Abstract
Upon stimulation of mature B cells, class switch recombination (CSR) can alter the specific immunoglobulin heavy chain constant region that is expressed. In a tissue culture cell line, we previously demonstrated that inhibition of late SV40 factor (LSF) family members enhanced IgM to IgA CSR. Here, isotype specificity of CSR regulation by LSF family members is addressed in primary mouse splenic B cells. First, we demonstrate that LBP-1a is the prevalent family member in B lymphocytes. Second, we demonstrate by ChIP that LBP-1a binds genomic sequences around mouse switch regions (S) in an isotype-specific manner, in accordance with computational predictions: binding is observed to Sμ and Sα, but not to the tested Sγ1, regions. Importantly, binding of LBP-1a is tightly regulated, with occupancy at genomic S regions dramatically decreasing following LPS stimulation. Finally, the consequence of DNA-binding by LBP-1a is determined using bone marrow chimeric mice in which LSF/LBP-1 activity is inhibited in hematopoietic lineages. Upon in vitro stimulation of such primary B-cells, CSR occurs with a higher efficiency to IgA, but not to IgG1. These results are supportive of a model whereby LBP-1a represses CSR in an isotype-specific manner via direct interaction with switch regions involved in the recombination.
Keywords: B cells, Immunoglobulins, Molecular Biology, Recombinant Viral Vectors
Introduction
Class switch recombination (CSR) involves genomic DNA recombination in mature B cells that alters expression of the immunoglobulin heavy (H) chain constant region. The consequence is diversification of antibody effector function while preserving antigen specificity. The nonhomologous DNA recombination occurs within or near switch (S) regions, which consist of tandemly repeated DNA motifs located upstream of each H chain constant coding region. Antigens, bacterial products, and cytokines induce switching, with each distinct combination of signals generating expression of specific isotype(s) [1–3]. Extracellular signaling drives switching to a particular isotype by activating transcription from a promoter located 5’ of the respective S region. Such germline transcription is required to facilitate accessibility of S region DNA for CSR [4;5]. The extent to which the concurrent, enhanced chromatin accessibility in S regions promotes CSR is unclear [5].
Although molecular mechanisms for CSR have been extensively studied, little is known about non-transcriptional regulatory pathways, and in particular, means to prevent inappropriate CSR. As the predominant sites of DNA recombination [4], the S regions themselves provide intriguing regulatory targets. Indeed, several proteins specifically recognize S regions and activate CSR [1;6;7]. Of these, NF-κB/p50 can promote isotype-specific CSR without influencing germline transcription [8;9].
As we demonstrated previously, another class of transcription factors, the Late SV40 Factor (LSF) family members, binds switch region sequences in vitro [10]. In the I.29μ B cell line, which undergoes IgM to IgA isotype switching in vitro, LSF or other family members represses CSR without altering levels of germline transcripts. This provided the first instance of non-transcriptional repression of CSR by a specific DNA-binding protein [10]. The LSF family of transcription factors includes three paralogs, which share a DNA recognition motif, CTGG N6 CTT/GG. The two ubiquitous members of the family, LSF and LBP-1a/b (leader binding protein-1a/b), are 72% identical in aa sequence and may be functionally redundant in many tissues [11].
Here we report that LBP-1a is the dominant family member expressed in mouse primary B lymphocytes. A high density of binding sites is predicted only in particular S regions. Consistent with these predictions, in resting B lymphocytes, LBP-1a specifically binds to particular S regions (e.g. Sα) but not demonstrably to others (e.g. Sγ1). Notably, LBP-1a is released from bound S regions after LPS stimulation. Finally, inhibition of LBP-1a function leads to enhanced expression of only specific isotypes (IgA but not IgG1) when mouse B lymphocytes are stimulated to undergo CSR in vitro, correlating with binding of LBP-1a to the relevant S region.
Results and Discussion
LBP-1a is the predominant LSF/LBP-1 family member in B-cells
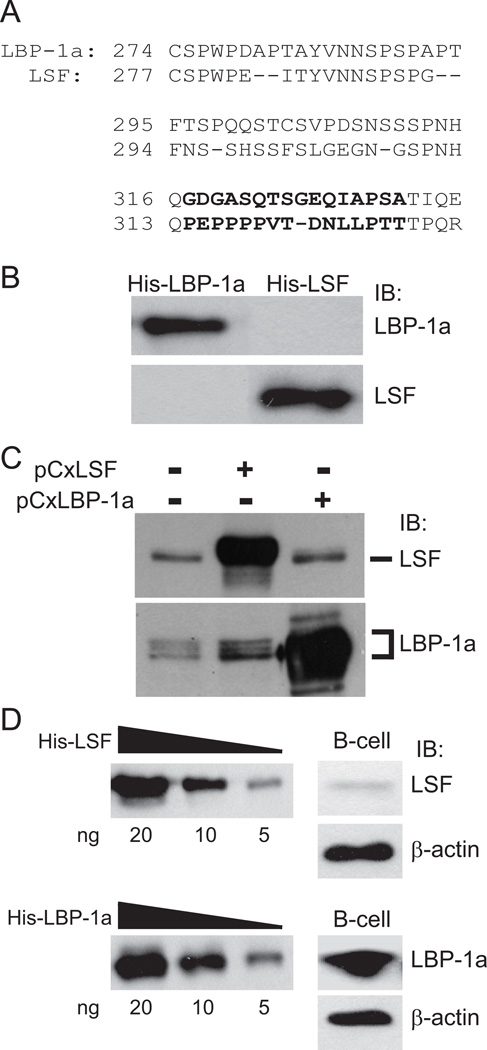
In order to distinguish LSF and LBP-1a, anti-peptide antibodies were produced against divergent regions (Fig. 1A). The antibody specificities and sensitivities were tested against recombinant proteins (Fig. 1B) and mammalian cell extracts in which each protein was independently overexpressed (Fig. 1C). In both instances, each antibody was highly specific for its cognate protein. The LBP-1a antibody recognizes multiple protein species from untransfected 293T extracts (Fig. 1C); due to alternative splicing, the cellular LBP-1a gene can encode two major products: LBP-1a and the larger LBP-1b [12]. LBP-1b is expressed in a tissue-specific manner [13], and is not evident in primary B cells (Fig. 1D).
Figure 1.
LBP-1a is the predominant LSF family member in primary B cells. A) Sequences of LBP-1a (aa 274–336) and LSF (aa 277–332) are aligned. Antipeptide antibodies were produced against nonconserved epitopes (bold). B) Ten ng of His-LBP-1a or His-LSF were subjected to SDS-PAGE and immunoblotted (IB) with the anti-peptide antibodies specific to LBP-1a or LSF, as indicated. C) Extracts (50 µg) from untransfected 293T cells, cells transfected with pCxLSF, or cells transfected with pCxLBP-1a were separated by SDS-PAGE through a 7.5% gel and immunoblotted (IB) with the antipeptide antibodies specific to LSF or LBP-1a, as indicated. D) Left panels: Decreasing levels of purified His-LSF or His-LBP-1a were immunoblotted with the respective antipeptide antibodies. Right panels: on the same gels, whole cell lysates from 20 × 106 splenic mouse B-cells in a single experiment were immunoblotted with antipeptide antibodies specific to either LSF (upper panels) or LBP-1a (lower panels), and subsequently with antibody against β-actin.
With these new reagents, we determined the relative abundance of LSF and LBP-1a in primary B-cells. By comparing the reactivity of each antibody against whole cell lysates from mouse splenic B-cells (Fig. 1D, right panels) to its reactivity against a standard curve of purified recombinant His-LSF or His-LBP-1a (Fig.1D, left panels), we concluded that primary mouse B cells contain at least 5-fold more LBP-1a (on the order of 104 molecules/cell) than LSF.
In primary, resting B-lymphocytes LBP-1a binds genomic DNA around particular S regions
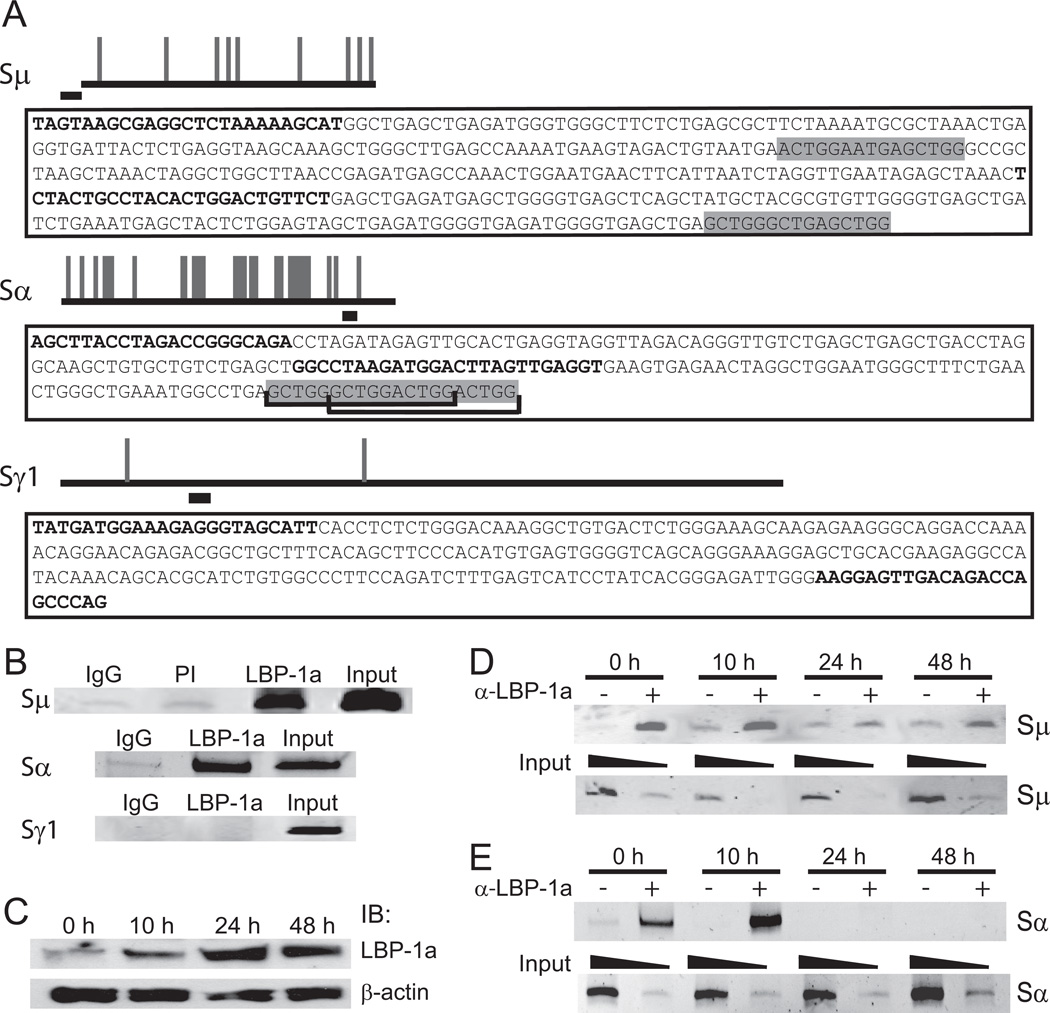
We previously demonstrated in vitro that recombinant LSF, as well as related proteins in mouse splenic cellular extracts, could bind specific sequences within Sμ, Sα, and Sε [10]. S regions can be clustered into two groups: Sμ, Sε, and Sα contain repeated pentameric sequences, whereas Sγ1, Sγ2a, Sγ2b, and Sγ3 contain 49–52 base-pair repeats [2;3]. We computationally analyzed the likelihood of LSF/LBP-1a binding sites within each S region. In some (Sμ, Sε, Sα, and Sγ3), numerous LSF/LBP-1a recognition sites were predicted; in others (Sγ1, Sγ2a, and Sγ2b), exceedingly few (Fig. 2A and Fig. S1A in supporting material). A similar pattern was evident when human S regions were analyzed (Fig. S2).
Figure 2.
LBP-1a binds to Sμ and Sα, but not Sγ1, regions in resting B cells and is released after LPS stimulation. A) Schematic of mouse immunoglobulin S region sequences with predicted LSF/LBP-1a binding sites. Long horizontal lines, to scale, represent each S region sequence; intersecting vertical lines represent predicted LSF/LBP-1a binding sites. Shorter horizontal lines below each S region represent regions amplified in ChIP assays. Amplicon sequences are in boxes below each S region schematic; PCR primers are in bold. Predicted LSF/LBP-1a binding sites within or flanking each amplicon are in shaded boxes. Brackets indicate overlapping sites. B) Isolated primary splenic murine B-lymphocytes were analyzed by ChIP with LBP-1a antiserum. Negative controls included non-specific rabbit IgG (IgG) and preimmune rabbit serum (PI). Input represents 1.25% of ChIP starting material. DNA were amplified with the primers in (A). Data are representative of at least 3 independent experiments. C) Whole cell extracts from 10 × 106 B-cells, stimulated with 50 µg/ml LPS for the indicated amounts of time, were immunoblotted (IB) with antibodies against LBP-1a or β-actin, as indicated. D–E) Resting B-cells were stimulated with 50 µg/ml LPS for the indicated times; LBP-1a occupancy at Sμ (D) and Sα (E) were analyzed as in (B). Upper panel: ChIP analysis with nonspecific rabbit IgG (−) or LBP-1a antiserum (+) at the indicated hours post-LPS stimulation. Lower panel: Amplication of input samples for each time point representing 0.125% (odd lanes) or 0.0125% (even lanes) of the ChIP starting material. Data are representative of 4 independent experiments.
We tested these predictions by ChIP assays in primary murine splenic B-lymphocytes, focusing on Sμ, which is operative in all CSR, and Sα and Sγ1, which are target switch regions of the two classes of repeats. Binding of LBP-1a, the more abundant LSF family member in B-cells (Fig. 1D), was monitored. Amplification of specific switch regions in the genomic DNA was analyzed by semi-quantitative PCR analysis (see Materials and Methods); the highly repetitive nature of these genomic sequences prevented design of real-time PCR primers. Binding in vivo was robust at both Sμ and Sα (Fig. 2B), where the amplicons were located at least 1.5 kb and 3.5 kb, respectively, downstream of transcriptional regulatory elements (the intronic promoters and the enhancer region), and either at the border of or well within S region sequences. However, as expected given the low density of predicted high affinity binding sites (roughly one per 3500 bp) LBP-1a binding was not detectable around Sγ1 in the region of the amplicon (Fig. 2B). Similarly, LBP-1a occupied Sε and Sγ3, the respective amplicons being at least 2.5 kb and 3.2 kb downstream of the intronic promoters, with minimal or no occupancy of Sγ2a and Sγ2b in the regions probed (Fig. S1B in supporting material). The experimental findings at all heavy chain gene loci are therefore consistent with predictions of LSF/LBP-1a binding sites within these S region sequences.
LPS stimulation of B-lymphocytes decreases binding of LBP-1a to S regions
Functionality of LBP-1a association with S regions in vivo was tested initially by assaying binding upon stimulation of primary B-lymphocytes. In previous studies, inherent DNA-binding activity of LSF/LBP-1a to S sites in mouse splenic extracts, assayed by EMSA, decreased after stimulation to undergo CSR [10]. Using the specific antibody (Fig. 1), levels of LBP-1a were measured in whole cell extracts of purified splenic B-lymphocytes stimulated by LPS for increasing amounts of time (Fig. 2C). Surprisingly, LBP-1a levels (and LSF protein levels; data not shown) substantially increased by 24 h after stimulation. In parallel LPS-stimulated cultures, we directly measured binding of LBP-1a to genomic S regions by ChIP. Occupancy at both Sμ (Fig. 2D) and Sα (Fig. 2E) was readily apparent at 0 and 10 h, but markedly decreased at 24 and 48 h post-stimulation. Thus, although protein levels increased at late time points after stimulation, binding decreased.
The decrease in DNA-binding with LPS stimulation is likely due to direct modification of LBP-1a. The closely related paralog, LSF, is directly targeted by mitogenic signal transduction pathways in multiple cell types, altering LSF DNA-binding potential, in part through modification of S291 [14;15]. LBP-1a contains an analogous serine; we hypothesize that LBP-1a is targeted downstream of cytokine signaling cascades in resting B cells, being modified to reduce binding to S regions.
The kinetics of release of LBP-1a from S region sequences following stimulation of B cells are consistent with LBP-1a inhibiting early events in CSR. Induction of AID mRNA and some germline transcripts occurs by 12 to 24 h post-stimulation [16;17], with Iα sterile transcripts [18], formation of R-loops [19], and alterations in histone acetylation by 48 h post-stimulation [20]. The inverse correlation between binding of LBP-1a and activation steps of CSR suggests that LBP-1a is a repressor.
LBP-1a-mediated repression of CSR to IgA, but not IgG1, in murine splenic B-cells
In a B cell line capable of undergoing CSR only from Sμ to Sα in vitro, LSF/LBP-1a did indeed repress CSR [10]. However, the question remained as to whether, in primary B cells, CSR in general would be subject to such inhibition, since all CSR in IgM-expressing B cells involves the Sμ switch region with which LSF/LBP-1a interacts, or if CSR only at certain heavy chain gene loci would be inhibited. To distinguish between these possibilities, we generated bone marrow chimeric mice in which a dominant negative form of LSF (LSFdn) was expressed in donor hematopoietic cells. LSF family members oligomerize with each other and bind DNA as obligate tetramers [11]. Two aa substitutions in the DNA-binding region of LSFdn prevent DNA-binding [21]; due to oligomerization this is a dominant phenotype that blocks all LSF family members [11]. This decrease in DNA-binding of endogenous LSF family members by LSFdn expression has previously been demonstrated in multiple studies, including in a murine B cell line [10].
Bone marrow cells from donor mice were transduced in vitro with retrovirus capable of expressing both LSFdn and GFP. Transplantation of these cells into lethally irradiated mice repopulated the recipient hematopoietic system. The resulting transduced, GFP-expressing splenic B cells were isolated and analyzed for CSR upon stimulation in vitro. Two control B cell populations were used. First, non-transduced cells in the donor bone marrow gave rise to GFP-negative splenic B cells in the same mouse, providing an ideal internal control for the GFP-positive, LSFdn-expressing cells (see Fig. S3 in supporting material for demonstration of GFP-negative versus GFP-positive populations). Second, other recipient mice were transplanted with donor bone marrow transduced with parental retrovirus, capable of expressing only GFP, in order to control for the possibility that transduction with any retrovirus would alter the efficiency of CSR.
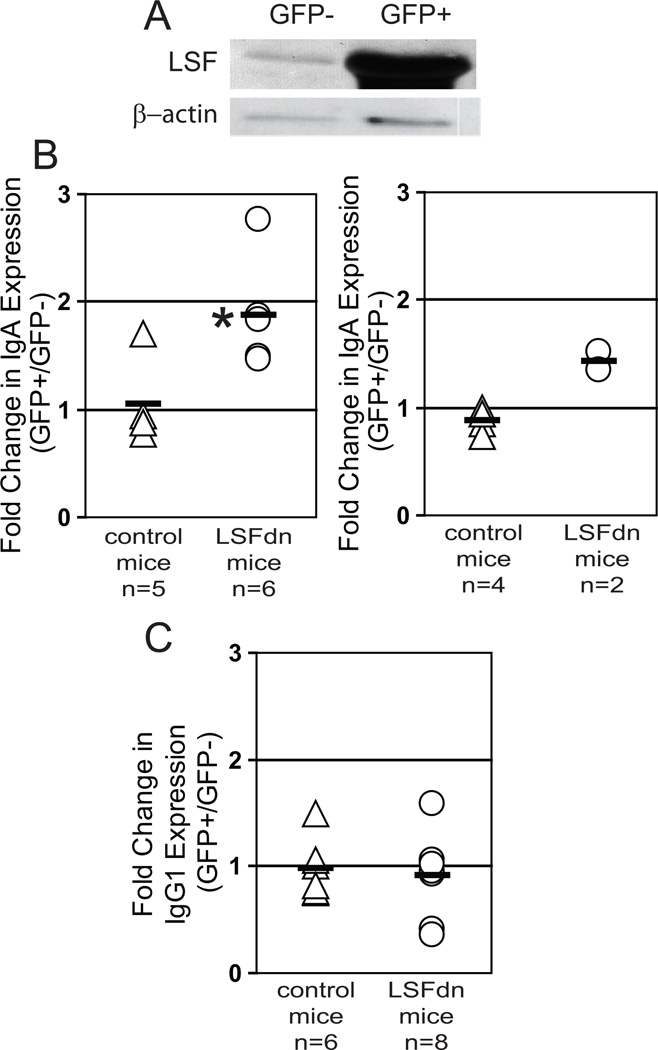
Expression of LSFdn was first validated in the LSFdn group of mice. Isolated GFP-positive and GFP-negative populations of primary splenic B-cells were harvested and immunoblotted with antibody specific to LSF. Endogenous murine LSF was detected in GFP- cells, whereas at least 50-fold higher levels of exogenous human LSFdn were detected in GFP+ cells from the same mouse (Fig. 3A). Considering relative expression levels of LBP-1a and LSF (Fig. 1D), LSFdn expression was also at least 10-fold higher than that of LBP-1a. Even equimolar concentrations, both in vitro and in vivo, are sufficient to dramatically suppress wild type DNA-binding of LSF family members [21;22].
Figure 3.
LSFdn expression in B-cells increases switching to IgA, but not to IgG1, upon induction of CSR. A) Isolated B-cells (5 × 106) from LSFdn chimeric mice were sorted into transduced (GFP+) and non-transduced (GFP−) populations based on GFP levels. Cell extracts from a single experiment were immunoblotted for LSF/LSFdn, using the LSF antipeptide antibody (Fig. 1), and for β-actin. B) Efficiency of IgM to IgA switching was determined for splenic B cells isolated from two groups of mice. Control mice (each indicated by a triangle) received bone marrow transduced with parental MSCV retrovirus. LSFdn mice (each indicated by a circle) received bone marrow transduced with MSCV-LSFdn retrovirus. Following stimulation in culture, splenic B cells were simultaneously analyzed for GFP and surface IgA expression. GFP expression levels separated transduced versus nontransduced populations. The fold change in IgA expression upon LSFdn expression, calculated as the ratio of the percentages of transduced cells vs. nontransduced cells that are IgA-positive, is plotted on the y-axis. Left panel: Cells were stimulated with LPS, IL-5, and TGF-β1 (control: n = 5; LSFdn: n = 6). Means (horizontal lines) and SEM were: 1.05 ± 0.17 for control mice, 1.88 ± 0.19 for LSFdn mice. *p=0.0048 (Student’s t-test). Right panel: Cells were stimulated with LPS and IL-4 (control: n = 4; LSFdn: n = 2) Means (horizontal lines) and SEM were: 0.88 ± 0.06 for control mice, 1.43 ± 0.09 for LSFdn mice. The probability that the values in these two data sets would be ordered by chance in this manner is 0.067 (Mann-Whitney U test). C) Splenic B cells derived from control and LSFdn mice characterized in (B) were analyzed for isotype switching to IgG1. Cells were stimulated with LPS and IL-4 (control: n = 6; LSFdn: n = 8). Cells were analyzed for GFP and IgG1 expression, and the fold change in IgG1 expression upon LSFdn expression calculated analogously to above (B). The means and SEM for the ratio of percentage IgG1-positive transduced cells to percentage IgG1-positive non-transduced cells were: 0.98 ± 0.11 for control mice and 0.92 ± 0.14 for LSFdn mice. The difference in means is not statistically significant.
To determine whether this expression of LSFdn affects the frequency of CSR, purified splenic B-cells from both chimeric LSFdn mice and chimeric control mice were stimulated to undergo CSR in vitro. Unlike results in the I.29μ B cell line, in the absence of stimulation, there was no detectable IgA or IgG1 expression in B cells isolated from LSFdn mice (data not shown). That alleviation of repression by LBP-1a is not sufficient to induce CSR in primary resting B cells in the absence of signaling is consistent with extensive analyses indicating that specific activation signals are absolutely required.
Due to the differences in density of LBP-1a occupancy at switch regions Sα vs. Sγ1 (Fig. 2), and to the relatively high efficiency of switching that can be induced in vitro to these two regions, we compared switching to IgA with that to IgG1 [2]. Cells were gated on the basis of GFP expression and surface levels of IgG1 and IgA expression after stimulation were monitored by flow cytometry (see Fig. S3 in supporting material for example). In initial experiments, comparison of the degree of CSR in BALB/cByJ B cells to that in the chimeric B cells indicated that CSR in the chimeric cells was less robust (approximately one fourth the levels of CSR), suggesting that bone marrow transplantation compromises the degree of CSR achievable in primary B cells in culture. We note that the depression in frequency of CSR in chimeric mouse B cells unfortunately precluded investigation of the inherently less efficient switching in vitro to isotypes other than IgA and IgG1.
By normalizing the percentage of transduced cells undergoing CSR to that of nontransduced cells in the same B cell population from the same mouse, we could most accurately determine the effect of LSFdn expression on CSR. The ratios for each individual mouse are plotted for expression of IgA and IgG1 (Fig. 3B and C, respectively), compiled from multiple independent experiments. These experiments included activation of B cells using multiple protocols; the effect of LSFdn on IgA CSR induced either by LPS, IL-5 and TGF-β1 (left panel) or by LPS and IL- 4 (right panel) are shown in Fig. 3B. Importantly, for chimeric control mice, splenic B cells switched expression from IgM to either IgG1 or IgA at the same frequencies, irrespective of whether or not they expressed GFP, resulting in a ratio for each isotype of 1.0. For the chimeric LSFdn-expressing mice, expression of IgG1 was also unaltered in the transduced cells (ratio of 0.94, Fig. 3C). In stark contrast, the percentage of LSFdn-expressing cells presenting surface IgA was significantly elevated (1.8-fold, P<0.005) compared to their non-transduced counterparts (Fig. 3B, left panel). A distinct cytokine cocktail appeared to result in a similar derepression of IgA expression by LSFdn (1.6-fold, P=0.067; Fig. 3B, right panel). These data demonstrate that LSF family members do not downregulate CSR in general, but only to certain isotypes (e.g. IgA).
How LBP-1a reduces CSR requires further investigation. Effects of LBP-1a, or other LSF family members, on CSR could potentially be either indirect or direct. Indirect effects might result, for instance, from LBP-1a affecting the degree of cell cycling. However, we view this possibility as unlikely for the following reasons. First, in a B cell line capable of undergoing CSR, expression of LSFdn, although similarly affecting CSR [10], did not detectably alter cell growth properties (data not shown). Second, since induction of IgG1 and of IgA in primary B cells both require multiple cell divisions [23;24], it is difficult to conceive of how indirect effects of LBP-1a would impact switching to one, but not to the other. Our combined findings that LBP-1a binds Sμ and Sα, but apparently not Sγ1, and that inhibition of LSF/LBP-1a increases levels of immunoglobulin class switching to IgA, but not to IgG1, instead suggest that LBP-1a inhibits CSR directly, as a consequence of LBP-1a binding S region sequences upstream of the respective heavy chain coding region.
Direct regulation of CSR by LBP-1a could result from either transcriptional or non-transcriptional mechanisms. Regulation of germline transcription by LBP-1a is unlikely, given that LSFdn did not alter the levels of germline transcripts in a B cell line in which it regulated CSR [10], and that the demonstrated binding of LBP-1a at the heavy chain loci is distant from the transcriptional regulatory regions. Alternative mechanisms for regulation of CSR by LBP-1a include, although are not limited to, inhibition of S-S synapsis, counteracting AID deamination or strand breakage, or reduction in chromatin accessibility. With regards to the latter, activation of CSR correlates with histone acetylation in S regions [5;25]. We note that LBP-1a may have the capability of inhibiting chromatin accessibility, in that the highly similar LSF interacts with multiple inhibitory chromatin modifying factors including histone deacetylases, Sin3A corepressor and the polycomb protein RING [11].
Concluding Remarks
The roles of specific switch region sequences in regulation of CSR remain to be fully elucidated, especially since the specific primary sequences of S regions appear unessential for the basic mechanism of CSR [4]. We hypothesize that specific repeat sequences mediate differential regulation of CSR, by providing specific binding sites for isotype-specific activators (e.g. NF-κB), and isotype-specific repressors (e.g. LBP-1a) to finetune appropriate CSR in response to the complex signaling to B-lymphocytes in vivo.
Materials and Methods
Recombinant protein purification
6X His-tagged LSF and LBP-1a were isolated using Ni-NTA nickel charged resin from bacteria transformed with pQE30LSF and pQE30LBP-1a, respectively (see supporting material for plasmid constructions). The concentrations of each recombinant protein were determined against a standard curve of purified BSA by SDS-PAGE, staining with Commassie-G250, densitometry, and ImageQuant analysis (version 1.2).
Antibody production and immunoblotting
Peptides corresponding to aa 317–332 of LBP-1a and 314–328 of LSF (bolded, Fig. 1A) were coupled to keyhole limpet hemocyanin. Antisera were produced in rabbits (Covance Research Products Inc.). Antibodies were purified using a HiTrap protein G column followed by affinity purification against peptide immobilized to SulfoLink coupling gel.
Purified B cells (5–20 × 106) were lysed in 2% SDS, 50 mM Tris HCl, pH 8.0, 0.1 mM DTT, 10% glycerol, subjected to SDS-PAGE, and immunoblotted with affinity-purified antibodies, followed by anti-β-actin (Sigma). 293T cells were maintained in DMEM plus 10% fetal bovine serum. Cells were transfected using lipofectamine (Invitrogen) and a total of 8 µg DNA, including pCXLBP-1a and pCXLBP-1c (LSF) [12] as indicated, per 10 cm dish. Nuclear extracts were prepared as described previously [26] prior to immunoblotting.
Bone marrow transplantation
Male BALB/cByJ mice (6–8 weeks) from The Jackson Laboratory were handled in accordance with the National Institutes of Health and institutional guidelines. Five days prior to bone marrow harvest from femurs, donor mice were injected i.p. with 5 mg of 5-fluorouracil. Marrow was subjected to density separation using Lympholyte-M and cultured in DMEM supplemented with 10% fetal bovine serum, 2 mM L-glutamine, antibiotics, 100 ng/ml SCF, 100 ng/ml flt-3 ligand and 50 ng/ml Thrombopoetin (R and D Systems) for 24 h prior to retroviral transduction.
Retroviral packaging cells were maintained in DMEM plus 10% fetal bovine serum. Following transfection with pCL-Eco [27] plus either MIGW [28] or MIGW-LSFdn (see supporting material for construction) using Fugene (Roche), viral supernatants were collected. Bone marrow cells were transduced 3 times for 24 h each with 80% viral supernatant, 20% bone marrow medium and 12 µg/ml polybrene.
Recipient mice were irradiated with 900 rads of gamma rays; 3 h later bone marrow from 1 donor (0.5 × 106 cells) was injected into the tail vein of one recipient mouse.
B cell isolation
Splenic B cells were isolated 8–10 wks after bone marrow transplantation, using negative selection [29]. Briefly, splenocytes were depleted of T cells using anti-Thy1.2 and rabbit complement. B-cells were isolated by density gradient centrifugation and cultured in RPMI plus 10% fetal bovine serum, 2 mM L-glutamine, 50 µM 2-ME, and antibiotics. The B cells were phenotyped as containing fewer than 1% T cells (B220-CD3+) or macrophages (B220-CD14+). For chimeric mice, cells were sorted by FACS into GFP-expressing and non-GFP-expressing populations. The gate for non-GFP expressing cells was set at minimal fluorescence, to eliminate contamination with cells expressing GFP at low levels. The gate for GFP-expressing cells was set above levels of maximal background fluorescence from BALB/cByJ splenocytes.
LBP-1a ChIP
LSF/LBP-1 binding sites were predicted with POSSUM (http://zlab.bu.edu/~mfrith/possum; score threshold 7, residue abundance range 1000, pseudocount 1) using the LSF binding matrix (accession number M00947) from TRANSFAC and mouse S region accession numbers Sμ: J00440-J00442, Sα: D11468, Sγ1: D78344.
ChIP assays were performed according to the Upstate Cell Signaling Solutions protocol. Briefly, 20 × 106 isolated, splenic B-cells were crosslinked in 1% formaldehyde for 10 min at 37°C, and quenched with 125 mM glycine. Cell lysates were sonicated to shear chromatin to around 500 bp; DNA was extracted from a fraction of the lysate for input controls. Precleared lysates were immunoprecipitated using 100 µl preimmune or immune rabbit serum or 5.0 µg normal rabbit IgG (Southern Biotech). Input and immunoprecipitated DNA were assayed for switch region sequences by PCR. Through amplification of increasing amounts of input DNA (e.g. Fig. 2D, 2E), a range of signals could be ascertained that was monotonically increasing with respect to the amount of input. In this manner, saturation of signals was avoided, and the results are semi-quantitative. Primer sequences included: Sμ as previously described [30]; Sα: AGCTTACCTAGACCGGGCAGAC and ACCTCAACTAAGTCCATCTTAGGCC; Sγ1: TATGATGGAAAGAGGGTAGCATT and CTGGGCTGGTCTGTCAACTCCTT.
CSR analysis
Isolated splenic B-cells were stimulated either 4 or 7 days with 25 µg/ml LPS and 25 ng/ml IL- 4 to assay IgG1 or IgA expression or with 25 µg/ml LPS, 25 ng/ml IL-5 and 1 ng/ml TGF-β1 to assay IgA expression. Surface IgG1 and IgA were measured by flow cytometry, using biotinylated IgG1 or IgA antibodies (1:500 dilution) and allophycocyanin-conjugated streptavidin (1:2000 dilution) in the presence of 2.4G2 anti-FcR antibody (BD Biosciences). The GFP-positive gate was set above the fluorescence levels of non-GFP-transduced B cells.. IgA- or IgG-positive gates were set above fluorescence levels of B-cells from chimeric mice stained either with biotinylated isotype control antibody (553923-BD Biosciences) followed by allophycocyanin-conjugated streptavidin, or allophycocyanin-conjugated streptavidin alone. Analyses were performed using FlowJo software. In calculating percentage of cells undergoing CSR, the percentage of IgA+ (or IgG1+) cells in the experimental sample were first adjusted by the percentage of IgA+ (or IgG1+) cells in the parallel isotype control analysis of the same cells. Samples in which the isotype control percentages were high relative to the experimental percentages (e.g. LSFdn mice, Fig. S3B in supporting material) were removed from consideration.
Supplementary Material
Acknowledgments
This work was supported by NIH R01 CA081157 to U.H., and NIH R01 AI29690 and NIH P01 AI60896 to T.L.R. K.J.R. was supported in part by T32 HL07501. We thank Quan Zhu for pQE30LSF, Robert Roeder for LBP-1a and LSF expression constructs, Luk Van Parijs for retroviral constructs, and Chun-Yan Bai and Essi Vulli for technical assistance.
Abbreviations
- AID
activation-induced deaminase
- CSR
class switch recombination
- LSF
Late SV40 Factor
- LBP-1a
Leader Binding Protein-1a
- S
switch region
Footnotes
Conflicts of Interest: The authors declare no financial or commercial conflict of interest.
References
- 1.Stavnezer J. Molecular processes that regulate class switching. Curr.Top.Microbiol.Immunol. 2000;245:127–168. doi: 10.1007/978-3-642-59641-4_6. [DOI] [PubMed] [Google Scholar]
- 2.Chaudhuri J, Alt FW. Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nat.Rev.Immunol. 2004;4:541–552. doi: 10.1038/nri1395. [DOI] [PubMed] [Google Scholar]
- 3.Pan-Hammarström Q, Zhao Y, Hammarström L. Class switch recombination: a comparison between mouse and human. Adv.Immunol. 2007;93:1–61. doi: 10.1016/S0065-2776(06)93001-6. [DOI] [PubMed] [Google Scholar]
- 4.Chaudhuri J, Basu U, Zarrin A, Yan C, Franco S, Perlot T, Vuong B, Wang J, Phan RT, Datta A, Manis J, Alt FW. Evolution of the immunoglobulin heavy chain class switch recombination mechanism. Adv.Immunol. 2007;94:157–214. doi: 10.1016/S0065-2776(06)94006-1. [DOI] [PubMed] [Google Scholar]
- 5.Yang SY, Schatz DG. Targeting of AID-mediated sequence diversification by cis-acting determinants. Adv.Immunol. 2007;94:109–125. doi: 10.1016/S0065-2776(06)94004-8. [DOI] [PubMed] [Google Scholar]
- 6.Kenter AL. Class switch recombination: an emerging mechanism. Curr.Top.Microbiol.Immunol. 2005;290:171–199. doi: 10.1007/3-540-26363-2_8. [DOI] [PubMed] [Google Scholar]
- 7.Honjo T, Kinoshita K, Muramatsu M. Molecular mechanism of class switch recombination: linkage with somatic hypermutation. Annu Rev Immunol. 2002;20:165–196. doi: 10.1146/annurev.immunol.20.090501.112049. [DOI] [PubMed] [Google Scholar]
- 8.Kenter AL, Wuerffel R, Dominguez C, Shanmugam A, Zhang H. Mapping of a functional recombination motif that defines isotype specificity for μ-->γ3 switch recombination implicates NF-κB p50 as the isotype-specific switching factor. J Exp.Med. 2004;199:617–627. doi: 10.1084/jem.20031935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang L, Wuerffel R, Kenter AL. NF-κB binds to the immunoglobulin Sγ3 region in vivo during class switch recombination. Eur.J Immunol. 2006;36:3315–3323. doi: 10.1002/eji.200636294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drouin EE, Schrader CE, Stavnezer J, Hansen U. The ubiquitously expressed DNA-binding protein Late SV40 Factor binds Ig switch regions and represses class switching to IgA. J Immunol. 2002;168:2847–2856. doi: 10.4049/jimmunol.168.6.2847. [DOI] [PubMed] [Google Scholar]
- 11.Veljkovic J, Hansen U. Lineage-specific and ubiquitous biological roles of the mammalian transcription factor LSF. Gene. 2004;343:23–40. doi: 10.1016/j.gene.2004.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoon J-B, Li G, Roeder RG. Characterization of a family of related cellular transcription factors which can modulate human immunodeficiency virus type 1 transcription in vitro. Mol Cell Biol. 1994;14:1776–1785. doi: 10.1128/mcb.14.3.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang HC, Chae JH, Lee YH, Park M-A, Shin JH, Kim S-H, Ye S-K, Cho YS, Fiering S, Kim CG. Erythroid cell-specific α-globin gene regulation by the CP2 transcription factor family. Mol Cell Biol. 2005;25:6005–6020. doi: 10.1128/MCB.25.14.6005-6020.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Volker JL, Rameh LE, Zhu Q, DeCaprio J, Hansen U. Mitogenic stimulation of resting T cells causes rapid phosphorylation of the transcription factor LSF and increased DNA-binding activity. Genes Dev. 1997;11:1435–1446. [Google Scholar]
- 15.Ylisastigui L, Kaur R, Johnson H, Volker J, He G, Hansen U, Margolis D. Mitogen-activated protein kinases regulate LSF occupancy at the human immunodeficiency virus type 1 promoter. J Virol. 2005;79:5952–5962. doi: 10.1128/JVI.79.10.5952-5962.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muramatsu M, Sankaranand VS, Anant S, Sugai M, Kinoshita K, Davidson NO, Honjo T. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J Biol.Chem. 1999;274:18470–18476. doi: 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]
- 17.Berton MT, Uhr JW, Vitetta ES. Synthesis of germ-line γ1 immunoglobulin heavy-chain transcripts in resting B cells: induction by interleukin 4 and inhibition by interferon γ. Proc Natl Acad Sci U.S.A. 1989;86:2829–2833. doi: 10.1073/pnas.86.8.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lebman DA, Lee FD, Coffman RL. Mechanism for transforming growth factor β and IL-2 enhancement of IgA expression in lipopolysaccharide-stimulated B cell cultures. J Immunol. 1990;144:952–959. [PubMed] [Google Scholar]
- 19.Yu K, Chedin F, Hsieh C-L, Wilson TE, Lieber MR. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat.Immunol. 2003;4:442–451. doi: 10.1038/ni919. [DOI] [PubMed] [Google Scholar]
- 20.Li Z, Luo Z, Scharff MD. Differential regulation of histone acetylation and generation of mutations in switch regions is associated with Ig class switching. Proc Natl Acad Sci U.S.A. 2004;101:15428–15433. doi: 10.1073/pnas.0406827101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shirra MK, Zhu Q, Huang H-C, Pallas D, Hansen U. One exon of the human LSF gene includes conserved regions involved in novel DNA-binding and dimerization motifs. Mol Cell Biol. 1994;14:5076–5087. doi: 10.1128/mcb.14.8.5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Powell CMH, Rudge TL, Zhu Q, Johnson LF, Hansen U. Inhibition of the mammalian transcription factor LSF induces S-phase-dependent apoptosis by downregulating thymidylate synthase expression. EMBO J. 2000;19:4665–4675. doi: 10.1093/emboj/19.17.4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hodgkin PD, HLee J-H, Lyons AB. B cell differentiation and isotype switching is related to division cycle number. J Exp.Med. 1996;184:277–281. doi: 10.1084/jem.184.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deenick EK, Hasbold J, Hodgkin PD. Switching to IgG3, IgG2b, and IgA is division linked and independent, revealing a stochastic framework for describing differentiation. J Immunol. 1999;163:4707–4714. [PubMed] [Google Scholar]
- 25.Kaminski DA, Stavnezer J. Stimuli that enhance IgA class switching increase histone 3 acetylation at Sα, but poorly stimulate sequential switching from IgG2b. Eur.J Immunol. 2007;37:240–251. doi: 10.1002/eji.200636645. [DOI] [PubMed] [Google Scholar]
- 26.Andrews NC, Faller DV. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 1991;19:2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naviaux RK, Costanzi E, Haas M, Verma IM. The pCL vector system: rapid production of helper-free, high-titer, recombinant retroviruses. J Virol. 1996;70:5701–5705. doi: 10.1128/jvi.70.8.5701-5705.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cherry SR, Biniszkiewicz D, van Parijs L, Baltimore D, Jaenisch R. Retroviral expression in embryonic stem cells and hematopoietic stem cells. Mol Cell Biol. 2000;20:7419–7426. doi: 10.1128/mcb.20.20.7419-7426.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rothstein TL, Wang JK, Panka DJ, Foote LC, Wang Z, Stanger B, Cui H, Ju ST, Marshak-Rothstein A. Protection against Fas-dependent Th1-mediated apoptosis by antigen receptor engagement in B cells. Nature. 1995;374:163–165. doi: 10.1038/374163a0. [DOI] [PubMed] [Google Scholar]
- 30.Nambu Y, Sugai M, Gonda H, Lee C-G, Katakai T, Agata Y, Yokota Y, Shimizu A. Transcription-coupled events associating with immunoglobulin switch region chromatin. Science. 2003;302:2137–2140. doi: 10.1126/science.1092481. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.