Research in the past decade has uncovered a new class of inherited neurodegenerative diseases, the polyglutamine (polyQ) expansion diseases (1). In each, the underlying mutation is an expansion of a CAG trinucleotide repeat that encodes polyQ in the respective disease proteins (Table 1). All are progressive, ultimately fatal disorders that typically begin in adulthood and progress over 10 to 30 years. The clinical features and pattern of neuronal degeneration differ among the diseases, yet increasing evidence suggests that polyQ diseases share important pathogenic features. In particular, abnormal protein conformation(s) promoted by polyQ expansion seem to be central to pathogenesis (2). PolyQ diseases thus join a growing group of neurodegenerative disorders, including Alzheimer's disease and many other dementias, in which abnormal protein folding and aggregation are implicated.
Table 1.
Polyglutamine diseases
| Disease | Protein | CAG repeat size
|
Evidence for aggregation in disease brain | |
|---|---|---|---|---|
| Normal | Disease | |||
| HD | Huntingtin | 6–35 | 38–180 | NI, neuropil aggregates |
| DRPLA | Atrophin | 6–35 | 49–88 | NI |
| SBMA | Androgen receptor | 9–36 | 38–65 | NI |
| SCA1 | Ataxin-1 | 6–44* | 39–82 | NI |
| SCA2 | Ataxin-2 | 15–31 | 34–64 | Cytoplasmic accumulation |
| SCA3/MJD | Ataxin-3 | 12–40 | 60–84 | NI |
| SCA6 | Calcium channel | 4–18 | 21–33 | Cytoplasmic accumulation |
| SCA7 | Ataxin-7 | 4–35 | 34–306 | NI |
NI, nuclear inclusions; DRPLA, dentatorubral-pallidoluysian atrophy; SBMA, spinobulbar muscular atrophy; MJD, Machado–Joseph disease.
In SCA1, normal alleles >21 repeats are interrupted by 1–3 CAT units, whereas disease alleles are pure CAG repeats.
Disease Mechanisms: An Unfolding Story
In polyQ diseases, as in these other disorders, fundamental questions remain unanswered. For example, why are neurons selectively vulnerable even though the mutant proteins are widely expressed? Why do certain populations of neurons degenerate whereas other neurons that also express the mutant protein do not? Do intranuclear and cytoplasmic aggregates of polyQ protein—a common pathologic hallmark of disease—contribute to pathogenesis or are they simply by-products of the disease process? Finally, what are the molecular pathways leading from conformational changes to neuronal dysfunction and cell death? Answers to these questions are critical for the successful development of therapies to treat these devastating disorders.
The basis of disease appears to be a novel, dominant toxic property conferred on the mutant protein. The threshold for this toxic property is a glutamine repeat of approximately 35 to 40 residues with one notable exception, the shorter disease repeat in spinocerebellar ataxia type 6 (SCA6). Longer repeats cause more severe disease, suggesting that the degree of toxicity is directly proportional to polyQ length.
Evidence is most consistent with a model of disease in which expansion increases the probability that polyQ will adopt a novel abnormal conformation. Evidence supporting an altered conformation includes antibodies that preferentially bind expanded polyQ; in vitro studies showing that mutant polyQ self-associates into amyloid-like fibrils; and studies in disease tissue, transfected cells, and animal models demonstrating that expanded polyQ proteins form insoluble intracellular inclusions. The fact that molecular chaperones and proteasome components localize to these inclusions, and that some chaperones can modulate polyQ aggregation and toxicity, further argues that these are diseases of abnormal conformation (2–9).
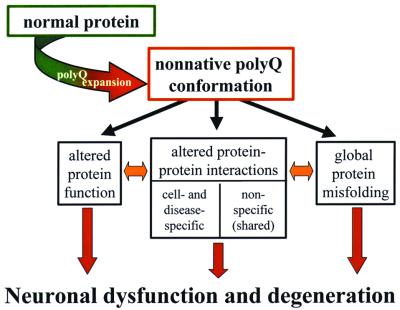
How this altered conformation leads to neurodegeneration is unclear. Fig. 1 illustrates several possible downstream consequences of a conformational change, some likely to be disease-specific and others shared among polyQ diseases. Among disease-specific effects, the intrinsic biologic activity of the protein could be altered by a conformational change in the polyQ domain (for example, expansions in the SCA6 protein may affect physiologic properties of this voltage-gated calcium channel). A second likely consequence is altered interactions of the mutant protein with its normal interacting partners and novel associations with still other proteins. Isolated, expanded polyQ fragments are intrinsically cytotoxic in a rather indiscriminant way, suggesting that the particular protein context of polyQ contributes to the selective vulnerability of certain neurons in each disease. Outside of polyQ, the disease proteins are otherwise dissimilar and thus certain changes in protein interactions will be unique to the individual disease proteins. But misfolded polyQ could also lead to novel or altered interactions with proteins that are common among the polyQ diseases. One intriguing group of candidate proteins are those that contain polyQ or glutamine-rich domains, because such domains are sufficient to recruit proteins into polyQ aggregates (10, 11). Mutant polyQ proteins might bind and even sequester glutamine-rich transcription factors, altering the expression of genes that are critical for neuronal function. A third possible consequence of polyQ expansion is global misfolding of the disease protein, resulting in disruption of normal protein homeostasis in neurons.
Figure 1.
Proposed model of early events in polyQ pathogenesis. Existing evidence suggests that CAG/polyQ expansion promotes the formation of a nonnative polyQ conformation. Shown are three major categories of downstream consequences that are not mutually exclusive and may be interdependent. How these or other consequences of a conformational change cause neuronal dysfunction and degeneration remains to be elucidated.
Although researchers are gaining insight into the molecular trigger of polyQ disease, the slow route to neuronal dysfunction and cell death is bound to be complex. A major challenge now is to identify which of the myriad changes occurring during chronic neurodegenerative disease contribute directly to pathogenesis and which are secondary phenomena. As described below, the use of genetically tractable animal models and the analysis of cell death pathways are two promising approaches.
Modeling Disease in Drosophila
To apply the power of genetics to the problem of polyQ neurodegeneration, these diseases have been recreated in the fruit fly Drosophila melanogaster (12, 13). Drosophila has many advantages for such study, including well advanced genetics and transgenic technologies. The rapid generation time of Drosophila is another benefit: Disease phenotypes that take years to develop in humans, and months in mice, can be modeled in days in the fly.
To take this approach, the human disease gene for one of the more common polyQ diseases, SCA3/Machado–Joseph disease, was introduced into the fly. The control and disease proteins had polyQ tracts of 27 and 78 residues, respectively. When expressed in Drosophila, the Q27 protein had no effect whereas the Q78 protein induced late-onset, progressive degeneration accompanied by the formation of nuclear inclusions (12). These studies demonstrate that the Drosophila model recapitulates fundamental characteristics of human polyQ disease with progressive neural degeneration and abnormal protein aggregation.
With such models in hand, Drosophila genetics now can be used to uncover mechanisms of degeneration and to define ways to slow neuronal loss. Toward this end, genetic screens can be performed to find genes that modulate neurodegeneration. In addition, candidate genes can be tested in the fly. For example, the human gene encoding Hsp70, a molecular chaperone that modulates protein folding, was introduced into Drosophila to ask whether raising Hsp70 levels can combat the toxic effects of polyQ. Indeed, coexpression of human Hsp70 ameliorated polyQ toxicity, suppressing degeneration (6). These studies, as well as genetic screens (7), indicate that manipulating specific molecular chaperones may be an effective therapeutic approach to neurodegenerative disease. In these ways, the fly can help pioneer new ways to understand and help prevent neurodegenerative disease in humans.
Mechanisms of Cell Death in Neurons
Neuronal cell loss characterizes many neurodegenerative diseases including polyQ diseases. The causal relationship between neuron loss and the onset of neurological dysfunction in polyQ disease is unclear, but the gradual and irreversible loss of specific neuronal subpopulations in Huntington's disease (HD) coincides with disease progression. Neuronal cell death is generally divided into two types, apoptotic and necrotic. Apoptotic cell death (programmed cell death) is a highly regulated process best characterized in Caenorhabditis elegans, where apoptotic execution is controlled by a linear molecular pathway involving the sequential actions of EGL-1, CED-9, CED-4, and CED-3 (14). Each of these molecules has multiple mammalian homologues. CED-3 is represented by Caspase-3 and 13 additional caspase family members, CED-4 by APAF-1 and several related molecules, CED-9 by antiapoptotic members of the Bcl-2 family including Bcl-XL and Bcl-2, and EGL-1 by proapoptotic Bcl-2 family members such as Bax and Bid (15).
Recent gene targeting studies have identified specific members of these families as key regulators of mammalian neuronal apoptosis, particularly during development. Bcl-XL is an important antiapoptotic factor and Bax, APAF-1, Caspase-9, and Caspase-3 are critical proapoptotic molecules in neurons (16). Apoptotic neuronal death typically occurs after activation of Caspase-3, an “effector” caspase. During nervous system development, Caspase-9, in concert with APAF-1, regulates Caspase-3 activation and neuronal apoptosis (16). At other developmental stages and in other cell types, Caspase-3 activation may be independent of Caspase-9 and may result from enzymatic cleavage by other upstream caspases such as Caspase-8. Interestingly, Caspase-8 activity has been reported to be increased in HD brain extracts, and both Caspase-8 and Caspase-1 have been implicated in cellular and animal models of polyglutamine-mediated neuronal degeneration (17, 18). These studies suggest that caspases play a role in the neuronal loss observed in HD, however the primacy of caspase activation to HD and other polyQ diseases is still uncertain.
Recent studies indicate that caspase-independent neuronal death pathways also exist. Autophagic cell death and neuronal death secondary to mitochondrial dysfunction can occur independently of caspase-dependent apoptosis (19). Both lysosomal-dependent autophagic death and mitochondrial-dependent “necrotic” neuronal death also have been observed in conjunction with apoptosis, and together, these processes may contribute to the neurodegeneration observed in polyQ and other neurodegenerative diseases. A key challenge now is to determine how and why these various death pathways are engaged in vulnerable neuronal subpopulations in polyQ diseases.
Footnotes
This paper is a summary of a session presented at the 11th annual symposium on Frontiers of Science, held November 11–13, 1999, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.210395797.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.210395797
References
- 1.Zoghbi H Y, Orr H T. Annu Rev Neurosci. 2000;23:217–247. doi: 10.1146/annurev.neuro.23.1.217. [DOI] [PubMed] [Google Scholar]
- 2.Perutz M F. Trends Biochem Sci. 1999;24:58–63. doi: 10.1016/s0968-0004(98)01350-4. [DOI] [PubMed] [Google Scholar]
- 3.Cummings C J, Mancini M A, Antalffy B, DeFranco D B, Orr H T, Zoghbi H Y. Nat Genet. 1998;19:148–154. doi: 10.1038/502. [DOI] [PubMed] [Google Scholar]
- 4.Chai Y, Koppenhafer S, Bonini N, Paulson H. J Neurosci. 1999;19:10338–10347. doi: 10.1523/JNEUROSCI.19-23-10338.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stenoien D L, Cummings C J, Adams H P, Mancini M G, Patel K, DeMartino G N, Marcelli M, Weigel N L, Mancini M A. Hum Molec Genet. 1999;8:731–741. doi: 10.1093/hmg/8.5.731. [DOI] [PubMed] [Google Scholar]
- 6.Warrick J M, Chan E, Gray-Board G, Paulson H L, Bonini N M. Nat Genet. 1999;23:425–428. doi: 10.1038/70532. [DOI] [PubMed] [Google Scholar]
- 7.Kazemi-Esfarjani P, Benzer S. Science. 2000;287:1837–1840. doi: 10.1126/science.287.5459.1837. [DOI] [PubMed] [Google Scholar]
- 8.Krobitsch S, Lindquist S. Proc Natl Acad Sci USA. 2000;97:1589–1594. doi: 10.1073/pnas.97.4.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Satyal S H, Schmidt E, Kitagawa K, Sondheimer N, Lindquist S, Kramer J M, Morimoto R I. Proc Natl Acad Sci USA. 2000;97:5750–5755. doi: 10.1073/pnas.100107297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perez M, Paulson H, Pendse S, Saionz S, Bonini N, Pittman R. J Cell Biol. 1998;143:1457–1470. doi: 10.1083/jcb.143.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kazantsev A, Preisinger E, Dranovsky A, Goldgaber D, Housman D. Proc Natl Acad Sci USA. 1999;96:11404–11409. doi: 10.1073/pnas.96.20.11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Warrick J M, Paulson H L, Gray-Board G, Bui Q T, Fischbeck K H, Pittman R N, Bonini N M. Cell. 1998;93:939–949. doi: 10.1016/s0092-8674(00)81200-3. [DOI] [PubMed] [Google Scholar]
- 13.Jackson G R, Salecker I, Dong X, Yao X, Arnheim N, Faber P W, MacDonald M E, Zipursky S L. Neuron. 1998;21:633–642. doi: 10.1016/s0896-6273(00)80573-5. [DOI] [PubMed] [Google Scholar]
- 14.Horvitz H R. Cancer Res. 1999;59:1701S–1706S. [PubMed] [Google Scholar]
- 15.Antonsson B, Martinou J-C. Exp Cell Res. 2000;256:50–57. doi: 10.1006/excr.2000.4839. [DOI] [PubMed] [Google Scholar]
- 16.Kuan C-Y, Roth K A, Flavell R A, Rakic P. Trends Neurosci. 2000;23:291–297. doi: 10.1016/s0166-2236(00)01581-2. [DOI] [PubMed] [Google Scholar]
- 17.Ona V O, Li M, Vonsattel J P G, Andrews L J, Khan S Q, Chung W M, Frey A S, Menon A S, Li X J, Stieg P E. Nature (London) 1999;399:263–267. doi: 10.1038/20446. [DOI] [PubMed] [Google Scholar]
- 18.Sánchez I, Xu C, Juo P, Kakizaka A, Blenis J, Yuan J. Neuron. 1999;22:623–633. doi: 10.1016/s0896-6273(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 19.Fiskum G, Murphy A N, Beal M F. J Cerebr Blood Flow Metab. 1999;19:351–369. doi: 10.1097/00004647-199904000-00001. [DOI] [PubMed] [Google Scholar]