SUMMARY
Tannerella forsythia is associated with subgingival biofilms in adult periodontitis, although the molecular mechanisms contributing to chronic inflammation and loss of periodontal bone remain unclear. We examined changes in the host transcriptional profiles during a T. forsythia infection using a murine calvarial model of inflammation and bone resorption. Tannerella forsythia was injected into the subcutaneous soft tissue over calvariae of BALB/c mice for 3 days, after which, the soft tissues and calvarial bones were excised. RNA was isolated and Murine GeneChip® array analysis of transcript profiles showed that 3226 genes were differentially expressed in the infected soft tissues (P < 0.05) and 2586 genes were differentially transcribed in calvarial bones after infection. Quantitative real-time reverse transcription-polymerase chain reaction analysis of transcription levels of selected genes corresponded well with the microarray results. Biological pathways significantly impacted by T. forsythia infection in calvarial bone and soft tissue included leukocyte transendothelial migration, cell adhesion molecules (immune system), extracellular matrix–receptor interaction, adherens junction, and antigen processing and presentation. Histologic examination revealed intense inflammation and increased osteoclasts in calvarias compared with controls. In conclusion, localized T. forsythia infection differentially induces transcription of a broad array of host genes, and the profiles differ between inflamed soft tissues and calvarial bone.
Keywords: calvarial bone, gene expression, microarray, Tannerella forsythia, tissue
INTRODUCTION
Periodontitis is a chronic immunoinflammatory infectious disease initiated by complex microbial subgingival biofilms resulting in destruction of periodontal tissues and resorption of alveolar bone. Tannerella forsythia, an oral, gram-negative, filament-shaped, non-pigmented, strict anaerobe, is an important member of pathogenic biofilms at sites of periodontal disease (Socransky et al., 1998; Tanner et al., 1998; Tanner & Izard, 2006). A strong association with chronic periodontitis has been shown for T. forsythia and Porphyromonas gingivalis (Haffajee et al., 2006). Several potential virulence factors of T. forsythia have been reported, including PrtH cysteine protease (Saito et al., 1997), forsythia-detaching factor (Nakajima et al., 2006), a sialidase (Ishikura et al., 2003), an apoptosis-inducing activity (Arakawa et al., 2000), a hemagglutinin (Murakami et al., 2002), alpha-D-glucosidase and N-acetyl-β-glucosaminidase (Hughes et al., 2003), production of methylglyoxal (Maiden et al., 2004), matrix metalloproteinase (MMP) -like enzyme karilysin (Karim et al., 2010), S-layer mediating hemagglutination, adhesion/invasion of epithelial cells, and murine subcutaneous abscess formation (Sabet et al., 2003). Tannerella forsythia also expresses a cell surface-associated and secreted protein BspA (Sharma et al., 1998), which has been recognized as an important virulence factor in inducing alveolar bone loss in mice (Sharma et al., 2005). Furthermore, BspA is an important modulator of host innate immune responses through activation of Toll-like receptor 2 in human gingival epithelial cells in cooperation with Toll-like receptor 1 (Onishi et al., 2008). In addition, twelve in vivo-induced genes (antigens) of T. forsythia were determined from sera of patients with periodontal disease using in vivo-induced antigen technology (Yoo et al., 2007). Recent reports indicate synergy between T. forsythia and Fusobacterium nucleatum in biofilm formation (Sharma et al., 2005). Moreover, P. gingivalis or its outer membrane vesicles enhance the attachment and invasion of T. forsythia to human oral epithelial cells (Inagaki et al., 2006), and growth of P. gingivalis is stimulated by cell extracts from T. forsythia (Yoneda et al., 2005). However, robust data on the in vivo role of potential virulence factors, as well as the broader aspects of the host response to virulence factors of T. forsythia in the periodontium remain to be defined.
The use of complementary DNA microarrays to survey transcriptional host responses after exposure to microbial pathogens has become a powerful approach to enhance understanding of the molecular basis of the host response to bacterial infections, which is critical for limiting tissue destruction. Host response profiling has identified transcripts uniquely affected by pathogens such as Listeria monocytogenes, Brucella abortus, and Mycobacterium tuberculosis (Cohen et al., 2000, Eskra et al., 2003, Rachman et al., 2006). Recent microarray studies have determined in vitro responses of host cells to challenge with P. gingivalis or its virulence components in primary human coronary artery endothelial cells and human aortic endothelial cells (Chou et al., 2005), gingival fibroblasts from healthy and inflammatory gingival tissues (Wang et al., 2003), gingival transcriptome patterns during induction and resolution of experimental gingivitis in humans (Offenbacher et al., 2009), and human gingival epithelial cells (Handfield et al., 2005). A recent gingival transcriptome study suggested that the microbial content of the periodontal pocket is a determinant of gene expression in the gingival tissues and provides new insights into the differential ability of periodontal species to elicit a local host response (Papapanou et al., 2009). In addition, gene ontology analysis of healthy and diseased gingival tissues from patients with advanced periodontitis identified 61 differentially expressed groups including apoptosis, antimicrobial immune response, and antigen presentation (Demmer et al., 2008). A recent review highlighted organism-specific transcriptional responses of gingival epithelial cell responses that correlated with the pathogenic potential of the real bacteria (Handfield et al., 2005). Nevertheless, there remains no report documenting T. forsythia induction of gene expression when the microorganism interacts with animal host cells in vivo. The aim of the present study was to define changes in the transcriptome of calvarial bone and overlying soft tissues in response to localized T. forsythia infection in mice using an established calvarial model of inflammation and bone resorption. We performed a genome-wide transcriptional analysis of the calvarial bone and overlying soft tissues isolated from T. forsythia-infected and sham-infected mice. Microarray data analysis identified several hundred altered probe sets, leading to subsequent identification of numerous pathways which were significantly changed in T. forsythia-infected mice.
METHODS
Mice
BALB/c female mice 8–10 weeks old (Harlan, Indianapolis, IN) were routinely acclimatized for at least 1 week before use. Mice were infected with T. forsythia ATCC 43037 cells as described below following isoflurane inhalation anesthesia. All mouse infection procedures were performed in accordance with the approved guidelines set forth by the Institutional Animal Care and Use Committee at the University of Kentucky (Lexington, KY).
Microorganism and mouse infection
Tannerella forsythia ATCC 43037 cells were grown in trypticase soy agar II basal media supplemented with yeast extract, phytone peptone, sheep blood (5%), and N-acetylmuramic acid for 3 days. Bacteria were scraped from the agar surface and resuspended in reduced transport fluid as described previously (Sharma et al., 2005). Tannerella forsythia were injected at 1.5 × 109 (n = 10 mice) into the soft tissues overlying the calvariae of the mice (Zubery et al., 1998). Bacterial infection, mouse euthanasia, collection, and preparation of calvarial bone and soft tissue for histology were performed as previously described (Zubery et al., 1998; Meka et al., 2010; Bakthavatchalu et al., 2010).
RNA isolation and mouse GeneChip hybridization
RNA isolation, assessment of quality of the RNA preparation, complementary RNA transcription, hybridization to the mouse GeneChip MG-MOE430A (Affymetrix, Santa Clara, CA), and scanning using an Affymetrix GCS 3000 7G Scanner were performed as previously described ((Meka et al., 2010; Bakthavatchalu et al., 2010).
Microarray data analysis
The T. forsythia microarray data were normalized, the dataset was evaluated by both unsupervised and supervised analyses, and hierarchal clustering analysis was performed, following which differences between the various treatment tissue classes, and determination of fold-change of significantly impacted genes were determined as previously described (Eisen et al., 1998; Li & Wong, 2001; Feezor et al., 2003; Handfield et al., 2005, 2008; Draghici et al., 2007, Khatri et al., 2007; Hasegawa et al., 2008; Bakthavatchalu et al., 2010; Meka et al., 2010).
Quantitative real-time reverse transcription-polymerase chain reaction analysis
Differential expression of selected genes, that showed significant variation in the microarray analyses, was confirmed by quantitative reverse trasncription–polymerase chain reaction (qRT-PCR) analysis as previously described (Piana et al., 2008; Bakthavatchalu et al., 2010; Meka et al., 2010). Seven representative upregulated genes based on a broad overview of the different functional category such as extracellular matrix, cell adhesion, cell proliferation, immune and defense responses, transport, and other category functions from both soft tissue and calvarial bone were evaluated. These genes were: pleiotrophin (Ptn), tumor necrosis factor receptor superfamily 19 (Tnfrsf19), periostin (Postn) from calvarial tissue and schafen (Slfn3), procollagen type I α1 (Col1a1), fibronectin (Fn), and matrix metalloproteinases 13 (Mmp13) from calvarial bone.
Calvarial bone histology
The mouse calvariae were fixed in 10% phosphate-buffered formalin, decalcified, embedded, sectioned, stained, and analysed for osteoclasts as previously described (Zubery et al., 1998; Bakthavatchalu et al., 2010; Meka et al., 2010).
Statistical analyses
Microarray data were analysed as described above. P-values of 0.05 or less were considered significant. The qRT-PCR data from two separate experiments were combined and results were expressed as means ± SD.
Microarray data accession numbers
The array results have been deposited in the GEO repository (http://www.ncbi.nlm.nih.gov/projects/geo/) under accession numbers GSE17110 and GSE20389.
RESULTS
Ontology of gene expression changes in murine calvarial soft tissue and bone
The mouse gene chip MOE430A contains 22,690 probe sets, with 17,395 and 17,274 probe sets (P < 0.001) providing positive readable signals to T. forsythia infection in calvarial soft tissue and bone, respectively. Significant differences were observed in mean gene expression levels of 3226 and 2586 probe sets in soft tissue and bone in response to infections (P < 0.05), respectively. Of the significantly regulated genes, 1528 were upregulated and 1698 were downregulated for soft tissue. In calvarial bone samples 1780 genes were upregulated and 806 were downregulated. The results of this initial gene profile analysis demonstrate that T. forsythia stimulated greater changes in the transcriptome of upregulated and downregulated genes in soft tissue compared with calvarial bone. The majority of genes with altered expression in calvarial soft tissue to T. forsythia infection were primarily associated with basic cellular functions [transcription, cell proliferation, cell cycle, transport, cell adhesion, extracellular matrix (ECM), apoptosis] for maintaining tissue integrity.
The ability of probe sets significant at P ≤ 0.05 to correctly identify differences between treatment groups was confirmed by leave-one-out cross-validation analysis (see Supporting Information, Tables S1 and S2). The significantly regulated probe sets were analysed by the PATHWAY EXPRESS tool as previously described (Draghici et al., 2007; Khatri et al., 2007). Pathways significantly impacted by T. forsythia at the P ≤ 0.05 level in bone and soft tissue types included: leukocyte transendothelial migration (LTM) (actin cytoskeleton, leukocyte) (Figs. 1 and 2), cell adhesion molecules (CAM) (immune system comprising antigen-presenting cells, T cells, and B cells) (Fig. 3), ECM–receptor interaction (ECM, integrin, VLA proteins, leukoproteins, cytoadhesin, focal adhesion, proteoglycan, glycoprotein) (Fig. 4), and focal adhesion (ECM–receptor, cytokine–cytokine receptor, actin cytoskeleton, cell proliferation, cell cycle, and three signaling pathway system) (Fig. 5). Table 1 shows soft tissue and calvarial bone pathways generated from this analysis that were predominantly affected in order of their impact factors. Tannerella forsythia significantly impacted 14 pathways in calvarial bone and 15 in soft tissue with an impact factor more than five. The high impact factors associated with these pathways predict that the effects of T. forsythia-induced gene expression changes in the bone or tissue will have a significant biological effect downstream. There are more than nine pathways, including LTM, CAM, focal adhesion, adherens junction, antigen processing and presentation, and phosphatidylinositol signaling system, that were significantly impacted or overlapped in both the calvarial bone and soft tissue (Table 1), and hence these were chosen for further study to compare and contrast the impact of T. forsythia on two distinct tissue types.
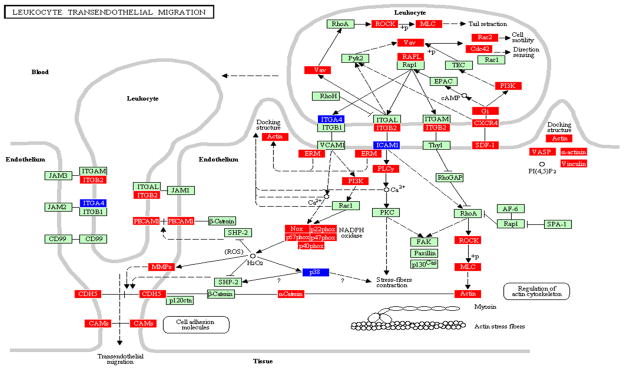
Figure 1.
Leukocyte transendothelial migration pathway containing genes differentially regulated by Tannerella forsythia in calvarial bone compared with sham-infected controls at P ≤ 0.05, adapted from PATHWAY EXPRESS and using the Kyoto Encyclopedia of Genes and Genomes nomenclature. Genes shown in red are upregulated, genes shown in blue are downregulated, and green indicates no change in gene expression at the P < 0.05 significance level. +p, phosphorylation event; −p, dephosphorylation event; ?, receptors that are yet to be identified; O, other molecule. An arrow indicates a molecular interaction resulting in transendothelial migration, leukocyte activation, regulation of actin cytoskeleton and a line without an arrowhead indicates a molecular interaction resulting in inhibition.
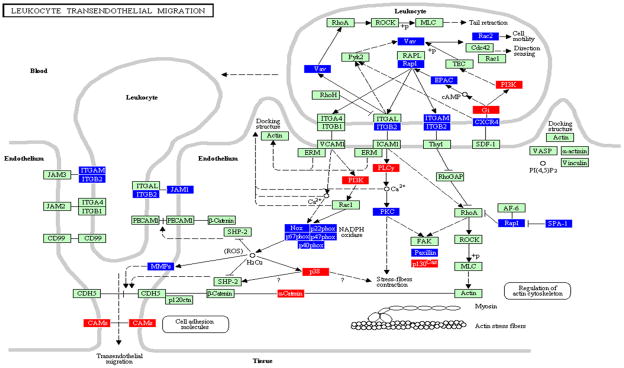
Figure 2.
Leukocyte transendothelial migration pathway containing genes differentially regulated by Tannerella forsythia in calvarial soft tissue compared with sham-infected controls at P ≤ 0.05, adapted from PATHWAY EXPRESS and using the Kyoto Encyclopedia of Genes and Genomes nomenclature. Red indicates induction, blue indicates repression, and green indicates no change in gene expression. +p, phosphorylation event; −p, dephosphorylation event; ?, receptors that are yet to be identified; O, other molecule. An arrow indicates a molecular interaction resulting in transendothelial migration, leukocyte activation, regulation of actin cytoskeleton and a line without an arrowhead indicates a molecular interaction resulting in inhibition.
Figure 3.
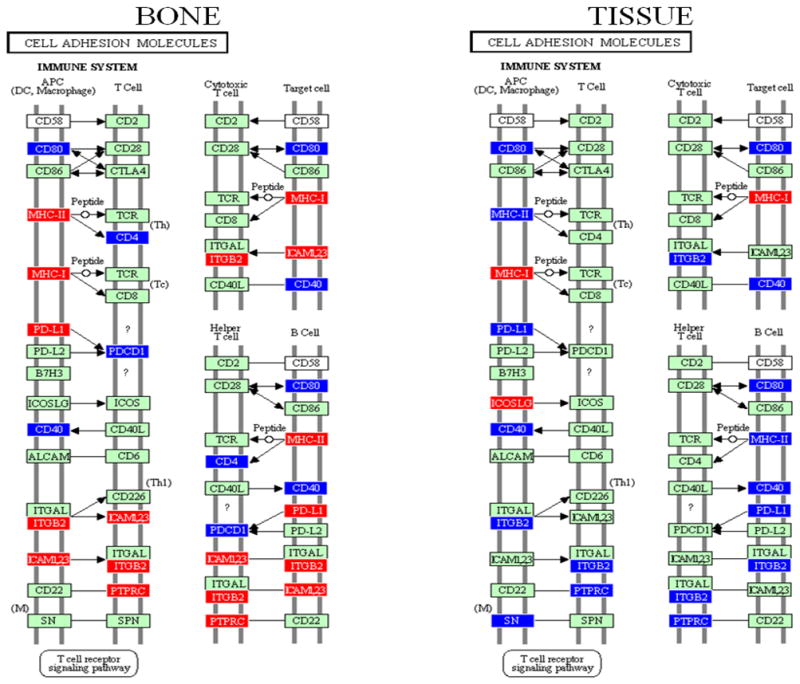
Cell adhesion molecules pathway (immune system) containing genes differentially regulated by Tannerella forsythia in calvarial bone and soft tissue compared with sham-infected controls at P ≤ 0.05, adapted from PATHWAY EXPRESS and using the Kyoto Encyclopedia of Genes and Genomes nomenclature. Red indicates induction, blue indicates repression, and green indicates no change in gene expression. An arrow indicates a molecular interaction resulting in activation of dendritic cells, macrophages, T cells (T-cell receptor signaling pathway), and a line without an arrowhead indicates a molecular interaction resulting in inhibition.
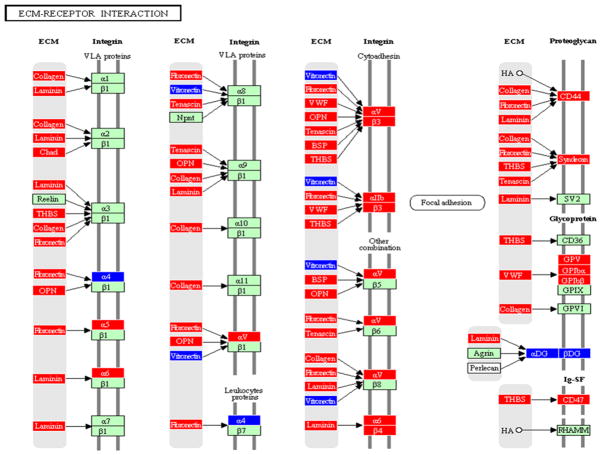
Figure 4.
Extracellular membrane receptor interaction pathway containing genes differentially regulated by Tannerella forsythia in calvarial bone compared with sham-infected controls at P ≤ 0.05, adapted from PATHWAY EXPRESS and using the Kyoto Encyclopedia of Genes and Genomes nomenclature. Red indicates induction, blue indicates repression, and green indicates no change in gene expression. An arrow indicates a molecular interaction resulting in extracellular matrix receptor activation, regulation of integrin (VLA proteins, leukoproteins, cytoadhesin, focal adhesion, proteoglycan, glycoprotein).
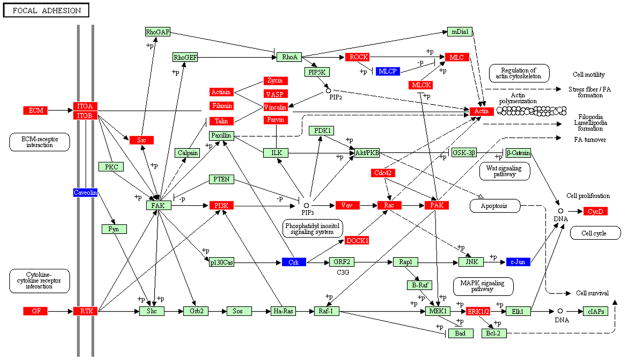
Figure 5.
Focal adhesion pathway containing genes differentially regulated by Tannerella forsythia in calvarial bone compared with sham-infected controls at P ≤ 0.05, adapted from PATHWAY EXPRESS and using the Kyoto Encyclopedia of Genes and Genomes nomenclature. Red indicates induction, blue indicates repression, and green indicates no change in gene expression. An arrow indicates a molecular interaction resulting in extracellular matrix-receptor interaction, cytokine–cytokine receptor interaction, regulation of actin cytoskeleton, cell proliferation, cell cycle, and three signaling pathway systems (phosphatidyl inositol, mitogen-activated protein kinase, Wnt).
Table 1.
Ontology analysis of calvarial bone and soft tissue gene pathways impacted by infection with Tannerella forsythia1
| Impacted pathway2 | Impact factor3 | No. of input genes/no.of pathway genes4 |
|---|---|---|
| Calvarial bone | ||
| Leukocyte transendothelial migration | 85.1 | 37/119 |
| Cell adhesion molecules | 79.3 | 30/159 |
| ECM–receptor interaction | 23.5 | 35/81 |
| Focal adhesion | 18.8 | 59/199 |
| DNA replication | 15.3 | 18/36 |
| Phosphatidylinositol signaling system | 14.5 | 14/75 |
| Adherens junction | 13.1 | 20/77 |
| Hematopoietic cell lineage | 11.7 | 29/86 |
| Regulation of actin cytoskeleton | 11.7 | 53/213 |
| Cell cycle | 9.7 | 32/124 |
| B-cell receptor signaling pathway | 9.1 | 19/71 |
| Antigen processing and presentation | 8.9 | 20/100 |
| Natural killer cell-mediated cytotoxicity | 5.7 | 22/109 |
| Jak-STAT signaling pathway | 5.7 | 31/157 |
| Calvarial soft tissue | ||
| Leukocyte transendothelial migration | 148.2 | 35/119 |
| Cell adhesion molecules | 136.7 | 31/159 |
| Adherens junction | 42.7 | 19/77 |
| Antigen processing and presentation | 37.1 | 11/100 |
| Phosphatidylinositol signaling system | 16.0 | 18/75 |
| Toll-like receptor signaling pathway | 9.8 | 33/101 |
| Natural killer cell-mediated cytotoxicity | 9.8 | 31/109 |
| B-cell receptor signaling pathway | 9.4 | 23/71 |
| Jak-STAT signaling pathway | 8.9 | 41/157 |
| Cytokine–cytokine receptor interaction | 8.5 | 63/249 |
| Focal adhesion | 6.8 | 50/199 |
| ErbB signaling pathway | 5.9 | 24/89 |
| p53 signaling pathway | 5.5 | 21/74 |
| Complement and coagulation cascades | 5.4 | 21/74 |
| VEGF signaling pathway | 5.4 | 21/76 |
The calvarial soft tissue and bone gene pathways were determined by PATHWAY EXPRESS (Draghici et al., 2007; Khatri et al., 2007).
Kyoto Encyclopedia of genes and genome pathways (http://www.genome.jp/kegg/).
The impact factor measures the pathways most affected by changes in gene expression in calvarial bone and soft tissue in response to T. forsythia by considering the proportion of differentially regulated genes, the perturbation factors of all pathway genes, and the propagation of these perturbations throughout the pathway (Draghici et al., 2007; Khatri et al., 2007). Only pathways with an impact factor greater than 5 are included in this table.
Number of regulated genes in a pathway/total number of genes currently mapped to this pathway.
Validation of microarray gene expression
Changes in T. forsythia-induced transcript expression levels of selected genes from the microarray studies were confirmed by qRT-PCR, including Ptn, Tnfrsf19, and Postn in calvarial tissue and Slfn3, Col1a1, Fn, and Mmp13 in calvarial bone using aliquots of the pooled RNA samples that were evaluated in the microarrays (see Supporting Information, Table S3). Transcripts of β-actin were used as an expression control and the qRT-PCR analyses were performed at least twice for each gene. The qRT-PCR results confirmed the microarray data and the selected upregulated genes in microarray showed corresponding increased expressions with qRT-PCR analysis, although some expression levels differed between the two techniques.
Inflammatory and immune response gene expression profiles
Since several studies of infection with oral pathogens have emphasized the chronic inflammatory nature of the challenge leading to localized tissue destruction, we also focused on examination of gene profiles related to inflammatory and immune responses. The proinflammatory cytokines, interleukin-1β (IL-1β), IL-6, tumor necrosis factor, and other cytokines such as IL-7, IL-15, IL-17, IL-18, IL-23 involved in induction of inflammation and calvarial bone resorption were not significantly altered in soft tissue and calvarial bone following T. forsythia infection. The majority of genes upregulated in calvarial bone during infection were associated with ECM, cell adhesion, cell proliferation, immune and defense responses, transport, and other category functions. The upregulated immune and defense response genes included multiple genes associated with the adaptive immune response regulation of humoral immunity, e.g. Igh-6, Igh, Igk-C, Igk-V8, Igj, Igh-VJ558, and Camp. Expression of the extracellular matrix protein gene chondroadherin (Chad), matrix metalloproteinases (Mmp3, Mmp13, Mmp9, Mmp14), several types of procollagen (Colla1), matrilin (Matn2), tissue inhibitor of metalloproteinase (Timp1, Timp2), various growth factors (Ctgf, Ltbp1), and especially periostin (Postn; three-fold) were upregulated. Increased expression of matrix metalloproteinases 13 (five-fold increase), 9 (11-fold increase), 8 (two-fold increase), 2 (two-fold increase), and 14 (two-fold increase), (Mmp13, Mmp8, Mmp2, Mmp9, Mmp14) and of cathepsins K (seven-fold increase), G (12-fold increase), E (three-fold increase), C (two-fold increase), H (two-fold increase), and Z (two-fold increase) genes showed upregulation in calvarial bone. In contrast, expression of several Mmp genes, including Mmp3 (0.3-fold decrease), Mmp8 (0.2-fold decrease), Mmp9 (0.2-fold decrease), Mmp10 (0.5-fold decrease), Mmp13 (0.2-fold decrease), Mmp14 (0.4-fold decrease), Mmp19 (0.3-fold decrease) and cathepsin genes, Ctsl (0.5 fold decrease), Ctsb (0.5-fold decrease), Ctsd (0.7-fold decrease), Ctss (0.3-fold decrease), and Ctsz (0.2-fold decrease), was downregulated in soft tissue. Probe sets representing tissue inhibitor of metalloproteinases Timp1 (five-fold increase) and Timp2 (three-fold increase) were modestly upregulated in calvarial bone and downregulated (Timp1, Timp2, Timp4) in soft tissue during T. forsythia infection.
Calvarial histology
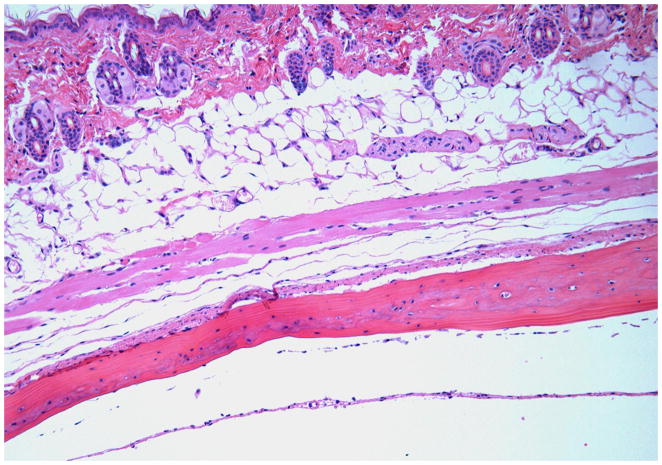
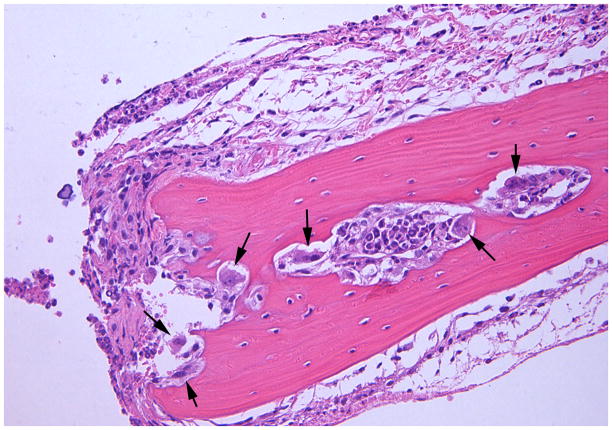
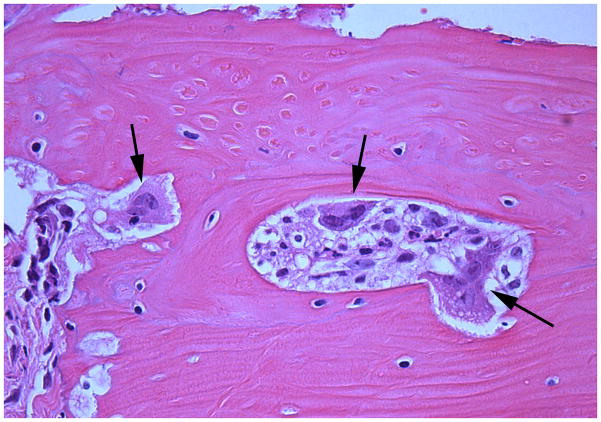
The sham-infected control mice showed a lack of edema and minimal inflammation in the soft tissue over the calvaria at the site of injection (Fig. 6A). Calvarial soft tissue swelling occurred at the injection site within 24 h of the first injection and was increased at 48 and 72 h in almost all of the mice injected with T. forsythia, but not in the sham-infected control mice. The T. forsythia-infected mice did not show local abscesses, ulceration of the overlying skin, or any evidence of spread of infection to neighboring sites. Histological examination of the calvarial sections revealed significant edema and an intense mixed inflammatory cell infiltrate consisting of aggregates of polymorphonuclear leukocytes, lymphocytes, and macrophages (Fig. 6B). Notably, prominent and increased numbers of osteoclasts were seen at the suture area on the inner aspect of the calvaria compared with sham-infected controls (data not shown). An increase in the size of the bone marrow spaces as a result of increased endosteal bone resorption was also noted. Activated osteoclasts were found along the entire suture area corresponding to the areas of bone resorption (Fig. 6C). Tannerella forsythia cells were not seen in Gram-stained calvarial sections from mice with moderate soft tissue inflammatory infiltrates.
Figure 6.
Effects of Tannerella forsythia local injection on mouse calvaria. Live T. forsythia bacteria (1.5 × 109) were injected once daily for 3 days into the subcutaneous tissues overlying the calvaria of mice. All photomicrographs are of sections stained with hematoxylin & eosin. (A) Lack of edema and inflammation in the calvarial soft tissue of a sham-infected control mouse (10 ×). (B) Numerous osteoclasts (black arrows) are seen throughout the inner aspects of the calvarial bone mainly in the suture area. (20 ×). (C) Activated osteoclasts at higher magnification (40 ×). Also note the increase in marrow space size, as a result of the increased bone resorption in (B, C).
DISCUSSION
We used a complementary DNA microarray to study the gene transcriptional profiles of host soft tissue and calvarial bone during a localized acute infectious challenge with the oral pathogen T. forsythia in BALB/c mice. Diverse functional classes of genes were altered by T. forsythia, suggesting that numerous cellular processes were transcriptionally modulated during the course of the infection.
The breadth of functional categories and gene families that were altered in response to the localized infection in calvarial soft tissue included transcription, cell proliferation, cell cycle, transport, cell adhesion, stress, apoptosis, defense response, ECM, and cell differentiation, indicating that T. forsythia elicits a multitude of specific gene expression changes during infection. In bone, functional categories and gene families that were altered in response to the localized infection included cell proliferation, cell cycle, transcription, transport, defense and immune response, apoptosis, and ECM. Similar functional categories were also altered following infection with other periodontal pathogens, P. gingivalis (Meka et al., 2010) and Treponema denticola (Bakthavatchalu et al., 2010) indicating a commonality of the molecular interactions following infection of these pathogens with host soft tissues and bone.
Tannerella forsythia infection induced robust transcriptional changes in ECM proteins in both calvarial bone and soft tissue. For example, expression of the genes for matrix metalloproteinases 13, 9, 2 and 14, (Mmp13, Mmp9, Mmp2, Mmp14) was upregulated in calvarial bone, whereas expression of tissue inhibitor of metalloproteinase (Timp1, Timp2, Timp3) was modestly upregulated during T. forsythia infection. Similar levels of upregulation of the expression of Mmp genes and Timp1, Timp2, and Timp3 genes were recorded in bone following Tr. denticola infection using this model (Bakthavatchalu et al., 2010), whereas P. gingivalis had no effect on the expression of Mmp and Timp genes (Meka et al., 2010). TIMPs may also serve as an early indicator of the acute-phase response. Furthermore, expression of several of the ECM proteins, including chondroadherin, procollagen types I, III, and V, fibronectin 1, fibromodulin, osteomodulin, and biglycan was elevated during T. forsythia infection as well as with Tr. denticola infection (Bakthavatchalu et al., 2010) but not following P. gingivalis infection (Meka et al., 2010). The changes we found in ECM protein transcriptional expression levels are in agreement with those induced robustly by Escherichia coli lipopolysaccharide inoculation in a canine model of oral infection (Higgins et al., 2003). The composition of the ECM is known to impact multiple cellular activities including differentiation, proliferation, and motility. Periostin (gene Postn; increased four-fold and two-fold in bone and soft tissue, respectively), is a protein secreted into ECM and its gene is widely expressed in various tissues. The periostin gene is highly expressed in the embryonic periosteum, periodontal ligaments, and placenta among other tissues. Moreover, Postn null mice exhibit incisor enamel defects and an early-onset periodontal disease-like phenotype, suggesting that periostin is critically required for maintenance of the periodontal ligament (Rios et al., 2005, 2008). Changes in periostin (ECM protein) expression in bone and tissue after Tr. denticola (Bakthavatchalu et al., 2010) and T. forsythia challenge but not P. gingivalis infection (Meka et al., 2010) in our model probably reflect its participation in the tissue remodeling of damaged calvarial bone and tissue after the acute T. forsythia or Tr. denticola infection. Damage caused by P. gingivalis infection, may be different from that induced by T. forsythia or Tr. denticola.
Tannerella forsythia has a number of virulence factors that can induce an inflammatory cascade involving proinflammatory cytokines, reactive oxygen species, and MMPs, and hence lead to the destruction of supportive soft and hard tissues around the teeth during periodontal disease. Several MMPs including MMP-2, MMP-3, MMP-8, and MMP-9 are increased in inflamed gingival tissue (Tervahartiala et al., 2000), and increased levels of MMP-8 and MMP-9 have been reported in gingival crevicular fluid during the active stages of periodontal disease (Kiili et al., 2002).
A significant difference between the calvarial bone and soft tissue samples was in the impact of infection on the LTM pathway in bone (33 genes increase; four genes decrease) including activation of Vav (signal transducer protein) which subsequently activates Rho-associated kinase, myosin light chain phosphorylation, resulting in the activation of actin (cytoskeleton) and α-cadherin. Moreover, leukocytes have a number of functions, including activation of endothelial cell signals, production of reactive oxygen species with subsequent activation of integrin β2, platelet endothelial cell adhesion molecule-1 (PECAM1), MMPs, CDH5 (cadherin 5), and CAMs. This LTM pathway is consistent with the major functional pathways and several of the genes (integrins, focal adhesion molecules, cadherins) upregulated during induction of experimental gingivitis in humans, which is consistent with the activation of the LTM pathway (Offenbacher et al., 2009). PECAM-1 is one of the most abundant proteins on the endothelial cell surface. It is expressed on the surface of platelets and leukocytes, and its expression on mononuclear infiltrates increases significantly with increasing size of infiltrates in the lesions of gingivitis and periodontitis (Gemmell et al., 1994). In contrast, in tissue more than 18 LTM genes were downregulated and seven genes were activated indicating a lesser interaction of T. forsythia in the soft tissues. These differences (LTM upregulation in bone and downregulation in soft tissue) are likely related to the differences in the types of cells in both types of tissues, given the clear difference in the composition of both tissues, i.e. there was an acute inflammatory infiltrate in the soft tissue, but not in the underlying bone. Importantly, the number (impact factor: bone 85 and tissue 148) and pattern of genes (bone: 37/119; tissue: 35/119) altered in the LTM pathway of bone samples derived from T. forsythia infections are quite different from the gene expression after Tr. denticola challenge (bone: impact factor 306; genes 14/119) (Bakthavatchalu et al., 2010). In contrast, the LTM pathway was not significantly affected in bone and soft tissue (impact factor: bone 18 and tissue 0) after P. gingivalis infection (Meka et al., 2010). Further studies will be required to determine why there are differences between bone and soft tissue levels of expression of LTM pathway members in response to these three pathogens.
Another major difference between the bone and tissue samples was in the impact of T. forsythia infection on the CAM pathway. In calvarial bone, T. forsythia upregulated several components of this immune system pathway including major histocompatibility complex class II (MHCII), MHCI, intercellular adhesion molecule 1 (ICAM-1), ICAM-2, and ICAM-3, and downregulated T cell, cytotoxic T cell, and T helper–B cell interacting molecules. This is consistent with recent descriptions of the expression of class I and II MHC antigens on oral mucosal epithelium and induction of activation of CD4+ T cells in response to bacterial challenge (Matsuyama et al., 2005). Similarly, Tr. denticola and P. gingivalis upregulated the expression of a range of genes in this pathway including, MHCII, MHCI, CD8, CD28, CD80, and CD40 ligand (Bakthavatchalu et al., 2010; Meka et al., 2010). In contrast to T. forsythia, both Tr. denticola and P. gingivalis infections downregulated expression of ICAM-1, -2, and -3 in tissues (Bakthavatchalu et al., 2010; Meka et al., 2010). Increased expression of ICAM-1 (a marker of endothelial dysfunction) has been found in gingival tissues from patients with periodontitis (Hayashi et al., 1994), and significant changes in soluble ICAM-1 levels of subjects with periodontitis were observed following treatment (Hannigan et al., 2004). ICAM-1 is required for cell adhesion in inflamed tissues (Tamai et al., 2005) and is also involved in the invasion of epithelial cells by P. gingivalis and Treponema medium (Tamai et al., 2005, 2007). In contrast to T. forsythia, only MHCI was upregulated in soft tissue and T cell, cytotoxic T cell, and T helper–B cell interacting molecules were downregulated following Tr. denticola infection.
Tannerella forsythia upregulated several components of the ECM–receptor pathway in calvarial bone including collagen, chondroadherin, fibronectin, osteopontin, bone sialoprotein, and thrombospondin. These components of the ECM–receptor pathway were not regulated by either Tr. denticola (Bakthavatchalu et al., 2010) or P. gingivalis infections (Meka et al., 2010) in this model. Collagen-I and Collagen-III are the major constituents of all periodontal tissues and are essential in establishing their structural and physiological integrity. Fibronectin (critical in regenerating periodontal tissues), osteopontin, and bone sialoprotein are non-collagenous glycoproteins, seem to have a unique distribution within the periodontium and accumulate predominantly at the hard tissue interfaces. As T. forsythia levels are higher in subgingival plaque samples from deep periodontal pockets (Tanner & Izard, 2006), they may induce the expression of several MMPs and cathepsins, which collectively can degrade ECM proteins in the periodontium and may contribute to the bone resorption and attachment loss observed in periodontal disease (Hannas et al., 2007). This level of T. forsythia-induced enhanced ECM–receptor interactions was not observed with infections by either P. gingivalis (Meka et al., 2010) or Tr. denticola (Bakthavatchalu et al., 2010). Similar observations in this pathway were not seen in the soft tissues after infection. These findings clearly suggest that T. forsythia, a potential critical member of pathogenic biofilms, may be an important contributor to the hard tissue destruction observed in periodontal disease.
The significant increase in osteoclast numbers and activity, dense inflammation, and full thickness calvarial bone resorption defects observed in response to T. forsythia infection were similar to the effects that we observed in the calvariae of mice following infection with other periodontal pathogens including P. gingivalis (Meka et al., 2010), Tr. denticola (Bakthavatchalu et al., 2010), Campylobacter rectus and F. nucleatum (Zubery et al., 1998), as well as with local injections of IL-1 (Boyce et al., 1989).
In conclusion, the present study reports findings from a comprehensive gene expression profile of inflamed soft tissue and calvarial bone that accompanied a localized, acute infection of mice with T. forsythia. Importantly, many of the most affected genes were related to biological processes, and specifically a large array of genes impacted ECM–receptor interaction in calvarial bone. Further studies are required to identify specific T. forsythia virulence factors that alter host gene expression. Furthermore, the substantive impact of T. forsythia in upregulation of host inflammatory/innate immune responses in this model supports the need for additional studies to explore the role of selected genes in the periodontal disease process, as well as the effect of T. forsythia on host response patterns as a component of a polymicrobial infection in periodontal disease.
Supplementary Material
Acknowledgments
We thank Drs. Arnold Stromberg, Cidambaram Srinivasan, Christopher Saunders, and Matt Hersh (Department of Statistics, University of Kentucky, Lexington, KY) for initial data management and microarray data analysis. We also thank Dr Quey-Chen and Ms Donna Wall (University of Kentucky, Lexington, KY; Microarray Core Facility) for microarray technical assistance. This study was supported by the National Institute of Dental and Craniofacial Research DE015720 to L.K., DE11111 to R.J.L., United States Public Health Services research grant U24 DE016509 (Robert Burne), and by a National Institute of Arthritis, Musculoskeletal and Skin Diseases grant (AR43510) to B.F.B.
Footnotes
Disclosures
The authors have no financial conflict of interest.
Table S1. Leave-one-out cross-validation – calvarial bone.
Table S2. Leave-one-out cross-validation – calvarial soft tissue.
Table S3. Comparison of expression of selected genes in calvarial tissue and calvariae (high dose) by microarray and real-time quantitative polymerase chain reaction.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors, any queries (other than missing material) should be directed to the corresponding author of the article.
References
- Arakawa S, Nakajima T, Ishikura H, Ichinose S, Ishikawa I, Tsuchida N. Novel apoptosis-inducing activity in Bacteroides forsythus: a comparative study with three serotypes of Actinobacillus actinomycetemcomitans. Infect Immun. 2000;68:4611–4615. doi: 10.1128/iai.68.8.4611-4615.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakthavatchalu V, Meka A, Sathishkumar S, et al. Molecular characterization of Treponema denticola infection-induced bone and tissue transcriptional profiles. Mol Oral Microbiol. 2010;25:1–15. doi: 10.1111/j.2041-1014.2010.00575.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce BF, Aufdemorte TB, Garrett IR, Yates AJ, Mundy GR. Effects of interleukin-1 on bone turnover in normal mice. Endocrinology. 1989;125:1142–1150. doi: 10.1210/endo-125-3-1142. [DOI] [PubMed] [Google Scholar]
- Chou HH, Yumoto H, Davey M, et al. Porphyromonas gingivalis fimbria-dependent activation of inflammatory genes in human aortic endothelial cells. Infect Immun. 2005;73:5367–5378. doi: 10.1128/IAI.73.9.5367-5378.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen P, Bouaboula M, Bellis M, et al. Monitoring cellular responses to Listeria monocytogenes with oligonucleotide arrays. J Biol Chem. 2000;275:11181–11190. doi: 10.1074/jbc.275.15.11181. [DOI] [PubMed] [Google Scholar]
- Demmer RT, Behle JH, Wolf DL, et al. Transcriptomes in healthy and diseased gingival tissues. J Periodontol. 2008;79:2112–2124. doi: 10.1902/jop.2008.080139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draghici S, Khatri P, Tarca AL, et al. A systems biology approach for pathway level analysis. Genome Res. 2007;17:1537–1545. doi: 10.1101/gr.6202607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskra L, Mathison A, Splitter G. Microarray analysis of mRNA levels from RAW264.7 macrophages infected with Brucella abortus. Infect Immun. 2003;71:1125–1133. doi: 10.1128/IAI.71.3.1125-1133.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feezor RJ, Oberholzer C, Baker HV, et al. Molecular characterization of the acute inflammatory response to infections with gram-negative versus gram-positive bacteria. Infect Immun. 2003;71:5803–5813. doi: 10.1128/IAI.71.10.5803-5813.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemmell E, Walsh LJ, Savage NW, Seymour GJ. Adhesion molecule expression in chronic inflammatory periodontal disease tissue. J Periodontal Res. 1994;29:46–53. doi: 10.1111/j.1600-0765.1994.tb01090.x. [DOI] [PubMed] [Google Scholar]
- Haffajee AD, Teles RP, Socransky SS. Association of Eubacterium nodatum and Treponema denticola with human periodontitis lesions. Oral Microbiol Immunol. 2006;21:269–282. doi: 10.1111/j.1399-302X.2006.00287.x. [DOI] [PubMed] [Google Scholar]
- Handfield M, Baker HV, Lamont RJ. Beyond good and evil in the oral cavity: insights into host–microbe relationships derived from transcriptional profiling of gingival cells. J Dent Res. 2008;87:203–223. doi: 10.1177/154405910808700302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handfield M, Mans JJ, Zheng G, et al. Distinct transcriptional profiles characterize oral epithelium–microbiota interactions. Cell Microbiol. 2005;7:811–823. doi: 10.1111/j.1462-5822.2005.00513.x. [DOI] [PubMed] [Google Scholar]
- Hannas AR, Pereira JC, Granjeiro JM, Tjaderhane L. The role of matrix metalloproteinases in the oral environment. Acta Odontol Scand. 2007;65:1–13. doi: 10.1080/00016350600963640. [DOI] [PubMed] [Google Scholar]
- Hannigan E, O’Connell DP, Hannigan A, Buckley LA. Soluble cell adhesion molecules in gingival crevicular fluid in periodontal health and disease. J Periodontol. 2004;75:546–550. doi: 10.1902/jop.2004.75.4.546. [DOI] [PubMed] [Google Scholar]
- Hasegawa Y, Tribble GD, Baker HV, Mans JJ, Handfield M, Lamont RJ. Role of Porphyromonas gingivalis SerB in gingival epithelial cell cytoskeletal remodeling and cytokine production. Infect Immun. 2008;76:2420–2427. doi: 10.1128/IAI.00156-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi J, Saito I, Ishikawa I, Miyasaka N. Effects of cytokines and periodontopathic bacteria on the leukocyte function-associated antigen 1/intercellular adhesion molecule 1 pathway in gingival fibroblasts in adult periodontitis. Infect Immun. 1994;62:5205–5212. doi: 10.1128/iai.62.12.5205-5212.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins MA, Berridge BR, Mills BJ, et al. Gene expression analysis of the acute phase response using a canine microarray. Toxicol Sci. 2003;74:470–484. doi: 10.1093/toxsci/kfg142. [DOI] [PubMed] [Google Scholar]
- Hughes CV, Malki G, Loo CY, Tanner AC, Ganeshkumar N. Cloning and expression of alpha-D-glucosidase and N-acetyl-β-glucosaminidase from the periodontal pathogen, Tannerella forsythensis (Bacteroides forsythus) Oral Microbiol Immunol. 2003;18:309–312. doi: 10.1034/j.1399-302x.2003.00091.x. [DOI] [PubMed] [Google Scholar]
- Inagaki S, Onishi S, Kuramitsu HK, Sharma A. Porphyromonas gingivalis vesicles enhance attachment, and the leucine-rich repeat BspA protein is required for invasion of epithelial cells by “Tannerella forsythia”. Infect Immun. 2006;74:5023–5028. doi: 10.1128/IAI.00062-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikura H, Arakawa S, Nakajima T, Tsuchida N, Ishikawa I. Cloning of the Tannerella forsythensis (Bacteroides forsythus) siaHI gene and purification of the sialidase enzyme. J Med Microbiol. 2003;52:1101–1107. doi: 10.1099/jmm.0.05349-0. [DOI] [PubMed] [Google Scholar]
- Karim AY, Kulczycka M, Kantyka T, et al. A novel matrix metalloprotease-like enzyme (karilysin) of the periodontal pathogen Tannerella forsythia ATCC 43037. Biol Chem. 2010;391:105–117. doi: 10.1515/BC.2010.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatri P, Voichita C, Kattan K, et al. Onto-Tools: new additions and improvements in 2006. Nucleic Acids Res. 2007;35:W206–211. doi: 10.1093/nar/gkm327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiili M, Cox SW, Chen HY, et al. Collagenase-2 (MMP-8) and collagenase-3 (MMP-13) in adult periodontitis: molecular forms and levels in gingival crevicular fluid and immunolocalisation in gingival tissue. J Clin Periodontol. 2002;29:224–232. doi: 10.1034/j.1600-051x.2002.290308.x. [DOI] [PubMed] [Google Scholar]
- Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiden MF, Pham C, Kashket S. Glucose toxicity effect and accumulation of methylglyoxal by the periodontal anaerobe Bacteroides forsythus. Anaerobe. 2004;10:27–32. doi: 10.1016/j.anaerobe.2003.12.001. [DOI] [PubMed] [Google Scholar]
- Matsuyama T, Kawai T, Izumi Y, Taubman MA. Expression of major histocompatibility complex class II and CD80 by gingival epithelial cells induces activation of CD4+ T cells in response to bacterial challenge. Infect Immun. 2005;73:1044–1051. doi: 10.1128/IAI.73.2.1044-1051.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meka A, Bakthavatchalu V, Sathishkumr S, et al. Porphyromonas gingivalis infection-induced tissue and bone transcriptional profiles. Mol Oral Microbiol. 2010;25:61–74. doi: 10.1111/j.2041-1014.2009.00555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y, Higuchi N, Nakamura H, Yoshimura F, Oppenheim FG. Bacteroides forsythus hemagglutinin is inhibited by N-acetylneuraminyllactose. Oral Microbiol Immunol. 2002;17:125–128. doi: 10.1046/j.0902-0055.2001.00093.x. [DOI] [PubMed] [Google Scholar]
- Nakajima T, Tomi N, Fukuyo Y, et al. Isolation and identification of a cytopathic activity in Tannerella forsythia. Biochem Biophys Res Commun. 2006;351:133–139. doi: 10.1016/j.bbrc.2006.10.012. [DOI] [PubMed] [Google Scholar]
- Offenbacher S, Barros SP, Paquette DW, et al. Gingival transcriptome patterns during induction and resolution of experimental gingivitis in humans. J Periodontol. 2009;80:1963–1982. doi: 10.1902/jop.2009.080645. [DOI] [PubMed] [Google Scholar]
- Onishi S, Honma K, Liang S, et al. Toll-like receptor 2-mediated interleukin-8 expression in gingival epithelial cells by the Tannerella forsythia leucine-rich repeat protein BspA. Infect Immun. 2008;76:198–205. doi: 10.1128/IAI.01139-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papapanou PN, Behle JM, Kebschull M, et al. Subgingival bacterial colonization profiles correlate with gingival tissue gene expression. BMC Microbiol. 2009;9:221. doi: 10.1186/1471-2180-9-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piana C, Wirth M, Gerbes S, Viernstein H, Gabor F, Toegel S. Validation of reference genes for qPCR studies on Caco-2 cell differentiation. Eur J Pharm Biopharm. 2008;69:1187–1192. doi: 10.1016/j.ejpb.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Rachman H, Kim N, Ulrichs T, et al. Critical role of methylglyoxal and AGE in mycobacteria-induced macrophage apoptosis and activation. PloS one. 2006;1:e29. doi: 10.1371/journal.pone.0000029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios H, Koushik SV, Wang H, et al. Periostin null mice exhibit dwarfism, incisor enamel defects, and an early-onset periodontal disease-like phenotype. Mol Cell Biol. 2005;25:11131–11144. doi: 10.1128/MCB.25.24.11131-11144.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios HF, Ma D, Xie Y, et al. Periostin is essential for the integrity and function of the periodontal ligament during occlusal loading in mice. J Periodontol. 2008;79:1480–1490. doi: 10.1902/jop.2008.070624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabet M, Lee SW, Nauman RK, Sims T, Um HS. The surface (S-) layer is a virulence factor of Bacteroides forsythus. Microbiology. 2003;149:3617–3627. doi: 10.1099/mic.0.26535-0. [DOI] [PubMed] [Google Scholar]
- Saito T, Ishihara K, Kato T, Okuda K. Cloning, expression, and sequencing of a protease gene from Bacteroides forsythus ATCC 43037 in Escherichia coli. Infect Immun. 1997;65:4888–4891. doi: 10.1128/iai.65.11.4888-4891.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Inagaki S, Honma K, Sfintescu C, Baker PJ, Evans RT. Tannerella forsythia-induced alveolar bone loss in mice involves leucine-rich-repeat BspA protein. J Dent Res. 2005;84:462–467. doi: 10.1177/154405910508400512. [DOI] [PubMed] [Google Scholar]
- Sharma A, Sojar HT, Glurich I, Honma K, Kuramitsu HK, Genco RJ. Cloning, expression, and sequencing of a cell surface antigen containing a leucine-rich repeat motif from Bacteroides forsythus ATCC 43037. Infect Immun. 1998;66:5703–5710. doi: 10.1128/iai.66.12.5703-5710.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- Tamai R, Asai Y, Kawabata A, Akisaka T, Ogawa T. Possible requirement of intercellular adhesion molecule-1 for invasion of gingival epithelial cells by Treponema medium. Can J Microbiol. 2007;53:1232–1238. doi: 10.1139/W07-094. [DOI] [PubMed] [Google Scholar]
- Tamai R, Asai Y, Ogawa T. Requirement for intercellular adhesion molecule 1 and caveolae in invasion of human oral epithelial cells by Porphyromonas gingivalis. Infect Immun. 2005;73:6290–6298. doi: 10.1128/IAI.73.10.6290-6298.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner A, Maiden MF, Macuch PJ, Murray LL, Kent RL., Jr Microbiota of health, gingivitis, and initial periodontitis. J Clin Periodontol. 1998;25:85–98. doi: 10.1111/j.1600-051x.1998.tb02414.x. [DOI] [PubMed] [Google Scholar]
- Tanner AC, Izard J. Tannerella forsythia, a periodontal pathogen entering the genomic era. Periodontology 2000. 2006;42:88–113. doi: 10.1111/j.1600-0757.2006.00184.x. [DOI] [PubMed] [Google Scholar]
- Tervahartiala T, Pirila E, Ceponis A, et al. The in vivo expression of the collagenolytic matrix metalloproteinases (MMP-2, -8, -13, and -14) and matrilysin (MMP-7) in adult and localized juvenile periodontitis. J Dent Res. 2000;79:1969–1977. doi: 10.1177/00220345000790120801. [DOI] [PubMed] [Google Scholar]
- Wang PL, Ohura K, Fujii T, et al. DNA microarray analysis of human gingival fibroblasts from healthy and inflammatory gingival tissues. Biochem Biophys Res Commun. 2003;305:970–973. doi: 10.1016/s0006-291x(03)00821-0. [DOI] [PubMed] [Google Scholar]
- Yoneda M, Yoshikane T, Motooka N, et al. Stimulation of growth of Porphyromonas gingivalis by cell extracts from Tannerella forsythia. J Periodontal Res. 2005;40:105–109. doi: 10.1111/j.1600-0765.2005.00774.x. [DOI] [PubMed] [Google Scholar]
- Yoo JY, Kim HC, Zhu W, et al. Identification of Tannerella forsythia antigens specifically expressed in patients with periodontal disease. FEMS Microbiol Lett. 2007;275:344–352. doi: 10.1111/j.1574-6968.2007.00906.x. [DOI] [PubMed] [Google Scholar]
- Zubery Y, Dunstan CR, Story BM, et al. Bone resorption caused by three periodontal pathogens in vivo in mice is mediated in part by prostaglandin. Infect Immun. 1998;66:4158–4162. doi: 10.1128/iai.66.9.4158-4162.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.