Sir, We read with interest the paper by Taioli et al. (2011) reporting eight different micromutations of PMP22 in patients with inherited demyelinating neuropathies. Small frameshift insertions/deletions (indels), nonsense nucleotide substitutions and splice-site mutations led to phenotypically and pathologically variable neuropathies. Patients with mutations causing a premature or delayed stop codon had a phenotype within the hereditary neuropathy with liability to pressure palsies (HNPP) spectrum. One patient with a splice-site mutation causing an in-frame skipping of exon 4 had a CMT1 phenotype. As the authors state, micromutations of PMP22 are considered very rare causes of Charcot–Marie–Tooth (CMT) in most populations and this certainly includes our experience of PMP22 analysis in cases with CMT from the UK. We presently report three novel PMP22 micromutations identified in a Greek cohort of patients with CMT that were associated with variable clinical phenotypes, ranging from severe CMT1 of the Dejerine–Sottas disease spectrum to HNPP. These novel cases, taken together with the findings of Taioli et al. (2011) illustrate that, despite some exceptions, in-frame indels in PMP22 usually lead to CMT1 of varying severity, whereas frameshift changes more often cause HNPP.
DNA was extracted from peripheral blood following standard procedures. Patients were previously found negative for the common duplication/deletion in chromosome 17 and for mutations in GJB1 and MPZ. The four coding exons (exons 2–5) of PMP22 with adjacent intronic sequences were amplified by polymerase chain reaction and sequenced on an Applied Biosystems 3730XL genetic analyser. Fragment analysis was performed with GeneMapper® software after running fragments on the 3730XL with a LIZ500 standard. RNA was obtained from peripheral blood using the Qiagen/PreAnalytix™ blood RNA system. Complementary DNA was synthesized using the Applied Biosystems high capacity complementary DNA reverse transcription kit. A 391 bp fragment of complementary DNA spanning the regions of interest in exons 4 and 5 was amplified using primers 5′-CCTGTCGATCATCTTCAGCA-3′ (within exon 4) and 5′-GGGATTTTGGGCTAGCTCTT-3′ (within the 3′-UTR; an 18-bp oligonucleotide was tagged to this primer). Subjects gave written informed consent for the performance of molecular genetic studies. The novel variants found were screened for in 450 chromosomes from Greek controls.
The novel micromutations in PMP22 identified included one in-frame duplication, one in-frame deletion and one splice-site mutation. Phenotypes were variable both between different mutations and within family members sharing the same mutation. The in-frame changes led to CMT1 of varying severity. The splice-site mutation, assumed by conceptual translation to cause exon skipping and frameshift with a premature stop codon, led to HNPP. Clinical features and nerve conduction studies of the patients are summarized in the Tables 1 and 2.
Table 1.
Clinical features of patients carrying novel PMP22 micromutations
| Family/ patient | PMP22 mutation | Exon/ intron | Predicted change at protein level | CMT type | FH | Gender | Age (years) | Age at onset | History | Examination findings |
|---|---|---|---|---|---|---|---|---|---|---|
| 1/III-1 | c.328_348dup21 | Exon 5 | p.Val110_Ile116 dup | CMT1 | AD | Male | 14 | 18 months | Onset with gait unsteadiness and falls; difficulty walking, support for climbing stairs; progressive. | Reduced tone, absent reflexes, wasted limbs and weakness, upper and lower limb sensory signs, pes cavus, scoliosis |
| 1/II-4 | c.328_348dup21 | Exon 5 | p.Val110_Ile116 dup | CMT1 | AD | Female | 38 | Adolescence | Onset with mild gait unsteadiness; difficulty walking, unable to wear high-heeled shoes. | Absent reflexes, peripheral lower limb weakness, lower limb sensory signs, pes cavus |
| 2/II-6 | c.296_301delCTGGAA | Exon 4 | p.Thr99_Gly100 del | Severe CMT1 (DSD) | Sporadic | Male | 22 | Infancy | Onset with delayed walking, gait unsteady and frequent falls; never walked independently; uses bilateral lower limb orthotics. | Absent reflexes, upper and lower limb atrophy and weakness, upper and lower limb sensory signs, scoliosis |
| 3 | c.79-2A > G | Intron 2 | p.Gln27_ Asn59 del_Glu60AsnfsX10 | HNPP | Isolated | Female | 37 | Adolescence | Onset with ulnar sensory neuropathy; transient positional sensory symptoms along right ulnar and both peroneal nerves | Reduced reflexes, pes cavus |
AD = autosomal dominant; de = deletion; DSD = Dejerine–Sottas disease; dup = duplication; FH = family history; fs = frameshift; HNPP = hereditary neuropathy with liability to pressure palsies.
Table 2.
Nerve conduction studies
| Family/patient | Age at testing (years) | Motor |
Sensory |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ulnar nerve |
Median nerve |
Peroneal nerve |
Ulnar nerve |
Median nerve |
Sural nerve |
|||||||||||
| DML (ms) | CMAP (mV) | CV (m/s) | DML (ms) | CMAP (mV) | CV (m/s) | DML (ms) | CMAP (mV) | CV (m/s) | SNAP (μV) | CV (m/s) | SNAP (μV) | CV (m/s) | SNAP (μV) | CV (m/s) | ||
| 1/III-1 | 4 | 5.9 | 0.4 | 14.3 | NA | NA | NA | Abs | Abs | Abs | NA | NA | NA | NA | Abs | Abs |
| 1/II-4 | 27 | 8.8 | 2.8 | 7.1 | NA | NA | NA | Abs | Abs | Abs | NA | NA | Abs | Abs | Abs | Abs |
| 2/II-6 | 9 | 40.0a | 0.4a | 2.3a | NA | NA | NA | Abs | Abs | Abs | Abs | Abs | Abs | Abs | Abs | Abs |
| 3 | 37 | 3.9 | 10.0 | 42.0/37.0b | 4.6 | 13.0 | 45.4 | 7.4 | 1.0 | 28.0 | 6.0 | 32.0 | 7.0 | 36.0 | 7.0 | 37.0 |
a Recorded using needle electrodes.
b Below/above elbow.
Abs = absent response; CMAP = compound muscle action potential; CV = conduction velocity; DML = distal motor latency; NA = records not available; SNAP = sensory nerve action potential.
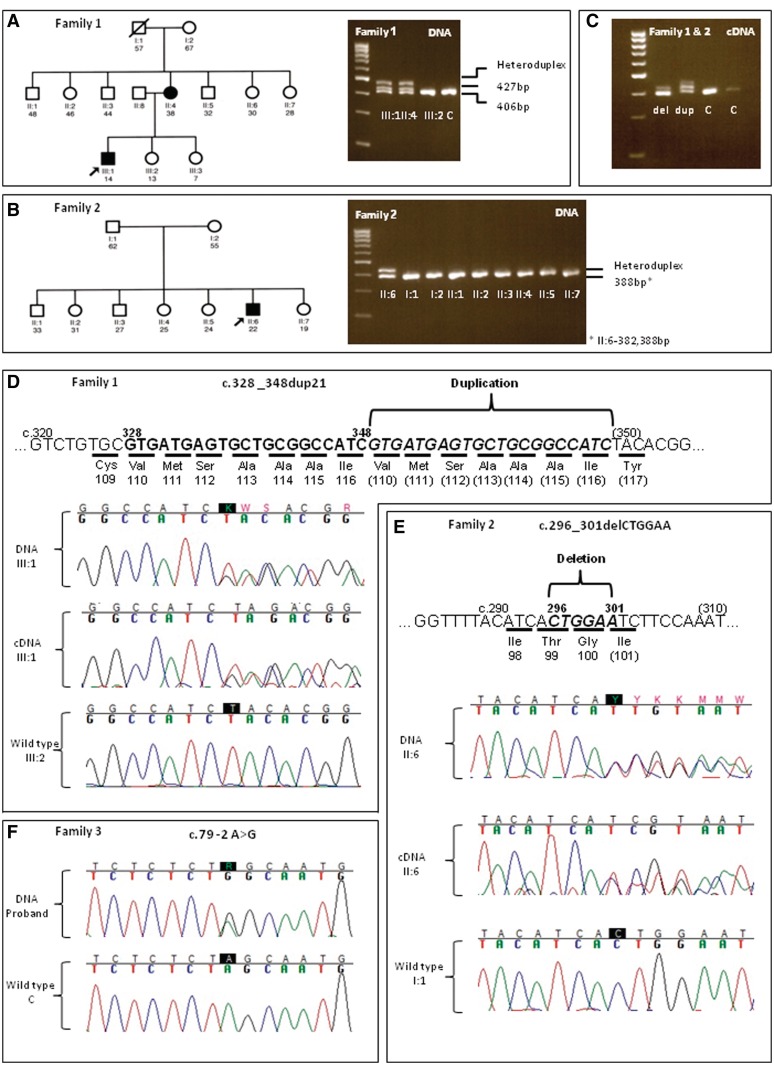
The first micromutation (Family 1) exhibiting autosomal dominant inheritance was an in-frame 21 bp duplication in exon 5 of PMP22 (c.328_348dup21, p.Val110_Ile116dup; Fig. 1D). Polymerase chain reaction amplification resulted in two fragments of 406 and 427 bp, which led to heteroduplex formation and three separate bands on agarose gel electrophoresis (Fig. 1A). Fluorescent polymerase chain reaction amplification and capillary electrophoresis confirmed that only two fragments of 406 and 427 bp were actually present (data not shown), further confirmed by sequencing. The mixed-base sequence caused by the duplication could be fully read in forward and reverse directions to distinguish the mutant and normal alleles (Fig. 1D). The sequencing was repeated in triplicate and using two different analysis software packages. Transcriptional analysis revealed the presence of two complementary DNA fragments of differing length, one wild-type and one bearing the duplication, which also resulted in heteroduplex formation (Fig. 1C), also confirmed by sequencing. The proband (III-1) was a 14-year-old male with CMT1. Disease onset was ∼18 months of age with gait unsteadiness and frequent falls. Mobility was moderately restricted throughout childhood and adolescence with gait disturbance and need of support for climbing stairs. Neurological examination revealed absent reflexes, lower and peripheral upper limb weakness and atrophy, upper and lower limb sensory signs, pes cavus and scoliosis. Nerve conduction studies showed severe conduction slowing (median motor conduction velocity 14.3 m/s). The mother of the proband (II-4) was also affected and carried the same mutation (Table 2). Her clinical phenotype was much milder compared with her son. She only had mild gait unsteadiness with onset in her teens. Neurological examination revealed absent reflexes, peripheral lower limb weakness, peripheral lower limb sensory signs and pes cavus. Despite the mild picture, she had severe slowing on nerve conduction studies with an ulnar motor conduction velocity of 7.1 m/s. Further details of the nerve conduction studies in the mother and son are shown in Table 2. An unaffected sister of the proband was free of the mutation (Fig. 1A), as were all control chromosomes. The remaining family members (Fig. 1A) were reportedly unaffected but DNA was not available for molecular studies. It is likely that the mutation arose de novo in the mother of the proband.
Figure 1.
Pedigrees, agarose gel electrophoresis and sequencing results of families with PMP22 micromutations. (A) Pedigree and DNA agarose gel electrophoresis of Family 1 with the c.328_348dup21 mutation; a third band visible in affected members is due to heteroduplex formation. (B) Pedigree and DNA agarose gel electrophoresis of Family 2 with the c.296_301delCTGGAA mutation; a second band visible in the proband is due to heteroduplex formation. (C) Complementary DNA (cDNA) agarose gel electrophoresis of probands from Families 1 and 2, as well as two controls; probands have extra bands due to heteroduplex formation. (D) Graphic representation of the c.328_348dup21 (p.Val110_116Iledup) mutation; DNA and complementary DNA sequencing results of the proband compared with the wild-type sequence. (E) Graphical representation of the c.296_301delCTGGAA (p.Thr99_Gly100del) mutation; DNA and complementary DNA sequencing results of the proband compared with the wild-type sequence. (F) DNA sequencing results of the c.79 -2A > G splice site mutation; proband compared with wild type sequence. del = deletion; dup = duplication.
The second micromutation (Family 2) was a sporadic in-frame 6-bp deletion in exon 4 of PMP22 (c.296_301delCTGGAA, p.Thr99_Gly100del; Fig. 1E). Polymerase chain reaction amplification resulted in two fragments of 388 and 382 bp, which led to heteroduplex formation and two separate bands on agarose gel electrophoresis (Fig. 1B). In this case, the 388 and 382-bp bands did not resolve well on the agarose gel, but fluorescent polymerase chain reaction showed the 6-bp deletion (data not shown), confirmed by sequencing. The mixed-base sequence caused by the deletion could be fully read in forward and reverse directions to distinguish the mutant and normal alleles (Fig. 1D). The sequencing was repeated in triplicate and analysed using two different software packages. Transcriptional analysis revealed the presence of two complementary DNA fragments of differing length, one wild-type and one bearing the deletion, which also resulted in heteroduplex formation (Fig. 1B), also confirmed by sequencing. The proband (II-6) was a 22-year-old male with severe CMT1 of the Dejerine–Sottas disease phenotype. Disease onset was in infancy with delayed walking, gait unsteadiness and frequent falls. During childhood and adolescence, mobility was severely restricted. Independent ambulation was never achieved and walking was only possible using orthotic devices and bilateral support. Neurological examination showed absent reflexes, upper and lower limb atrophy and weakness, upper and lower limb sensory dysfunction and scoliosis. Nerve conduction studies showed severe slowing with an ulnar motor conduction velocity of 2.3 m/s (Table 2). A sural nerve biopsy performed at 7 years of age showed severe reduction of myelinated fibres with complete loss of large and medium diameter fibres. Schwann cells were dispersed among the smaller diameter fibres with disproportionally thin myelin sheaths, suggestive of remyelination and consistent with a hypomyelinating neuropathy. This case was shown to be de novo dominant. No other family members were affected or carried the mutation (Fig. 1B). The variant was not found in control chromosomes.
The third micromutation (Family 3) was an isolated 3′ splice-site nucleotide substitution in the intronic sequence between exons 2 and 3 (c.79 -2 A > G; Fig. 1F). Conceptual translation predicted the skipping of exon 3 and a frameshift leading to a stop codon 10 amino acids downstream (p.Gln27_Asn59 del_Glu60AsnfsX10). RNA was not available for the transcriptional analysis. The proband was a 37-year-old female with HNPP and symptom onset in adolescence with an ulnar sensory neuropathy. Her clinical picture was mild. She had transient positional sensory symptoms along a right ulnar and a bilateral peroneal nerve distribution. Examination revealed reduced tendon reflexes and pes cavus. On nerve conduction study, she had median motor conduction velocity of 45.4 m/s, right ulnar motor conduction velocity of 37.0 and 42.0 m/s above and below the elbow, respectively, and peroneal motor conduction velocity of 28.0 m/s (Table 2). This was an isolated case. No other family members were reported affected but DNA was not available for molecular studies. The variant was not found in control chromosomes.
The first two novel PMP22 micromutations identified, the 21-bp duplication and the 6-bp deletion, cause in-frame changes in PMP22. As shown by transcriptional analysis, the mutant alleles are expressed in both cases. The patients had CMT1 of varying severity, ranging from mild CMT1 to severe CMT1 of the Dejerine–Sottas disease type. Among the micromutations reported by Taioli et al. (2011), the c.179 -2A > G splice-site mutation (Ekici et al., 2000; Park et al., 2006), which according to transcriptional analysis led to an in-frame deletion and a stable transcript, was the only one leading to a CMT1 rather than HNPP phenotype. Cases with in-frame deletions in PMP22 from previous reports have also led to a CMT1 phenotype (Silander et al., 1998; Mersiyanova et al., 2000; Sambuughin et al., 2003). The above taken together illustrate that in-frame indels in PMP22 usually lead to a CMT1 phenotype.
The two novel indels reported result in the duplication of amino acids 110–116 and the deletion of amino acids 99–100, respectively. In both cases, the amino acids lie in the third transmembrane domain of PMP22. The majority of PMP22 point mutations located in the second and third transmembrane domain of the protein cause a CMT1 phenotype (Posciotta et al., 2009). It is believed that a toxic gain of function leading to impaired intracellular trafficking and protein aggregation underlies the pathogenesis in such cases. Mutant PMP22 can also form heterodimers with the wild-type protein. Different mutations can differ in the degree of protein aggregation thus affecting the severity of the clinical phenotype (Houlden and Reilly, 2006). However, the clinical severity can also vary among individuals with the same mutation. Within the family with the 21-bp duplication, the severity of the neuropathy varied significantly, with the son having a relatively severe phenotype and the mother only mild CMT1. This remarkable intrafamilial variability that can be observed with PMP22 mutations was specifically noted by Taioli et al. (2011), and has also been commented on by other authors (Russo et al., 2011). It is further emphasized by the present report.
The third novel PMP22 micromutation identified was a c. 79 -2 A > G substitution in intron 2, disturbing the 3′ splice-site. This is expected to cause a deletion of exon 3 followed by a frameshift and a premature stop codon 10 amino acids downstream. It should result, therefore, in a severely truncated protein or most likely lead to degradation at the RNA level by nonsense-mediated messenger RNA decay, as expected by any frameshift mutation causing a premature stop codon located more than 55 bp from a downstream exon–intron boundary (Nagy and Maquat, 1998). Unfortunately, we did not have RNA to confirm this by transcriptional analysis. Such mutations, thought to result in haploinsufficiency, are usually associated with HNPP (Houlden and Reilly, 2006), as was the case in our patient. There are previously reported frameshift mutations in PMP22 causing CMT1 and even Dejerine–Sottas disease rather than HNPP (Ionasescu et al., 1997; Numakura et al., 2002; Choi et al., 2004). However, these mutations cause premature stop codons towards the end of the penultimate or within the last exon leading to messenger RNA that is expected to be immune to nonsense-mediated decay and therefore, unlikely to cause haploinsufficiency. Nevertheless, exceptions exist and frameshift mutations with similar characteristics have also been reported to lead to an HNPP phenotype (Taroni et al., 1995; Taioli et al., 2011). Taioli et al. (2011) also reported three frameshift mutations causing delayed stop codons and leading to HNPP. Although no transcriptional data were presented for these patients, the possibility of messenger RNA instability leading to haploinsufficiency was raised. However, previous cases with frameshift mutations causing delayed stopped codons and a CMT1 phenotype have also been reported (Ikegami et al., 1998; Lenssen et al., 1998). Therefore, although the stability of the messenger RNA transcript is likely to determine the emergence of a CMT1 or HNPP phenotype, it is not straightforward to predict this stability based purely on sequence data. It will be important in the future to analyse skin biopsies in patients with PMP22 mutations to quantify PMP22 expression in myelinated dermal fibres by real-time polymerase chain reaction and immunoelectron microscopy to confirm these results (Li, 2005).
In conclusion, the present report confirms the identification of PMP22 micromutations, which have a higher prevalence in the Greco-Roman population. Through the study of three novel variants, our report emphasizes the variability of phenotypes that can be associated with different PMP22 micromutations, as noted by Taioli et al. (2011), and taken together with the findings of that study illustrates that, despite some exceptions, in-frame indels usually lead to CMT1 of varying severity, whereas frameshift changes more often cause HNPP.
Funding
The Medical Research Council (MRC) (HH fellowship and project grant); The Wellcome Trust; The French Muscular Dystrophy Association (AFM); The Muscular Dystrophy Association; The National Organisation for Rare Disorders (NORD) and the Department of Health's National Institute for Heath Research (NIHR) Biomedical Research Centres (BRC) funding scheme to UCLH/UCL.
Acknowledgements
We are grateful to the patients and families for their help with this work.
References
- Choi BO, Lee MS, Shin SH, Hwang JH, Choi KG, Kim WK, et al. Mutational analysis of PMP22, MPZ, GJB1, EGR2 and NEFL in Korean Charcot-Marie-Tooth neuropathy patients. Hum Mutat. 2004;24:185–6. doi: 10.1002/humu.9261. [DOI] [PubMed] [Google Scholar]
- Ekici AB, Schweitzer D, Park O, Lorek D, Rautenstrauss B, Kruger G, et al. Charcot-Marie-Tooth disease and related peripheral neuropathies: novel mutations in the peripheral myelin genes connexin 32 (Cx32), peripheral myelin protein 22 (PMP22) and peripheral myelin protein zero (MPZ) Neurogenetics. 2000;3:49–50. doi: 10.1007/pl00022981. [DOI] [PubMed] [Google Scholar]
- Houlden H, Reilly MM. Molecular genetics of autosomal-dominant demyelinating Charcot-Marie-Tooth disease. Neuromolecular Med. 2006;8:43–62. doi: 10.1385/nmm:8:1-2:43. [DOI] [PubMed] [Google Scholar]
- Ikegami T, Ikeda H, Aoyama M, Matsuki T, Imota T, Fukuuchi Y, et al. Novel mutations of the peripheral myelin protein 22 gene in two pedigrees with Dejerine-Sottas disease. Hum Genet. 1998;102:294–8. doi: 10.1007/s004390050694. [DOI] [PubMed] [Google Scholar]
- Ionasescu VV, Searby CC, Ionasescu R, Reisin R, Ruggieri V, Arberas C. Severe Charcot-Marie-Tooth neuropathy type 1A with 1-base pair deletion and frameshift mutation in the peripheral myelin protein 22 gene. Muscle Nerve. 1997;20:1308–10. doi: 10.1002/(sici)1097-4598(199710)20:10<1308::aid-mus14>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Lenssen PP, Gabreëls-Festen AA, Valentijn LJ, Jongen PJ, van Beersum SE, van Engelen BG, et al. Hereditary neuropathy with liability to pressure palsies . Phenotypic differences between patients with the common deletion and a PMP22 frame shift mutation. Brain. 1998;121:1451–8. doi: 10.1093/brain/121.8.1451. [DOI] [PubMed] [Google Scholar]
- Li J, Bai Y, Ghandour K, Qin P, Grandis M, Trostinskaia A, et al. Skin biopsies in myelin-related neuropathies: bringing molecular pathology to the bedside. Brain. 2005;128(Pt 5):1168–77. doi: 10.1093/brain/awh483. [DOI] [PubMed] [Google Scholar]
- Mersiyanova IV, Ismailov SM, Polyakov AV, Fedotov VP, Nelis E, Löfgren A, et al. Screening for mutations in the peripheral myelin genes PMP22, MPZ and Cx32 (GJB1) in Russian Charcot-Marie-Tooth neuropathy patients. Hum Mutat. 2000;15:340–7. doi: 10.1002/(SICI)1098-1004(200004)15:4<340::AID-HUMU6>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Nagy E, Maquat LE. A rule for termination-codon position with intron-containing genes: when nonsense affects RNA abundance. Trends Biochem Sci. 1998;23:198–9. doi: 10.1016/s0968-0004(98)01208-0. [DOI] [PubMed] [Google Scholar]
- Numakura C, Lin C, Ikegami T, Guldberg P, Hayasaka K. Molecular analysis in Japanese patients with Charcot-Marie-Tooth disease: DGGE analysis for PMP22, MPZ,andCx32/GJB1 mutations. Hum Mutat. 2002;20:392–8. doi: 10.1002/humu.10134. [DOI] [PubMed] [Google Scholar]
- Park HK, Kim BJ, Sung DH, Ki CS, Kim JW. Mutation analysis of the PMP22, MPZ, EGR2, LITAF and GJB1 gene in Korean patients with Charcot-Marie-Tooth neuropathy type 1. Clin Genet. 2006;70:253–6. doi: 10.1111/j.1399-0004.2006.00669.x. [DOI] [PubMed] [Google Scholar]
- Pisciotta C, Manganelli F, Iodice R, Bellone E, Geroldi A, Volpi N, et al. Two families with novel PMP22 point mutations: genotype-phenotype correlation. J Peripher Nerv Syst. 2009;14:208–12. doi: 10.1111/j.1529-8027.2009.00235.x. [DOI] [PubMed] [Google Scholar]
- Russo M, Laurá M, Polke JM, Davis MB, Blake J, Brandner S, et al. Variable phenotypes are associated with PMP22 missense mutations. Neuromuscul Disord. 2011;21:106–14. doi: 10.1016/j.nmd.2010.11.011. [DOI] [PubMed] [Google Scholar]
- Sambuughin N, de Bantel A, McWilliams S, Sivakumar K. Deafness and CMT disease associated with a novel four amino acid deletion in the PMP22 gene. Neurology. 2003;60:506–8. doi: 10.1212/01.wnl.0000044048.27971.fc. [DOI] [PubMed] [Google Scholar]
- Silander K, Meretoja P, Juvonen V, Ignatius J, Pihko H, Saarinen A, et al. Spectrum of mutations in Finnish patients with Charcot-Marie-Tooth disease and related neuropathies. Hum Mutat. 1998;12:59–68. doi: 10.1002/(SICI)1098-1004(1998)12:1<59::AID-HUMU9>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Taioli F, Cabrini I, Cavallaro T, Acler M, Fabrizi GM. Inherited demyelinating neuropathies with micromutations of peripheral myelin protein 22 gene. Brain. 2011;134:608–17. doi: 10.1093/brain/awq374. [DOI] [PubMed] [Google Scholar]
- Taroni F, Botti S, Sghirlanzoni A, Botteon G, DiDonato S, Pareyson D. A nonsense mutation in the PMP22 gene in hereditary neuropathy with liability to pressure palsies (HNPP) not associated with the 17p11.2 deletion. Am J Hum Genet. 1995;57:A229. [Google Scholar]