Abstract
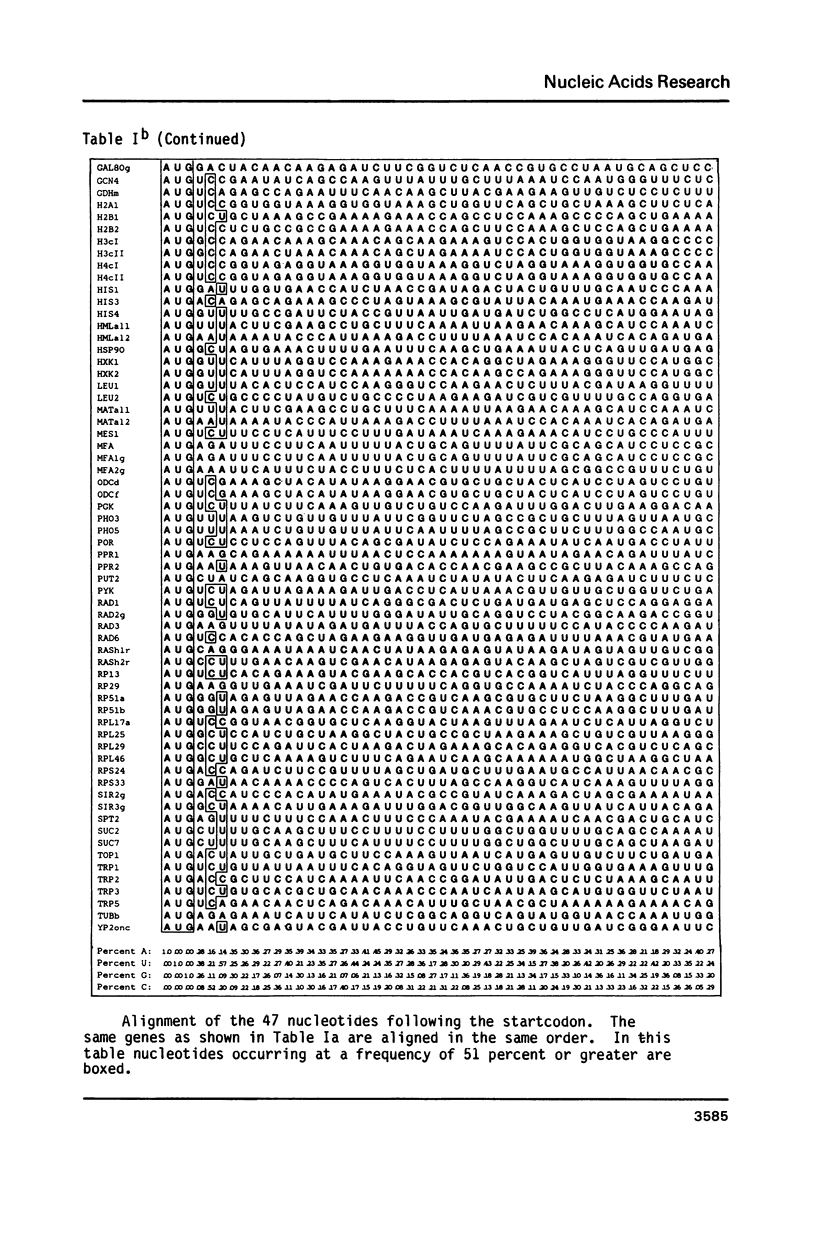
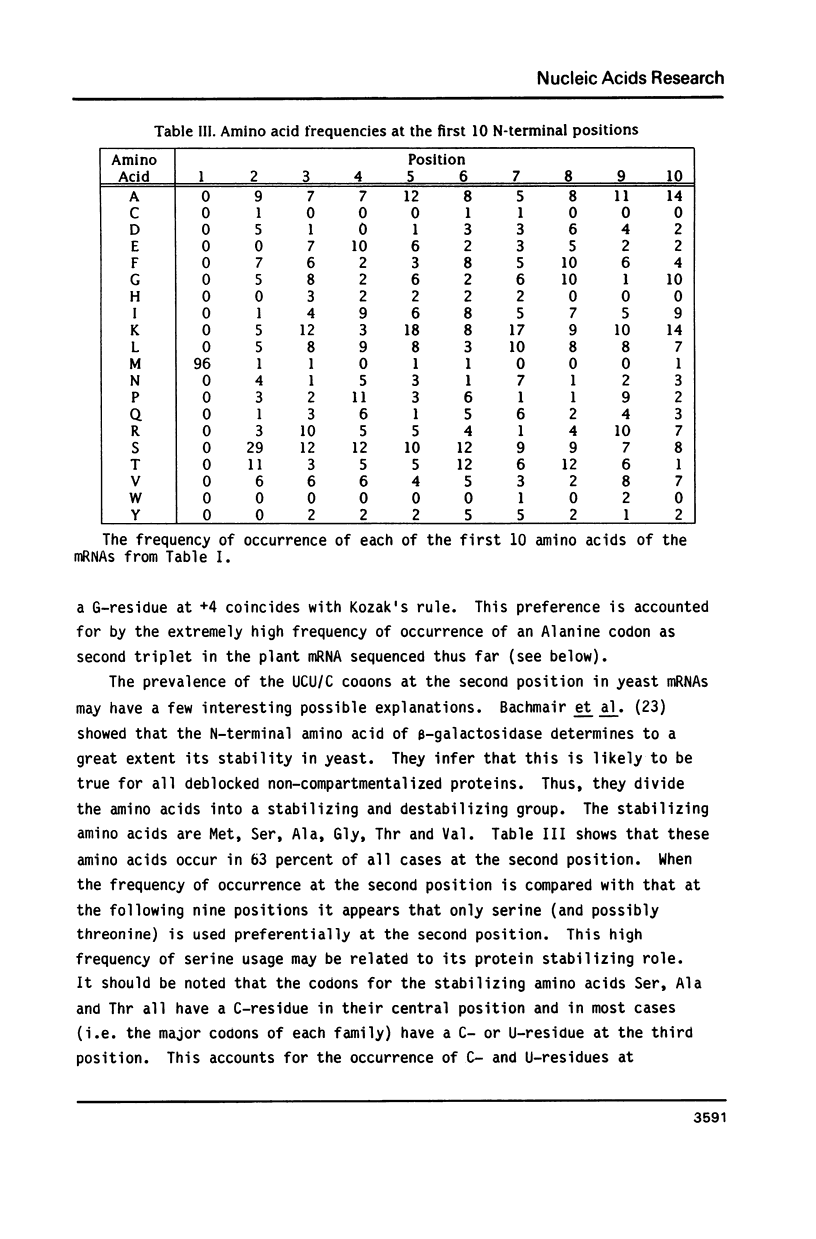
The nucleotide sequence of the translation initiation regions of 96 Saccharomyces cerevisiae mRNAs was compiled and compared. The entire 5' untranslated sequence of most mRNAs is very rich in A-residues. G-residues are underrepresented in the untranslated region. The AUG startcodon context appeared to be distinctly different from that of animal mRNAs, although an A-residue at -3 also occurs very frequently (81 percent) in yeast mRNAs. The prevailing codon 3' adjacent to the AUG is the UCU serine codon. All these features are more extreme in the highly expressed genes. Fifty percent of all highly expressed genes use the UCU serine codon as second triplet. In this group G-residues are completely absent in the 7 bases preceding the startcodon and an A-residue occurs at position -1 and -3 at a frequency of 89 percent and 100 percent, respectively. The abundance of A-residues throughout the leader suggests that unstructured mRNA is required for efficient translation initiation in yeast. The consensus sequence for the AUG context in highly expressed genes can be summarized as follows: (Sequence: see text).
Full text
PDF










Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmair A., Finley D., Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986 Oct 10;234(4773):179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- Hsu Y. P., Schimmel P. Yeast LEU1. Repression of mRNA levels by leucine and relationship of 5'-noncoding region to that of LEU2. J Biol Chem. 1984 Mar 25;259(6):3714–3719. [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of yeast transfer RNAs and the occurrence of the respective codons in protein genes. Differences in synonymous codon choice patterns of yeast and Escherichia coli with reference to the abundance of isoaccepting transfer RNAs. J Mol Biol. 1982 Jul 15;158(4):573–597. doi: 10.1016/0022-2836(82)90250-9. [DOI] [PubMed] [Google Scholar]
- Johnston M., Davis R. W. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Aug;4(8):1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kniskern P. J., Hagopian A., Montgomery D. L., Burke P., Dunn N. R., Hofmann K. J., Miller W. J., Ellis R. W. Unusually high-level expression of a foreign gene (hepatitis B virus core antigen) in Saccharomyces cerevisiae. Gene. 1986;46(1):135–141. doi: 10.1016/0378-1119(86)90177-0. [DOI] [PubMed] [Google Scholar]
- Kozak M. Comparison of initiation of protein synthesis in procaryotes, eucaryotes, and organelles. Microbiol Rev. 1983 Mar;47(1):1–45. doi: 10.1128/mr.47.1.1-45.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 1984 Jan 25;12(2):857–872. doi: 10.1093/nar/12.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Evaluation of the "scanning model" for initiation of protein synthesis in eucaryotes. Cell. 1980 Nov;22(1 Pt 1):7–8. doi: 10.1016/0092-8674(80)90148-8. [DOI] [PubMed] [Google Scholar]
- Kozak M. Mechanism of mRNA recognition by eukaryotic ribosomes during initiation of protein synthesis. Curr Top Microbiol Immunol. 1981;93:81–123. doi: 10.1007/978-3-642-68123-3_5. [DOI] [PubMed] [Google Scholar]
- Kozak M. Point mutations close to the AUG initiator codon affect the efficiency of translation of rat preproinsulin in vivo. Nature. 1984 Mar 15;308(5956):241–246. doi: 10.1038/308241a0. [DOI] [PubMed] [Google Scholar]
- Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986 Jan 31;44(2):283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- Kozak M. Possible role of flanking nucleotides in recognition of the AUG initiator codon by eukaryotic ribosomes. Nucleic Acids Res. 1981 Oct 24;9(20):5233–5252. doi: 10.1093/nar/9.20.5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Selection of initiation sites by eucaryotic ribosomes: effect of inserting AUG triplets upstream from the coding sequence for preproinsulin. Nucleic Acids Res. 1984 May 11;12(9):3873–3893. doi: 10.1093/nar/12.9.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C. C., Simonsen C. C., Levinson A. D. Initiation of translation at internal AUG codons in mammalian cells. Nature. 1984 May 3;309(5963):82–85. doi: 10.1038/309082a0. [DOI] [PubMed] [Google Scholar]
- McNeil J. B., Smith M. Saccharomyces cerevisiae CYC1 mRNA 5'-end positioning: analysis by in vitro mutagenesis, using synthetic duplexes with random mismatch base pairs. Mol Cell Biol. 1985 Dec;5(12):3545–3551. doi: 10.1128/mcb.5.12.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart J. W., Sherman F., Shipman N. A., Jackson M. Identification and mutational relocation of the AUG codon initiating translation of iso-1-cytochrome c in yeast. J Biol Chem. 1971 Dec 25;246(24):7429–7445. [PubMed] [Google Scholar]
- Valenzuela P., Gray P., Quiroga M., Zaldivar J., Goodman H. M., Rutter W. J. Nucleotide sequence of the gene coding for the major protein of hepatitis B virus surface antigen. Nature. 1979 Aug 30;280(5725):815–819. doi: 10.1038/280815a0. [DOI] [PubMed] [Google Scholar]
- Zalkin H., Paluh J. L., van Cleemput M., Moye W. S., Yanofsky C. Nucleotide sequence of Saccharomyces cerevisiae genes TRP2 and TRP3 encoding bifunctional anthranilate synthase: indole-3-glycerol phosphate synthase. J Biol Chem. 1984 Mar 25;259(6):3985–3992. [PubMed] [Google Scholar]
- Zitomer R. S., Walthall D. A., Rymond B. C., Hollenberg C. P. Saccharomyces cerevisiae ribosomes recognize non-AUG initiation codons. Mol Cell Biol. 1984 Jul;4(7):1191–1197. doi: 10.1128/mcb.4.7.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]



