Abstract
Protein phosphorylation plays a critical role in the signaling pathways regulating water and solute transport in the distal renal tubule (i.e., renal collecting duct). A central mediator in this process is the antidiuretic peptide hormone arginine vasopressin, which regulates a number of transport proteins including water channel aquaporin-2 and urea transporters (UT-A1 and UT-A3). Within the past few years, tandem mass spectrometry-based proteomics has played a pivotal role in revealing global changes in the phosphoproteome in response to vasopressin signaling in the renal collecting duct. This type of large-scale ‘shotgun’ approach has resulted in an exponential increase in the number of phosphoproteins known to be regulated by vasopressin and has expanded on the established signaling mechanisms and kinase pathways regulating collecting duct physiology. This article will provide a brief background on vasopressin action, will highlight a number of recent quantitative phosphoproteomic studies in both native rat kidney and cultured collecting duct cells, and will conclude with a perspective focused on emerging trends in the field of phosphoproteomics.
Keywords: aquaporin, collecting duct, kidney, LC-MS/MS, tandem mass spectrometry, vasopressin, water transport
Vasopressin signaling in the renal collecting duct
Regulation of water excretion by the kidney is critical for maintaining the proper tonicity of body fluids. The peptide hormone arginine vasopressin (AVP) is the key regulator of renal water excretion. When AVP levels are high (the ‘antidiuretic’ state), more water is reabsorbed by the kidney, which produces concentrated urine. When AVP levels are low (the ‘diuretic’ state), a larger volume of water is excreted by the kidney, which produces diluted urine. Dysregulation of AVP signaling features prominently in disorders such as diabetes insipidus, syndromes of inappropriate antidiuretic hormone hypersecretion and congestive heart failure (reviewed by Nielsen et al. [1]).
The entire cascade of known signaling events begins with the release of AVP from the posterior pituitary in response to an elevation in blood osmolality above the normal range (290–294 mosmol/kg H2O). AVP binding to the V2 receptor (a Gs protein-coupled receptor) on the basolateral membrane of collecting duct epithelial cells triggers activation of two adenylyl cyclases (types III and VI) [2,3] and a subsequent rise in intracellular cAMP. One of the main targets of cAMP is protein kinase (PK)A, which has been shown to phosphorylate a serine residue, Ser256, in the C-terminal tail of the membrane water channel aquaporin-2 (AQP2) [4]. This phosphorylation event is critical for trafficking of vesicles containing AQP2 to the apical plasma membrane [5–8], which increases the osmotic water permeability of the collecting duct epithelium and allows water to be reabsorbed back into the body. This entire process occurs quite rapidly, within a few minutes of AVP binding to its receptor. AVP also regulates AQP2 via a long-term process, namely regulation of AQP2 gene transcription (reviewed in [9]), which may be partly PKA dependent. A number of recent review articles offer a more detailed description of AVP action in the collecting duct [10,11].
Although the exact signaling mechanisms governing AQP2 trafficking are incompletely understood, it is clear that phosphorylation plays a prominent role. Within the past few years, investigators have turned to tandem mass spectrometry (MS/MS)-based phosphoproteomics in order to elucidate these unknown phosphorylation events.
The emerging field of phosphoproteomics
Phosphoproteomics comprises any large-scale identification or characterization of the protein phosphorylation state of a given proteome (reviewed in [12,13]). Although the field is less than 10 years old, a number of recent technological advances have led to a rapid improvement in both the quality and quantity of phosphoproteomic data.
Phosphopeptide enrichment strategies such as immobilized metal affinity chromatography (IMAC) and metal oxide affinity chromatography (MOAC) have allowed investigators to overcome the low stoichiometry and ion-suppression effects routinely associated with analysis of phosphopeptides by mass spectrometry (MS). There have also been rapid advances in MS instrumentation, including the development of newer, more powerful hybrid instruments capable of extremely high mass accuracy and greater sensitivity, as well as the development of newer fragmentation methods (e.g., electron transfer dissociation [14]). Finally, a dramatic expansion in software programs tailored to searching, filtering and manipulating phosphoproteomic data has resulted in higher-quality datasets with very low numbers of predicted false positives (often <1% as determined by false discovery rate estimation). Many of these programs are written as ad hoc software that is often shared with other laboratories.
A number of recent phosphoproteomic studies [15–21] have capitalized on these advancements, many reporting identification and quantitation of hundreds or thousands of phosphorylation events from a single sample. A recent paper by Swaney et al. reported a staggering 10,844 phosphorylation sites from human embryonic stem cells [22].
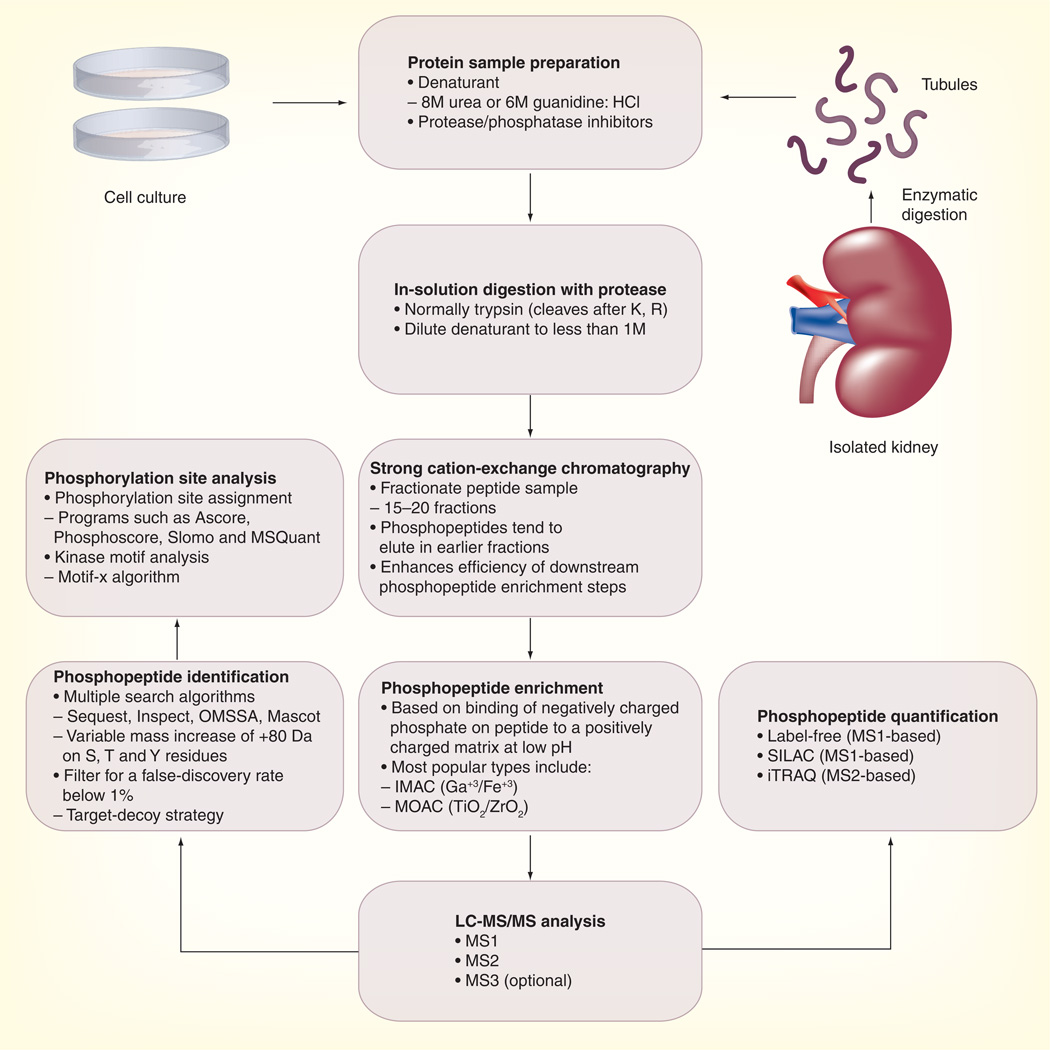
A typical large-scale phosphoproteomic workflow as used in our laboratory is presented in Figure 1. Proteins from either cell culture or isolated tissue are denatured in sample buffer containing 8M urea or 6M guanidine with added inhibitors to prevent protein degradation and phosphate loss. The sample is then digested with a site-specific protease (normally trypsin) that cleaves the polypeptide chain after arginine and lysine residues. The peptide sample is then subjected to strong cation-exchange chromatography, which separates the sample into multiple fractions and enhances recovery of phosphopeptides. Phosphopeptides are further enriched using IMAC or MOAC approaches. IMAC and TiO2-MOAC preferentially enrich for multiply phosphorylated peptides while ZrO2 tends to enrich monophosphorylated peptides. A recent review by Dunn et al. provides a detailed explanation regarding the differences in binding affinities among the various IMAC and MOAC resins [23]. The phosphopeptide sample is then subjected to liquid chromatography-MS/MS (LC-MS/MS) analysis. The MS2 and MS3 fragmentation spectra that are generated are then compared with protein sequence databases using multiple programs to identify the peptides. Software such as Sequest [24], OMSSA [25] and Mascot [26] are based on pattern matching, while Inspect [27] includes a de novo sequence tagging component for more efficient spectral identification. Those phosphopeptides that pass stringent false discovery rate filtering are then analyzed by various ‘site-assignment’ programs [28–31] to identify the most likely site(s) of phosphorylation (i.e., which particular S, T or Y residues are modified). Various quantification strategies including label-free methods [19,32], stable isotope labeling by amino acids in cell culture (SILAC) [33] and isobaric tags for relative and absolute quantitation (iTRAQ) [34] can be utilized to quantify changes in phosphopeptide abundances between samples. Please see Grimsrud et al. for a detailed summary of these quantitative proteomic strategies [12].
Figure 1. Typical workflow for a large-scale phosphoproteomics experiment.
This workflow combines a number of different approaches that have been used to successfully identify and quantify phosphopeptides from the kidney collecting duct.
HCl: Hydrochloride; IMAC: Immobilized metal affinity chromatography; iTRAQ: Isobaric tags for relative and absolute quantitation; LC-MS/MS: Liquid chromatography-tandem mass spectrometry; MOAC: Metal oxide affinity chromatography; MS: Mass spectrometry; SILAC: Stable isotope labeling by amino acids in cell culture.
Phosphoproteomic studies of renal collecting duct
Hoffert et al. were the first to utilize a quantitative phosphoproteomic approach to analyze the response of the collecting duct to AVP [19]. Isolated inner medullary collecting ducts from rats were incubated in the presence or absence of 1 nM (deamino-Cys1, D-Arg8)-vasopressin (dDAVP; a V2-receptor-specific analog of vasopressin) for 10 min and then processed for LC-MS/MS analysis utilizing a strategy similar to that shown in Figure 1. A total of 714 phosphorylation sites on 223 proteins were identified (data available at [101]), including four phosphorylation sites in the C-terminus of the water channel AQP2, one of the main molecular targets of vasopressin. One site, Ser-256, had been previously identified as being critical for AQP2 trafficking to the apical cell membrane [35], while the other three sites (Ser-261, Ser-264 and Ser-269) were previously unidentified. Label-free LC-MS/MS quantification of reconstructed MS1 precursor ion intensities demonstrated that two of these sites are reciprocally regulated: phosphorylation at Ser-256 increases while phosphorylation at Ser-261 decreases in the presence of AVP. Subsequent studies have examined the phosphorylation kinetics and physiological roles of these newly identified phosphosites. Immunoblotting with newly generated phospho-specific antibodies against each site demonstrated that phosphorylation at Ser-264 and Ser-269 also increase with dDAVP and that prior phosphorylation at Ser-256 is a requirement for phosphorylation at these downstream residues [4]. Phosphorylation at Ser-269 is believed to play a role in retention at the apical membrane [4] through a reduction in the rate of endocytosis and decreased interaction with a number of proendocytotic proteins including heat-shock protein 70, heat-shock cognate 70, dynamin and clathrin [36]. Ser-261 phosphorylation may be involved in regulating polyubiquitylation and proteasomal degradation of AQP2 [37]. The kinase(s) responsible for phosphorylating AQP2 at these sites remain unknown.
A subsequent large-scale phosphoproteomic study by Bansal et al. also analyzed the AVP response in isolated rat inner medullary collecting ducts [38], but with a number of important modifications designed to increase the sensitivity of the analysis: an upstream strong cation-exchange chromatography step was added, which stratified the peptide sample and increased the recovery of phosphopeptides after IMAC; samples were run on a higher sensitivity mass spectrometer, the Orbitrap XL™ (Thermo Scientific); and spectra were searched with multiple algorithms (Sequest, Inspect and OMSSA). As a result of these advances, over 2700 unique phosphopeptides were identified (data available at [102]). The authors also performed label-free MS quantitation, which identified 29 phosphopeptides that changed significantly with AVP treatment (p < 0.05; n = 3 unfractionated biological replicates). A major finding from this study was the identification of three phosphorylation sites on urea transporters UT-A1 and UT-A3, all of which were shown by MS to increase in abundance in response to AVP. AVP-mediated phosphorylation of UT-A1 is believed to increase the permeability of the collecting duct to urea, possibly by affecting membrane trafficking [39]. Immunoblotting with phospho-specific antibodies to Ser-84 and Ser-486 of rat UT-A1 confirmed responsiveness to AVP and involvement of PKA. Another phosphoprotein that was increased with AVP was β-catenin (Ctnnb1), a protein with both structural and signaling roles that has been proposed to play a role in regulation of AQP2 [40]. Other phosphorylation sites that increased with AVP that have potential roles in the regulation of water and/or urea transporter activity included sites in protein kinase PCTAIRE-3 (Pctk3) and plexin domain containing-2 (Plxdc2). Phosphorylation sites decreased with AVP were present in septin-9 (Sept9), ArfGAP with FG repeats 1 (Agfg1) and epsin-3 (Epn3).
A recent study by Rinschen et al. measured AVP-responsive phosphoproteins in a SILAC-labeled mouse cortical collecting duct cell line (mpkCCD) [41]. Cells labeled with heavy or light amino acids were exposed to either dDAVP (0.1 nM) or vehicle control for 30 min and processed for LC-MS/MS analysis. Of the 2884 phosphopeptides that were quantified, 273 increased and 254 decreased with AVP. The threshold for change was based on the results of preliminary experiments in which both heavy and light samples were vehicle treated (data are available at [103]) Identification of upregulated phosphorylation sites on calmodulin-dependent kinase II pointed to a potential role for the Ca+2-calmodulin pathway in the response of the collecting duct to AVP. This finding was consistent with prior studies demonstrating that vasopressin triggers both intracellular calcium mobilization [42] and calmodulin-dependent phosphorylation of myosin regulatory light chain protein by myosin light chain kinase [43], which are both critical for AQP2 trafficking. In order to assess which kinase pathways may be regulated by vasopressin, the authors performed a global analysis of phosphorylation motifs (i.e., the amino acid sequences surrounding the phosphorylation site) for phosphopeptides regulated by AVP. This analysis was performed using the motif-x algorithm [44], which generates sequence logos representing amino acid sequences that are significantly over-represented in input datasets compared with background. For phosphopeptides increased by AVP, there was an over-representation of so-called ‘basophilic’ phosphorylation motifs, which contained positively charged amino acids arginine and lysine prior to the site of phosphorylation (i.e., R- [R/K]-X- [S/T]*, where X is any amino acid and * indicates the site of phosphorylation). These basophilic motifs are associated with phosphorylation by the AGC class of kinases, which includes members such as PKA, PKC, PKG and some calmodulin-dependent kinases. For phosphopeptides decreased by AVP, there was an over-representation of ‘proline-directed’ motifs, which contain a single proline residue at position +1 (i.e., X-X- [S/T]*-P-X) and are associated with phosphorylation by MAP kinases and cyclin-dependent kinases. Thus, these results reveal that AVP signaling is associated with inactivation of one or more proline-directed kinases. A similar profile was demonstrated in a recent analysis of the AVP response in the medullary thick ascending limb of rat kidney [45].
Expert commentary
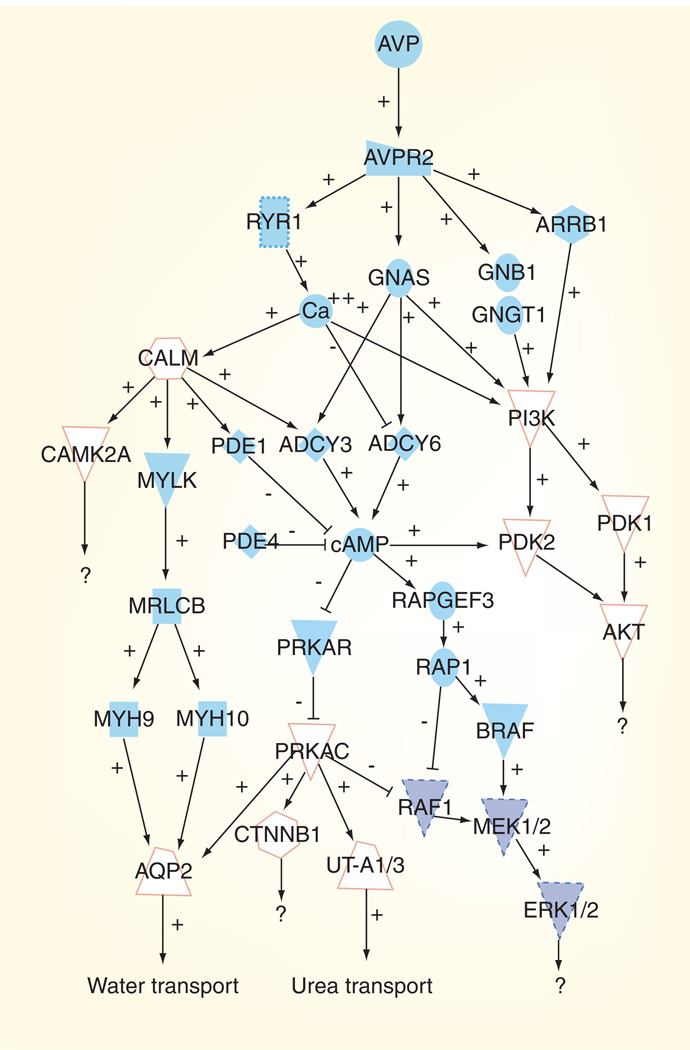
The field of phosphoproteomics has had a tremendous impact on the study of vasopressin signaling in the mammalian collecting duct. The studies summarized in this article have dramatically expanded the number of known vasopressin-regulated phosphoproteins. This new knowledge has resulted in the expansion of established signaling mechanisms and kinase pathways regulated by vasopressin. Figure 2 shows an abbreviated view of the vasopressin signaling network. From both classical physiology as well as phosphoproteomic approaches, it is clear that vasopressin regulates phosphorylation of AQP2 and urea transporters at multiple sites, thereby increasing the permeability of the collecting duct to water and urea, respectively. Newly identified phosphorylation sites in these proteins have spurred tremendous research interest within the vasopressin/aquaporin communities. These phosphorylation events are probably mediated predominantly through PKA. However, phosphoproteomic analysis has also confirmed that vasopressin activates the Ca+2-calmodulin pathway and inhibits signaling through MAP kinases. Vasopressin is also known to upregulate the AKT pathway [46]. Thus, it is clear that vasopressin can regulate multiple signaling pathways in addition to PKA, although the downstream effectors of these signaling events largely remain unknown. Further exploration of these additional pathways will be required to fully understand their relevance to vasopressin signaling.
Figure 2. The vasopressin signaling network in the kidney collecting duct.
Pathways that are upregulated by arginine vasopressin are indicated by the white shapes with an orange outline. Pathways that are downregulated by arginine vasopressin are indicated by the filled dark blue shapes with a dashed outline.
Five-year view
Where is the field of phosphoproteomics headed in the near future? A number of current trends are likely to continue: a gradual shift in focus from readily tractable biological models (cell culture, yeast and bacteria) to more physiologically and pathophysiologically motivated studies in native tissue and whole-animal models; increased use of versatile peptide labeling techniques such as iTRAQ; increased use of newer, more powerful hybrid mass spectrometers; and consolidation of computational approaches in order to create integrated, refined phosphoproteomics analysis software packages. The last objective is particularly challenging because separations and MS technologies are changing very rapidly, making it difficult to keep up with the state-of-the-art in terms of data formats and dataset characteristics.
As the amount of phosphoproteomic data increases, researchers will continue to use this valuable information to construct more accurate models of cell signaling networks. This type of analysis usually includes detailed dynamic data acquired across multiple timepoints (as provided by iTRAQ or SILAC labeling) that can be used for temporal and spatial clustering of the various components of a signaling pathway [47,48]. Ideally, this phosphorylation data would be mapped to the specific kinase(s)/phosphatase(s) involved, utilizing a combination of motif analysis, kinase/phosphatase assays and other traditional biochemical approaches. Network modeling represents a promising tool for understanding which proteins would make the most effective drug targets and could lead to a deeper understanding of obscure diseases that have not been well characterized at the molecular level.
One topic that deserves special attention in the years ahead would be the issue of ‘quality’ versus ‘quantity’ in phosphoproteomics. Although the total number of phosphopeptides identified in some large-scale studies is impressive, too often comprehensive analysis seems to be the overriding goal, rather than a deeper understanding of the physiological mechanisms. Thus, researchers may need to shift their investigative focus to examining the functional relevance of individual sites. This issue is all the more pressing given a recent estimate that as many as 50% of phosphorylation sites detected in phosphoproteomic screens are not conserved across species and are therefore less likely to play important functional roles [49]. A recent study in kidney cell culture by Hallows et al. highlights the benefits of a ‘targeted’ phosphoproteomic approach [50]. By using a combination of immunoprecipitation followed by MS/MS, the authors identified four novel phosphorylation sites on Nedd4–2, an E3 ubiquitin ligase that regulates the degradation of the collecting duct epithelial sodium channel ENaC. Through site-directed mutagenesis and classic electrophysiological measurements, they further demonstrated that these sites are the target of the MAP kinase JNK1 and that phosphorylation at these sites is important for inhibition of ENaC activity. In the growing realm of large-scale ‘systems’ biology, it is critical that these classical ‘reductionist’ approaches to molecular and cellular physiology are not overlooked, as these techniques offer an unparalleled way of sharpening our understanding of defined aspects of a biological system. It is also important to realize that neither a systems approach nor a reductionist approach alone is likely to provide an optimal path to understanding the complex interaction of phosphorylation events governing a physiological process.
Key issues.
Protein phosphorylation plays a critical role in the molecular pathways governing vasopressin-mediated control of water and solute transport in the kidney collecting duct.
Phosphoproteomic methodologies have allowed unprecedented identification of protein targets and signaling pathways that are regulated by vasopressin.
Phosphoproteomic methodologies can be adapted to virtually any biological system to address pertinent physiological questions.
A fraction of the phosphorylation sites identified in large-scale studies may not be biologically relevant and must be analyzed further by using traditional biochemical, cell and molecular biological techniques.
Acknowledgments
This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
Footnotes
Financial & competing interests disclosure
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
• of interest
•• of considerable interest
- 1.Nielsen S, Frokiaer J, Marples D, et al. Aquaporins in the kidney: from molecules to medicine. Physiol. Rev. 2002;82:205–244. doi: 10.1152/physrev.00024.2001. [DOI] [PubMed] [Google Scholar]
- 2.Hoffert JD, Chou CL, Fenton RA, Knepper MA. Calmodulin is required for vasopressin-stimulated increase in cyclic AMP production in inner medullary collecting duct. J. Biol. Chem. 2005;280:13624–13630. doi: 10.1074/jbc.M500040200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strait KA, Stricklett PK, Chapman M, Kohan DE. Characterization of vasopressin-responsive collecting duct adenylyl cyclases in the mouse. Am. J. Physiol. Renal Physiol. 2010;298:F859–F867. doi: 10.1152/ajprenal.00109.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoffert JD, Fenton RA, Moeller HB, et al. Vasopressin-stimulated increase in phosphorylation at ser-269 potentiates plasma membrane retention of aquaporin-2. J. Biol. Chem. 2008;283:24617–24627. doi: 10.1074/jbc.M803074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nielsen S, Chou CL, Marples D, et al. Vasopressin increases water permeability of kidney collecting duct by inducing translocation of aquaporin-CD water channels to plasma membrane. Proc. Natl Acad. Sci. USA. 1995;92:1013–1017. doi: 10.1073/pnas.92.4.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nunes P, Hasler U, McKee M, et al. A fluorimetry-based ssYFP secretion assay to monitor vasopressin-induced exocytosis in LLC-PK1 cells expressing aquaporin-2. Am. J. Physiol. Cell Physiol. 2008;295:C1476–C1487. doi: 10.1152/ajpcell.00344.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuwahara M, Fushimi K, Terada Y, et al. cAMP-dependent phosphorylation stimulates water permeability of aquaporin-collecting duct water channel protein expressed in Xenopus oocytes. J. Biol. Chem. 1995;270:10384–10387. doi: 10.1074/jbc.270.18.10384. [DOI] [PubMed] [Google Scholar]
- 8.Kamsteeg EJ, Heijnen I, van Os CH, Deen PM. The subcellular localization of an aquaporin-2 tetramer depends on the stoichiometry of phosphorylated and nonphosphorylated monomers. J. Cell Biol. 2000;151:919–930. doi: 10.1083/jcb.151.4.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hasler U, Leroy V, Martin PY, Feraille E. Aquaporin-2 abundance in the renal collecting duct: new insights from cultured cell models. Am. J. Physiol. Renal Physiol. 2009;297:F10–F18. doi: 10.1152/ajprenal.00053.2009. [DOI] [PubMed] [Google Scholar]
- 10.Hoffert JD, Chou CL, Knepper MA. Aquaporin-2 in the ‘-omics’ era. J. Biol. Chem. 2009;284:14683–14687. doi: 10.1074/jbc.R900006200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Noda Y, Sohara E, Ohta E, Sasaki S. Aquaporins in kidney pathophysiology. Nat. Rev. Nephrol. 2010;6:168–178. doi: 10.1038/nrneph.2009.231. [DOI] [PubMed] [Google Scholar]
- 12. Grimsrud PA, Swaney DL, Wenger CD, Beauchene NA, Coon JJ. Phosphoproteomics for the masses. ACS Chem. Biol. 2010;5:105–119. doi: 10.1021/cb900277e. • A very informative and current review of the field of phosphoproteomics.
- 13. Hoffert JD, Knepper MA. Taking aim at shotgun phosphoproteomics. Anal. Biochem. 2008;375:1–10. doi: 10.1016/j.ab.2007.11.023. • Summarizes a number of different large-scale methodologies that have been used to study aquaporin-2 biology.
- 14.Mikesh LM, Ueberheide B, Chi A, et al. The utility of ETD mass spectrometry in proteomic analysis. Biochim. Biophys. Acta. 2006;1764:1811–1822. doi: 10.1016/j.bbapap.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Villen J, Beausoleil SA, Gerber SA, Gygi SP. Large-scale phosphorylation analysis of mouse liver. Proc. Natl Acad. Sci. USA. 2007;104:1488–1493. doi: 10.1073/pnas.0609836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li X, Gerber SA, Rudner AD, et al. Large-scale phosphorylation analysis of α-factor-arrested Saccharomyces cerevisiae. J. Proteome Res. 2007;6:1190–1197. doi: 10.1021/pr060559j. [DOI] [PubMed] [Google Scholar]
- 17.Beausoleil SA, Jedrychowski M, Schwartz D, et al. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc. Natl Acad. Sci. USA. 2004;101:12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bodenmiller B, Mueller LN, Mueller M, Domon B, Aebersold R. Reproducible isolation of distinct, overlapping segments of the phosphoproteome. Nat. Methods. 2007;4:231–237. doi: 10.1038/nmeth1005. [DOI] [PubMed] [Google Scholar]
- 19. Hoffert JD, Pisitkun T, Wang G, Shen RF, Knepper MA. Quantitative phosphoproteomics of vasopressin-sensitive renal cells: regulation of aquaporin-2 phosphorylation at two sites. Proc. Natl Acad. Sci. USA. 2006;103:7159–7164. doi: 10.1073/pnas.0600895103. •• The first quantitative phosphoproteomic study of the kidney collecting duct.
- 20.Gruhler A, Olsen JV, Mohammed S, et al. Quantitative phosphoproteomics applied to the yeast pheromone signaling pathway. Mol. Cell Proteomics. 2005;4:310–327. doi: 10.1074/mcp.M400219-MCP200. [DOI] [PubMed] [Google Scholar]
- 21.Olsen JV, Blagoev B, Gnad F, et al. Global in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 22.Swaney DL, Wenger CD, Thomson JA, Coon JJ. Human embryonic stem cell phosphoproteome revealed by electron transfer dissociation tandem mass spectrometry. Proc. Natl Acad. Sci. USA. 2009;106:995–1000. doi: 10.1073/pnas.0811964106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dunn JD, Reid GE, Bruening ML. Techniques for phosphopeptide enrichment prior to analysis by mass spectrometry. Mass Spectrom. Rev. 2010;29:29–54. doi: 10.1002/mas.20219. [DOI] [PubMed] [Google Scholar]
- 24.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 25.Geer LY, Markey SP, Kowalak JA, et al. Open mass spectrometry search algorithm. J. Proteome Res. 2004;3:958–964. doi: 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
- 26.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 27.Tanner S, Shu H, Frank A, et al. InsPecT: identification of posttranslationally modified peptides from tandem mass spectra. Anal. Chem. 2005;77:4626–4639. doi: 10.1021/ac050102d. [DOI] [PubMed] [Google Scholar]
- 28.Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol. 2006;24:1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- 29.Ruttenberg BE, Pisitkun T, Knepper MA, Hoffert JD. PhosphoScore: an open-source phosphorylation site assignment tool for MSn data. J. Proteome Res. 2008;7:3054–3059. doi: 10.1021/pr800169k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mortensen P, Gouw JW, Olsen JV, et al. MSQuant, an open source platform for mass spectrometry-based quantitative proteomics. J. Proteome Res. 2010;9:393–403. doi: 10.1021/pr900721e. [DOI] [PubMed] [Google Scholar]
- 31.Bailey CM, Sweet SM, Cunningham DL, et al. SLoMo: automated site localization of modifications from ETD/ECD mass spectra. J. Proteome Res. 2009;8:1965–1971. doi: 10.1021/pr800917p. [DOI] [PubMed] [Google Scholar]
- 32.Wang G, Wu WW, Zeng W, Chou CL, Shen RF. Label-free protein quantification using LC-coupled ion trap or FT mass spectrometry: reproducibility, linearity, and application with complex proteomes. J. Proteome Res. 2006;5:1214–1223. doi: 10.1021/pr050406g. [DOI] [PubMed] [Google Scholar]
- 33.Ong SE, Blagoev B, Kratchmarova I, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 34.Ross PL, Huang YN, Marchese JN, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 35.Fushimi K, Sasaki S, Marumo F. Phosphorylation of serine 256 is required for cAMP-dependent regulatory exocytosis of the aquaporin-2 water channel. J. Biol. Chem. 1997;272:14800–14804. doi: 10.1074/jbc.272.23.14800. [DOI] [PubMed] [Google Scholar]
- 36.Moeller HB, Praetorius J, Rutzler MR, Fenton RA. Phosphorylation of aquaporin-2 regulates its endocytosis and protein-protein interactions. Proc. Natl Acad. Sci. USA. 2010;107:424–429. doi: 10.1073/pnas.0910683107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nedvetsky PI, Tabor V, Tamma G, et al. Reciprocal regulation of aquaporin-2 abundance and degradation by protein kinase A and p38-MAP kinase. J. Am. Soc. Nephrol. 2010;21(10):1645–1656. doi: 10.1681/ASN.2009111190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bansal AD, Hoffert JD, Pisitkun T, et al. Phosphoproteomic profiling reveals vasopressin-regulated phosphorylation sites in collecting duct. J. Am. Soc. Nephrol. 2010;21:303–315. doi: 10.1681/ASN.2009070728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blount MA, Mistry AC, Frohlich O, et al. Phosphorylation of UT-A1 urea transporter at serines 486 and 499 is important for vasopressin-regulated activity and membrane accumulation. Am. J. Physiol. Renal Physiol. 2008;295:F295–F299. doi: 10.1152/ajprenal.00102.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nielsen J, Hoffert JD, Knepper MA, et al. Proteomic analysis of lithium-induced nephrogenic diabetes insipidus: mechanisms for aquaporin 2 down-regulation and cellular proliferation. Proc. Natl Acad. Sci. USA. 2008;105:3634–3639. doi: 10.1073/pnas.0800001105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rinschen MM, Yu MJ, Wang G, et al. Quantitative phosphoproteomic analysis reveals vasopressin V2-receptor-dependent signaling pathways in renal collecting duct cells. Proc. Natl Acad. Sci. USA. 2010;107:3882–3887. doi: 10.1073/pnas.0910646107. •• A recent quantitative phosphoproteomic study that effectively utilizes stable isotope labeling with amino acids in cell culture in cultured collecting duct cells and incorporates large-scale profiling of phosphorylation motifs.
- 42.Yip KP. Coupling of vasopressin-induced intracellular Ca2+ mobilization and apical exocytosis in perfused rat kidney collecting duct. J. Physiol. 2002;538:891–899. doi: 10.1113/jphysiol.2001.012606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chou CL, Christensen BM, Frische S, et al. Non-muscle myosin II and myosin light chain kinase are downstream targets for vasopressin signaling in the renal collecting duct. J. Biol. Chem. 2004;279:49026–49035. doi: 10.1074/jbc.M408565200. [DOI] [PubMed] [Google Scholar]
- 44.Schwartz D, Gygi SP. An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat. Biotechnol. 2005;23:1391–1398. doi: 10.1038/nbt1146. [DOI] [PubMed] [Google Scholar]
- 45.Gunaratne R, Braucht DW, Rinschen MM, et al. Quantitative phosphoproteomic analysis reveals cAMP/vasopressin-dependent signaling pathways in native renal thick ascending limb cells. Proc. Natl Acad. Sci. USA. 2010;107:15653–15658. doi: 10.1073/pnas.1007424107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pisitkun T, Jacob V, Schleicher SM, et al. Akt and ERK1/2 pathways are components of the vasopressin signaling network in rat native IMCD. Am. J. Physiol. Renal Physiol. 2008;295:F1030–F1043. doi: 10.1152/ajprenal.90339.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y, Wolf-Yadlin A, Ross PL, et al. Time-resolved mass spectrometry of tyrosine phosphorylation sites in the epidermal growth factor receptor signaling network reveals dynamic modules. Mol. Cell Proteomics. 2005;4:1240–1250. doi: 10.1074/mcp.M500089-MCP200. [DOI] [PubMed] [Google Scholar]
- 48.Naegle KM, Gymrek M, Joughin BA, et al. PTMScout: a web resource for analysis of high-throughput post-translational proteomic studies. Mol. Cell Proteomics. 2010;9(11):2558–2570. doi: 10.1074/mcp.M110.001206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Levy ED, Landry CR, Michnick SW. Cell signaling. Signaling through cooperation. Science. 2010;328:983–984. doi: 10.1126/science.1190993. [DOI] [PubMed] [Google Scholar]
- 50.Hallows KR, Bhalla V, Oyster NM, et al. Phosphopeptide screen uncovers novel phosphorylation sites of Nedd4–2 that potentiate its inhibition of the epithelial Na+ channel. J. Biol. Chem. 2010;285:21671–21678. doi: 10.1074/jbc.M109.084731. [DOI] [PMC free article] [PubMed] [Google Scholar]
Websites
- 101.Collecting Duct Phosphoprotein Database. http://dir.nhlbi.nih.gov/papers/lkem/cdpd.
- 102.IMCD Phosphoproteome Database. http://dir.nhlbi.nih.gov/papers/lkem/mpkccdprot.
- 103.mpkCCD Cell Phosphoproteomic Database. http://dir.nhlbi.nih.gov/papers/lkem/cdpd_private.