Abstract
Background
CREB (cAMP-response element binding protein) is the prototypical signal-regulated transcription factor. In neurons, it is the target of the synaptic activity-induced nuclear calcium-calcium/calmodulin dependent protein kinase (CaMK) IV signaling pathway that controls the expression of genes important for acquired neuroprotection as well as other long-lasting adaptive processes in the nervous system. The function of CREB as a transcriptional activator is controlled by its phosphorylation on serine 133, which can be catalyzed by CaMKIV and leads to the recruitment of the co-activator, CREB binding protein (CBP). Activation of CBP function by nuclear calcium-CaMKIV signaling is a second regulatory step required for CREB/CBP-mediated transcription.
Results
Here we used recombinant adeno-associated virus (rAAV) to increase the levels of wild type CREB or to overexpress a mutant version of CREB (mCREB) containing a serine to alanine mutation at position amino acid 133 in mouse hippocampal neurons. Increasing the levels of CREB was sufficient to boost neuroprotective activity even under basal conditions (i.e., in the absence of stimulation of synaptic activity). In contrast, overexpression of mCREB increased cell death. The ratio of phospho(serine 133)CREB to CREB immunoreactivity in unstimulated hippocampal neurons was similar for endogenous CREB and overexpressed wild type CREB and, as expected, dramatically reduced for overexpressed mCREB. A gene expression analysis revealed that increased expression of CREB but not that of mCREB in hippocampal neurons led to elevated expression levels of bdnf as well as that of several members of a previously characterized set of Activity-regulated Inhibitor of Death (AID) genes, which include atf3, btg2, gadd45β, and gadd45γ.
Conclusions
Our findings indicate that the expression levels of wild type CREB are a critical determinant of the ability of hippocampal neurons to survive harmful conditions. Increasing the levels of wild type CREB can, even without inducing synaptic activity, increase pro-survival gene expression and strengthen the neurons’ neuroprotective shield. The observed degradation of CREB protein following NMDA treatment of hippocampal neurons suggests that the known CREB shut-off associated with extrasynaptic NMDA receptor-induced excitotoxicity is followed by CREB proteolysis.
Keywords: CREB, Neuroprotection, Hippocampal neurons, Nuclear calcium, CaMKIV, Gene expression, Viral gene delivery, Recombinant adeno-associated virus
Background
In neurons, the transcription factor CREB and its co-activator CBP are central players in synaptic activity-driven gene transcription that contributes to the process through which neurons convert signals from the environment into genomic responses required for long-lasting adaptations [1-3]. Acquired neuroprotection is a well-studied adaptive response in which neurons that have undergone periods of synaptic activity are rendered more resistant to harmful conditions [2,4-11]. This built-up of a neuroprotective shield is initiated by calcium entry though synaptic NMDA receptors and requires calcium transient invading the cell nucleus [8-12]. Nuclear calcium acting principally via nuclear CaMKIV leads to phosphorylation of CREB on its activator site serine 133, allowing CREB to form a complex with CBP [13]. The second regulatory step required for induction of CREB/CBP-mediated transcription is the activation of CBP function by nuclear calcium-CaMKIV signaling [14-16]. Transcriptome analyses have identified a set of neuroprotective genes that are induced by synaptic activity and controlled by nuclear calcium signaling [9,10]. Several members of this group of genes, which were termed Activity-regulated Inhibitor of Death (AID) genes, are known or putative CREB target genes [10,11]. In addition to the ability of CREB to mediate the process that leads to ‘added-on’ survival activity upon synaptic stimulation, the presence of CREB seems to be required for the health of the neurons even under conditions of basal synaptic activity. Mice that lack CREB (and its close relative cAMP response element modulator, CREM) exhibit widespread cell death in the brain [17], and the reduction of functional CREB by means of expression of inhibitors of CREB can severely compromise the well being of neuron [18-20]. Given that physiological expression levels of functional CREB appear to be required cell survival we reasoned that increasing the levels of CREB in hippocampal neurons may enhance neuroprotection even under basal conditions. Here we have tested this hypothesis and found that indeed overexpression of wild type CREB, even without inducing synaptic activity, increases the expression of several AID genes and renders hippocampal neurons more resistant to cell death inducing conditions. Consistent with the importance of having physiological levels of functional CREB available at promoter regions for the maintenance of cell vitality, expression of mCREB increased cell death.
Results and discussion
Expression and serine 133 phosphorylation of wild type and mutant CREB
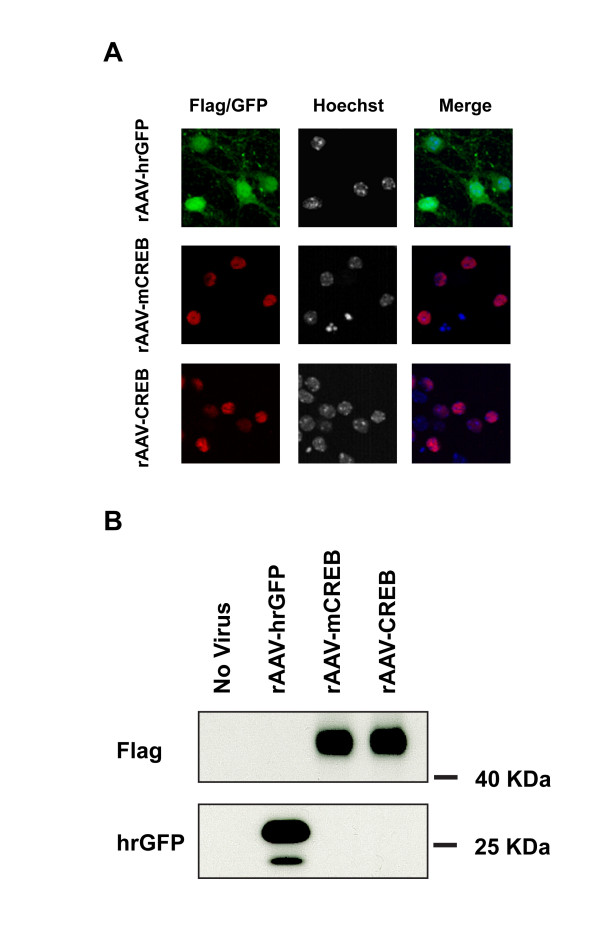
To investigate the relationship between cellular CREB levels and neuronal survival activity, we infected primary mouse hippocampal neurons with a recombinant adeno-associated virus containing an expression cassette for wild type rat CREB, a mutant version of CREB (mCREB) containing a serine to alanine mutation at position amino acid 133, or hrGFP (humanized Renilla reniformis green fluorescent protein). Expression of all three proteins following infection of mouse hippocampal neurons with rAAV-CREB, rAAV-mCREB, or rAAV-hrGFP was readily detectable immunocytochemically or GFP fluorescence in 80 to 95% of the viable cells (Figure 1A). The correct size proteins were also detected in immunoblots (Figure 1B; note that due to the fusion of the exogenously expressed CREB and mCREB to a triple Flag tag, the size of the exogenously expressed CREB and mCREB is slightly larger than that of the endogenous CREB).
Figure 1.
Immunocytochemical (A) and immunoblot analyses (B) of hippocampal neurons infected with rAAVs containing expression cassettes for hrGFP, CREB or mCREB. CREB and mCREB contain triple Flag epitope tags and were detected with M2 Flag antibody; hrGFP was detected with an antibody to hrGFP. Cell nuclei were counterstained with Hoechst 33258. Representative images are shown. Scale bar, 10 μm.
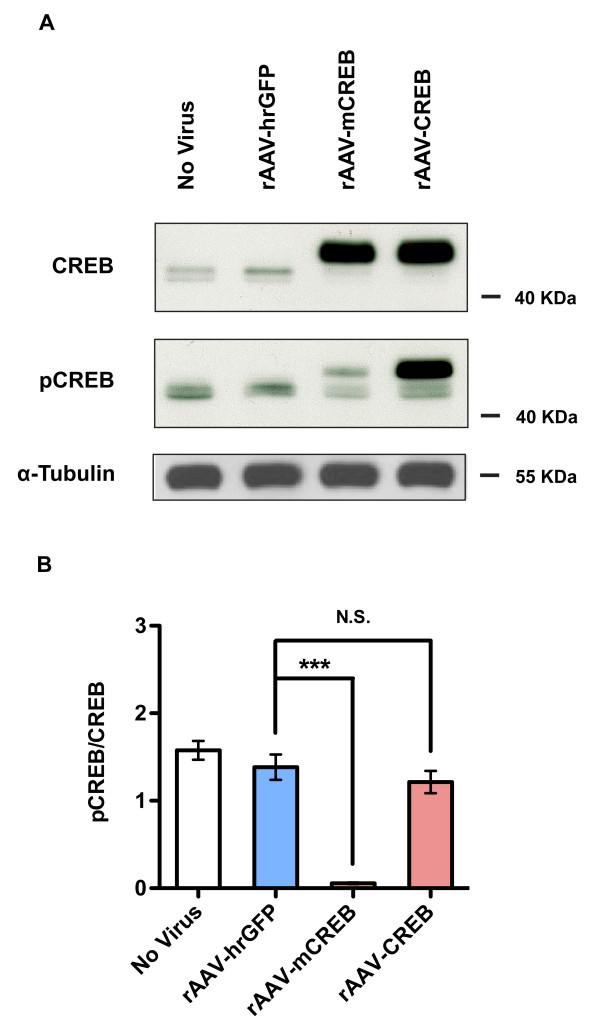
Analysis of the phosphorylation of CREB on serine 133 in unstimulated hippocampal neurons using phospho(serine133)CREB-specific antibodies revealed very weak signals for the endogenous CREB protein in all samples analyzed (Figure 2A). We detected much stronger phospho(serine133)CREB immunoreactivity for the overexpressed CREB protein, while, as expected, low levels of phospho(serine133)CREB immunoreactivity were obtained for mCREB. The ratio of phospho(serine133)CREB to CREB immunoreactivity was similar for the endogenous CREB and the overexpressed wild type CREB, but, in contrast, was dramatically reduced for overexpressed mCREB (Figure 2B). This suggests that the strong phospho(serine133)CREB signal obtained using lysates from hippocampal neurons infected with rAAV-CREB reflects the large amount of CREB protein present in the cells, which compared to the endogenous CREB appears to contain a similar fraction of the serine 133-phosphorylated form. However, we cannot rule out the possibility that the phospho(serine133)CREB-specific antibody solution contains a small fraction of antibodies that bind to CREB even when it is not phosphorylated on serine 133. In this scenario, the phospho(serine133)CREB immunoreactivity detected in samples from rAAV-CREB infected neurons may not indicate the presence of a subset of serine133-phosphorylated CREB molecules but it is merely the result of the binding to CREB of the fraction of the antibody solution that can interact with the non-phosphorylated epitope. The observation that mCREB yielded a very weak signal with the phospho(serine133)CREB-specific antibody solution does not argue against this possibility because the mutation of serine 133 not only abolished any possible phosphorylation event at amino acid 133 but may also alter the epitope thereby reducing the binding to CREB of those antibodies (within the solution of the phospho(serine133)CREB-specific antibodies) that recognize CREB not phosphorylated on serine 133. The antibody used to detect total CREB levels was generated using a recombinant N-terminal fragment of CREB as an antigen.
Figure 2.
(A) Immunoblot analysis of expression and phosphorylation on serine 133 of endogenous and overexpressed CREB in uninfected hippocampal neurons and in neurons infected with rAAV-hrGFP, rAAV-CREB and rAAV-mCREB. CREB expression and CREB phosphorylation on serine 133 (pCREB) was assessed using antibodies to CREB and antibodies specific for the serine 133 phosphorylated form of CREB, respectively. (B) Quantitative analysis of the levels of CREB and CREB phosphorylated on serine 133 (pCREB). In uninfected hippocampal neurons and in neurons infected with rAAV-hrGFP, the ratio of pCREB to CREB (pCREB/CREB) was calculated for endogenous CREB; for hippocampal neurons infected with rAAV-CREB and rAAV-mCREB, the ratios were determined for overexpressed CREB and mCREB. Bars represent means ± SEM (n = 3). Statistical significance was determined by One-way ANOVA, Bonferroni post hoc test; statistically significant differences are indicated with an asterisk (***p < 0.001 versus rAAV-hrGFP; N.S., not significant).
Elevated CREB level protects against apoptosis and excitotoxicity
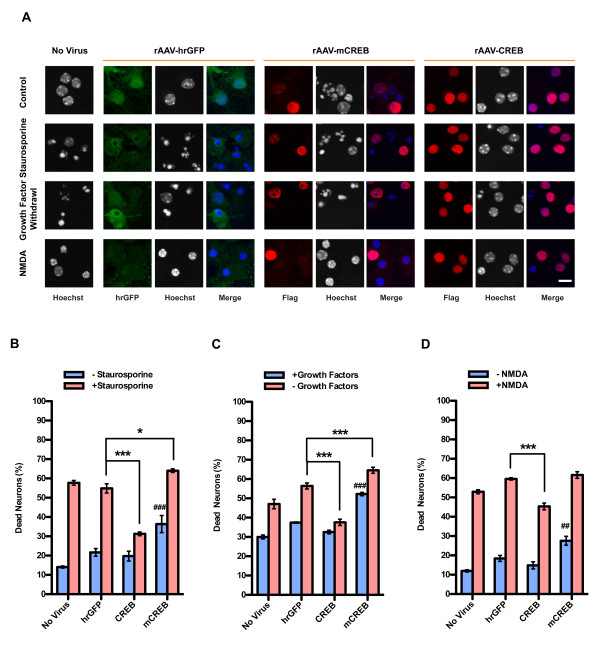
In order to investigate a possible neuroprotective effect of elevating intracellular CREB levels, hippocampal neurons were infected with rAAV-mCREB, rAAV-CREB, rAAV-hrGFP on day in vitro (DIV) 4. Six days post-infection, the neurons were challenged using three different types of cell death-inducing conditions. We treated neurons with a low concentration of staurosporine and growth factor withdrawal to induce apoptosis, and bath application of 20 μM NMDA to trigger excitotoxicity [10,11]. We found that in all three types of cell death assays, the stimulus-induced increase in the percentage of dead neurons was smaller in hippocampal cultures infected with rAAV-CREB compared to the uninfected or rAAV-hrGFP infected control (Figure 3A-D). This indicates that increasing the levels of CREB in hippocampal neurons renders the cells more resistant to apoptosis and excitotoxicity. In contrast, overexpression of mCREB failed to provide neuroprotection and, instead, increased the basal death rates (Figure 3A-D). Because mCREB can bind to the CRE [21], it competes with endogenous CREB for docking sites within gene regulatory regions. This may explain the cell death-promoting activity of mCREB, consistent with the hypothesis that a reduction below physiological levels of functional CREB available at target gene promoters renders neurons more susceptible for harmful conditions.
Figure 3.
Increasing levels of CREB protects against apoptosis and excitotoxicity. (A) Analysis of cell death in uninfected hippocampal neurons and in hippocampal neurons infected with rAAVs-hrGFP, rAAV-CREB or rAAV-mCREB. CREB and mCREB were detected with the M2 Flag antibody; hrGFP was detected with an antibody to hrGFP. Cell death was determined by analyzing Hoechst stained nuclei with shrunken cell body and large round chromatin clumps. For mCREB expression we obtained very weak Flag immunoreactivity in the death-inducing conditions, which is most likely due to compromised cell health following mCREB expression. Representative images are shown. Scale bar, 10 μm. Quantitative analysis of cell death induced by 10 nM staurosporine (B), growth factor withdrawal (C), or bath application of 20 μM NMDA (D). Bars represent means ± SEM (n = 4). Statistical significance was determined by Two-way ANOVA, Bonferroni post hoc test; statistically significant differences are indicated (***p < 0.001, **p < 0.01, *p < 0.05 compared to rAAV-hrGFP infected neurons in groups induced to undergo cell death; ###p < 0.001, ##p < 0.01 compared to rAAV-hrGFP infected neurons in untreated groups).
NMDA-induced CREB phosphorylation, CREB shut-off, and CREB degradation
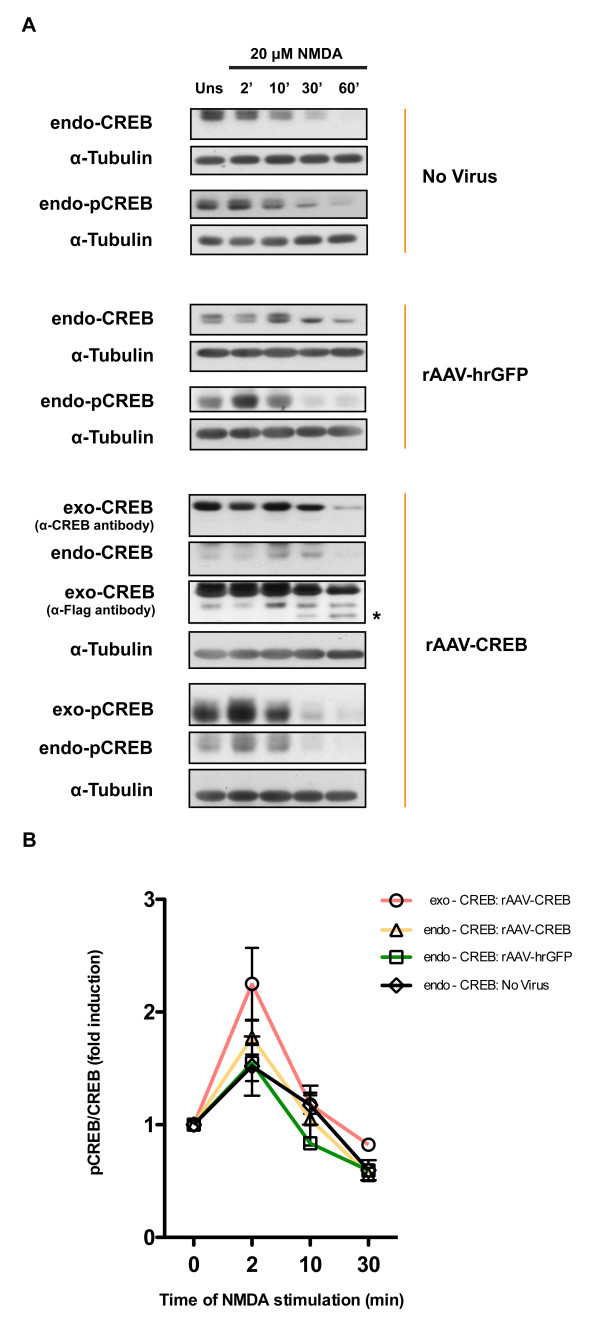
Increasing the levels of CREB expression in hippocampal neurons did not change the dynamics of CREB phosphorylation on serine 133 after exposure to 20 μM NMDA. We found that the ratio of phospho(serine133)CREB immunoreactivity (pCREB) to CREB immunoreactivity increased very transiently after stimulation in uninfected hippocampal neurons and in neurons infected with rAAV-CREB and rAAV-hrGFP (Figure 4A, B). Peak levels of the pCREB/CREB immunoreactivities for both endogenously and exogenously expressed CREB were observed at 2 min after stimulation, which is followed by a rapid shut-off of CREB phosphorylation on serine 133 (Figure 4A, B). Because we observed significant degradation of CREB at 60 min after NMDA treatment (Figure 4A), a meaningful ratio of pCREB to CREB immunoreactivities cannot be determined for this time point. The degradation of CREB protein after induction of excitotoxicity, which is already detectable 30 min after exposure to NMDA, appears to start within the amino-terminus of CREB because degradation products of exogenously expressed CREB, Flag-tagged at the carboxyl-terminus, are detected with antibodies to Flag but not with antibodies to CREB that recognize an amino-terminal epitope (Figure 4A, CREB degradation products are indicated by an asterisk). It is tempting to speculate that CREB degradation, which may be catalyzed by the calcium dependent protease, calpain, is causally involved in the cell death process. Thus, excitotoxicity triggered by the stimulation of extrasynaptic NMDA receptors leads in a first phase to a rapid CREB shut-off (i.e., dephosphorylation of phospho(serine133) of CREB) [7], which is followed by the degradation of CREB protein. Overexpression of CREB delays the excitotoxicity-associated depletion of CREB from the neurons, which likely contributes to neuroprotection.
Figure 4.
(A) Immunoblot analysis of expression and phosphorylation on serine 133 of endogenous CREB (endo-CREB; endo-pCREB) and overexpressed CREB (exo-CREB; exo-pCREB) in uninfected hippocampal neurons and in neurons infected with rAAV-hrGFP or rAAV-CREB at the indicated times after treatment with 20 μM NMDA. Expression of endogenous and overexpressed CREB was analyzed with an antibody to CREB; overexpressed CREB was also detected using an anti-Flag antibody. CREB phosphorylation on serine 133 (pCREB) was assessed using antibodies specific for the serine 133 phosphorylated form of CREB. CREB degradation products are indicated by an asterisk. (B) Quantitative analysis of the pCREB/CREB ratios of the immunoblot analyses shown in (A). Means ± SEM are given (n = 4).
CREB regulation of survival gene expression
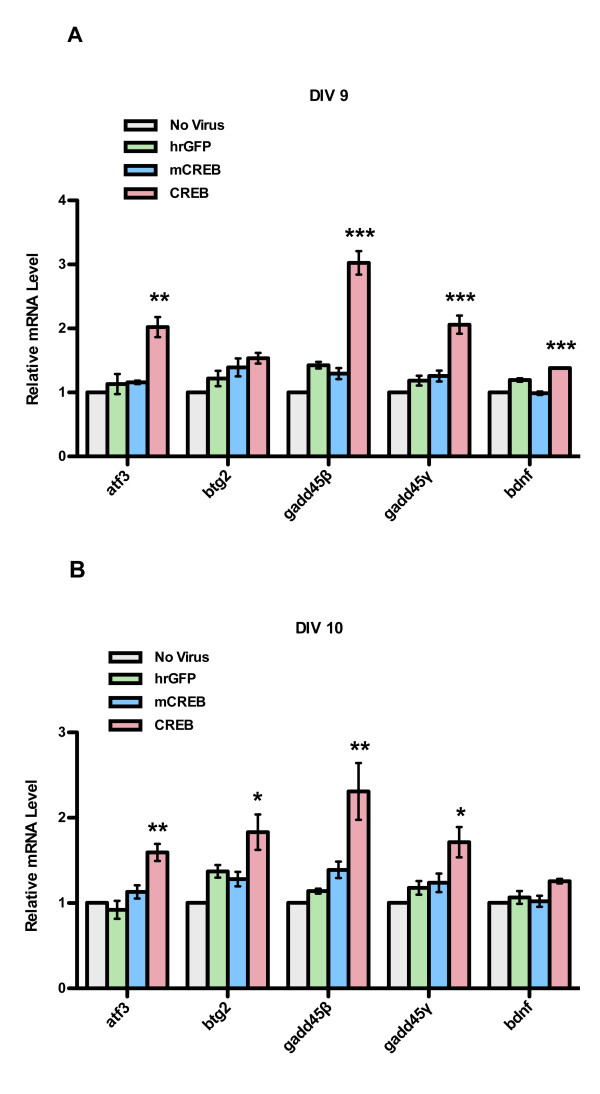
In a previous study we have identified a set of synaptic activity- and nuclear calcium-regulated neuroprotective genes, which were termed Activity-regulated Inhibitor of Death (AID) genes [10] Several AID genes are known or putative CREB target genes [10,11]. We therefore investigated the possibility that the observed neuroprotection following infection with rAAV-CREB is associated with an increase in AID gene expression. Compared to uninfected hippocampal neurons and neurons infected with rAAV-hrGFP or rAAV-mCREB, we detected a 2- to 3- fold increase in the expression of the AID genes, atf3btg2gadd45β, and gadd45γ after 5 and/or 6 days post infection (i.e., DIV 9 or DIV 10) with rAAV-CREB (Figure 5A, B). We also observed a slight but significant increase in the bdnf expression 5 days after infection with rAAV-CREB (Figure 5A). These findings suggest that neuroprotection afforded by elevated CREB levels may be the result of increased expression of neuroprotective genes.
Figure 5.
QRT-PCR analysis ofatf3,btg2,gadd45β, andgadd45γandbdnfexpression in uninfected hippocampal neurons and in neurons infected with rAAV-hrGFP, rAAV-CREB and rAAV-mCREB. Gene expression analyses were done on DIV 9 (A) and DIV 10 (B), which correspond to five days and six days, respectively, post-infection. Bars represent SEM (n = 3). Statistical significance was determined by One-way ANOVA, Bonferroni post hoc test, statistically significant differences are indicated with an asterisk (***p < 0.001, **p < 0.01, *p < 0.05).
Conclusions
Our results indicate that increasing cellular CREB levels can enhance the ability of hippocampal neurons to survive neurotoxic stimuli. This form of CREB-mediated neuroprotection is being built up under conditions of basal neuronal activity and is associated with an increase of several survival promoting genes.
Methods
Hippocampal cell culture
Hippocampal neurons from newborn C57Black6 mice were prepared as described previously [9,22,23]. Neurons were plated onto either 12 mm glass coverslips or 35 mm dishes at a density between 400 and 600 cells per mm2 and cultured in Neurobasal media (Invitrogen, Gaithersburg, MD, USA) containing 1% rat serum and B27 (Invitrogen), and penicillin and streptomycin (Sigma). On days in vitro (DIV) 3, 2.4 μM cytosine D-arabinofuranoside (Sigma) was added to each dish to prevent proliferation of non-neuronal cells. At DIV 8, medium was replaced with transfection medium (TM) [22] which consists of a salt-glucose-glycine (SGG) solution and minimum Eagle’s medium (9:1; v/v) with sodium selenite 10 μg/ml, insulin 15 μg/ml, transferring 8.25 μg/ml, and penicillin-streptomycin 0.5%. SGG includes (in mM): NaCl 114, NaHCO3 26, KCl 6.3, MgCl2 1, CaCl2 2, Hepes 10, glycine 1, glucose 30, sodium pyruvate 0.5, Phenol Red 0.2%. All the experiments were done after a culturing period of 10 to 13 days during which hippocampal neurons develop a rich network of processes, express function NMDA-type and AMPA/kainite-type glutamate receptors, and form synaptic contacts [7,24].
Recombinant adeno-associated viruses
The vectors used to construct and package rAAVs have been described previously [9,10,25]. The recombinant virus for the expression of the humanized Renilla reniformis green fluorescent (hrGFP) was generated as described in previous work [9]. A recombinant adeno-associated virus vector containing 1 kbp cytomegalovirus enhancer (CMV)/chicken β actin hybrid promoter (CBA) was used to express CREB and mCREB. Both rAAV-CREB and rAAV-mCREB contain triple Flag epitope tag. All the vectors were generated by standard molecular biology techniques and verified by sequencing. Viral particles were produced and purified as described previously [9,10,26,27]. For viral infection, neurons were infected with 1011 rAAV particles/μL at DIV 4. Infection efficiencies were determined immunocytochemically at DIV 9 or 10 by using antibodies to the Flag tag, or by analyzing the fluorescence of hrGFP; they ranged from 80%-95% of the viable neurons.
Immunocytochemistry
Mouse hippocampal neurons were fixed with 4% paraformaldehyde for 15 min and permeabilized with 0.3% Triton X 100 in PBS for 10 min. PFA was blocked by 10 min incubation in 1.25 M Glycine. After washing in PBS, the cells were blocked with blocking buffer (2% BSA, 10% normal goat serum in PBS) at room temperature for 1 h. The cells were exposed to primary antibody (anti-Flag M2 Flag antibody, Sigma, MO, US) in dilution buffer (2% BSA, 0.1% Triton X 100 in PBS) at 4°C overnight, washed in PBST (0.1% Tween 20 in PBS) and exposed to secondary antibodies in dilution buffer. After washing with PBST, the cells were incubated with Hoechst 33258 for 5 min, washed in PBS and mounted with mowiol and glycerol mounting medium, and dried at room temperature overnight. The cells were examined by fluorescent microscopy using a Leica SP2 confocal microscope (Leica, Wetzlar, Germany).
Immunoblot analysis
Hippocampal neurons incubated under appropriate conditions were washes with ice-cold PBS and cell lysates were prepared. The lysates were mixed with 5 × Laemmli sample buffer and boiled for 5 min. The proteins were resolved on 10% SDS-polyacrylamide gels and transferred to nitrocellulose (NC) membranes. The blots were blocked with Tris-buffered saline with Tween-20 (TBST; 20 mM Tris–HCl [PH 7.6], 0.136 M NaCl and 0.5% Tween-20 [vol./vol.]) containing 5% skim milk at room temperature for 1 h followed by incubation of primary antibody in TBST containing 5.0% BSA at 4°C overnight. Expression of CREB and phospho-CREB (pCREB) were measured by immunoblotting using the antibodies to the CREB (rabbit monoclonal antibody; Cell Signaling Technology, MA, USA) and pCREB (rabbit polyclonal antibody; Upstate, MA, USA); hrGFP expression levels were detected using antibody to the hrGFP (rabbit polyclonal antibody; Stratagene); tubulin (mouse monoclonal antibody; Sigma, MO, US) was chosen as the loading control. After washing with TBST, goat-anti-rabbit or mouse HRP conjugated IgG (Promega, WI, US) were added for 1 h at room temperature. Blots were then washes with TBST and exposed to X-ray film. The blots were quantitatively analyzed using ImageJ (http://rsb.info.nih.gov/ij/). All results are given as means ± SEM; statistical significance was determined by One-way ANOVA, Bonferroni post hoc test.
Quantitative reverse transcriptase PCR
To determine the mRNA levels of the potential pro-survival genes regulated by CREB, QRT-PCR was performed using real-time TaqMan technology with a sequence detection system model 7300 Real Time PCR System (Applied Biosystems, Foster City, CA, USA). Total RNA was extracted using RNeasy kit (Qiagen GmbH, Germany) with additional on-column DNase I digestion during RNA purification. cDNA was generated from 1.5 μg of total RNA using High Capacity cDNA Reverse Transcription kit (Applied Biosystems). QRT-PCR was carried out using TaqMan Universal PCR Master Mix (Applied Biosystems). The following TaqMan gene expression assays were used in this study: atf3 (Mm00476032), bdnf (Mm00432069), btg2 (Mm00476162), gadd45β (Mm00435123), gadd45γ (Mm00442225), gusb (Mm00446953_m1). The thermal cycling conditions comprised 10 min at 95°C, and 45 cycles of 15 s for denaturation at 95°C and 60 s for annealing and extension at 60°C. The expression levels of the target mRNA were normalized to the relative ratio of the expression of gusb mRNA. Each QRT-PCR assay was performed three times. All results are given as means ± SEM; statistical significance was determined by One-way ANOVA, Bonferroni post hoc test.
Assessment of cell death
The induction and analysis of NMDA induced neuronal cell death assay was done as described with slight changes [11,27,28]. Briefly, the cells were treated with 20 μM NMDA for 10 min at 37°C, washed three times with TM and incubated at 37°C for 20 h. The percentage of dead cells was determined by analyzing Hoechst 33258 stained nuclei, and the percentage of dead cells was determined. The Staurosporine induced and growth factor withdrawal induced apoptosis assay have been described previously [9-11]. Briefly, 36 h after staurosporine (10 nM) exposure, or 72 h after keeping hippocampal neurons in TM medium minus the growth and trophic factors, all in the presence of 1 μM tetrodotoxin (TTX, Tocris Bioscience), cell death was assessed by determining the percentage of hippocampal neurons with shrunken cell body and large round chromatin clumps. All the cell death assays were done at DIV 10-13, at least 20 visual fields from each coverslip (corresponding to 1500–2000 cells per coverslip) were counted. All results are given as means ± SEM; statistical significance was determined by Two-way ANOVA, Bonferroni post hoc test.
Abbreviations
atf3: Activating transcription factor 3; btg2: B-cell translocation gene 2; gadd45β: Growth arrest and DNA-damage-inducible, beta; gadd45γ: Growth arrest and DNA-damage-inducible, gamma; bdnf: Brain-derived neurotrophic factor; CaMKIV: Calcium/calmodulin-dependent protein kinase 4; CBP: CREB binding protein; CREM: cAMP response element modulator.
Authors’ contributions
HB conceived the study and participated in its design. YWT designed and performed the cell death assay, the immunoblot experiments, and the quantitative PCR assay. SJZ generated the rAAV constructs and viruses. TH carried out part of the gene expression analysis. HB and YWT wrote the manuscript. All authors approved the final manuscript.
Contributor Information
Yan-Wei Tan, Email: tan@nbio.uni-heidelberg.de.
Sheng-Jia Zhang, Email: sheng-jia.zhang@ntnu.no.
Tina Hoffmann, Email: Taina19@gmx.de.
Hilmar Bading, Email: Hilmar.Bading@uni-hd.de.
Acknowledgements
We would like to thank Iris Bünzli-Ehret for preparing hippocampal cultures. This work was supported by the Alexander von Humboldt-Foundation (Wolf-gang-Paul-Prize to HB), the ERC Advanced Grant (HB), German Ministry of Education and Research (BMBF, 01GQ1003A), the EU Project GRIPANNT, the EU Network of Excellence NeuroNE, the Sonderforschungsbereich (SFB) 488 and 636, and the German Ministry of Education and Research (BMBF, 01GQ1003A). HB is a member of the Excellence Cluster CellNetworks at Heidelberg University.
References
- Hagenston AM, Bading H. Calcium Signaling in Synapse-to-Nucleus Communication. Cold Spring Harb Perspect Biol. 2011;3:a004564. doi: 10.1101/cshperspect.a004564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardingham GE, Bading H. Synaptic versus extrasynaptic NMDA receptor signalling: implications for neurodegenerative disorders. Nat Rev Neurosci. 2010;11:682–696. doi: 10.1038/nrn2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer PL, Greenberg ME. From synapse to nucleus: calcium-dependent gene transcription in the control of synapse development and function. Neuron. 2008;59:846–860. doi: 10.1016/j.neuron.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Ghosh A, Ginty DD, Bading H, Greenberg ME. Calcium regulation of gene expression in neuronal cells. J Neurobiol. 1994;25:294–303. doi: 10.1002/neu.480250309. [DOI] [PubMed] [Google Scholar]
- Mao Z, Bonni A, Xia F, Nadal-Vicens M, Greenberg ME. Neuronal activity-dependent cell survival mediated by transcription factor MEF2. Science. 1999;286:785–790. doi: 10.1126/science.286.5440.785. [DOI] [PubMed] [Google Scholar]
- Bonni A, Brunet A, West AE, Datta SR, Takasu MA, Greenberg ME. Cell survival promoted by the Ras-MAPK signaling pathway by transcription-dependent and -independent mechanisms. Science. 1999;286:1358–1362. doi: 10.1126/science.286.5443.1358. [DOI] [PubMed] [Google Scholar]
- Hardingham GE, Fukunaga Y, Bading H. Extrasynaptic NMDARs oppose synaptic NMDARs by triggering CREB shut-off and cell death pathways. Nat Neurosci. 2002;5:405–414. doi: 10.1038/nn835. [DOI] [PubMed] [Google Scholar]
- Papadia S, Stevenson P, Hardingham NR, Bading H, Hardingham GE. Nuclear Ca2+ and the cAMP response element-binding protein family mediate a late phase of activity-dependent neuroprotection. J Neurosci. 2005;25:4279–4287. doi: 10.1523/JNEUROSCI.5019-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SJ, Steijaert MN, Lau D, Schutz G, Delucinge-Vivier C, Descombes P, Bading H. Decoding NMDA receptor signaling: identification of genomic programs specifying neuronal survival and death. Neuron. 2007;53:549–562. doi: 10.1016/j.neuron.2007.01.025. [DOI] [PubMed] [Google Scholar]
- Zhang SJ, Zou M, Lu L, Lau D, Ditzel DA, Delucinge-Vivier C, Aso Y, Descombes P, Bading H. Nuclear calcium signaling controls expression of a large gene pool: identification of a gene program for acquired neuroprotection induced by synaptic activity. PLoS Genet. 2009;5:e1000604. doi: 10.1371/journal.pgen.1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SJ, Buchthal B, Lau D, Hayer S, Dick O, Schwaninger M, Veltkamp R, Zou M, Weiss U, Bading H. A signaling cascade of nuclear calcium-CREB-ATF3 activated by synaptic NMDA receptors defines a gene repression module that protects against extrasynaptic NMDA receptor-induced neuronal cell death and ischemic brain damage. J Neurosci. 2011;31:4978–4990. doi: 10.1523/JNEUROSCI.2672-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardingham GE, Bading H. Coupling of extrasynaptic NMDA receptors to a CREB shut-off pathway is developmentally regulated. Biochim Biophys Acta. 2002;1600:148–153. doi: 10.1016/S1570-9639(02)00455-7. [DOI] [PubMed] [Google Scholar]
- Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol. 2001;2:599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- Chawla S, Hardingham GE, Quinn DR, Bading H. CBP: a signal-regulated transcriptional coactivator controlled by nuclear calcium and CaM kinase IV. Science. 1998;281:1505–1509. doi: 10.1126/science.281.5382.1505. [DOI] [PubMed] [Google Scholar]
- Hardingham GE, Chawla S, Cruzalegui FH, Bading H. Control of recruitment and transcription-activating function of CBP determines gene regulation by NMDA receptors and L-type calcium channels. Neuron. 1999;22:789–798. doi: 10.1016/S0896-6273(00)80737-0. [DOI] [PubMed] [Google Scholar]
- Impey S, Fong AL, Wang Y, Cardinaux JR, Fass DM, Obrietan K, Wayman GA, Storm DR, Soderling TR, Goodman RH. Phosphorylation of CBP mediates transcriptional activation by neural activity and CaM kinase IV. Neuron. 2002;34:235–244. doi: 10.1016/S0896-6273(02)00654-2. [DOI] [PubMed] [Google Scholar]
- Mantamadiotis T, Lemberger T, Bleckmann SC, Kern H, Kretz O, Martin Villalba A, Tronche F, Kellendonk C, Gau D, Kapfhammer J. et al. Disruption of CREB function in brain leads to neurodegeneration. Nat Genet. 2002;31:47–54. doi: 10.1038/ng882. [DOI] [PubMed] [Google Scholar]
- Jaworski J, Mioduszewska B, Sanchez-Capelo A, Figiel I, Habas A, Gozdz A, Proszynski T, Hetman M, Mallet J, Kaczmarek L. Inducible cAMP early repressor, an endogenous antagonist of cAMP responsive element-binding protein, evokes neuronal apoptosis in vitro. J Neurosci. 2003;23:4519–4526. doi: 10.1523/JNEUROSCI.23-11-04519.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jancic D, Lopez de Armentia M, Valor LM, Olivares R, Barco A. Inhibition of cAMP response element-binding protein reduces neuronal excitability and plasticity, and triggers neurodegeneration. Cereb Cortex. 2009;19:2535–2547. doi: 10.1093/cercor/bhp004. [DOI] [PubMed] [Google Scholar]
- Ao H, Ko SW, Zhuo M. CREB activity maintains the survival of cingulate cortical pyramidal neurons in the adult mouse brain. Mol Pain. 2006;2:15. doi: 10.1186/1744-8069-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez GA, Montminy MR. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell. 1989;59:675–680. doi: 10.1016/0092-8674(89)90013-5. [DOI] [PubMed] [Google Scholar]
- Bading H, Greenberg ME. Stimulation of protein tyrosine phosphorylation by NMDA receptor activation. Science. 1991;253:912–914. doi: 10.1126/science.1715095. [DOI] [PubMed] [Google Scholar]
- Bading H, Ginty DD, Greenberg ME. Regulation of gene expression in hippocampal neurons by distinct calcium signaling pathways. Science. 1993;260:181–186. doi: 10.1126/science.8097060. [DOI] [PubMed] [Google Scholar]
- Bading H, Segal MM, Sucher NJ, Dudek H, Lipton SA, Greenberg ME. N-methyl-D-aspartate receptors are critical for mediating the effects of glutamate on intracellular calcium concentration and immediate early gene expression in cultured hippocampal neurons. Neuroscience. 1995;64:653–664. doi: 10.1016/0306-4522(94)00462-E. [DOI] [PubMed] [Google Scholar]
- Klugmann M, Symes CW, Leichtlein CB, Klaussner BK, Dunning J, Fong D, Young D, During MJ. AAV-mediated hippocampal expression of short and long Homer 1 proteins differentially affect cognition and seizure activity in adult rats. Mol Cell Neurosci. 2005;28:347–360. doi: 10.1016/j.mcn.2004.10.002. [DOI] [PubMed] [Google Scholar]
- Lau D, Bading H. Synaptic activity-mediated suppression of p53 and induction of nuclear calcium-regulated neuroprotective genes promote survival through inhibition of mitochondrial permeability transition. J Neurosci. 2009;29:4420–4429. doi: 10.1523/JNEUROSCI.0802-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick O, Bading H. Synaptic activity and nuclear calcium signaling protect hippocampal neurons from death signal-associated nuclear translocation of FoxO3a induced by extrasynaptic N-methyl-D-aspartate receptors. J Biol Chem. 2010;285:19354–19361. doi: 10.1074/jbc.M110.127654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtson CP, Dick O, Bading H. A quantitative method to assess extrasynaptic NMDA receptor function in the protective effect of synaptic activity against neurotoxicity. BMC Neurosci. 2008;9:11. doi: 10.1186/1471-2202-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]