016/j.dnarep.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pursell ZF, Isoz I, Lundstrom EB, Johansson E, Kunkel TA. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science. 2007;317:127–130. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Asturias FJ, Cheung IK, Sabouri N, Chilkova O, Wepplo D, Johansson E. Structure of Saccharomyces cerevisiae DNA polymerase epsilon by cryo-electron microscopy. Nat Struct Mol Biol. 2006;13:35–43. doi: 10.1038/nsmb1040. [DOI] [PubMed] [Google Scholar]
- 14.Pursell ZF, Isoz I, Lundstrom EB, Johansson E, Kunkel TA. Regulation of B family DNA polymerase fidelity by a conserved active site residue: characterization of M644W, M644L and M644F mutants of yeast DNA polymerase epsilon. Nucleic Acids Res. 2007;35:3076–3086. doi: 10.1093/nar/gkm132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pavlov YI, Newlon CS, Kunkel TA. Yeast origins establish a strand bias for replicational mutagenesis. Mol Cell. 2002;10:207–213. doi: 10.1016/s1097-2765(02)00567-1. [DOI] [PubMed] [Google Scholar]
- 16.Morrison A, Bell JB, Kunkel TA, Sugino A. Eukaryotic DNA polymerase amino acid sequence required for 3′----5′ exonuclease activity. Proc Natl Acad Sci U S A. 1991;88:9473–9477. doi: 10.1073/pnas.88.21.9473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldstein AL, McCusker JH. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 18.Shcherbakova PV, Kunkel TA. Mutator phenotypes conferred by MLH1 overexpression and by heterozygosity for mlh1 mutations. Mol Cell Biol. 1999;19:3177–3183. doi: 10.1128/mcb.19.4.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tapper DP, Clayton DA. Altered mobility of polydeoxyribonucleotides in high resolution polyacrylamide gels due to removal of terminal phosphates. Nucleic Acids Res. 1981;9:6787–6794. doi: 10.1093/nar/9.24.6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Derbyshire V, Freemont PS, Sanderson MR, Beese L, Friedman JM, Joyce CM, Steitz TA. Genetic and crystallographic studies of the 3′,5′-exonucleolytic site of DNA polymerase I. Science. 1988;240:199–201. doi: 10.1126/science.2832946. [DOI] [PubMed] [Google Scholar]
- 21.Derbyshire V, Grindley ND, Joyce CM. The 3′-5′ exonuclease of DNA polymerase I of Escherichia coli: contribution of each amino acid at the active site to the reaction. EMBO J. 1991;10:17–24. doi: 10.1002/j.1460-2075.1991.tb07916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lopes M, Cotta-Ramusino C, Pellicioli A, Liberi G, Plevani P, Muzi-Falconi M, Newlon CS, Foiani M. The DNA replication checkpoint response stabilizes stalled replication forks. Nature. 2001;412:557–561. doi: 10.1038/35087613. [DOI] [PubMed] [Google Scholar]
- 23.Reichard P. Interactions between deoxyribonucleotide and DNA synthesis. Annu Rev Biochem. 1988;57:349–374. doi: 10.1146/annurev.bi.57.070188.002025. [DOI] [PubMed] [Google Scholar]
- 24.Lazzaro F, Novarina D, Amara F, Watt DL, Stone JE, Costanzo V, Burgers PM, Kunkel TA, Plevani P, Muzi-Falconi M. RNase H and Postreplication Repair Protect Cells from Ribonucleotides Incorporated in DNA. Mol Cell. 2012;45:99–110. doi: 10.1016/j.molcel.2011.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim N, Huang SN, Williams JS, Li YC, Clark AB, Cho JE, Kunkel TA, Pommier Y, Jinks-Robertson S. Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I. Science. 2011;332:1561–1564. doi: 10.1126/science.1205016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nick McElhinny SA, Kissling GE, Kunkel TA. Differential correction of lagging-strand replication errors made by DNA polymerases {alpha} and {delta} Proc Natl Acad Sci U S A. 2010;107:21070–21075. doi: 10.1073/pnas.1013048107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morrison A, Sugino A. The 3′-->5′ exonucleases of both DNA polymerases delta and epsilon participate in correcting errors of DNA replication in Saccharomyces cerevisiae. Mol Gen Genet. 1994;242:289–296. doi: 10.1007/BF00280418. [DOI] [PubMed] [Google Scholar]
- 28.Shcherbakova PV, Pavlov YI, Chilkova O, Rogozin IB, Johansson E, Kunkel TA. Unique error signature of the four-subunit yeast DNA polymerase epsilon. J Biol Chem. 2003;278:43770–43780. doi: 10.1074/jbc.M306893200. [DOI] [PubMed] [Google Scholar]
- 29.Johnson KA. The kinetic and chemical mechanism of high-fidelity DNA polymerases. Biochim Biophys Acta. 2010;1804:1041–1048. doi: 10.1016/j.bbapap.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beckman RA, Loeb LA. Multi-stage proofreading in DNA replication. Q Rev Biophys. 1993;26:225–331. doi: 10.1017/s0033583500002869. [DOI] [PubMed] [Google Scholar]
- 31.Reha-Krantz LJ. DNA polymerase proofreading: Multiple roles maintain genome stability. Biochim Biophys Acta. 2010;1804:1049–1063. doi: 10.1016/j.bbapap.2009.06.012. [DOI] [PubMed] [Google Scholar]
- 32.Baker TA, Kornberg A. DNA replication. 2. W.H. Freeman and Company; New York, NY: 1991. [Google Scholar]
- 33.Clark AB, Kunkel TA. The importance of being DNA. Cell Cycle. 2010;9:4422–4424. doi: 10.4161/cc.9.22.14052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ninio J. Kinetic amplification of enzyme discrimination. Biochimie. 1975;57:587–595. doi: 10.1016/s0300-9084(75)80139-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.