Abstract
A dynamic balance between stem cell maintenance and differentiation paces generation of post-mitotic progeny during normal development and maintenance of homeostasis. Recent studies show that Notch plays a key role in regulating the identity of neuroepithelial stem cells, which generate terminally differentiated neurons that populate the adult optic lobe via the intermediate progenitor cell type called neuroblast. Thus, understanding how Notch controls neuroepithelial cell maintenance and neuroblast formation will provide critical insight into the intricate regulation of stem cell function during tissue morphogenesis. Here, we showed that a low level of Notch signaling functions to maintain the neuroepithelial cell identity by suppressing the expression of pointedP1 gene through the transcriptional repressor Anterior open. Increased Notch signaling, which coincides with transient cell cycle arrest but precedes the expression of PointedP1 in cells near the medial edge of neuroepithelia, defines transitioning neuroepithelial cells that are in the process of acquiring the neuroblast identity. Transient up-regulation of Notch signaling in transitioning neuroepithelial cells decreases their sensitivity to PointedP1 and prevents them from becoming converted into neuroblasts prematurely. Down-regulation of Notch signaling combined with a high level of PointedP1 trigger a synchronous conversion from transitioning neuroepithelial cells to immature neuroblasts at the medial edge of neuroepithelia. Thus, changes in Notch signaling orchestrate a dynamic balance between maintenance and conversion of neuroepithelial cells during optic lobe neurogenesis.
Introduction
During mammalian cortical neurogenesis, neural stem cells initially divide symmetrically to expand their population and then divide asymmetrically to produce layer-specific neurons (Kriegstein and Alvarez-Buylla, 2009). Thus, a dynamic balance between stem cell maintenance versus stem cell differentiation directly impinges on the pace of generating post-mitotic progeny in a developing tissue, but the underlying mechanisms remain virtually unknown. Neuroepithelial cells in the larval optic lobe first divide symmetrically to expand their population during first and second larval instar and become progressively converted into neuroblasts that divide asymmetrically to generate neurons in the third larval instar (Egger et al., 2007). A recent study demonstrates that the mechanisms that regulate symmetric expansion of neuroepithelial cells and their subsequent conversion into neuroblasts appear to be distinct (Ngo et al., 2010). Thus, elucidating the mechanisms that controls conversion of neuroepithelial cells into neuroblasts will contribute critical insight into regulation of the balance between stem cell maintenance and differentiation during tissue morphogenesis.
Recent studies have shown that Notch plays a central role in regulating the identity of neuroepithelial stem cells in the developing larval optic lobe (Egger et al., 2010; Ngo et al., 2010; Reddy et al., 2010; Yasugi et al., 2010; Orihara-Ono et al., 2011; Wang et al., 2011). Counterintuitively, while down-regulation of Notch signaling is necessary and sufficient to convert neuroepithelial cells into neuroblasts, the expression of Notch reporter transgenes becomes up-regulated prior to the conversion. One study suggests that activation of the EGF receptor triggers increased Notch signaling and proposes that Notch and EGF signaling function cooperatively to assure the directional progression of conversion in neuroepithelia (Yasugi et al., 2010). However, how Notch and EGF might function in concert to regulate conversion of neuroepithelia cannot be fully understood until several fundamental questions are addressed. What are the functional properties of the intermediate cell types during conversion of neuroepithelia into neuroblasts? What is the molecular basis by which Notch maintains the identity of neuroepithelial cells? What purpose does up-regulation of Notch signaling serve in neuroepithelial cells prior to their conversion into neuroblasts?
Consistent with the requirement of Notch signaling in maintaining neuroepithelial cell identity, its ligand Delta is detected throughout most neuroepithelia (Egger et al., 2010; Ngo et al., 2010; Reddy et al., 2010; Yasugi et al., 2010; Orihara-Ono et al., 2011; Wang et al., 2011). While reducing the function of Delta throughout neuroepithelia leads to premature formation of neuroblasts, removing or over-expressing Delta in the mosaic clone located near the medial edge of neuroepithelia results in both accelerating and inhibiting formation of neuroblasts (Egger et al., 2010; Reddy et al., 2010; Yasugi et al., 2010; Orihara-Ono et al., 2011; Wang et al., 2011). These data strongly suggest an intricate spatial regulation of Notch signaling by Delta in neuroepithelial cells, but the mechanisms are unknown. Delta can regulate the output of Notch signaling via two distinct mechanisms (Sprinzak et al., 2010; del Álamo et al., 2011). During trans-activation, Delta activates Notch signaling in the adjacent cell and the level of Delta directly correlates with the level of Notch signaling output. During cis-inhibition, however, above the threshold level of Delta inactivates Notch signaling in the same cell, so the level of Delta inversely correlates with the output of Notch signaling (Miller et al., 2009). cis-inhibition of Notch signaling functions to sharply define the boundary between cells that show activated Notch signaling and the adjacent cells that lack activated Notch signaling.
From a genetic screen, we identified mutations in two genes required for activation of Notch signaling that led to the entire swath of neuroepithelial cells prematurely differentiating into neuroblasts. We focus our study on elucidating the mechanisms by which Notch signaling regulates maintenance and conversion of neuroepithelia in the outer proliferation center. We show that a low level of Notch signaling maintains the identity of neuroepithelial cells by suppressing the function of the pointedP1 (pntP1) gene via an Anterior open (Aop; also known as Yan)-dependent mechanism. An increase in Notch signaling functionally defines transitioning neuroepithelial cells near the medial edge of neuroepithelia, which are in the process of converting into future neuroblasts. Transient up-regulation of Notch signaling in transitioning neuroepithelial cells raises the threshold of response to PntP1 and functions to pace progressive conversion of the entire swath of neuroepithelia into immature neuroblasts in a medial-to-lateral orientation. The combination of abrupt down-regulation of Notch signaling and a high level of PntP1 triggers a synchronous conversion from transitioning neuroepithelial cells to immature neuroblasts. Our data strongly suggest that changes in Notch signaling dynamically balance maintenance and conversion of neuroepithelial cells during larval optic lobe morphogenesis.
Results
Identification of genes required for proper development of optic lobe neuroepithelia
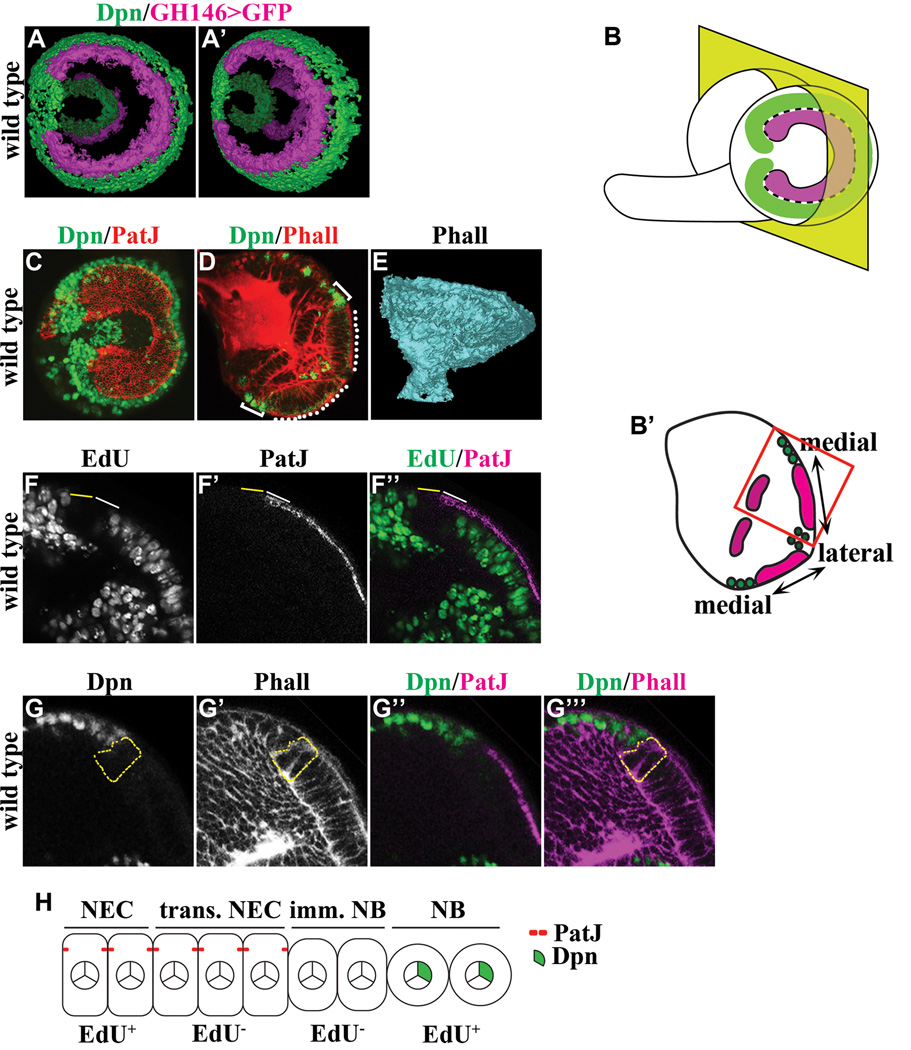
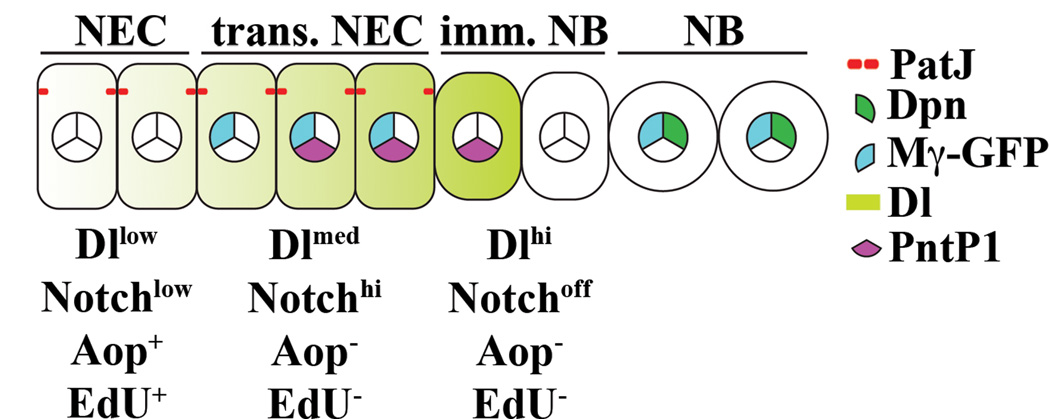
Defining the functional characteristics of epithelial-shape cells at the medial edge of neuroepithelia and the adjacent round-shape cells is pivotal for investigating the mechanisms that regulate conversion of neuroepithelial cells into neuroblasts (Figure 1A–E; Movie S1). Absence of EdU incorporation following a 3-hour pulse labeling in epithelial-shape cells at the medial edge of neuroepithelia and their adjacent round-shape cells indicate that they are functionally distinct from the rest of neuroepithelia and neuroblasts (Figure 1F-F’’). The epithelial-shape cells at the medial edge of neuroepithelia expressed PatJ but lacked Dpn and were named transitioning neuroepithelial cells (Figure 1F–G’’’). The round-shape cells immediately adjacent to transitioning neuroepithelial cells expressed neither PatJ nor Dpn and were named immature neuroblasts (Figure 1G-G’’’). Thus, conversion of neuroepithelia into neuroblasts occurs in the following sequence: neuroepithelial cells -> transitioning neuroepithelial cells -> immature neuroblasts -> neuroblasts (Figure 1H).
Figure 1. Identification of intermediate cell types during differentiation of neuroepithelial cells into neuroblasts.
(A–A’) Three-dimensionally reconstructed views of the outer and inner proliferation center in the third larval instar optic lobe. The expression of GH146-Gal4 marks neuroepithelia in the outer (bright colored) and inner (faded colored) proliferation center whereas expression of Deadpan (Dpn) marks neuroblasts.
(B–B’) Visualization of the developing larval optic lobe. (B) A lateral projection view of a wild type larval optic lobe shows the overall morphology of the outer proliferation center, which consists of a C-shape swath of neuroepithelia (magenta) surrounded by neuroblasts (green). The medial edge of neuroepithelia separating neuroepithelial cells and neuroblasts is indicated by the black & white dotted line. (B’) A dorsoventral single confocal optical section of a wild type larval brain (as indicated by the yellow plane in B) reveals neuroepithelia in the inner and outer proliferation center. In this view, optic lobe neuroblasts flank the medial edge of neuroepithelia in the outer proliferation center. The higher magnification image of neuroepithelia and neuroblasts boxed in red is used in subsequent figures to illustrate the effects of removing or increasing the function of a gene.
(C) The lateral projection view of a wild type larval optic lobe stained for PatJ and Dpn reveals the swath of neuroepithelia flanked by neuroblasts.
(D) A dorsoventral single confocal optical section of a wild type larval brain stained for Phalloidin (Phall) and Dpn shows neuroepithelia in the outer proliferation center (white dotted line) flanked by neuroblasts (white bracket).
(E) A three-dimensionally reconstructed model of a wild type optic lobe stained for Phall reveals the axon bundles that constitute the neuropil.
(F–F’’) Differentiating neuroepithelial cells and immature neuroblasts are transiently arrested in cell cycle. Transitioning neuroepithelial cells (solid white line) located at the medial edge of neuroepithelia and the adjacent immature neuroblasts (solid yellow line) do not incorporate EdU following a 3-hour pulse labeling. The area shown corresponds to the red box in B’.
(G–G’’’) Immature neuroblasts do not maintain the epithelial cell morphology and do not express the neuroblast marker. Immature neuroblasts (outlined in dotted yellow line) located immediately adjacent to transitioning neuroepithelial cells lack expression of PatJ and Dpn. The area shown corresponds to the red box in B’.
(H) A cartoon summarizes the intermediate cell types during conversion of neuroepithelia into neuroblasts. NEC: neuroepithelial cells; trans. NEC: transitioning neuroepithelial cells; imm. NB: immature neuroblast; NB: neuroblast.
To gain insight into the mechanisms that regulate differentiation of neuroepithelia, we screened for zygotic lethal mutations induced by transposable P-element or ethyl methane sulfonate (EMS) that led to aberrant morphology in the third instar larval optic lobe. By using expression of the PCNA::3XEmGFP transgene (kindly provided by Dr. R. Duronio) to rapidly visualize all proliferating cells in the larval optic lobe, we identified three unique categories of mutants (Figure S1). The neuroepithelia in the third instar optic lobe absent mutant larvae was extremely small despite continuous bodily growth overall. We never detected expansion of neuroepithelia and formation of neuroblasts in the second or the third instar optic lobe absent mutant larval optic lobe (Figure S1C–D; data not presented). Furthermore, we did not observe formation of the axonal bundle in the third instar optic lobe absent mutant optic lobe (Figure S1E). Together, these data indicate that the optic lobe absent mutations perturbs initiation of proliferation in the post-embryonic optic lobe neuroepithelia. We mapped these mutations to the connector enhancer of KSR (cnk) and corkscrew (csw) genes, which have been shown to function in receptor tyrosine kinase signaling (Perkins et al., 1992; Van Vactor et al., 1996; Therrien et al., 1998; Kolch, 2005). Consistent with a recently published study, removal of the EGF function also leads to the "optic lobe absent " phenotype (Yasugi et al., 2010). Thus, the receptor tyrosine kinase signaling cascade plays a critical role in initiating neuroepithelial cell proliferation in the developing larval optic lobe.
The neuroepithelia in the third instar optic lobe expanded mutant larvae became overly expanded and partially overlapped. We detected continuous expansion of neuroepithelia and formation of neuroblasts in the second and the third instar optic lobe expanded mutant larvae (Figure S1F–G; data not presented). In addition, we observed formation of the funnel-shape neuropil in the third instar optic lobe expanded mutant larval optic lobe. Together, these data indicate that the optic lobe expanded mutation does not perturb proliferation of the optic lobe neuroepithelia and neuroblasts. We mapped this mutation to the fat gene, which encodes a critical activator of hippo signaling and regulates tissue growth (Zhao et al., 2010). Our result is consistent with a recent study showing that fat-hippo signaling triggers transient cell cycle arrest in cells near the medial edge of neuroepithelia and inactivation of hippo signaling delays conversion of neuroepithelial cells into neuroblasts (Reddy et al., 2010).
The third instar optic lobe prematurely lost mutant larvae contained a swath of neuroblasts in the place of neuroepithelia and a fragmented neuropil in the larval optic lobe (Figure S1I–K). This category of mutants consisted of two mutations mapped to the o-fucosyltransferase 1 (o-fut1) and anterior pharynx defective 1 (aph-1) genes, which encode critical activator proteins of Notch signaling (Francis et al., 2002; Okajima and Irvine, 2002; Bray, 2006). The o-fut12834 mutation leads to a single nucleotide substitution disrupting the splicing acceptor site in the first intron of the o-fut1 gene (Figure S1L). Neuroblasts in o-fut12834/2834 or o-fut12834/Df mutant brains showed similar cytosolic accumulation of the Notch protein (Figure S1M–N’). This result strongly suggests that the o-fut12834 mutation is a strong hypomorphic mutant allele of the o-fut1 gene. The aph-15072 mutation leads to substitution of glycine with arginine in the first transmembrane domain of the Aph-1 protein (Figure S1O). Since the optic lobe prematurely lost phenotype was completely penetrant in aph-15072/5072 and aph-15072/Df mutant larvae, we conclude that aph-15072 is a strongly hypomorphic mutant allele of the aph-1 gene. We next tested if the “optic lobe prematurely lost” phenotype observed in o-fut1 or aph-1 mutant larvae might be due to inactivation of Notch signaling. While the Notch reporter E(spl)mγ-GFP co-localized with Deadpan (Dpn) in many neuroblasts in the second instar o-fut1 or aph-1 mutant larval brain, E(spl)mγ-GFP completely diminished from neuroblasts in the third instar mutant larval brain (Figure S1P–S). Coincidentally, neuroepithelia was present in the second instar o-fut1 or aph-1 mutant larvae, but became lost in the third instar mutant larvae (Figure S1P–S). Together, these results support our hypothesis that inactivation of Notch signaling leads to premature conversion of neuroepithelia into neuroblasts.
Notch signaling becomes up-regulated in transitioning neuroepithelial cells but down-regulated in immature neuroblasts
We tested directly if Notch is required for maintenance of neuroepithelia by inducing the wild type or Notch mutant neuroepithelial cell clone 24 hours after hatching. Most neuroepithelial cells in the wild type clone adopted the neuroblast fate as indicated by expression of Dpn and located medially from remaining neuroepithelia on the surface of the optic lobe (Figure S2A-A’; N = 9 clones). Although neuroepithelial cells in 85.7% of Notch mutant clones also assumed the neuroblast fate, these cells delaminated inward away from the rest of neuroepithelia and located deep in the developing medulla (Figure S2B-B’; N = 21 clones). These data indicate that Notch functions cell autonomously to maintain the identity of neuroepithelial cells. We next tested if down-regulation of Notch signaling is necessary for conversion of neuroepithelial cells into neuroblasts by assessing the identity of cells in the neuroepithelial cell clone over-expressing a UAS-Notchintra transgene. Control neuroepithelial cells lacking expression of Notchintra expanded medially and differentiated into neuroblasts synchronously with cells located outside of the clone (Figure S2C-C’; N = 10). In contrast, neuroepithelial cells that show constitutively activated Notch signaling expanded beyond the medial edge of neuroepithelia and became surrounded by neuroblasts located outside of the clone (Figure S2D-D’; N = 8). Thus, our data are consistent with recent studies showing that Notch acts cell autonomously to maintain neuroepithelial cell identity and down-regulation of Notch signaling is necessary for conversion of neuroepithelial cells into neuroblasts.
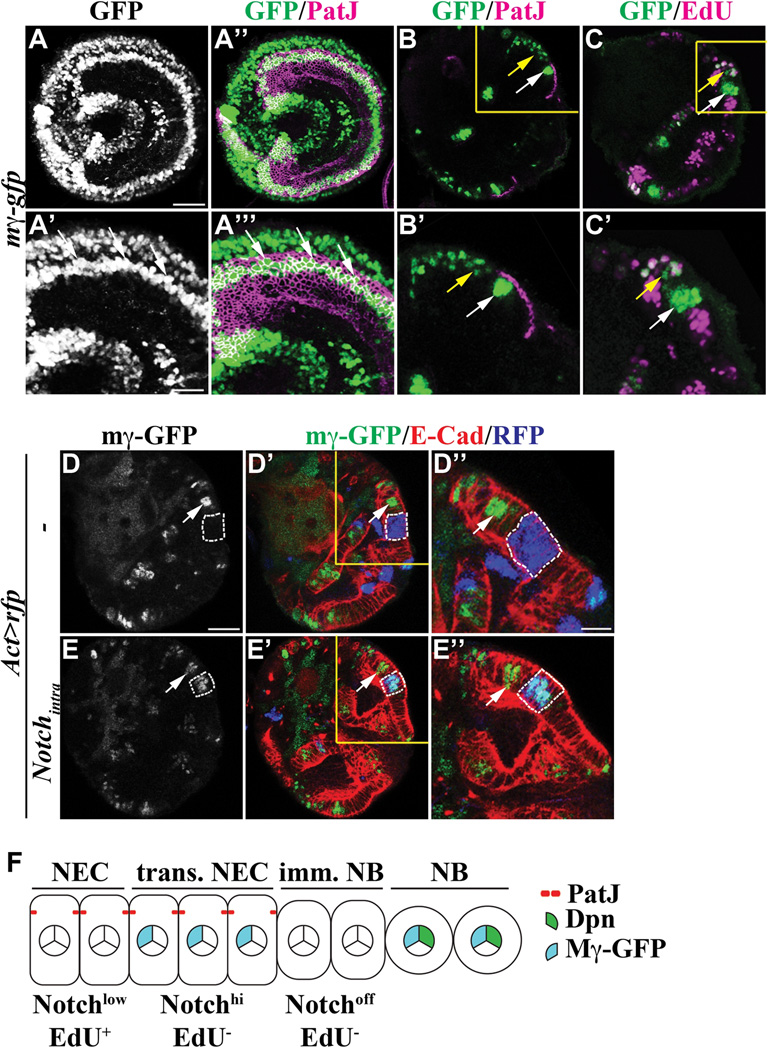
Defining the spatial pattern of Notch signaling will be key toward elucidating the mechanisms by which Notch regulates conversion of neuroepithelia into neuroblasts. We examined expression of the Notch reporter E(spl)mγ-GFP and detected GFP expression only in cells located at and near the medial edge of neuroepithelia (Figure 2A-A’’’). This result led us to hypothesize that Notch signaling becomes up-regulated in transitioning neuroepithelial cells but down-regulated in immature neuroblasts. Indeed, E(spl)mγ-GFP co-localized with PatJ across two-to-three neuroepithelial cells located at the medial edge of neuroepithelia, but was undetectable in the adjacent PatJ− immature neuroblast (Figure 2B-B’; N = 8). In addition, we did not detect any EdU incorporation in the neuroepithelial cells that expressed E(spl)mγ-GFP following a 3-hour pulse labeling (Figures 2C-C’ and S3A-A’). These data indicate that Notch signaling becomes transiently up-regulated in transitioning neuroepithelial cells, but abruptly down-regulated in immature neuroblasts (Figure 2F).
Figure 2. Changes in Notch signaling coincide with conversion of neuroepithelia.
(A–A’’’) Notch signaling becomes drastically increased in neuroepithelial cells near the medial edge of neuroepithelia. (A and A’’) E(spl)mγ-GFP expression co-localizes with PatJ in neuroepithelial cells near the medial edge of neuroepithelia. The scale bar = 20 µm. The higher magnification image of the cells expressing E(spl)mγ-GFP (white arrow) is shown in A’ and A’’’. The scale bar = 10 µm.
(B–C’) Notch signaling becomes drastically increased in transitioning neuroepithelial cells. (B-B’) E(spl)mγ-GFP co-localizes with PatJ in transitioning neuroepithelial cells (white arrow), but is undetectable in adjacent immature neuroblasts. The higher magnification image of the boxed area is shown in B’. (C-C’) E(spl)mγ-GFP does not co-localize with EdU incorporation in neuroepithelia (white arrow) following a 3-hour pulse labeling. However, E(spl)mγ-GFP co-localizes with EdU incorporated into neuroblasts (yellow arrow). The higher magnification image of the boxed area is shown in C’.
(D–E’’) Notch signaling appears to be low throughout most of neuroepithelia. (D-D’’) Cells in the RFP-marked control clone (outlined in dotted white line) located laterally from transitioning neuroepithelial cells (white arrow) lack expression of E(spl)mγ-GFP. The clone was induced in larvae carrying the E(spl)mγ-GFP transgene 72 hours after hatching and the phenotype was assessed 24 hours after clone induction. The scale bar = 20 µm. The higher magnification image of the boxed area is shown in D’’. The scale bar = 10 µm. (E-E’) Cells over-expressing Notchintra in the clone (outlined in dotted white line) located laterally from transitioning neuroepithelial cells (white arrow) show expression of E(spl)mγ-GFP. The higher magnification image of the boxed area is shown in E’’.
(F) A cartoon summarizes changes in Notch signaling during conversion of neuroepithelial cells into neuroblasts. NEC: neuroepithelial cells; trans. NEC: transitioning neuroepithelial cells; imm. NB: immature neuroblast; NB: neuroblast.
The spatial expression pattern of E(spl)mγ-GFP appeared to contradict with the requirement of Notch signaling for the maintenance of neuroepithelial cells. One possibility might be that cells located laterally from transitioning neuroepithelial cells show a too low level of activated Notch to trigger expression of the E(spl)mγ-GFP transgene. We tested this hypothesis by over-expressing Notchintra for 24 hours in the RFP-marked neuroepithelial cell clone in larvae that carry the E(spl)mγ-GFP transgene. Pulsed-expression of Notchintra induced cell autonomous expression of E(spl)mγ-GFP in all RFP-marked clones located laterally from transitioning neuroepithelial cells (Figure 2D–E’’; N = 8). Thus, neuroepithelial cells located laterally from the medial edge of neuroepithelia show a very low level of Notch signaling that is not sufficient to induce E(spl)mγ-GFP expression under physiological conditions. Hence, maintenance of neuroepithelial cell identity requires a low level of Notch signaling (Figure 2F).
Delta regulates changes in Notch signaling during conversion of neuroepithelia into neuroblasts
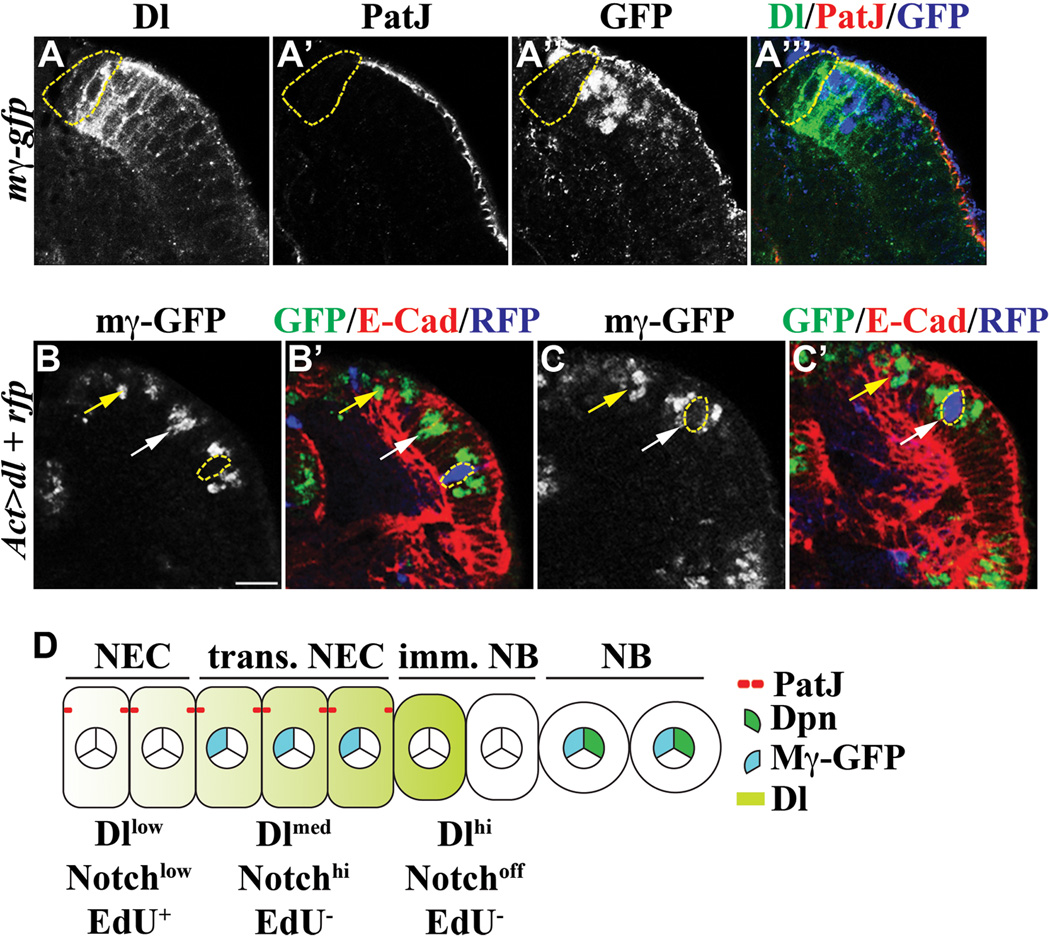
In order to investigate how Delta might spatially regulate Notch signaling in neuroepithelial cells, we first co-localized the endogenous Delta protein with PatJ and E(spl)mγ-GFP in neuroepithelial cells. While low Delta expression was detected throughout neuroepithelia lacking E(spl)mγ-GFP, moderate Delta expression co-localized with E(spl)mγ-GFP in transitioning neuroepithelial cells (Figure 3A-A’’’). Most intriguingly, Delta expression peaked in immature neuroblasts immediately adjacent to transitioning neuroepithelial cells, but became undetectable in the rest of immature neuroblasts (Figure 3A-A’’’). Thus, the expression profile of Delta closely resembles the change in Notch signaling throughout neuroepithelia. We next examined how Delta regulates the dynamic change in Notch signaling in neuroepithelia by transiently over-expressing a UAS-Delta transgene for 24 hours in the neuroepithelial cell clone in larvae carrying the E(spl)mγ-GFP transgene. Over-expression of Delta in the RFP-marked clone located laterally from transitioning neuroepithelial cells led to aberrant expression of E(spl)mγ-GFP cell non-autonomously, (Figure 3B-B’; N = 6). Since previous studies show that reducing function of Delta throughout neuroepithelia triggers premature formation of neuroblasts, our result suggests that the low level of Delta expression trans-activates Notch signaling during maintenance of neuroepithelia. In contrast, over-expression of Delta strikingly abolished E(spl)mγ-GFP expression cell autonomously in the clone located in transitioning neuroepithelial cells, indicating that Delta can cis-inhibit Notch signaling during conversion of neuroepithelia into neuroblasts (Figure 3C-C’; N = 10). This result also suggests that the peak level of Delta expression in immature neuroblasts likely abrogates Notch signaling in the adjacent transitioning neuroepithelial cells. Thus, we propose that trans-activation of a low level of Notch signaling by Delta contributes to maintenance of neuroepithelia whereas cis-inhibition of Notch signaling by Delta triggers the conversion from transitioning neuroepithelial cells to immature neuroblasts (Figure 3D).
Figure 3. Delta regulates changes in Notch signaling in neuroepithelia.
(A–A’’’) The expression pattern of Delta mostly correlates with changes in Notch signaling. Delta is expressed in a low level throughout neuroepithelia, but rises dramatically in transitioning neuroepithelial cells marked by the expression of E(spl)mγ-GFP. Delta expression peaks in the immature neuroblast immediately adjacent to transitioning neuroepithelial cells (outlined in dotted yellow line).
(B–C’) Delta regulates changes in Notch signaling. (B-B’) Transient over-expression of Delta in the RFP-marked clone (outlined in dotted yellow line) located laterally from transitioning neuroepithelial cells (white arrow) activates cell non-autonomous expression of E(spl)mγ-GFP. The UAS-Delta transgene was driven by Actin-Gal4 for 24 hours in the neuroepithelial cell clone in larvae carrying the E(spl)mγ-GFP transgene. The yellow arrow indicates optic lobe neuroblasts. The scale bar = 10 µm. (C-C’) Transient over-expression of Delta in the RFP-marked genetic clone (outlined in dotted yellow line) located in transitioning neuroepithelial cells (white arrow) triggers cell autonomous inhibition of E(spl)mγ-GFP expression. The yellow arrow indicates neuroblasts.
(D) A cartoon summarizes the expression pattern of Delta expression and the Notch activity during conversion of neuroepithelial cells into neuroblasts. NEC: neuroepithelial cells; trans. NEC: transitioning neuroepithelial cells; imm. NB: immature neuroblast; NB: neuroblast.
Notch maintains the identity of neuroepithelial cells via Aop
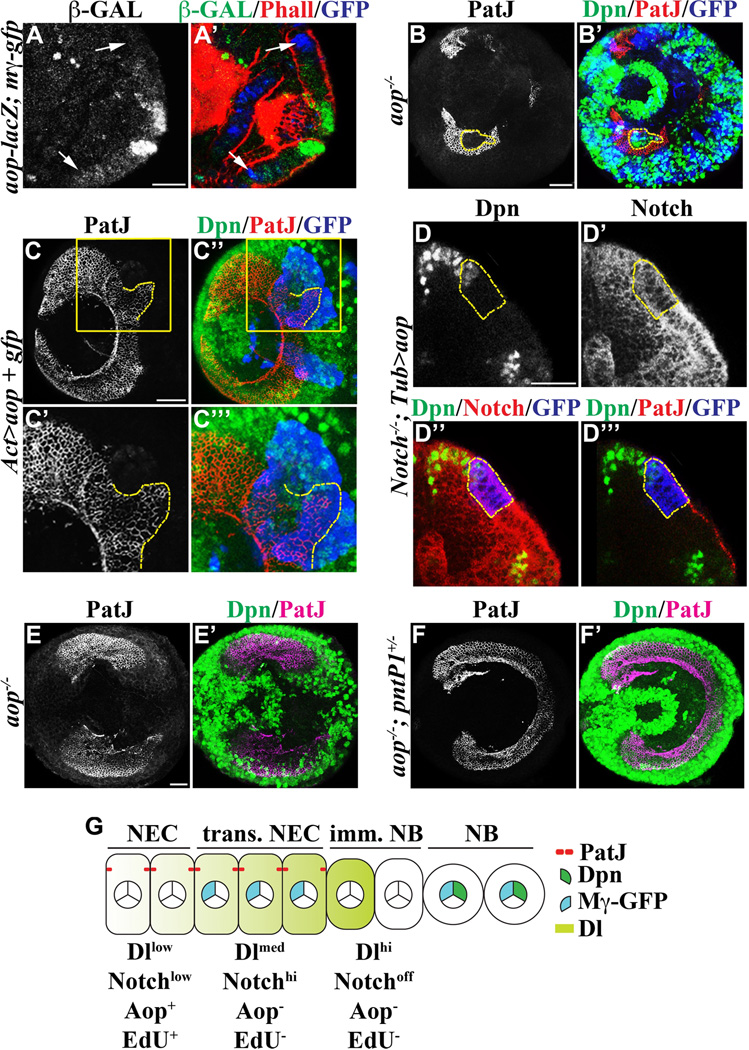
We took a candidate gene approach to test if aop might act downstream of Notch during maintenance of the neuroepithelial cell identity for the following reasons. First, the synchrony of neuroepithelial cell conversion into neuroblasts across the entire neuroepithelial swath resembles the morphogenic furrow sweeping across the larval eye-antennal imaginal disc (Hsiung and Moses, 2002; Doroquez and Rebay, 2006). Notch plays an important role in regulating eye morphogenesis by directly activating the aop gene (Rohrbaugh et al., 2002). Second, a recent study shows that Notch regulates maintenance of adult muscle progenitor cells by directly activating aop gene transcription (Rohrbaugh et al., 2002; Krejcí et al., 2009). We attempted to investigate if Aop is expressed in neuroepithelia by using a specific antibody, but failed to detect any specific expression patterns presumably due to a very low level of the endogenous Aop protein (data not presented). As an alternative approach, we analyzed expression of an aop-lacZ reporter transgene whose expression pattern largely mimics endogenous Aop in the developing eye disc (Rohrbaugh et al., 2002). We detected a low level of Aop-lacZ expression throughout most neuroepithelia in a non-overlapping pattern with E(spl)mγ-GFP (Figure 4A-A’; N = 5). Thus, we conclude that aop is expressed in neuroepithelia located laterally from transitioning neuroepithelial cells.
Figure 4. Aop functions downstream of Notch signaling to maintain the identity of neuroepithelial cells.
(A–A’) Aop is likely expressed in a low level in neuroepithelia. A low level of Aop-lacZ can be detected throughout neuroepithelia located laterally from transitioning neuroepithelial cells (white arrows). The scale bar = 20 µm.
(B–B’) Aop is necessary for maintaining the identity of neuroepithelial cells. Neuroepithelial cells in the GFP-marked aop mutant mosaic clones become prematurely converted into neuroblasts perturbing the swath of neuroepithelia in the outer proliferation center. The scale bar = 20 µm. The dotted yellow line outlines an aop mutant clone surrounded by wild type neuroepithelial cells.
(C–C’’’) Aop is sufficient to promote the identity of neuroepithelial cells. Neuroepithelial cells over-expressing aop (outlined in dotted yellow line) in the GFP-marked genetic clone expanded beyond the medial edge of neuroepithelia and became surrounded by neuroblasts located outside of the clone. However, many neuroepithelial cells in the clone marked by GFP eventually become converted into immature neuroblasts as indicated by the absence of PatJ and Dpn. A few neuroblasts can also been seen in the clone. The scale bar = 20 µm. The higher magnification image of the boxed area is shown in C’ and C’’’.
(D–D’’’) Over-expression of Aop suppresses premature conversion of neuroepithelial cells into neuroblasts in the Notch mutant clone. Ectopic expression of Aop allows Notch mutant neuroepithelial cells marked by the expression of GFP (outlined in dotted yellow line) to maintain their identity and undergo conversion into neuroblasts synchronously with neuroepithelial cells outside of the clone. Immunofluorescent staining using an antibody specific for the intracellular domain of the Notch protein confirms the Notch mutant clone. The scale bar = 20 µm.
(E–F’) Aop maintains the identity of neuroepithelial cells by repressing the pntP1 gene. (E-E’) Neuroepithelial cells in the sensitized aop1/yan1 mutant larval optic lobe become prematurely converted into neuroblasts perturbing the continuity of the neuroepithelial swath. The scale bar = 20 µm. (F-F’) Reduced function of pntP1 prevents premature conversion of neuroepithelial cells into neuroblasts and restores a continuous swath of neuroepithelia.
(G) A cartoon summarizes the expression pattern of Aop during conversion of neuroepithelial cells into neuroblasts. NEC: neuroepithelial cells; trans. NEC: transitioning neuroepithelial cells; imm. NB: immature neuroblast; NB: neuroblast.
We combined loss- and gain-of-function analyses to determine the function of aop in neuroepithelial cells. We induced the GFP-marked mosaic clone derived from aop mutant neuroepithelial cells and assessed the identity of cells within the clone. While wild type cells located directly adjacent to the clone maintained expression of the neuroepithelial cell marker PatJ, all cells within the aop mutant clone expressed the neuroblast marker Dpn (Figure 4B-B’). The high efficiency in aop mutant clone induction led to widespread premature formation of neuroblasts throughout neuroepithelia, indicating that aop is necessary for maintenance of neuroepithelia (Figure 4B-B’). We next tested if down-regulation of aop is necessary for formation of optic lobe neuroblasts by over-expressing a UAS-aop transgene in the neuroepithelial cell clone. Neuroepithelial cells in the control clone assumed the neuroblast identity synchronously with the cells outside of the clone (Figure 2E). In contrast, neuroepithelial cells in the clone over-expressing aop expanded beyond the medial edge of neuroepithelia and were surrounded by neuroblasts outside of the clone (Figure 4C-C’’’; N = 11). Importantly, the aop over-expressing clone also contained many immature neuroblasts and some neuroblasts, leading us to conclude that expression of Aop significantly delays but does not prevent conversion of neuroepithelial cells into neuroblasts (Figure 4C-C’’’; N = 11). Taken together, these data indicate that Aop is necessary for maintenance of neuroepithelia and down-regulation of Aop promotes efficient formation of neuroblasts.
We next examined if aop functions downstream of Notch to maintain neuroepithelial cell identity. In the absence of Notch function, neuroepithelial cells became converted into neuroblasts prematurely and delaminated inward from the rest of wild type neuroepithelia (Figure S2D). In contrast, over-expression of aop prevented Notch mutant neuroepithelia from becoming converted into neuroblasts prematurely and allowed them to undergo conversion into neuroblasts as the adjacent neuroepithelial cells outside of the clone (Figure 4D-D’’’; 82%, N = 11). Thus, Notch maintains the neuroepithelial cell identity via an Aop-dependent mechanism, but the conversion from neuroepithelial cells into neuroblasts occurs via an Aop-insensitive mechanism.
Transcriptional repression of the pntP1 gene is a well established mechanism by which Aop regulates various cell responses in the context of development (Hsiung and Moses, 2002; Doroquez and Rebay, 2006). Thus, we hypothesized that premature conversion of neuroepithelial cells into neuroblasts in the aop mutant optic lobe might be due to increased pntP1 function. We tested this hypothesis in the sensitized aop1/yan1 mutant genetic background, in which 66.7% of the mutant larvae show premature formation of neuroblasts interrupting the neuroepithelia swath in the outer proliferation center (Figure 4E-E’; N = 12). In contrast, heterozygosity of the pntP1-specific mutant allele (pntΔ33) prevented premature conversion of neuroepithelial cells into neuroblasts and restored continuity in the neuroepithelia swath in 100% of aop1/yan1 mutant larvae (Figure 4F-F’; N = 12). Thus, we conclude that Aop maintains the identity of neuroepithelial cells by repressing pntP1 (Figure 4G).
Up-regulation of Notch signaling in transitioning neuroepithelial cells increases the threshold of response to PntP1
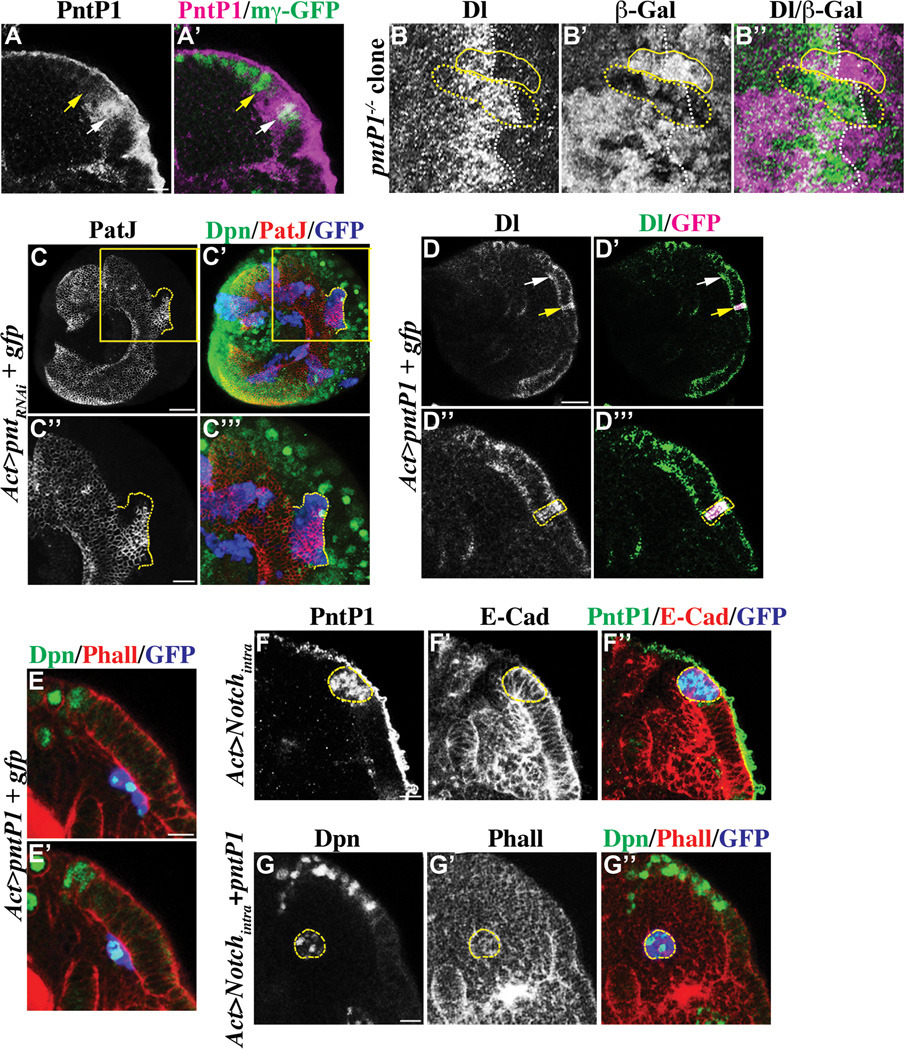
Since decreased pntP1 function suppressed premature formation of neuroblasts in the aop mutant optic lobe, we tested if pntP1 regulates conversion of neuroepithelial cells into neuroblasts. We first determined the expression of pntP1 by immunostaining and observed endogenous PntP1 largely co-localizing with E(spl)mγ-GFP in transitioning neuroepithelial cells but also is detectable in immature neuroblasts (Figure 5A-A’). The expression pattern of PntP1 led us to propose that pntP1 regulates the conversion of transitioning neuroepithelial cells into immature neuroblasts. We tested this hypothesis by generating negatively marked mosaic clones derived from pntΔ33 (pntP1-specific allele) mutant neuroepithelial cells and assessed the identity of cells within the clone spanning across the medial edge of neuroepithelia by using Delta as the marker. Cells in the wild type twin-spot clone showed abrupt down-regulation of Delta synchronously with cells outside of the clone (Figure 5B-B’’). In contrast, multiple rows of cells in the pntP1 mutant clone maintained robust expression of Delta despite locating beyond the medial edge of neuroepithelia as determined by sparse Delta expression in the surrounding cells located outside of the clone (Figure 5B-B’’). This result indicates that pntP1 is necessary for a timely conversion of transitioning neuroepithelial cells into immature neuroblasts, but additional parallel signaling mechanisms must also exist as Delta expression eventually became down-regulated.
Figure 5. PntP1 promotes conversion of transitioning neuroepithelial cells into immature neuroblasts.
(A–A’) PntP1 becomes transiently expressed in transitioning neuroepithelial cells. The expression of PntP1 largely co-localizes with E(spl)mγ-GFP in transitioning neuroepithelial cells (white arrow), but is undetectable in neuroblasts (yellow arrow). Please note that the antibody against PntP1 shows non-specific background staining on the surface of the brain. The scale bar = 10 µm.
(B–B’’) pntP1 is required for efficient conversion of neuroepithelia into neuroblasts. The conversion of neuroepithelial cells into neuroblasts is significant delayed in the negatively marked pntP1 mutant mosaic clone (outline in dotted yellow line) as indicated by prolonged expression of a high level of Delta. In contrast, neuroepithelial cells in the wild type control twin-spot clone (outlined in solid yellow line) show down-regulation Delta expression (white dotted line) and undergo conversion into neuroblasts synchronously with the surrounding cells outside of the clone.
(C–C’’’) The pnt gene is necessary for conversion of neuroepithelial cells into neuroblasts. Simultaneously reducing the function of pntP1 and pntP2 prevents neuroepithelial cells in the GFP-marked mosaic clone (dotted yellow line) from becoming converted into immature neuroblasts. The scale bar = 20 µm. The higher magnification image of the boxed area is shown in C’’ and C’’’. The scale bar = 10 µm.
(D–E’) pntP1 is sufficient to induce conversion of neuroepithelial cells into neuroblasts. (D-D’’’) Transient over-expression of pntP1 leads to dramatic up-regulation of Delta in neuroepithelial cells in the GFP-marked genetic clone (yellow arrow). The white arrow indicates transitioning neuroepithelial cells. The scale bar = 20 µm. The higher magnification image of neuroepithelia containing the clone (outlined in dotted yellow line) is shown in D’’ and D’’’. (E-E’) Neuroepithelial cells over-expressing pntP1 marked by expression of GFP delaminate inward away from the rest of neuroepithelia, and become converted into neuroblasts prematurely. A superficial optical section is shown in E and a distal optical section is shown in E’. The scale bar = 10 µm.
(F–G’’) Constitutively activated Notch signaling de-sensitizes neuroepithelial cells from PntP1. (F-F’’) Neuroepithelial cells over-expressing Notchintra marked by expression of GFP (outlined in dotted yellow line) accumulate at the medial edge of neuroepithelia. These neuroepithelial cells express PntP1, but do not become converted into neuroblasts. The scale bar = 10 µm. (G-G’’) Expression of PntP1 triggers neuroepithelial cells over-expressing Notchintra marked by GFP (outlined in yellow) to undergo conversion into neuroblasts. The scale bar = 10 µm.
The pnt locus encodes at least two protein isoforms P1 and P2 and PntP2 can compete with Aop for binding to the promoter of various target genes including pntP1 (Hsiung and Moses, 2002; Doroquez and Rebay, 2006). However, we could not directly test if pntP2 regulates differentiation of neuroepithelia because the pntP2-specific pntΔ78 mutant allele failed to complement the pntΔ33 mutant allele in our hands. Instead, we over-expressed a UAS-pntRNAi transgene that targets the common coding region for all pnt transcripts in the neuroepithelial cell clone and assessed the fate of cells within the GFP-marked clone located medially from the edge of neuroepithelia. Ten or more rows of neuroepithelial cells in 77.8 % of the pnt mutant clones expanded beyond the medial edge of neuroepithelia, and became surrounded by neuroblasts located outside of the clone (Figure 5C-C’’’; N = 9). Thus, simultaneously reducing all transcripts from the pnt locus prevented conversion of neuroepithelial cells more efficiently than removing the function of pntP1 alone. Thus, a successful conversion from transitioning neuroepithelial cells to neuroblasts likely requires both pntP1 and pntP2.
We extended our analyses to test if over-expression of pntP1 is sufficient to prematurely convert neuroepithelial cells into neuroblasts by transiently over-expressing a UAS-pntP1 transgene in the GFP-marked neuroepithelial cell clone. We then assessed whether neuroepithelia located laterally from transitioning neuroepithelial cells might show up-regulation of Delta. While wild type cells located immediately surrounding the clone showed a very low level of Delta expression, cells within the clone showed dramatic elevated expression of Delta (Figure 5D-D’’’; N = 16). We prolonged over-expression of pntP1 and assessed the identity of the cells within the GFP-marked clone. Neuroepithelial cells in the clone became prematurely converted into neuroblasts and delaminated inward away from the rest of neuroepithelia (Figure 5E-E’; N = 12). Thus, over-expression of pntP1 is sufficient to trigger premature conversion of neuroepithelial cells into neuroblasts.
Since the expression of pntP1 is critical for converting transitioning neuroepithelial cells into neuroblasts, we tested if constitutively activated Notch signaling prevents formation of neuroblasts by repressing pntP1 expression. We over-expressed Notchintra in the neuroepithelial cell clone and then assessed the identity of cells within the GFP-marked clone located at the edge of neuroepithelia. Unexpectedly, most cells in the clone resembled transitioning neuroepithelial cells in that they maintained the epithelial cell morphology and expressed PntP1 (Figure 5F-F’’; N = 10). This result was surprising since over-expression of pntP1 efficiently convert neuroepithelial cells into neuroblasts prematurely. One possible reason that neuroepithelial cells showing constitutively activated Notch signaling fail to become converted into neuroblasts despite expressing PntP1 is that their threshold of response to PntP1 might not have been reached. We tested this hypothesis by co-expressing Notchintra and pntP1 in the GFP-marked clone derived from neuroepithelial cells. Consistent with our hypothesis, neuroepithelial cells in 80% of the clones assumed the neuroblast fate as indicated by the absence of the epithelial cell morphology and expression of Dpn (Figure 5G-G’’; N = 15). Taken together, we conclude that transient activation of Notch signaling in transitioning neuroepithelial cells raises their threshold of response to PntP1.
Discussion
A deregulated conversion of neuroepithelial cells into neuroblasts perturbs formation of the neuronal network and will almost certainly lead to visual impairment of the adult fly. Thus, a dynamic balance between neuroepithelial cell maintenance and differentiation plays a pivotal role during morphogenesis of the optic lobe. In this study, we provide evidence that changes in Notch signaling regulate the dynamic balance between maintenance of neuroepithelial cells and formation of neuroblasts (Figure 6). A low level of Notch signaling maintains the neuroepithelial cell identity by triggering Aop-dependent repression of the pntP1 gene. Transient up-regulation of Notch signaling in transitioning neuroepithelial cells raises their threshold of response to PntP1 preventing them from precociously converting into immature neuroblasts. Finally, abrupt down-regulation of Notch signaling together with a high level of PntP1 trigger the conversion from transitioning neuroepithelial cells into immature neuroblasts at the medial edge of neuroepithelia. Thus, interplay between changes in Notch signaling and transient up-regulation of pntP1 orchestrates synchronous and progressive formation of neuroblasts in a medial-to-lateral orientation across the entire neuroepithelial swath.
Figure 6. A model summarizing changes in Notch signaling during conversion of neuroepithelial cells into neuroblasts.
Maintenance of optic lobe neuroepithelia
Lack of Notch reporter transgene expression throughout neuroepithelia located laterally from transitioning neuroepithelial cells has been perplexing in light of recent studies reporting that Notch signaling is necessary for maintenance of their identity. One possibility might be that these Notch reporter transgenes including E(spl)mγ-GFP might not contain all necessary regulatory response elements to respond to Notch signaling in most neuroepithelial cells. Alternatively, the level of Notch signaling might simply be too low to activate the expression of the Notch reporter transgene. We favor the second hypothesis for the following reasons. Since over-expression of Notchintra is sufficient to trigger robust cell autonomous expression of E(spl)mγ-GFP in neuroepithelia located laterally from transitioning neuroepithelial cells, this transgene does contain all necessary regulatory elements to respond to a high level of Notch signaling. Furthermore, the Notch ligand Delta is expressed in a low level throughout neuroepithelia located laterally from transitioning neuroepithelial cells and Delta likely functions to trans-activate Notch signaling in these cells. In the context of trans-activation of Notch signaling by Delta, the level of the ligand correlates with the level of signaling output (Sprinzak et al., 2010; del Álamo et al., 2011). Taken together, we conclude that maintenance of the neuroepithelial cell identity requires a low level of Notch signaling.
We propose that Notch maintains the identity of neuroepithelial cells by activating Aop-dependent repression of the pntP1 gene. The Suppressor of Hairless protein, which is necessary for activating transcription of Notch targets genes, directly binds to the promoter of the aop gene (Rohrbaugh et al., 2002; Krejcí et al., 2009). Furthermore, removing the Notch or aop function triggered premature conversion of neuroepithelia into neuroblasts whereas over-expressing Notch or aop prevented conversion of neuroepithelial cells into neuroblasts. Most importantly, over-expression of aop suppressed premature differentiation of Notch mutant neuroepithelial cells. Finally, heterozygosity of the pntP1 gene completely suppressed premature conversion of neuroepithelial cells into neuroblasts in a hypomorphic aop mutant genetic background. These data lead us to conclude that Notch signaling maintains the identity of neuroepithelial cells by activating an Aop-dependent repression of pntP1. In the future, analyses of Notch and pntP1 double mutants will be necessary to confirm this regulatory mechanism.
The conversion from transitioning neuroepithelial cells to immature neuroblasts
Down-regulation of Notch signaling is necessary for formation of neuroblasts, so transient up-regulation of Notch signaling in transitioning neuroepithelial cells appears rather counterproductive. One possibility might be that up-regulation of Notch signaling paces the conversion from transitioning neuroepithelial cells into neuroblasts by increasing their threshold of response to PntP1. Consistent with this hypothesis, constitutively activated Notch signaling prevented transitioning neuroepithelial cells from becoming converted into neuroblasts despite expressing PntP1. This hypothesis was further supported by co-expression of pntP1 overcoming the blockade by constitutively activated Notch signaling and restoring conversion of transitioning neuroepithelial cells into neuroblasts. Thus, we propose that up-regulation of Notch signaling in transitioning neuroepithelial cells raises their threshold of response to PntP1 and functions to prevent them from becoming converted into immature neuroblasts precociously. Such an elaborated mechanism only permits transitioning neuroepithelial cells expressing the highest level of PntP1 to convert into immature neuroblasts. This mechanism is consistent with a recent study reporting that the EGF ligand is processed and secreted by cells near the medial edge of the optic lobe neuroepithelia (Yasugi et al., 2010). As a result of simple diffusion, transitioning neuroepithelial cells at the medial edge of neuroepithelia will be exposed to the highest level of the EGF ligand and will express the highest level of PntP1. As such, EGF signaling likely creates a vector field establishing the directionality of conversion from neuroepithelial cells into neuroblasts whereas Notch signaling refines the functional output of EGF signaling by raising the threshold response to PntP1.
Many important questions arise from this highly plausible mechanism by which the interplay between Notch and EGF signaling paces synchronous conversion of neuroepithelial cells into neuroblasts one row at a time. This model will predict that immature neuroblasts immediately adjacent to transitioning neuroepithelial cells should secrete the processed EGF ligand. However, the antibody specific for the Rhomboid (Rho) protease required for proteolytic activation of the EGF protein is currently unavailable and a genomic fragment encompassing the rho-1 locus tagged with YFP did not show detectable expression in the larval optic lobe (Weng and Lee, unpublished observation). Alternatively, a recent study shows that pntP1 is a direct target of Notch in vivo (Krejcí et al., 2009). Thus, up-regulation of Notch signaling might directly activate transcription of the pntP1 gene in transitioning neuroepithelial cells. Since Notch signaling becomes abruptly down-regulated at the medial edge of neuroepithelia, it is highly possible that the threshold of response to PntP1 also becomes lowered in the same cells. Thus, the pre-existing level of PntP1 protein will likely be sufficient to trigger the conversion from transitioning neuroepithelial cells into immature neuroblasts. More experiments including identification of the cell type from which the processed EGF ligand is released and a direct test to confirm the role of EGF signaling during conversion of neuroepithelia into neuroblasts will be key to distinguish these two possible mechanisms.
Other signaling mechanisms that affect differentiation of neuroepithelial cells
The fat-hippo signaling mechanism controls tissue growth by regulating proliferation and cell death and promotes timely differentiation of optic lobe neuroepithelial cells (Reddy et al., 2010; Zhao et al., 2010). While inactivation of fat-hippo signaling delays conversion of neuroepithelia into neuroblasts, removal of the downstream effecter yorkie only accelerates the conversion near the medial edge of the optic lobe neuroepithelia (Reddy et al., 2010). Thus, fat-hippo signaling likely functions as a gatekeeper to prevent over-growth of optic lobe neuroepithelia by triggering transient cell cycle arrest. Intriguingly, transient cell cycle arrest precedes increased Notch signaling in transitioning neuroepithelial cells. Detailed studies in the future will be necessary to determine if activation of the fat-hippo signaling might contribute to increased Notch signaling in transitioning neuroepithelial cells.
Experimental Procedure
Fly Genetics and Transgenes
o-fut12834, aph-15072 and fat3477 alleles were recovered from EMS mutagenesis following a standard protocol. Drosophila cultures were kept at 25oC on standard cornmeal food. Other mutant alleles and transgenes used in this study include pcna-3XEmgfp (R. Duronio), o-fut14R6 (Sasamura et al., 2007), E(spl)mγ-gfp (Wech et al., 1999), UAS-Notchintra (Chung and Struhl, 2001), aop-lacZ (Rohrbaugh et al., 2002), hs-flp; FRT40A, tub-Gal80; tub-Gal4, UAS-mCD8-GFP (Izergina et al., 2009), FRT19A, tub-Gal80, hs-flp; UAS-mcd8::GFP (Bayraktar et al., 2010), rho-1–YFP (Yogev et al., 2010). The following stocks were obtained from the Bloomington Stock Center: tub-Gal4, hs-flp(F38), act-FRT-CD2-FRT-Gal4, UAS-GFP, act-FRT-CD2-FRT-Gal4, UAS-mRFP, UAS-mCD8-GFP, tub-GAL80ts,GH146-Gal4, UAS-delta, UAS-aop.WT, UAS-pntP1, N55e11, FRT19A, aopyan1, aop1, aph-1D35, pntΔ33, cswG0170, cnkE2083, cnkk16314, ftG-rv.
Immunofluorescent microscopy and antibodies
Larval brains were dissected in Schneider’s medium and fixed in PBS containing 0.3% Triton X-100 (PBS-T) containing 4% Formaldehyde for 23 min at room temperature; quickly washed in PBS-T; incubated in primary antibodies diluted in PBS-T for 3-4 hrs at the room temperature or overnight at 4°C. After quick washing with PBS-T, the specimen was incubated in secondary antibodies in PBS-T at the room temperature or overnight at 4°C. After quick washing in PBS-T, the specimen was equilibrated in Prolong Gold (Molecular Probe). Antibodies used in this study include rat Dpn (1:1), guinea pig Dpn (1:2500, J. Skeath), rabbit PatJ (1:2000, H. Bellen), rabbit PntP1 (1:1000, J. Skeath), L’(sc) (A. Carmena), rat α-tubulin (1:100, Serotec), rabbit phosphorylated-histone H3 (1:1000, Upstate), rabbit β-Gal (1:1000, ICN/Cappel), mouse β-Gal (1:100, Sigma), chicken GFP (1:2000, Aves Lab), rabbit RFP (1:100, Rockland), mouse BrdU (1:100, BD Biosciences), mouse Elav (DSHB), mouse dE-Cad (DSHB), mouse Notchintra (1:100, DSHB) and mouse Delta (1:100, DSHB). Secondary antibodies were from Molecular Probes and Jackson Labs (details are available upon request). To stain cortical actin, rhodamine Phalloidin or Alexa 488 Phalloidin (Molecular Probes) was used as 1:100 in PBS-T. The images were acquired on a Leica SP5 scanning confocal microscope. Three-dimensional reconstruction of the fly larval optic lobe was performed by using the Mimics software (Materialise).
EdU staining
Larvae were aged for 72 hr after hatching and were pulse labeled for 3 hrs by feeding on the Kankel-White media containing 50 µg/ml EdU (5-ethynyl-2’deoxyuridine) (Daul et al., 2010). Half of the larvae were processed for staining immediately following the pulse; remaining larvae were transferred to standard media for a 12 hr EdU-free chase. Larvae were dissected and processed for antibody staining. Incorporated EdU was detected by Click-iT fluorescent dye azide reaction as described in the Click-iT product literature (Invitrogen).
Supplementary Material
Acknowledgement
We thank Dr. Chris Doe in whose lab the optic lobe defective mutants were isolated when C.-Y. Lee was a post-doctoral fellow. We thank Kristin Wildermuth for the technical assistance in the isolation of these mutants and Dr. M. Rolls for the UAS-3xEmGFP vector. We thank Drs. H. Bellen, S. Bray, A. Carmena, C. Doe, R. Duronio, Z.-C. Lai, R. Lehmann, K. Matsuno, H. Reichert, B. Shilo, J. Skeath and G. Struhl for fly stocks and antibody reagents. We thank the Bloomington Drosophila Stock Center and Vienna Drosophila RNAi Center for fly stocks. We thank Drs. I. Maillard and Y. Yamashita and the members of the Lee lab for reading the manuscript and providing critical comments. M.W. was supported by a Rackham graduate school predoctoral fellowship from the University of Michigan. C.-Y.L was initially supported by a Damon Runyon postdoctoral fellowship, and is currently supported by the Burroughs Wellcome Fund Career Award in the Biomedical Sciences (1006160.01), a Sontag Foundation Distinguished Scientist Award and a NIH grant (GM092818).
Reference
- Bayraktar OA, Boone JQ, Drummond ML, Doe CQ. Drosophila type II neuroblast lineages keep Prospero levels low to generate large clones that contribute to the adult brain central complex. Neural Dev. 2010;5:26. doi: 10.1186/1749-8104-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- Chung HM, Struhl G. Nicastrin is required for Presenilin-mediated transmembrane cleavage in Drosophila. Nat. Cell Biol. 2001;3:1129–1132. doi: 10.1038/ncb1201-1129. [DOI] [PubMed] [Google Scholar]
- Daul AL, Komori H, Lee C-Y. EdU (5-ethynyl-2'-deoxyuridine) labeling of Drosophila mitotic neuroblasts. Cold Spring Harb Protoc. 2010;2010 doi: 10.1101/pdb.prot5461. pdb.prot5461. [DOI] [PubMed] [Google Scholar]
- del Álamo D, Rouault H, Schweisguth F. Mechanism and significance of cis-inhibition in Notch signalling. Curr Biol. 2011;21:R40–R47. doi: 10.1016/j.cub.2010.10.034. [DOI] [PubMed] [Google Scholar]
- Doroquez DB, Rebay I. Signal Integration During Development: Mechanisms of EGFR and Notch Pathway Function and Cross-Talk. Crit Rev Biochem Mol Biol. 2006;41:339–385. doi: 10.1080/10409230600914344. [DOI] [PubMed] [Google Scholar]
- Egger B, Boone JQ, Stevens N, Brand AH, Doe C. Regulation of spindle orientation and neural stem cell fate in the Drosophila optic lobe. Neural Dev. 2007;2:1. doi: 10.1186/1749-8104-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger B, Gold KS, Brand AH. Notch regulates the switch from symmetric to asymmetric neural stem cell division in the Drosophila optic lobe. Development. 2010;137:2981–2987. doi: 10.1242/dev.051250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis R, McGrath G, Zhang J, Ruddy DA, Sym M, Apfeld J, Nicoll M, Maxwell M, Hai B, Ellis MC, Parks AL, Xu W, Li J, Gurney M, Myers RL, Himes CS, Hiebsch R, Ruble C, Nye JS, Curtis D. aph-1 and pen-2 are required for Notch pathway signaling, gamma-secretase cleavage of betaAPP, and presenilin protein accumulation. Dev Cell. 3:85–97. doi: 10.1016/s1534-5807(02)00189-2. [DOI] [PubMed] [Google Scholar]
- Hsiung F, Moses K. Retinal development in Drosophila: specifying the first neuron. Hum Mol Genet. 2002 May 15;11(10):1207–1214. doi: 10.1093/hmg/11.10.1207. Review. Hum Mol Genet: 1207-1214. [DOI] [PubMed] [Google Scholar]
- Izergina N, Balmer J, Bello B, Reichert H. Postembryonic development of transit amplifying neuroblast lineages in the Drosophila brain. Neural Dev. 2009;11:44. doi: 10.1186/1749-8104-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolch W. Coordinating ERK/MAPK signalling through scaffolds and inhibitors. Nat Rev Mol Cell Biol. 2005;6:827–837. doi: 10.1038/nrm1743. [DOI] [PubMed] [Google Scholar]
- Krejcí A, Bernard F, Housden BE, Collins S, Bray SJ. Direct response to Notch activation: signaling crosstalk and incoherent logic. Sci Signal 2:ra1. 2009 doi: 10.1126/scisignal.2000140. [DOI] [PubMed] [Google Scholar]
- Kriegstein A, Alvarez-Buylla A. The glial nature of embryonic and adult neural stem cells. Annu Rev Neurosci. 2009;32:149–184. doi: 10.1146/annurev.neuro.051508.135600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AC, Lyons EL, Herman TG. cis-Inhibition of Notch by endogenous Delta biases the outcome of lateral inhibition. Curr Biol. 2009;19:1378–1378. doi: 10.1016/j.cub.2009.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo KT, Wang J, Junker M, Kriz S, Vo G, Asem B, Olson JM, Banerjee U, Hartenstein V. Concomitant requirement for Notch and Jak/Stat signaling during neuro-epithelial differentiation in the Drosophila optic lobe. Dev Biol. 2010;346:284–295. doi: 10.1016/j.ydbio.2010.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okajima T, Irvine KD. Regulation of notch signaling by o-linked fucose. Cell. 2002;111:893–904. doi: 10.1016/s0092-8674(02)01114-5. [DOI] [PubMed] [Google Scholar]
- Orihara-Ono M, Toriya M, Nakao K, Okano H. Downregulation of Notch mediates the seamless transition of individual Drosophila neuroepithelial progenitors into optic medullar neuroblasts during prolonged G1. Dev Biol. 2011;351:163–175. doi: 10.1016/j.ydbio.2010.12.044. [DOI] [PubMed] [Google Scholar]
- Perkins LA, Larsen I, Perrimon N. corkscrew encodes a putative protein tyrosine phosphatase that functions to transduce the terminal signal from the receptor tyrosine kinase torso. Cell. 1992;70:225–236. doi: 10.1016/0092-8674(92)90098-w. [DOI] [PubMed] [Google Scholar]
- Reddy BVVG, Rauskolb C, Irvine KD. Influence of Fat-Hippo and Notch signaling on the proliferation and differentiation of Drosophila optic neuroepithelia. Development. 2010;137:2397–2408. doi: 10.1242/dev.050013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrbaugh M, Ramos E, Nguyen D, Price M, Wen Y, Lai Z-C. Notch Activation of yan Expression Is Antagonized by RTK/Pointed Signaling in the Drosophila Eye. Current Biology. 2002;12:576–581. doi: 10.1016/s0960-9822(02)00743-1. [DOI] [PubMed] [Google Scholar]
- Sasamura T, Ishikawa HO, Sasaki N, Higashi S, Kanai M, Nakao S, Ayukawa T, Aigaki T, Noda K, Miyoshi E, Taniguchi N, Matsuno K. The O-fucosyltransferase O-fut1 is an extracellular component that is essential for the constitutive endocytic trafficking of Notch in Drosophila. Development. 2007;134:1347–1356. doi: 10.1242/dev.02811. [DOI] [PubMed] [Google Scholar]
- Sprinzak D, Lakhanpal A, LeBon L, Santat LA, Fontes ME, Anderson GA, Garcia-Ojalvo J, Elowitz MB. Cis-interactions between Notch and Delta generate mutually exclusive signalling states. Nature. 2010;465:86–90. doi: 10.1038/nature08959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therrien M, Wong AM, Rubin GM. CNK, a RAF-binding multidomain protein required for RAS signaling. Cell. 1998;95:343–353. doi: 10.1016/s0092-8674(00)81766-3. [DOI] [PubMed] [Google Scholar]
- Van Vactor D, O'Reilly AM, Neel BG. Genetic analysis of protein tyrosine phosphatases. Curr Opin Genet Dev. 1996;8:112–126. doi: 10.1016/s0959-437x(98)80070-1. [DOI] [PubMed] [Google Scholar]
- Wang W, Liu W, Wang Y, Zhou L, Tang X, Luo H. Notch signaling regulates neuroepithelial stem cell maintenance and neuroblast formation in Drosophila optic lobe development. Dev Biol. 2011;350:414–428. doi: 10.1016/j.ydbio.2010.12.002. [DOI] [PubMed] [Google Scholar]
- Wech I, Bray S, Delidakis C, Preiss A. Distinct expression patterns of different enhancer of split bHLH genes during embryogenesis of Drosophila melanogaster. Dev Genes Evol. 1999;209:370–375. doi: 10.1007/s004270050266. [DOI] [PubMed] [Google Scholar]
- Yasugi T, Sugie A, Umetsu D, Tabata T. Coordinated sequential action of EGFR and Notch signaling pathways regulates proneural wave progression in the Drosophila optic lobe. Development. 2010;137:3193–3203. doi: 10.1242/dev.048058. [DOI] [PubMed] [Google Scholar]
- Yogev S, Schejter ED, Shilo BZ. Polarized secretion of Drosophila EGFR ligand from photoreceptor neurons is controlled by ER localization of the ligand-processing machinery. PLoS Biol. 2010:8. doi: 10.1371/journal.pbio.1000505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Guan K-L. Hippo signaling at a glance. J Cell Sci. 2010;123:4001–4006. doi: 10.1242/jcs.069070. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.