Summary
Most mutations in cancer genomes are thought to be acquired after the initiating event, which may cause genomic instability, driving clonal evolution. However, for acute myeloid leukemia (AML), normal karyotypes are common, and genomic instability is unusual. To better understand clonal evolution in AML, we sequenced the genomes of AML samples with a known initiating event (PML-RARA) vs. normal karyotype AML samples, and the exomes of hematopoietic stem/progenitor cells (HSPCs) from healthy people. Collectively, the data suggest that most of the mutations found in AML genomes are actually random events that occurred in HSPCs before they acquired the initiating mutation; the mutational history of that cell is “captured” as the clone expands. In many cases, only one or two additional, cooperating mutations are needed to generate the malignant founding clone. Cells from the founding clone can acquire additional cooperating mutations, yielding subclones that can contribute to disease progression and/or relapse.
Introduction
The molecular pathogenesis of acute myeloid leukemia (AML) has not yet been completely defined. Recurrent chromosomal structural variations (e.g., t(15;17), t(8;21), inv(16), t(9;21), t(9;11), del5, del7, etc.) are established diagnostic and prognostic markers, suggesting that acquired genetic abnormalities play an essential role in leukemogenesis (Betz and Hess, 2010). However, nearly 50% of AML cases have a normal karyotype (NK), and many of these lack recurrent structural abnormalities, even with high density comparative genomic hybridization (CGH) or single nucleotide polymorphisms (SNP) arrays (Bullinger et al., 2010; Suela et al., 2007; Walter et al., 2009)). Targeted sequencing efforts have identified several mutations that carry diagnostic and prognostic information, including mutations in FLT3, NPM1, KIT, CEBPA and TET2 (reviewed in (Bacher et al., 2010; Stirewalt and Radich, 2003)). The advent of massively parallel sequencing enabled the discovery of recurrent mutations in DNMT3A (Ley et al., 2010; Yamashita et al., 2010) and IDH1 (Mardis et al., 2009). Despite these efforts, more than 25% of AML patients carry no mutations in the known leukemia-associated genes (Shen et al., 2011). Furthermore, defining the molecular consequences of recurring mutations (e.g. whether a mutation is an initiating or a cooperating event) has been challenging.
PML-RARA is one of the best-characterized leukemia-initiating mutations. This fusion gene results from t(15;17)(q22;21), and is associated exclusively with acute promyelocytic leukemia (APL, FAB M3 AML) (Allford et al., 1999; Rowley et al., 1977). Its expression is diagnostic of this single type of leukemia with unique clinical features (Sanz et al., 2009). Its presence predicts a near universal therapeutic response to a targeted agent, all-trans retinoic acid (ATRA), which is abrogated by mutations that inhibit ATRA binding to PML-RARA (Imaizumi et al., 1998; Larson and Le Beau, 2011; Takayama et al., 2001). Early myeloid expression of PML-RARA results in leukemia with promyelocytic features in multiple mouse models of the disease, although long latency (which can be shortened by radiation, alkylator treatment, or FLT3 ITD co-expression), suggests that PML-RARA requires cooperating events to cause leukemia (Funk et al., 2008; Kelly et al., 2002; Kogan, 2007; Sohal et al., 2003; Walter et al., 2004).
In this study, we sequenced the genomes of 24 AML cases. We chose to compare 12 genomes from patients with FAB M3 AML (where the initiating event is known) to 12 genomes from patients with AML without maturation (FAB M1) with normal cytogenetics, where the initiating event is less clear for most patients. In this and previous studies, we have demonstrated that AML genomes generally contain hundreds of mutations, that the total number of mutations per AML genome is related to the age of the patient, and that nearly all AML cells in the samples contain all of the mutations (although very few of these mutations are recurrent in AML or other malignancies) (Ding et al., 2012; Ley et al., 2008; Link et al., 2011; Mardis et al., 2009; Welch et al., 2011b). We show here that clonally derived hematopoietic cells from normal individuals also accumulate mutations as a function of age. This suggests that most of the mutations present in AML genomes were already present in the hematopoietic cell that was ‘transformed’ by the initiating mutation; nearly all of these preexisting mutations are probably benign and irrelevant for pathogenesis.
Consistent with this hypothesis, we observed that M1 and M3 genomes have similar numbers of total mutations, and that M1 genomes contain unique mutations (e.g., DNMT3A, NPM1, IDH1, or TET2) that almost never occur in M3 cases. In addition, there are many mutations that are shared between the two subtypes (e.g., FLT3 ITD), suggesting that these mutations can cooperate with a variety of initiating mutations. Because the data is comprehensive for all 24 genomes, it also allows us to estimate the minimum number of recurring mutations that may be responsible for the pathogenesis of AML.
Results
Whole genome sequencing of 24 AML samples
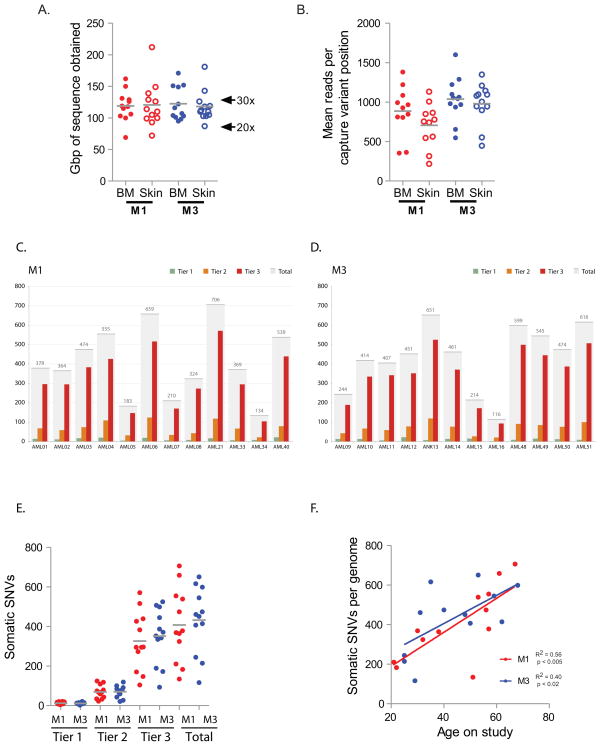
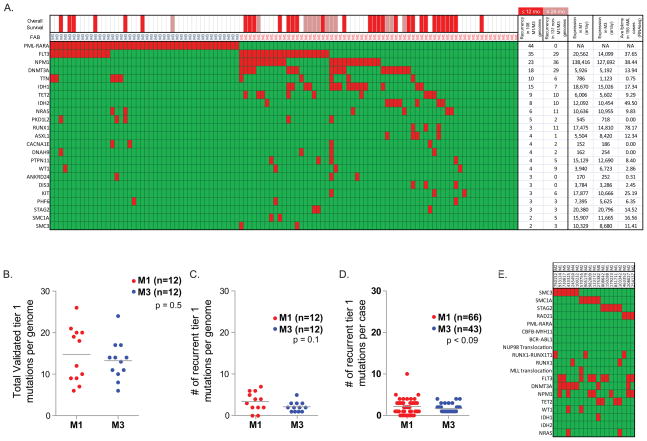
We subjected 12 cases of NK M1 AML and 12 cases of t(15;17)-positive M3 AML to whole genome sequencing (WGS) (case descriptions provided in Supplemental Information, summarized in Supplemental Table 1 and Supplemental Figure 1). To identify somatic, AML-associated mutations, we subjected both the bone marrow (leukemic tissue) and skin (normal tissue) to WGS (average haploid coverage 28×, Figure 1A and Supplemental Table 2); the mutations in the AML1 and AML2 genomes have been previously reported and deposited in dbGaP (Ding et al., 2012; Ley et al., 2008; Mardis et al., 2009). They are included in this study for ease of reference. Because of the prevalence of false positive calls in WGS (between 20% – 50%, depending on the stringency of type I errors tolerated), we validated all single nucleotide variants (SNVs), small insertions and deletions (indels), and structural variants (SVs) identified in tiers 1, 2, or 3 (which contain the non-repetitive portion of the genome; see ((Mardis et al., 2009) for definitions of tiers) using patient-specific custom NimbleGen capture arrays, followed by Illumina sequencing (Figure 1B). All subsequent analysis relies on these validated data, and not on the primary genome discovery sequence. An average coverage of 972 reads per somatic variant was obtained at validation. We observed a higher validation frequency in tier 1 than in tier 2 and 3 (mean frequency 0.5 vs. 0.35 and 0.29, p < 0.002 and p < 0.0001 respectively, Supplemental Table 2), which may reflect the lower GC content and increased uniqueness of tier 1. Numbers of mutations and validation frequencies were similar across all tiers in M1 vs. M3 genomes (Figure 1C – E and Supplemental Table 2). These data yielded a total of 10,563 validated somatic variants for the 24 genomes (average 440/genome) (Supplemental Tables 2, 3, and 4). Indels were less frequent than SNVs in M1 and M3 AML (M1: average 19.1 per genome, range 4–45; M3: average 20.3 per genome, range 4–56), and occurred proportionally less often in tier 1 (average 1.4 per genome, range 0–4). Of these total somatic mutations, 319 occurred in the coding regions of 287 unique genes; an average of 10 somatic mutations had translational consequences in each genome (Supplemental Table 3). A fusion event between PML and RARA was identified in all 12 M3 AML cases (Supplemental Table 5). Only 43 additional structural variants (translocations, insertions, deletions, and inversions) were identified (median 2 per genome, range 0–8, Supplemental Table 5). The t(15;17) breakpoints occurred in exon 5 and introns 3 and 5 of PML, and within RARA intron 2 in all cases, as expected; two cases were associated with large deletions that are predicted to result in PML-RARA expression without reciprocal RARA-PML expression (Supplemental Figure 2). None of the other structural variations found by WGS were identified with metaphase cytogenetics; most are predicted to be cryptic when examined by routine cytogenetics. The others may exist in minor leukemic subclones that were not otherwise detected, or in cells that did not expand during preparation for cytogenetic studies.
Figure 1. Coverage and number of validated single nucleotide variants (SNVs) by tier per genome.
A. Gigabase pairs of sequence obtained for each genome assessed by whole genome sequencing. B. Mean number of reads obtained for each variant assessed per genome during validation. C and D. Number of tier 1, 2, 3, and total variants validated in each sample. E. Total number of validated SNVs by tier in M1 AML (red) versus M3 AML (blue) cases. F. Total number of validated SNVs per genome in non-redundant regions (tier 1, 2, and 3) versus age of patient; M1 AML (red), M3 AML (blue). The R2 for the combined 24 cases is 0.5, p < 0.0001.
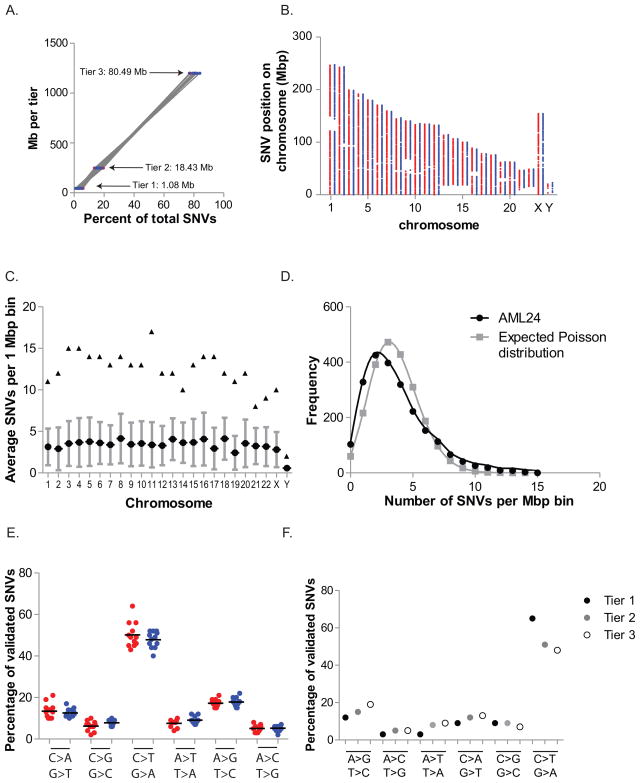
The total number of validated SNVs per genome increased in proportion to patient age, in both M1 and M3 AML genomes (Figure 1F). We also observed that somatic AML mutations were widely distributed throughout the genome; in all 24 cases, the number of SNVs in each tier was directly proportional to the amount of the genome present in that tier (SNVs per Mb: tier 1: 0.264 ± 0.024; tier 2: 0.283 ± 0.026; tier 3: 0.283 ± 0.024, p < 0.91) (Figure 2A) and these mutations were distributed across all chromosomes (Figure 2B). However, with this sample size, modestly increased mutational frequency within small regions of the genome cannot be excluded (Figure 2C and D).
Figure 2. Genomic distribution of somatic variants.
A. Percentage of validated SNVs in each tier versus the number of genomic megabases occupied by that tier; M1 (red), M3 (blue). Average R2 M1: 0.999 (range: 0.998–1.000); M3: 0.999 (range: 0.998–1.000). B. Distribution of validated SNVs throughout the genome. Each point represents the genomic position of an SNV: X axis represents chromosomes in numerical order; Y axis represents base pair position on each chromosome in megabase pairs; M1 AML (red), M3 AML (blue). The large discontinuous regions represent regions of the reference genome without defined sequence. C. Average (circle), standard deviation (bars), and maximal (triangles) number of SNVs per megabase by chromosome in all 24 cases. D. Frequency of SNVs per megabase of non-tier 4 genomic space compared with the expected Poisson distribution if SNVs were randomly distributed across non-tier 4 space. E. Mutational spectrum of validated SNVs. Percentage of validated SNVs that occur in non-redundant regions of the genome (tiers 1, 2, and 3) with indicated nucleotide changes in M1 AML (red) and M3 AML (blue). F. Mutational spectrum as defined by tier of each event (tier 1, genic; tier 2, highly conserved and/or with regulatory potential; tier 3, unique in the genome, and not in tier 1 or tier 2, see Mardis et. al. 2009 for details).
The mutational spectrum was dominated by C>T/A>G transitions, and was similar for M1 and M3 cases (Figure 2E). Tier 1 mutations favored C>T/A>G transitions, probably due to an increased proportion of methylated cytosine nucleotides in tier 1, which is associated with increased susceptibility to deamination and subsequent transition mutations (Figure 2F) (Pfeifer, 2006).
For this study, skin samples were obtained when the patients presented with leukemia. As expected, we observed low-level contamination of leukemic variants in the skin samples, which was proportional to the peripheral WBC count at the time of skin collection (Supplemental Figure 3A–C). M3 AML cases usually had very low leukemia contamination in the skin (often completely absent), although we observed two outlier cases. There was no correlation between disproportionally high skin contamination and clinical history (e.g. leukemia cutis, gum hypertrophy, pulmonary hemorrhage, or intracranial hemorrhage).
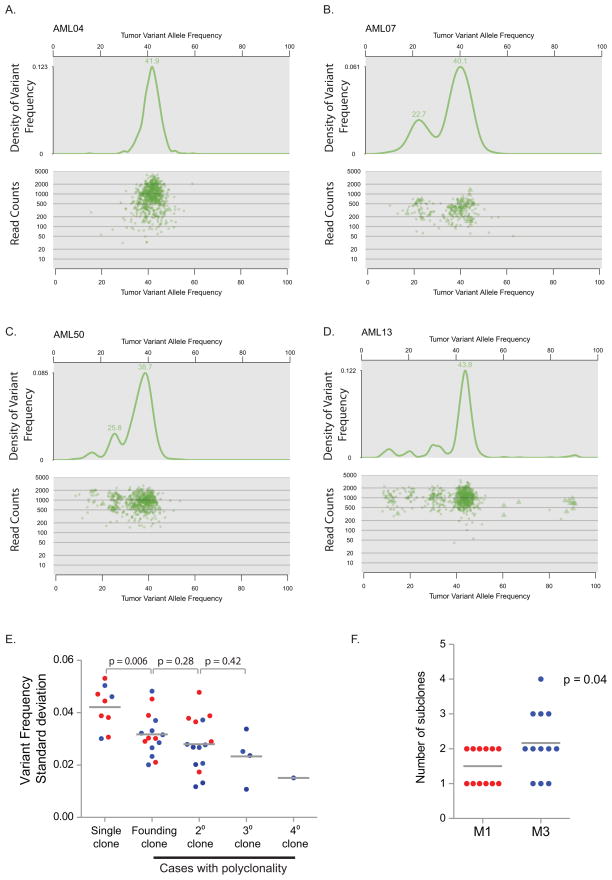
Clusters of mutations with similar variant allele frequencies within individual cases provide evidence of a single founding clone in both M1 and M3 samples (Ding et al., 2012). Using kernel density analysis, we identified 1–4 clusters of mutations (representing the founding clone in all cases, with or without additional subclones derived from the founding clone) in each genome (Figure 3A–D); the clusters were independent of the number of read counts for each SNV. In cases with subclones, the number of variants specific to each subclone was relatively small (an average of 40.4 SNVs per subclone (range 6 – 110)); SNVs in subclones represented only 14% of the total SNVs per genome (range 2% – 33%). Because each genome’s founding clone contains heterozygous SNVs that appear to be present in nearly all of the cells in the sample, subclones must also contain all of the SNVs in the founding clone. For example, the average variant frequency of heterozygous SNVs in the founding clone of AML13 (Figure 3D) is 44%, suggesting that 88% of the cells in the sample contain these heterozygous mutations; therefore, the subclones with average variant allele frequencies of 12%, 22%, and 32% must also contain the mutations found in the founding clone. Genomes with two or more mutation clusters displayed a similar standard deviation of variant frequencies within separate clusters (Figure 3E). However, genomes with a single cluster tended to have an increased standard deviation of the variant frequency within that cluster, suggesting that overlapping subclones may exist in these samples that are below this level of resolution. There was a trend towards more subclones in M3 cases (M1 average 1.5 clones/genome; M3 average 2.2 clones/genome; p = 0.04) (Figure 3F).
Figure 3. Clonality assessment in NK M1 AML and t(15;17)-positive M3 AML.
A–D. Clusters of variants that occur with similar frequencies identify AML founding clones and subclones. Upper panels: probability density function using kernel density estimation of data in lower panels. The graphed line shows the relative probability of each variant allele frequency in the total sample. Lower panels: for each SNV, the frequency of the variant allele within the bone marrow sample is plotted versus the total number of sequencing reads covering the corresponding nucleotide position. Triangles indicate SNVs on the X chromosome. E. The standard deviation of the variant frequency for each cluster in all 24 cases: M1 AML (red); M3 AML (blue). F. Number of clusters identified by kernel density estimation in M1 AML (red) and M3 AML (blue).
Several pieces of data suggest that most of the mutations in the founding clone may be random events that preexisted in the hematopoietic cell that acquired the initiating mutation: 1) the presence of hundreds of mutations in the founding clones of all AML genomes (which is far greater than the number expected to be biologically relevant), 2) their presence in nearly all of the AML cells in the sample, and 3) the correlation of the number of total mutations with the patient’s age. We expect that an AML-initiating mutation would provide a growth advantage for an HSPC, and that the preexisting, random mutations in that HSPC would therefore be “captured” in all its progeny when that cell clonally expands. Even though the preexisting mutations would not be pathogenetically relevant, they would be present in all of the cells in the founding clone, and appear as somatic mutations when the AML sample is sequenced. Although this hypothesis cannot be proven directly with current technologies, it strongly suggests that HSPCs derived from healthy people may acquire random, benign mutations as a function of age. This issue is addressed in the next section.
Identification of background mutations in normal human hematopoietic cells
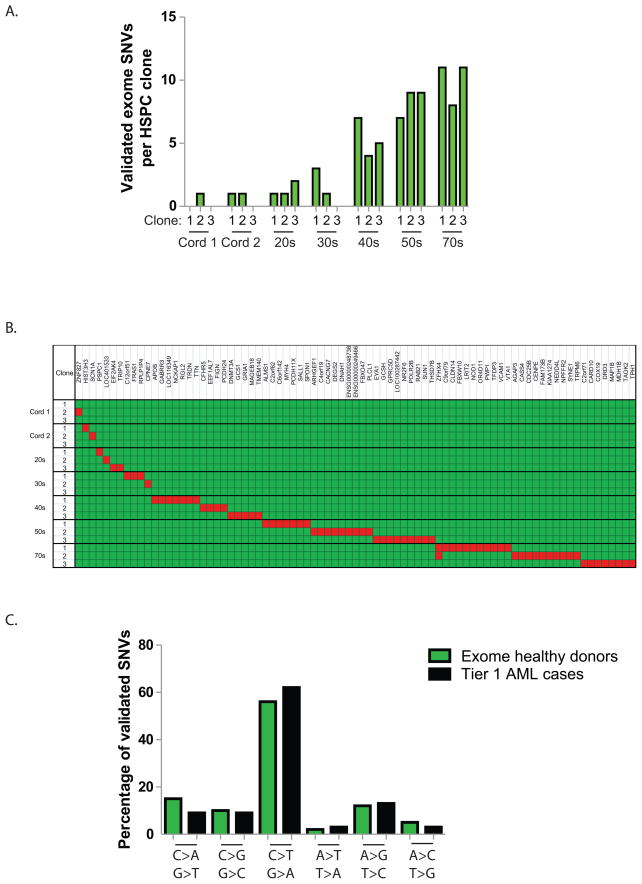
To determine whether normal human hematopoietic cells accumulate mutations over time, we performed exome sequencing on the progeny of three different HSPCs from seven healthy individuals of different ages, and compared these sequences to those of normal peripheral blood from the same donors (which are thought to have contributions from ~1,000 hematopoietic stem cells at any given time) (Abkowitz et al., 1996; Abkowitz et al., 2000; Catlin et al., 2011). Details of the experiments are provided in the Methods section, and Supplemental Figure 6. We focused on mutations with variant allele frequencies of 50% (+/− 2 SDs), which strongly suggests that they are present in all cells in the sample; mutations that are not present in all cells may have arisen during the outgrowth of the HSPCs in culture, and would be artifacts of the ex vivo expansion. The number of validated mutations in each clonally derived sample was lowest in the cord blood samples, and increased as a function of age (Figure 4A); in adult volunteers, the number of mutations detected in each exome was similar to that detected in AML patients of the same age. Each of the mutations was distinct for each of the clones (Figure 4B and Supplemental Table 6). The mutational spectrum was very similar to that of AML samples (Figure 4C); both have mostly transitions, suggesting that they may represent mutations caused by deamination of methylcytosine residues. Collectively, these data suggest that normal, self-renewing hematopoietic stem cells accumulate random background mutations as a function of age. These background mutations would therefore be present in all cells present in an AML founding clone, would increase with age, and would generally be irrelevant for AML pathogenesis.
Figure 4. Background mutations in normal HSPCs accumulate as a function of age.
A. Number of validated SNVs per exome identified in each of 3 clones that were derived from individual hematopoietic stem/progenitor cells from 7 anonymous, healthy donors with normal blood counts. B. Clustering of mutations by clone. The Y-axis represents the analyzed clones from each donor. The X-axis represents the genes with mutations in each clone (red). Note that the two clones with ZFHX4 variants contained unique mutations (Q1660K vs. P3497A). C. Mutational spectrum in normal donor exomes (81 validated variants) vs. 24 tier 1 AML genomes (343 validated variants).
Identification of genes with recurring mutations
To better identify the recurrently mutated genes in the sequenced AML genomes, we extended the analysis to additional AML cases. A liquid-phase capture approach followed by Illumina sequencing was used to assess the mutational status of 284 of the tier 1 mutated genes in an additional 53 M1 and 31 M3 cases (Supplemental Figure 1D and Supplemental Table 7). We also assessed the mutational status of 53 genes previously reported to be mutated in AML (see Supplemental Methods). Forty-one of these cases did not have a matched normal sample.
Among the 108 genomes assessed, we identified 23 genes containing mutations with translational consequences in at least 3 independent samples (Figure 5A). In the 24 cases subjected to WGS, we observed an average of ~14.5 total tier 1 mutations per genome (M1: 14.8 +/− 1.9; M3: 13.3 +/− 1.4; p = 0.5, Figure 5B). Of those, there was an average of ~3 recurrent mutations in each M1 genome (3.4 +/− 0.7, range 0–7) vs. 2 recurrent mutations in each M3 genome (2.2 +/− 0.3, range 1–5, p = 0.1, including PML-RARA, Figure 5C). Among the 108 total genomes assessed, there were an average of ~2 recurrently mutated genes per M1 sample (M1: 2.2 +/− 0.2, range 0–10) vs. ~2 genes per M3 sample (1.7 +/− 0.1, range 1–4; p < 0.09, including PML-RARA, Figure 5D).
Figure 5. Recurrently mutated genes in M1 and M3 AML.
A. 108 total cases of M1 and M3 AML were screened for somatic variants in the 284 genes identified by WGS and in 53 genes previously reported to be mutated in AML. Mutations with translational consequences that occur in at least three unique genomes are represented in red. FAB subtype indicated above graph. Overall survival is indicated by red (≤ 12 months), pink (12–24 months), and white (> 24 months) bars above graph. B. The total number of validated genic variants per genome identified by WGS in 12 NK M1 AML cases (red) and 12 M3 AML cases (blue), including PML-RARA. C. The total number of recurrently mutated genes per genome identified in 12 NK M1 AML cases (red) and 12 M3 AML cases (blue), including PML-RARA. Only mutations with translational consequences were assessed. D. Total number of recurrently mutated genes per genome identified in 65 M1 AML cases (red) and 43 M3 AML cases (blue), including PML-RARA. Only mutations with translational consequences were assessed. E. Somatic variants in cohesin complex genes (SMC3, SMC1A, STAG2, and RAD21) identified in 199 cases of AML (red). Additional mutations with translational consequences in highly recurrent genes are also shown for each genome (red).
Patterns of recurrent mutations in M1 vs. M3 AML genomes
We identified 9 recurrently mutated genes found in both the M1 and M3 genomes (FLT3, TTN, NRAS, PKD1L2, CACNA1E, DNAH9, WT1, ANKRD24, and PHF6) (Figure 5A). Mutations in these genes may represent cooperating events that are capable of interacting with several different initiating mutations.
Thirteen genes were recurrently mutated only in M1 genomes (NPM1, DNMT3A, IDH1, TET2, IDH2, RUNX1, ASXL1, PTPN11, DIS3, KIT, SMC1A, SMC3, and STAG2) (Figure 5A), suggesting that they might be involved with AML initiation. Of this group, six had mutation frequencies that were statistically higher in M1 vs. M3 patients (NPM1: p < 0.0001, IDH1: p < 0.0001, IDH2: p < 0.01, TET2: p < 0.001, DNMT3A: p < 0.0001, ASXL1: p < 0.03).
Mutations in three genes (NPM1, DNMT3A, and IDH1) coexisted only in M1 genomes. These genomes contained additional mutations that were also found in M3 genomes. Recurring mutations that co-occur with PML-RARA are likely to be cooperating mutations, rather than initiating mutations. This further suggests that mutations in NPM1, DNMT3A, and IDH1, are more likely to be initiating, rather than cooperating, events in M1 AML genomes.
We observed non-random associations of NPM1, DNMT3A, IDH1 and FLT3 mutations in M1 AML cases, as follows: NPM1+DNMT3A, p < 0.028; NPM1+IDH1, p < 0.011; NPM1+FLT3, p < 0.014; DNMT3A+IDH1, p < 0.001; DNMT3A+FLT3, p < 0.04, similar to our previous report and to others (Supplemental Figure 7) (Ley et al., 2010; Markova et al., 2011; Shen et al., 2011).
We extended these findings by examining mutational frequency data from whole genome or exome sequencing studies of 131 additional AML cases that included all other FAB subtypes from the TCGA AML study (Figure 5A and Supplemental Table 3). Most of the genes (20/23) were recurrently mutated in these additional samples, suggesting that these mutations may participate in the pathogenesis of other AML subtypes, as expected. Some genes with low mutation frequencies in the M1 and M3 cases were not recurrently mutated in the 131 additional cases (e.g. ANKRD24, DIS3, and PML-RARA), but none of the differences were statistically significant.
Three of the recurrently mutated genes, STAG2, SMC3, and SMC1A, are members of the cohesin complex, which is a tetrameric structure that also includes RAD21; this complex is involved in sister chromatid separation during anaphase, and CTCF-mediated chromatin topologic constraints (Carretero et al., 2010; Millau and Gaudreau, 2011). STAG2 was recently found to be deleted in an AML genome (Walter et al., 2009) and in cancer cell lines (Rocquain et al., 2010; Solomon et al., 2011). We therefore assessed all four genes for mutations in a larger set of 183 AML samples subjected to exome or whole genome sequencing as a part of The Cancer Genome Atlas (TCGA) study of AML (T.J. Ley and R.K. Wilson, unpublished). We identified mutations in all four cohesin complex genes, while the related genes STAG1 and STAG3 were not mutated (Figure 5E). These mutations were not detected in any M3 genomes. Of the 19 mutations identified in cohesin complex genes, 11 had loss-of-function consequences (nonsense, splice site, or gene deletions). Two cohesin complex genes are found on the X chromosome, and mutations in those genes were identified in 6 male patients. Cohesin complex gene mutations were not associated with chromosomal instability (Solomon et al., 2011); 12 of the cases with cohesin mutations had normal cytogenetics, 6 had 3 or fewer cytogenetic abnormalities, and only 1 had complex cytogenetics.
We examined the expression of all of the recurrently mutated genes using expression array data for all of the M1 and M3 cases available in our dataset (Figure 5A and Supplemental Table 3). The expression values of all probe sets on these arrays are normalized to an average chip mean of 1,500. Most of the recurrently mutated genes are highly expressed in both M1 and M3 AML cases. Several of the genes had very low expression levels (e.g. TTN, PKD1L2, ANKRD24, CACNA1E, and DNAH9) on the Affymetrix U133 Plus 2 array. Using readcounts from RNA-Seq data obtained from 155 AML cases from the TCGA study of AML, we were able to show that spliced transcripts from TTN and ANKRD24 are detected in virtually all AML samples tested, while PKD1L2, CACNA1E and DNAH9 transcripts were absent in all (or nearly all) of the same samples. We suggest that mutations in genes that are not detectably expressed are less likely to be relevant for pathogenesis.
We identified only two germline variants that were likely to be relevant for AML pathogenesis: WT1 R430* in AML15 (age 25), and PTPN11 Y197* in AML9 (age 25). These data are presented in the Supplemental Information and Supplemental Table 8.
Integration of co-existing variants within individual patients
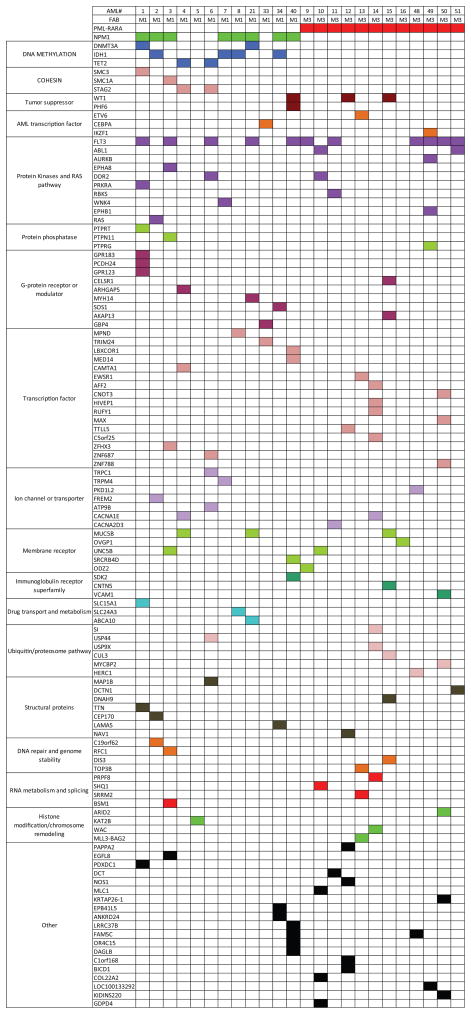
Mutations that may be relevant for AML pathogenesis are integrated for all 24 fully sequenced patients in Table 1. This analysis identified at least 2 likely AML driver mutations (in genes that are recurrently mutated in AML and/or other cancer types, and expressed in the patient’s own AML sample) in 23 of 24 cases, and at least 4 likely drivers in 15 of 24 cases. This outcome is consistent with our predicted statistical power from this experimental design and sample size (see Supplemental Methods). These mutations are organized into currently annotated pathways (Figure 6), which reveal potential relationships among genes that are infrequently mutated in this set of cases.
Table 1.
Patient-specific mutations that may contribute to AML pathogenesis
| UPN | WGS# | FAB | Age at Dx | Structural Variants1 | Clones | OS (months) | Mutated in AML2, expressed in patient3 | Mutated in AML2, not expressed in patient4 | Mutated in other cancer types5, expressed in patient3 | Inherited mutation in AML gene, expressed in patient3 |
|---|---|---|---|---|---|---|---|---|---|---|
| 933124 | AML1 | M1 | 57 | 3 | 1 | 24.6 | NPM1, FLT3, SMC3, DNMT3A, GPR183, TTN | PCDH24, SLC15A1, PTPRT, GPR123 | PDXDC1, PRKRA 6 | |
| 807970 | AML2 | M1 | 38 | 0 | 2 | 54.9 | NPM1, IDH1, NRAS, CEP170, C19orf62 | FREM2 | ||
| 123172 | AML3 | M1 | 56 | 0 | 2 | 53.9 | NPM1, FLT3, SMC1A, PTPN11, ZFHX3, UNC5B | EPHA8, EGFL8 | BMS1, RFC1 | |
| 831711 | AML4 | M1 | 57 | 0 | 1 | 53.4 | STAG2, TET2 | MUC5B, CACNA1E | CAMTA1, ARHGAP5 | |
| 849660 | AML5 | M1 | 22 | 5 | 1 | 27 | KAT2B | |||
| 808642 | AML6 | M1 | 61 | 5 | 2 | 15.5 | FLT3, TET2 | ZNF687, MAP1B, TRPC1 | DDR2, ATP9B, USP44 | |
| 509754 | AML7 | M1 | 21 | 8 | 2 | 73.9 | NPM1, IDH1 | TRPM4, WNK4 | ||
| 327733 | AML8 | M1 | 32 | 3 | 2 | 56.3 | NPM1, IDH1, SLC24A3, MPND | |||
| 224143 | AML21 | M1 | 67 | 1 | 1 | 0.8 | NPM1, FLT3, DNMT3A | MYH14, MUC5B, MUC5B, ABCA10 | ||
| 545259 | AML33 | M1 | 30 | 0 | 1 | 33.7 | CEBPA | GBP4 | TRIM24 | |
| 548327 | AML34 | M1 | 51 | 2 | 2 | 31.9 | NPM1, IDH1 | LAMA5, EPB41L5, ANKRD24 | SOS1 | |
| 804168 | AML40 | M1 | 53 | 1 | 1 | 30 | NPM1, FLT3, WT1, PHF6, LRRC37B | SDK2, SRCRB4D, FAM5C, LBXCOR1, OR4C15 | MED14, DAGLB | |
| 709968 | AML9 | M3 | 25 | 2 | 2 | 71.7 | PML-RARA 6, FLT3 | ODZ2 | PTPN11 Y197* | |
| 863018 | AML10 | M3 | 62 | 4 | 2 | 69.4 | PML-RARA 6, UNC5B, COL11A2, ABL1 | GDPD4 | SHQ1, DDR2, MLC1 6 | |
| 478908 | AML11 | M3 | 50 | 1 | 2 | 35.5 | PML-RARA 6, FLT3, RBKS | DCT | CACNA2D3 | |
| 344551 | AML12 | M3 | 48 | 3 | 1 | 69.3 | PML-RARA 6, WT1, NAV1, NOS1 | C1orf168 | TTLL5, PAPPA2, BICD1 | |
| 673778 | AML13 | M3 | 53 | 3 | 4 | 76.1 | PML-RARA 6, ETV6, SRRM2 | TOP3B | EWSR1, MLL3-BAG2 6 | |
| 321258 | AML14 | M3 | 31 | 1 | 3 | 64.3 | PML-RARA 6, WAC, HIVEP1, C5orf25 | CACNA1E, SI | AFF2, USP9X, RUFY1, PRPF8 | |
| 758168 | AML15 | M3 | 25 | 5 | 1 | 49.8 | PML-RARA 6, AKAP13, DIS3 | DNAH9, CNTN5, MUC5B | CUL3, CELSR1 | WT1 R430* 7 |
| 455499 | AML16 | M3 | 29 | 3 | 2 | 37.1 | PML-RARA 6 | OVGP1 | ||
| 202127 | AML48 | M3 | 68 | 1 | 1 | 1 | PML-RARA 6, FLT3, HERC1 | PKD1L2, FAM5C | ||
| 529205 | AML49 | M3 | 59 | 2 | 2 | 31.3 | PML-RARA 6, FLT3 | EPHB1, LOC100133292 | AURKB, IKZF1, PTPRG | |
| 501944 | AML50 | M3 | 40 | 1 | 3 | 73.6 | PML-RARA 6 | KRTAP26-1, ZNF788 | MYCBP2, MAX, VCAM1, ARID2, KIDINS220, CNOT3 | |
| 943309 | AML51 | M3 | 35 | 1 | 3 | 63.1 | PML-RARA 6, FLT3 | DCTN1 |
Details regarding structural variants in Supplemental Table 5.
At least 4 separate cases of AML with somatic mutations in this gene within 222 cases analyzed.
Gene expression ≥ 800 by Affymetrix U133 Plus2 array (chip mean 1,500).
Gene expression < 800 by Affymetrix U133 Plus2 array (chip mean 1,500).
At least 4 separate cases with somatic mutations in this gene in Cosmic databases.
Structural variant
Absent in ESP cohort (N = 4540).
Figure 6. Pathway analysis of key mutations in the 24 sequenced genomes.
Mutations identified in Table 1 were individually curated in Pubmed for biological function. Functional categories represented by 2 or fewer mutated genes were excluded. Mutations in NPM1, DNA methylation, and cohesin genes occurred only in the M1 cases, suggesting that they may substitute for PML-RARA as initiating events, or do not productively cooperate with PML-RARA. Mutations in ubiquitin pathway genes occurred more often in M3 AML. Genes representing other functional categories were mutated in both M1 and M3 AML cases.
Discussion
In this report, we describe the comprehensive sequencing of the entire genome of 24 carefully selected acute myeloid leukemia cases. Paired-end sequencing of the entire tumor and normal skin genomes was performed for each case, along with independent validation of all putative mutations identified in the non-repetitive part of the human genome. We compared the genomes of patients with a known initiating mutation (PML-RARA) with genomes that have less known about the initiating events (AML without maturation-FAB M1 with normal karyotype). Both AML subtypes have approximately the same number of overall mutations in their genomes, and both are associated with relative genomic stability (in all cases we observed a small number of total SNVs, indels, and SVs, compared with most solid tumors). The SNVs were distributed across the entire genome, and the mutational spectrum of SNVs was virtually identical between the two AML subtypes. By comparing these two subtypes of AML, we were able to distinguish the mutations that are likely to be initiating events in the cytogenetically normal cases, vs. the cooperating events that are common to both subtypes; this analysis suggests that mutations in NPM1, DNMT3A, and IDH1 may act as major initiating mutations in M1 AML.
We found a small number of mutations with translational consequences (e.g. non-synonymous mutations in tier 1) in each of the 24 genomes: the average was ~11 for M1 AML, and ~10 for M3 AML (including the PML-RARA fusion event). Only a portion of these mutations were found to be expressed in the patient’s sample and recurrent in other M1/M3 AML genomes, yielding a median number of 3 recurring mutations in M1 genomes, vs. 2 in M3 genomes (including the PML-RARA fusion). Additional mutations are likely to contribute to pathogenesis (such as mutations that occur with < 3% frequency in AML or that occur in solid tumor-associated genes), although their biological significance in AML is less certain (Table 1 and Figure 6).
These data, since they were derived from completely sequenced genomes, allow us to estimate the minimum number of genic mutations that may be required for AML pathogenesis. We observed one M3 AML genome in which the only detected recurrent AML or cancer-associated tier 1 somatic mutations were PML-RARA and FLT3 ITD (AML9, Table 1 and Figure 6). In mice, this combination of mutations can cooperate to accelerate the onset (or development) of AML with many of the features of acute promyelocytic leukemia (Kelly et al., 2002; Sohal et al., 2003). Furthermore, by sequencing an APL genome derived from a mouse expressing human PML-RARA, we recently defined a recurring, spontaneous mutation in the Jak1 gene at position V657F (Wartman et al., 2011); this mutation that is identical to that found in some human patients with B lymphoblastic leukemia (Flex et al., 2008; Harvey et al., 2010; Mullighan et al., 2009), and a patient with M3 AML (Jeong et al., 2008). When this Jak1 mutation was coexpressed with PML-RARA in mice, the animals developed an explosive, oligoclonal APL syndrome within 4 weeks, strongly suggesting that these two mutations were sufficient to produce the disease (Wartman et al., 2011). These data support the idea that as few as two key somatic mutations may cause AML in some patients.
We observed that human hematopoietic stem/progenitor cells (HSPCs) from healthy volunteers each contain private mutations that accumulate as a function of age; we calculated that 0.13 +/− 0.02 exonic mutations are acquired per year of life (Figure 4A). Current models of human hematopoietic stem cells (HSCs) suggest that one cell division occurs every 25–50 weeks (Catlin et al., 2011). Experimental methods of interrogating human HSC turnover are, however, somewhat limited. Murine HSC division rates are better characterized, and the data suggests that murine HSCs fluidly transition between two pools, one cycling every 2–6 weeks and a second cycling every 20–50 weeks (Foudi et al., 2009; Takizawa et al., 2008). DNA replication fidelity rates have been measured between 0.06 – 1.5 × 10−9, depending on cell type (germ cells having the highest fidelity and cells grown in tissue culture have the lowest) (Gundry and Vijg, 2012; Lynch, 2010). Therefore, if hematopoietic stem/progenitor cells (HSPCs) have a fidelity rate of 0.78 × 10−9 mutations per genomic base pair per cell division (mid way between the fidelity of germ cells and cells in tissue culture), and undergo 2–20 divisions per year, we would predict that they would acquire between 0.069 – 0.858 exonic mutations per year. The mutation rate we have measured is within the expected parameters, which are based on our current understanding of replication fidelity and HSPC cell division rates. These data provide a plausible explanation for the large number of unique background mutations in each AML genome.
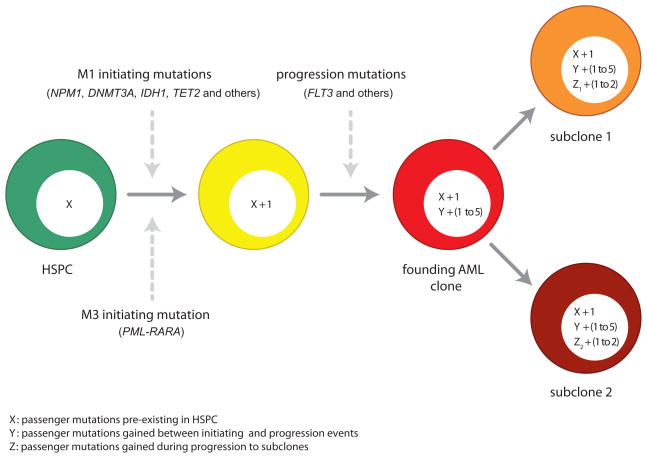
These data are consistent with previously described models of oncogenesis (Figure 7) (Armitage and Doll, 1957; Calabrese et al., 2004; Loeb et al., 1974; Nowell, 1976; Tomlinson et al., 1996). In the first step of leukemogenesis, a driver mutation (e.g. PML-RARA or NPM1) occurs within the context of an HSPC that already contains hundreds of random, benign mutations that have accumulated over time. This mutation confers an advantage to this cell, and as a consequence, all of the random background mutations within its genome (passengers) are “captured” and are carried forward as the clone expands. When an additional driver mutation (e.g. FLT3 ITD) occurs within a cell in an expanding clone, any additional passenger mutations that have accumulated in that cell (since acquisition of the first driver mutation) also will be captured. Eventually, these cells become the “founding” clone that is detected at presentation, which contains only a few drivers, but many passengers that were captured at initiation, and a few additional passengers that were captured as cooperating events were added. Each progression event (and/or events that contribute to subclone outgrowth) probably yields a small cluster of new mutations, of which only one or two may be relevant for clonal outgrowth. Our data suggests that the number of additional passengers added with progression events is typically much smaller than the number of passengers captured with the initiating event (which accumulated over the lifetime of the founding cell).
Figure 7. Integrated model for the origin of driver and passenger mutations during AML evolution.
Hematopoietic stem/progenitor cells (HSPCs, shown in green) are long-lived cells that accumulate random, benign background mutations as a function of age. Based on the fact that HSPCs from normal, healthy donors accumulate cell-specific, age-dependent mutations, and that the total mutational burden in AML genomes is related to age, we suggest that the largest numbers of mutations in AML genomes are variants that were preexisting in the cell that received the initiating event. “X” represents these background mutations in the HSPCs, and we estimate that X may range between 100 and 1000 events, depending on age. The initiating mutations, which are drivers, are different for M1 AML cases and M3 AML cases. The initiating event provides an advantage for the affected cell and clonal expansion ensues, so that all of the preexisting mutations are “captured” by cloning. Each progression event gives the expanding clone an additional advantage; based on the data in this paper, we suggest that 1–5 events contribute to progression in most cases of AML. Each progression mutation is expected to capture all mutations that occurred between the initiating event and the progression event (designated as “Y” in the yellow cell). This number would be affected by the time between these events, and the mutation rate in these cells. Although this number is unknown at present, the analysis of clonal progression of secondary AML (Walter et al., 2012) suggests that each progression event may capture dozens to hundreds of mutations. Clonal outgrowth of cells with appropriate progression events results in AML, which is dominated by the “founding” AML clone, designated in red. When this clone is sequenced, the driver mutations (1–6 events) and all associated passenger mutations (hundreds of events) would all be present in all cells within the sample, which is what we observe. Subclones arise from the founding AML clone by acquiring a small number of additional mutations that confer an advantage to that cell, along with any additional background mutations that may have occurred in the interim (represented as “Z”). In this study, the average number of mutations captured in subclones was 40 (range 6–110).
The 13 recurrently mutated genes that are found only in M1 samples may be involved in the initiation of M1 AML. Mutations in NPM1, DNMT3A, and IDH1 occur only rarely in M3 genomes (Ley et al., 2010; Lin et al., 2011; Shen et al., 2011), and co-occur more often than can be explained by chance (Ley et al., 2010; Markova et al., 2011; Shen et al., 2011). This pattern suggests that mutations in these genes may sometimes cooperate to initiate AML, and that this is likely to be a central AML pathway, around which rare and private mutations can coalesce. No patterns of cooperation could be discerned between the other recurrently mutated genes that are M1-specific, except for mutations in the genes that encode members of the cohesin complex, which will be discussed below.
Nine recurrently mutated genes were detected both in M1 and M3 genomes, suggesting that these mutations may cooperate with a variety of initiating events to produce the disease. By far the most frequently mutated gene in this set is FLT3, where internal tandem duplications are known to cooperate with other initiating events in mice to produce disease progression, but do not initiate leukemia on their own (Kelly et al., 2002; Li et al., 2008; Sallmyr et al., 2008). Although several groups have suggested that an activated tyrosine kinase may be an essential feature of all AML cases (Gilliland, 2002; Ishikawa et al., 2009; Pedersen-Bjergaard et al., 2008; Zheng et al., 2009), we identified mutations in alternative tyrosine kinase genes in only 1 of the 13 completely sequenced genomes that lacked a FLT3 mutation (AML10, which contained both ABL1 P937L and DDR2 M291I). In the entire set of 108 cases, we observed mutations in tyrosine kinase genes in only 5 of the 69 genomes that lacked a FLT3 mutation (these 4 additional cases had somatic variants in KIT D816V and D816Y, JAK2 V617F, and DDR2 G222R). We had previously addressed this possibility by sequencing the expressed tyrosine kinases in AML cases (Tomasson et al., 2008), however that study was limited by the fact that only targeted gene sequencing was performed. Here, where whole genomes were sequenced, we are more confident that a second mutated tyrosine kinase gene is not required for most cases of AML. However, it is possible that mutations in protein phosphatases and G-protein receptor/modulators might substitute for kinase mutations in some cases (Figure 6).
We identified recurring mutations in the genes that encode all four members of the cohesin complex (STAG2, SMC3, RAD21, and SMC1A), a set of proteins that assemble into a ring structure that encircles sister chromatids during metaphase, maintaining their polarity during mitosis (Carretero et al., 2010; Millau and Gaudreau, 2011). The cohesin complex mutations occurred in ~10% of non-M3 AML cases and co-occurred with NPM1, DNMT3A, TET2, or RUNX1 mutations in 17/19 cases. Eleven of these cohesin mutations were predicted to cause loss-of-function for the affected gene; STAG2 and SMC1A are both on the X chromosome, and null mutations in these genes are predicted to create complete protein deficiency for the three male patients with these mutations. All of the mutations were mutually exclusive of one another, suggesting that an alteration of only one component of the complex may be sufficient to disrupt or alter the entire complex. Recently, loss-of-function mutations in the STAG2 gene were shown to be associated with aneuploidy in cancer cell lines derived from solid tumors (Solomon et al., 2011); however, only one of the 19 AML genomes with cohesin complex mutations was aneuploid. Although we do not yet understand the mechanisms by which these mutations may contribute to AML pathogenesis, the discovery of recurrent mutations in this important structure represents yet another new pathway that is undoubtedly relevant for this disease.
Although we have focused this report on recurrently mutated genes, we have also identified 15 recurrently mutated non-genic regions (e.g. mutations occur within 100 base pairs of one another) within these 24 completely sequenced genomes. We previously reported one identical mutation in two AML genomes, falling in a conserved but unannotated portion of the genome (Mardis et al., 2009). None of the 15 recurrent mutations identified here was associated with altered expression of its nearest neighbor gene, and none occurred within 10,000 base pairs of an annotated, untranslated RNA (data not shown). As additional AML genomes are sequenced, the identification of significantly mutated regions may suggest functional properties for these unannotated parts of the genome (e.g. cryptic genes or transcribed regions, or regulatory domains).
In summary, we have been able to provide new insights into the origin and evolution of AML mutations with whole genome sequencing. Although many AML mutations have been discovered by sequencing more limited parts of the genome, candidate gene sequencing and/or exome sequencing cannot provide enough information to confidently predict the full spectrum of mutations required for disease in an individual patient, or to determine the global genomic character of a disease. Our data suggest that most of the somatic events in AML genomes appear to be random, preexisting, background mutations in the hematopoietic cell that acquired the key initiating mutation. This event “captures” the mutational history of this cell as it evolves to become the founding clone at presentation (we have recently reported a similar phenomenon when fibroblasts are cloned via reprogramming to become induced pluripotent stem cells (Young et al., 2012)). Only a tiny fraction of the total mutations in each AML genome are therefore likely to be relevant for pathogenesis, for disease classification, and for targeted therapy. The comprehensive analysis of AML genomes, while technically complex, has therefore suggested that AML is not a disease caused by hundreds of mutations, but only a few. These data may have implications for the origins of mutations in other cancer types as well.
Experimental Procedures
Whole genome sequencing and somatic mutation discovery
Whole genome sequence data were generated using Illumina GA and Illumina GA IIx instruments (Ley et al., 2008; Mardis et al., 2009). Paired-end reads 50–100 bp in length were aligned to NCBI 36 using Maq (v0.6.8 or v0.7.1) or BWA (v0.5.5). Target haploid reference coverage was a minimum of 25X for tumor and 20X for normal. Sample specific variants were called using MAQ (v0.6.8) or Samtools (r320, r453, or r599). Somatic SNVs were identified using SomaticSniper as previously described (Ding et al., 2012; Larson and Dooling, 2011). Somatic indels were detected as previously described (Ding et al., 2012). We used Pindel in both tumor and normal and selected only tier 1 Pindel predictions regardless of filtering (could be germline or somatic) for validation.
Somatic structural variants were identified as previously described using BreakDancer; structural variant predictions were not filtered based on assembled sequence data from breakpoint flanking regions (Chen et al., 2009; Ding et al., 2012). Further detail provided in Supplemental Methods.
All sequence variants for the AML tumor samples from 24 cases have been submitted to dbGaP (accession number phs000159.v3.p2).
Capture validation and analysis
Capture validation and analysis were performed as previously described (Ding et al., 2012).
Mutation cluster analysis (clonality)
Tumor clonality estimates were determined as described previously (Ding et al., 2012). Briefly, supporting readcounts from custom capture validation data for somatic mutations in regions of diploid copy number (2N) with a minimum reference coverage >100× were input into a custom R function that drew a kernel density estimate (KDE) plot and estimated the number and relative composition of clones in these tumors.
Extension sequencing and analysis
Nimblegen Liquid phase custom capture reagents were used to extend putative variant calls from the initial M1/M3 discovery data. These variants were extended on an additional set of samples including 53 M1 and 31 M3 cases. The target regions included 522 genes, 9 non-coding regions that had mutations in more than one discovery sample, and 24 identity single nucleotide polymorphism sites, and additional genes with ambiguous mutations. This final design space was 4.6Mb. DNA Libraries were constructed using ligation of Illumina adaptors to sheared whole genome amplified DNA. A Solid Phase Reversible Immobilization (SPRI) bead cleanup procedure was conducted to select size fractions between 300 and 500bp. Hybridizations were performed using a customized version of the Nimblegen EZ exome protocol. Barcoding adaptors were used to enable post-capture multiplexing of 15 samples per lane on the sequencing instrument. qPCR was used to determine the quantity of captured library necessary for loading on a single lane of an Illumina Hi-Seq 2000 (2 × 100bp) PE sequences in order to produce ~2 Gbp of sequence/sample.
Differential mutation frequency analysis across subtypes
Expression quartiles were defined using the average, highest expressed probe set from Affymetrix U133 Plus2 results for 5 independent flow sorted CD34+ bone marrow samples and 52 unique M1-AML bone marrow samples and have been previously published; Geo accession number GSE12662 (Payton et al., 2009).
RNA-seq-based expression analysis
RNA-seq data for 155 TCGA AML tumor samples consisting of of 50 bp paired-end reads, were extracted from BAM into FASTQ format using Picard 1.31 (http://picard.sourceforge.net/) and subsequently aligned using the Tophat 1.1.4 release (Trapnell et al., 2009). Cufflinks 0.9.2 (Trapnell et al., 2010) was used to estimate expression levels for all known UCSC genes and transcript isoforms. The resulting transcript abundance estimates are expressed in fragments per kilobase of exon per million fragments mapped (FPKM), as proposed by Mortazavi et al (Mortazavi et al., 2008).
Exome sequencing of normal volunteer samples
Flow sorted CD34+ CD38− Lineage− cells were expanded on AFT024 stromal cells. DNA from individual colonies was prepared, captured with Agilent SureSelect Human All Exon v2 kit, and sequenced using similar methods as we have published (Ding et al., 2012; Mardis et al., 2009; Welch et al., 2011b). Experimental details are described in Supplemental Information.
Statistical Analysis
Students T-test, Wilcoxon comparison, ANOVA, log-rank, and hypergeometric distributions were used to compare results from two cohorts (Prism, GraphPad, La Jolla, California, and Excel, Microsoft, Seattle, Washington). The expected Poisson distribution was generated using WinPepi (Abramson, 2011).
Supplementary Material
Highlights.
Normal HSPCs contain random background mutations that increase with aging
The total number of mutations in AML genomes increases with age
AML genomes contain hundreds of mutations, but very few are recurrent
Most AML mutations are probably background events in HSPCs, “captured” by cloning
Acknowledgments
We thank the Analysis Pipeline group for developing the automated sequence analysis pipelines; the LIMS group for developing tools and software to manage samples and sequencing; and the Systems group for providing the IT infrastructure and HPC solutions required for sequencing and analysis. We thank William S. Schierding and Nathan D. Dees for their help on tumor clonality analysis, Dr. David Spencer for help with pathway analysis, and Joshua McMichael for help with figure preparation. We also thank The Cancer Genome Atlas for allowing us to use unpublished data for this study, the Washington University Cancer Genome Initiative for their support, and the Siteman Cancer Center Flow Cytometry Core, which provided cell sorting service and is supported in part by an NCI Cancer Center Support Grant #P30 CA91842. This work was funded by grants to Richard K. Wilson from Washington University in St. Louis and the National Human Genome Research Institute (NHGRI U54 HG003079), and grants to John S. Welch (K99 HL103975) and Timothy J. Ley from the National Cancer Institute (PO1 CA101937) and the Barnes-Jewish Hospital Foundation (00335-0505-02).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abkowitz JL, Catlin SN, Guttorp P. Evidence that hematopoiesis may be a stochastic process in vivo. Nat Med. 1996;2:190–197. doi: 10.1038/nm0296-190. [DOI] [PubMed] [Google Scholar]
- Abkowitz JL, Golinelli D, Harrison DE, Guttorp P. In vivo kinetics of murine hemopoietic stem cells. Blood. 2000;96:3399–3405. [PubMed] [Google Scholar]
- Abramson JH. WINPEPI updated: computer programs for epidemiologists, and their teaching potential. Epidemiol Perspect Innov. 2011;8:1. doi: 10.1186/1742-5573-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allford S, Grimwade D, Langabeer S, Duprez E, Saurin A, Chatters S, Walker H, Roberts P, Rogers J, Bain B, et al. Identification of the t(15;17) in AML FAB types other than M3: evaluation of the role of molecular screening for the PML/RARalpha rearrangement in newly diagnosed AML. The Medical Research Council (MRC) Adult Leukaemia Working Party. Br J Haematol. 1999;105:198–207. [PubMed] [Google Scholar]
- Armitage P, Doll R. A two-stage theory of carcinogenesis in relation to the age distribution of human cancer. Br J Cancer. 1957;11:161–169. doi: 10.1038/bjc.1957.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacher U, Schnittger S, Haferlach T. Molecular genetics in acute myeloid leukemia. Curr Opin Oncol. 2010;22:646–655. doi: 10.1097/CCO.0b013e32833ed806. [DOI] [PubMed] [Google Scholar]
- Betz BL, Hess JL. Acute myeloid leukemia diagnosis in the 21st century. Arch Pathol Lab Med. 2010;134:1427–1433. doi: 10.5858/2010-0245-RA.1. [DOI] [PubMed] [Google Scholar]
- Bullinger L, Kronke J, Schon C, Radtke I, Urlbauer K, Botzenhardt U, Gaidzik V, Cario A, Senger C, Schlenk RF, et al. Identification of acquired copy number alterations and uniparental disomies in cytogenetically normal acute myeloid leukemia using high-resolution single-nucleotide polymorphism analysis. Leukemia. 2010;24:438–449. doi: 10.1038/leu.2009.263. [DOI] [PubMed] [Google Scholar]
- Calabrese P, Tavare S, Shibata D. Pretumor progression: clonal evolution of human stem cell populations. Am J Pathol. 2004;164:1337–1346. doi: 10.1016/S0002-9440(10)63220-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carretero M, Remeseiro S, Losada A. Cohesin ties up the genome. Curr Opin Cell Biol. 2010;22:781–787. doi: 10.1016/j.ceb.2010.07.004. [DOI] [PubMed] [Google Scholar]
- Catlin SN, Busque L, Gale RE, Guttorp P, Abkowitz JL. The replication rate of human hematopoietic stem cells in vivo. Blood. 2011;117:4460–4466. doi: 10.1182/blood-2010-08-303537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Wallis JW, McLellan MD, Larson DE, Kalicki JM, Pohl CS, McGrath SD, Wendl MC, Zhang Q, Locke DP, et al. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat Methods. 2009;6:677–681. doi: 10.1038/nmeth.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, Ritchey JK, Young MA, Lamprecht T, McLellan MD, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481:506–510. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flex E, Petrangeli V, Stella L, Chiaretti S, Hornakova T, Knoops L, Ariola C, Fodale V, Clappier E, Paoloni F, et al. Somatically acquired JAK1 mutations in adult acute lymphoblastic leukemia. J Exp Med. 2008;205:751–758. doi: 10.1084/jem.20072182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foudi A, Hochedlinger K, Van Buren D, Schindler JW, Jaenisch R, Carey V, Hock H. Analysis of histone 2B-GFP retention reveals slowly cycling hematopoietic stem cells. Nat Biotechnol. 2009;27:84–90. doi: 10.1038/nbt.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk RK, Maxwell TJ, Izumi M, Edwin D, Kreisel F, Ley TJ, Cheverud JM, Graubert TA. Quantitative trait loci associated with susceptibility to therapy-related acute murine promyelocytic leukemia in hCG-PML/RARA transgenic mice. Blood. 2008;112:1434–1442. doi: 10.1182/blood-2008-01-132084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilliland DG. Molecular genetics of human leukemias: new insights into therapy. Semin Hematol. 2002;39:6–11. doi: 10.1053/shem.2002.36921. [DOI] [PubMed] [Google Scholar]
- Gundry M, Vijg J. Direct mutation analysis by high-throughput sequencing: from germline to low-abundant, somatic variants. Mutat Res. 2012;729:1–15. doi: 10.1016/j.mrfmmm.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey RC, Mullighan CG, Chen IM, Wharton W, Mikhail FM, Carroll AJ, Kang H, Liu W, Dobbin KK, Smith MA, et al. Rearrangement of CRLF2 is associated with mutation of JAK kinases, alteration of IKZF1, Hispanic/Latino ethnicity, and a poor outcome in pediatric B-progenitor acute lymphoblastic leukemia. Blood. 2010;115:5312–5321. doi: 10.1182/blood-2009-09-245944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi M, Suzuki H, Yoshinari M, Sato A, Saito T, Sugawara A, Tsuchiya S, Hatae Y, Fujimoto T, Kakizuka A, et al. Mutations in the E-domain of RAR portion of the PML/RAR chimeric gene may confer clinical resistance to all-trans retinoic acid in acute promyelocytic leukemia. Blood. 1998;92:374–382. [PubMed] [Google Scholar]
- Ishikawa Y, Kiyoi H, Tsujimura A, Miyawaki S, Miyazaki Y, Kuriyama K, Tomonaga M, Naoe T. Comprehensive analysis of cooperative gene mutations between class I and class II in de novo acute myeloid leukemia. Eur J Haematol. 2009;83:90–98. doi: 10.1111/j.1600-0609.2009.01261.x. [DOI] [PubMed] [Google Scholar]
- Jeong EG, Kim MS, Nam HK, Min CK, Lee S, Chung YJ, Yoo NJ, Lee SH. Somatic mutations of JAK1 and JAK3 in acute leukemias and solid cancers. Clin Cancer Res. 2008;14:3716–3721. doi: 10.1158/1078-0432.CCR-07-4839. [DOI] [PubMed] [Google Scholar]
- Kelly LM, Kutok JL, Williams IR, Boulton CL, Amaral SM, Curley DP, Ley TJ, Gilliland DG. PML/RARalpha and FLT3-ITD induce an APL-like disease in a mouse model. Proc Natl Acad Sci U S A. 2002;99:8283–8288. doi: 10.1073/pnas.122233699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogan SC. Mouse models of acute promyelocytic leukemia. Curr Top Microbiol Immunol. 2007;313:3–29. doi: 10.1007/978-3-540-34594-7_2. [DOI] [PubMed] [Google Scholar]
- Larson DE, Harris CC, Chen K, Koboldt DC, Abbott TE, Dooling DJ, Ley TJ, Mardis ER, Wilson RK, Ding L. SomaticSniper: identification of somatic point mutations in whole genome sequencing data. Bioinformatics. 2011;28:311–317. doi: 10.1093/bioinformatics/btr665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson RA, Le Beau MM. Prognosis and therapy when acute promyelocytic leukemia and other “good risk” acute myeloid leukemias occur as a therapy-related myeloid neoplasm. Mediterr J Hematol Infect Dis. 2011;3:e2011032. doi: 10.4084/MJHID.2011.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, Kandoth C, Payton JE, Baty J, Welch J, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley TJ, Mardis ER, Ding L, Fulton B, McLellan MD, Chen K, Dooling D, Dunford-Shore BH, McGrath S, Hickenbotham M, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Piloto O, Nguyen HB, Greenberg K, Takamiya K, Racke F, Huso D, Small D. Knock-in of an internal tandem duplication mutation into murine FLT3 confers myeloproliferative disease in a mouse model. Blood. 2008;111:3849–3858. doi: 10.1182/blood-2007-08-109942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Yao DM, Qian J, Chen Q, Qian W, Li Y, Yang J, Wang CZ, Chai HY, Qian Z, et al. Recurrent DNMT3A R882 Mutations in Chinese Patients with Acute Myeloid Leukemia and Myelodysplastic Syndrome. PLoS One. 2011;6:e26906. doi: 10.1371/journal.pone.0026906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link DC, Schuettpelz LG, Shen D, Wang J, Walter MJ, Kulkarni S, Payton JE, Ivanovich J, Goodfellow PJ, Le Beau M, et al. Identification of a novel TP53 cancer susceptibility mutation through whole-genome sequencing of a patient with therapy-related AML. JAMA. 2011;305:1568–1576. doi: 10.1001/jama.2011.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb LA, Springgate CF, Battula N. Errors in DNA replication as a basis of malignant changes. Cancer Res. 1974;34:2311–2321. [PubMed] [Google Scholar]
- Lynch M. Evolution of the mutation rate. Trends Genet. 2010;26:345–352. doi: 10.1016/j.tig.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, Koboldt DC, Fulton RS, Delehaunty KD, McGrath SD, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markova J, Michkova P, Burckova K, Brezinova J, Michalova K, Dohnalova A, Maaloufova JS, Soukup P, Vitek A, Cetkovsky P, et al. Prognostic impact of DNMT3A mutations in patients with intermediate cytogenetic risk profile acute myeloid leukemia. Eur J Haematol. 2011;88:128–135. doi: 10.1111/j.1600-0609.2011.01716.x. [DOI] [PubMed] [Google Scholar]
- McCormick SR, McCormick MJ, Grutkoski PS, Ducker GS, Banerji N, Higgins RR, Mendiola JR, Reinartz JJ. FLT3 mutations at diagnosis and relapse in acute myeloid leukemia: cytogenetic and pathologic correlations, including cuplike blast morphology. Arch Pathol Lab Med. 2010;134:1143–1151. doi: 10.5858/2009-0292-OA.1. [DOI] [PubMed] [Google Scholar]
- Millau JF, Gaudreau L. CTCF, cohesin, and histone variants: connecting the genome. Biochem Cell Biol. 2011;89:505–513. doi: 10.1139/o11-052. [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Mullighan CG, Zhang J, Harvey RC, Collins-Underwood JR, Schulman BA, Phillips LA, Tasian SK, Loh ML, Su X, Liu W, et al. JAK mutations in high-risk childhood acute lymphoblastic leukemia. Proc Natl Acad Sci U S A. 2009;106:9414–9418. doi: 10.1073/pnas.0811761106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- Payton JE, Grieselhuber NR, Chang LW, Murakami M, Geiss GK, Link DC, Nagarajan R, Watson MA, Ley TJ. High throughput digital quantification of mRNA abundance in primary human acute myeloid leukemia samples. J Clin Invest. 2009;119:1714–1726. doi: 10.1172/JCI38248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen-Bjergaard J, Andersen MK, Andersen MT, Christiansen DH. Genetics of therapy-related myelodysplasia and acute myeloid leukemia. Leukemia. 2008;22:240–248. doi: 10.1038/sj.leu.2405078. [DOI] [PubMed] [Google Scholar]
- Pfeifer GP. Mutagenesis at methylated CpG sequences. Curr Top Microbiol Immunol. 2006;301:259–281. doi: 10.1007/3-540-31390-7_10. [DOI] [PubMed] [Google Scholar]
- Rego EM, Wang ZG, Peruzzi D, He LZ, Cordon-Cardo C, Pandolfi PP. Role of promyelocytic leukemia (PML) protein in tumor suppression. J Exp Med. 2001;193:521–529. doi: 10.1084/jem.193.4.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocquain J, Gelsi-Boyer V, Adelaide J, Murati A, Carbuccia N, Vey N, Birnbaum D, Mozziconacci MJ, Chaffanet M. Alteration of cohesin genes in myeloid diseases. Am J Hematol. 2010;85:717–719. doi: 10.1002/ajh.21798. [DOI] [PubMed] [Google Scholar]
- Rowley JD, Golomb HM, Vardiman J, Fukuhara S, Dougherty C, Potter D. Further evidence for a non-random chromosomal abnormality in acute promyelocytic leukemia. Int J Cancer. 1977;20:869–872. doi: 10.1002/ijc.2910200608. [DOI] [PubMed] [Google Scholar]
- Sallmyr A, Fan J, Datta K, Kim KT, Grosu D, Shapiro P, Small D, Rassool F. Internal tandem duplication of FLT3 (FLT3/ITD) induces increased ROS production, DNA damage, and misrepair: implications for poor prognosis in AML. Blood. 2008;111:3173–3182. doi: 10.1182/blood-2007-05-092510. [DOI] [PubMed] [Google Scholar]
- Sanz MA, Grimwade D, Tallman MS, Lowenberg B, Fenaux P, Estey EH, Naoe T, Lengfelder E, Buchner T, Dohner H, et al. Management of acute promyelocytic leukemia: recommendations from an expert panel on behalf of the European LeukemiaNet. Blood. 2009;113:1875–1891. doi: 10.1182/blood-2008-04-150250. [DOI] [PubMed] [Google Scholar]
- Shen Y, Zhu YM, Fan X, Shi JY, Wang QR, Yan XJ, Gu ZH, Wang YY, Chen B, Jiang CL, et al. Gene mutation patterns and their prognostic impact in a cohort of 1185 patients with acute myeloid leukemia. Blood. 2011;118:5593–5603. doi: 10.1182/blood-2011-03-343988. [DOI] [PubMed] [Google Scholar]
- Shih LY, Huang CF, Wu JH, Wang PN, Lin TL, Dunn P, Chou MC, Kuo MC, Tang CC. Heterogeneous patterns of FLT3 Asp(835) mutations in relapsed de novo acute myeloid leukemia: a comparative analysis of 120 paired diagnostic and relapse bone marrow samples. Clin Cancer Res. 2004;10:1326–1332. doi: 10.1158/1078-0432.ccr-0835-03. [DOI] [PubMed] [Google Scholar]
- Sohal J, Phan VT, Chan PV, Davis EM, Patel B, Kelly LM, Abrams TJ, O’Farrell AM, Gilliland DG, Le Beau MM, et al. A model of APL with FLT3 mutation is responsive to retinoic acid and a receptor tyrosine kinase inhibitor, SU11657. Blood. 2003;101:3188–3197. doi: 10.1182/blood-2002-06-1800. [DOI] [PubMed] [Google Scholar]
- Solomon DA, Kim T, Diaz-Martinez LA, Fair J, Elkahloun AG, Harris BT, Toretsky JA, Rosenberg SA, Shukla N, Ladanyi M, et al. Mutational inactivation of STAG2 causes aneuploidy in human cancer. Science. 2011;333:1039–1043. doi: 10.1126/science.1203619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirewalt DL, Radich JP. The role of FLT3 in haematopoietic malignancies. Nat Rev Cancer. 2003;3:650–665. doi: 10.1038/nrc1169. [DOI] [PubMed] [Google Scholar]
- Suela J, Alvarez S, Cigudosa JC. DNA profiling by arrayCGH in acute myeloid leukemia and myelodysplastic syndromes. Cytogenet Genome Res. 2007;118:304–309. doi: 10.1159/000108314. [DOI] [PubMed] [Google Scholar]
- Takayama N, Kizaki M, Hida T, Kinjo K, Ikeda Y. Novel mutation in the PML/RARalpha chimeric gene exhibits dramatically decreased ligand-binding activity and confers acquired resistance to retinoic acid in acute promyelocytic leukemia. Exp Hematol. 2001;29:864–872. doi: 10.1016/s0301-472x(01)00651-8. [DOI] [PubMed] [Google Scholar]
- Takizawa H, Regoes RR, Boddupalli CS, Bonhoeffer S, Manz MG. Dynamic variation in cycling of hematopoietic stem cells in steady state and inflammation. J Exp Med. 2008;208:273–284. doi: 10.1084/jem.20101643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasson MH, Xiang Z, Walgren R, Zhao Y, Kasai Y, Miner T, Ries RE, Lubman O, Fremont DH, McLellan MD, et al. Somatic mutations and germline sequence variants in the expressed tyrosine kinase genes of patients with de novo acute myeloid leukemia. Blood. 2008;111:4797–4808. doi: 10.1182/blood-2007-09-113027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson IP, Novelli MR, Bodmer WF. The mutation rate and cancer. Proc Natl Acad Sci U S A. 1996;93:14800–14803. doi: 10.1073/pnas.93.25.14800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter MJ, Park JS, Lau SK, Li X, Lane AA, Nagarajan R, Shannon WD, Ley TJ. Expression profiling of murine acute promyelocytic leukemia cells reveals multiple model-dependent progression signatures. Mol Cell Biol. 2004;24:10882–10893. doi: 10.1128/MCB.24.24.10882-10893.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter MJ, Payton JE, Ries RE, Shannon WD, Deshmukh H, Zhao Y, Baty J, Heath S, Westervelt P, Watson MA, et al. Acquired copy number alterations in adult acute myeloid leukemia genomes. Proc Natl Acad Sci U S A. 2009;106:12950–12955. doi: 10.1073/pnas.0903091106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter MJ, Shen D, Ding L, Shao J, Koboldt DC, Chen K, Larson DE, McLellan MD, Dooling D, Abbott R, et al. Clonal Architecture of Secondary Acute Myeloid Leukemia. New England Journal of Medicine. 2012;366:1090–1098. doi: 10.1056/NEJMoa1106968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wartman LD, Larson DE, Xiang Z, Ding L, Chen K, Lin L, Cahan P, Klco JM, Welch JS, Li C, et al. Sequencing a mouse acute promyelocytic leukemia genome reveals genetic events relevant for disease progression. J Clin Invest. 2011;121:1445–1455. doi: 10.1172/JCI45284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch JS, Klco JM, Varghese N, Nagarajan R, Ley TJ. Rara haploinsufficiency modestly influences the phenotype of acute promyelocytic leukemia in mice. Blood. 2011a;117:2460–2468. doi: 10.1182/blood-2010-08-300087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch JS, Westervelt P, Ding L, Larson DE, Klco JM, Kulkarni S, Wallis J, Chen K, Payton JE, Fulton RS, et al. Use of whole-genome sequencing to diagnose a cryptic fusion oncogene. JAMA. 2011b;305:1577–1584. doi: 10.1001/jama.2011.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita Y, Yuan J, Suetake I, Suzuki H, Ishikawa Y, Choi YL, Ueno T, Soda M, Hamada T, Haruta H, et al. Array-based genomic resequencing of human leukemia. Oncogene. 2010;29:3723–3731. doi: 10.1038/onc.2010.117. [DOI] [PubMed] [Google Scholar]
- Young MA, Larson DE, Sun CW, George DR, Ding L, Miller CA, Lin L, Pawlik KM, Chen K, Fan X, et al. Background mutations in parental cells account for most of the genetic heterogeneity of induced pluripotent stem cells. Cell Stem Cell. 2012;10:570–582. doi: 10.1016/j.stem.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Oancea C, Henschler R, Ruthardt M. Cooperation between constitutively activated c-Kit signaling and leukemogenic transcription factors in the determination of the leukemic phenotype in murine hematopoietic stem cells. Int J Oncol. 2009;34:1521–1531. doi: 10.3892/ijo_00000281. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.