SUMMARY
Adult-onset neurodegenerative disorders are disabling and often fatal diseases of the nervous system whose underlying mechanisms of cell death remain, in most instances, unknown. Defects in mitochondrial respiration had previously been proposed to contribute to the occurrence of many, if not all of the most common neurodegenerative disorders. However, the discovery of genes mutated in hereditary forms of these enigmatic diseases has additionally suggested defects in mitochondrial dynamics. Such disturbances can lead to changes in mitochondrial trafficking, in interorganellar communication, and in mitochondrial quality control. These new mechanisms by which mitochondria may also be linked to neurodegeneration will likely have far-reaching implications for our understanding of the pathophysiology and treatment of adult-onset neurodegenerative disorders.
INTRODUCTION
Adult-onset neurodegenerative diseases are a large group of heterogeneous disorders characterized by the relatively selective death of neuronal subtypes. In most cases, they arise for unknown reasons, and are relentlessly progressive. Age is the most consistent and robust risk factor, and thus the number of patients is expected to increase dramatically in the years to come, especially in industrialized countries. For instance, the number of cases of Alzheimer’s disease (AD) and other dementias, including Lewy body disease and frontotemporal dementia, was estimated by the World Health Organization in 2005 at almost 25 million individuals worldwide, with ~5 million new cases annually, and is projected to more than double by 2025. Existing approved medicines provide only symptomatic relief, and their chronic use is often associated with deleterious side effects; none appear to modify the natural course of the diseases. Clearly, the development of effective therapies is hindered by our limited knowledge of the molecular mechanisms underlying these conditions.
Despite the phenotypic diversity of neurodegenerative disorders, insights gained in the last decade into their pathophysiology, especially through genetics, has begun to reveal some underlying themes. These include disturbances in cellular quality control mechanisms (e.g. endoplasmic reticulum [ER] stress, defects in proteasomal and autophagic function, accumulation and/or aggregation of misfolded proteins), oxidative stress, neuroinflammation, and impaired subcellular trafficking. Another pathogenic theme that has come to prominence, and which is the focus of this review, is the role of impaired mitochondrial function, not only as it pertains to defects in mitochondrial energy production, but also to mitochondrial dynamics (i.e. organellar shape, size, distribution, movement, and anchorage), communication with other organelles, and turnover.
Of necessity, we have limited our discussion to a subset of neurodegenerative disorders (Table 1), focusing on those that best illustrate our central points. We recognize that this selection introduces a bias, yet the diseases we have chosen encompass the vast majority of patients afflicted with neurodegenerative disease, and thus should provide a faithful picture of the state of affairs regarding the role of mitochondria in neurodegeneration.
Table 1.
| Name | Frequency per 100,000 |
Presentation | Typical age of on set (yr.) |
Main clin ical manifestations | Main neuropathological features |
|---|---|---|---|---|---|
|
Alzheimer’s Diseas
(AD) |
1600 | >90% sporadc <10% famlal |
>60 (often younger for familial cases) |
Cognitive impairment primarily featuring memory problems (e.g. trouble in remembering recent events, the names of people and things). As the disease progresses, language (e.g. inability to recall vocabulary), perceptual skills, attention, constructive abilities, orientation, problem solving and functional ability difficulties also arise, as w ell as behavioral and neuropsychiatric changes, including w andering, irritability and labile a f fec ts . |
Gross cerebral cortex atrophy ( particularly in the temporal, parietal, and parts of the frontal lobes, and in the cingulate gyrus) due to a loss of neurons and synapses. These chnages are associated w ith amyloid plaques and neurofibrillary tangles. |
|
Amyotrophic latera
sclerosis (ALS) |
1 to 3 | >90% sporadic <10% familial |
~55 (often younger for familial cases) |
Muscle w asting and w eakness, and increased muscle tone. |
Loss of cortical and spinal motor neurons; degeneration of corticospinal track; multiple forms of pertinacious inclusions; gliosis. |
|
Charcot-Marie-Tooth
disease (CMT) |
~40 | Familial. Autosomal Dominant, Recessive, X-linked |
5-25 (sometimes >30) |
Progres sive disorder of the peripheral nerves giving rise to weakness, muscle w asting, and sensory loss, predominantly in the feet and legs, but also in the hands and arms in advanced stages. Often the first manifestation is difficulty in w alking. |
Degenerative changes are seen in the peripheral nerves, w here a depletion of large myelinated motor and sensory fibers is observed. Spared fibers show damaged axons and myelin sheaths, with the distal part of the nerve often more affected than the proximal part. In some forms of CMT, the affected nerve may be enlarged and show “onion-bulb” formation of Schw ann and fibroblast cells. |
|
Huntington’s diseas
(HD) |
3 to 7 | Familial. Autosomal Domnant. CAG tridnucleotide expansions in the huntingtin gene |
40-50 Of note, the greater the number of CAG repeats , the earlier the onset |
Often begins with personality changes (e.g. irritability) and mood disturbances (e.g. depression) follow ed by abnormal movements of a choreic nature, primarily of the face and fingers. As the disease progresses, chorea spreads, athetoid and dystonic monuments appear, and intellectual functions decline, giving r is e to a dementia. |
Gross atrophy of caudate nucleus and putamen accompanied w ith mild frontal and temporal atrophy. The most salient neurodegenerative changes invoke a loss of medium-size spiny neurons in the striatopallidal striatonigral pathways assoc iated with striatal gliosis |
|
Hereditary spastic
parapares is (HSP) |
4 to 6 | Familial. Autosomal Dominant, Recessive, X-linked |
<35 or 40-60 | Difficulty in w alking and poor balance are often the first signs. Progressive increased muscle tone, brisk reflexes, muscle w eakness, bladder disturbances, and paresthesia are part of the core manifestations. Depending of the genetic form, ataxia, dementia, abormal movements, visual dysfunction, epilepsy or even extraneurological signs may be observed. |
Axonal degeneration primarily in the corticospinal tracts and the fasciculus gracilis, and to a lesser extent in the spinocerebellar tract. Loss of anterior spinal horn is observed in some cases. Dorsal root ganglia, posterior roots, and peripheral nerves are normal. |
| Optic atrophy (OA) | 2 to 10 | Familial. Autosomal Dominant, Recessive, X-linked, Mitochondrially inherited |
18-25 | Progressive bilateral visual loss. Central vision affected prior to peripheral vision. |
Degeneration of the retinal ganglion cell bodies and axonal pathways up to the lateral geniculate nuclei. |
|
Parkinson ’s disease
(PD) |
e 160 | >90% sporadic <10% familial |
~60 (often younger for familial cases) |
Tremor; slowness of movements; stiffness; poor balance. As the disease progresses, non-motor manifestations arise, including dementia, constipation, sleep disturbances, and orthostatic hypotension. |
Loss of pigmented neurons in ventral midbr ain ( e.g. substantia nigra pars compacta) and other pigmented nuclei (e.g. locus ceruleus, dorsal motor nucleus of the vagus); intraneuronal Lewy body inclusions; gliosis. |
|
Spinocerebellar
ataxias (SCA) |
1 to 4 | Familial. Autosomal Dominant, Recessive, X-linked. ~30 different gene mutations, but a CAG trinucleotide expansion (in different genes) is found in several forms |
<10 to >60 | Progres sive incoordination of gait, and often associated w ith poor coordination of hands, speech, and eye movements. Dementia, movement disorders such as parkinsonism, myoclonus, seizures, retinal degeneration and optic atrophy, and peripheral neuropathy are observed in some forms of SCA. |
Degeneration of the spinal cord and the cerebellum, as well as many nuclei of the basal ganglia and the brainstem. |
CAN GENETICS SHED LIGHT ONTO THE MITOCHONDRIAL LINK IN NEURODEGENERATIVE DISORDERS?
Many of the prominent adult-onset neurodegenerative disorders, such as AD, Parkinson’s disease (PD), and amyotrophic lateral sclerosis (ALS), are primarily sporadic, i.e. occurring in the absence of any genetic linkage. However, in rare instances they can be inherited. The phenotypes of both the sporadic and familial forms of these diseases are essentially indistinguishable, implying that they might share common underlying mechanisms. We believe that this similarity justifies the analysis of rare genetic forms of a common sporadic disorder, as it could well illuminate the pathogenesis of both. Moreover, the familial counterparts of all of the common sporadic neurodegenerative disorders are due to mutations not just in a single gene, but in multiple distinct and often ostensibly dissimilar genes. This apparent genetic heterogeneity associated with specific syndromes should not come as a surprise, since thus far the taxonomy of neurodegenerative disorders rests on clinical, biochemical, and neuropathological criteria, lumping under the same label diseases that merely look alike. Nonetheless, this striking situation raises the possibility that however disparate these genes may appear to be, the functions of the respective gene products might intersect in common pathways. Furthermore, the observations that mutations in a specific gene can give rise to more than one distinct clinical phenotype (Chen et al., 2004; Elden et al., 2010; Moreira et al., 2004; Pulst et al., 1996) suggest that while the disease classification scheme is useful clinically, it may be equally helpful to view different neurodegenerative disorders as reflecting different, and perhaps more nuanced, expressions of shared, fundamental underlying problems.
In the last several years, 188 separate genetic loci have been associated with inherited forms of the eight adult-onset neurodegenerative syndromes that we have selected (AD, ALS, CMT, HSP, Huntington’s disease [HD], optic atrophy [OA], PD, and SCA [Table 1]), and 104 genes have been identified (Table 2; see also Online Mendelian Inheritance in Man [http://www.ncbi.nlm.nih.gov/omim]). In connection to the topic of this review, it is worth noting that of the 104 identified genes, at least 36 have some type of association to mitochondrial function, either directly (i.e. proteins in known mitochondrial biochemical pathways and structure; 24 genes) or indirectly (i.e. proteins that are not necessarily targeted to mitochondria, but that affect them secondarily, such as those associated with the communication between mitochondria and the ER; 12 genes) (Table 3). The fraction of mitochondrial-resident gene products associated with neurodegenerative disorders (24/104, or ~23%) is well above the proportion expected by mere chance alone (~8%, i.e. ~1,600 genes encoding mitochondrial proteins/~20,000 total protein-coding genes), suggesting a predilection for defects in these organelles to be associated with late-onset neurodegenerative disorders. Based on the above discussion, let us start our journey through mitochondria and see where the path of human genetics leads us.
Table 2.
| Type | Inh. | Gene | Chromosome |
|---|---|---|---|
| Alzheimer disease (AD) | |||
| AD1 | AD | APP | 21q21.3 |
| AD2 | ? | APOE | 19q 13.32 |
| AD3 | AD | PSEN1 | 14q24.2 |
| AD4 | AD | PSEN2 | 1q4213 |
| AD5 | ? | ? | 12p11-q13 |
| AD6 | ? | ? | 10q 24 |
| AD 7 | ? | ? | 10p13 |
| AD8 | ? | ? | 20p |
| AD9 | ? | ? | 19p13.2 |
| AD10 | ? | ? | 7q36 |
| AD11 | ? | ? | 9p21.3 |
| AD12 | ? | ? | 8p12-q22 |
| AD13 | ? | ? | 1q21 |
| AD14 | ? | ? | 1q25 |
| AD15 | ? | ? | 3q22-q24 |
| AD16 | XL | ? | Xq 21.3 |
| Amyotrophic lateral sclerosis ( ALS) | |||
| ALS1 | AD | SOD1 | 21q 22.11 |
| ALS2 (J) | AR | ALS2 | 2q33.1 |
| ALS3 | AD | ? | 18q 21 |
| ALS4 (J) | AD | SETX | 9q34.13 |
| ALS5 | AR | ? | 15q15-q21 |
| ALS6 | AR | FUS | 16p11.2 |
| ALS7 | AD | ? | 20p13 |
| ALS8 | AD | VAPB | 20q 13.32 |
| ALS9 | AD | ANG | 14q11.2 |
| ALS10 | AD | TARDBP | 1p36.22 |
| ALS11 | AD | FIG4 | 6q21 |
| ALS12 | AD/AR | OPTN | 10p13 |
| ALS13 | AD | ATXN2 | 12q 24.12 |
| ? | AR | SPG11 | 15q21.1 |
| ? | AD | VCP | 9p13.3 |
| Charcot-Marie-Tooth disease ( CMT) | |||
| CMT1A | AD | PM P22 | 17p12 |
| CMT1B | AD | MPZ | 1q23.3 |
| CMT1C | AD | LITAF | 16p13.13 |
| CMT1D | AD | EGR2 | 10q 21.3 |
| CMT1F | AD | NEFL | 8p21.2 |
| CMT2A1 | AD | KIF1B | 1p36.22 |
| CM T2A2 | AD | MFN2 | 1p36.22 |
| CM T2B | AD | RAB7A | 3q21.3 |
| CMT2B1 | AR | LMNA | 1q22 |
| CM T2B2 | AR | MED25 | 19q 13.33 |
| CM T2C | AD | TR PV4 | 12q 24.11 |
| CM T2D | AD | GARS | 7p14.3 |
| CM T2E | AD | NEFL | 8p21.2 |
| CM T2F | AD/AR | HSPB1 | 7q11.23 |
| CM T2G | AD | ? | 12q12-q13.3 |
| CM T2H | AR | ? | 8q21 3 |
| CM T2I | AD | MPZ | 1q23.3 |
| CM T2J | AD | MPZ | 1q23.3 |
| CM T2K | AD | GDAP1 | 8q21.11 |
| CM T2L | AD | HSPB8 | 12q 24.23 |
| CM T2M | AD | DN M2 | 19p13.2 |
| CM T2N | AD | AARS | 16q 22.1 |
| CM T4A | AR | GDAP1 | 8q21.11 |
| CMT4B1 | AR | MTMR2 | 11q 21 |
| CM T4B2 | AR | SBF2 | 11p15.4 |
| CM T4C | AR | SH 3TC2 | 5q32 |
| CM T4D | AR | NDRG1 | 8q24.22 |
| CM T4E | AD/AR | EGR2 | 10q 21.3 |
| CM T4F | AR | PR X | 19q 13.2 |
| CM T4G | AR | ? | 10q 23.2 |
| CM T4H | AR | FGD4 | 12p1121 |
| CM T4J | AR | F G4 | 6q21 |
| CM TDIA | AD | ? | 10q24.1-q25.1 |
| CM TDIB | AD | DN M2 | 19p13.2 |
| CM TDIC | AD | YARS | 1p35.1 |
| CM TX1 | XL | GJB1 | Xq 13 1 |
| Huntington disease (HD) | |||
| HD | AD | HTT | 4p16 3 |
| Hereditary spastic paraplegia ( HSP) | |||
| SPG1 | XL | L1CAM | Xq 28 |
| SPG2 | XL | PLP1 | Xq 22 |
| SPG3A | AD | ATL1 | 14q 22 1 |
| SPG4 | AD | SPAST | 2p22 3 |
| SPG5A | AR | CYP7B1 | 8q12 3 |
| SPG6 | AD | NIPA1 | 15q112 |
| SPG7 | AR | SPG7 | 16q 24 3 |
| SPG8 | AD | KIAA0196 | 8q24.13 |
| SPG9 | AD | ? | 10q 23.3-q24.1 |
| SPG10 | AD | KIF5A | 12q 13.3 |
| SPG11 | AR | SPG11 | 15q21.1 |
| SPG12 | AD | ? | 19q13.11-q 13.13 |
| SPG13 | AD | HSPD1 | 2q33.1 |
| SPG14 | AR | ? | 3q27-q28 |
| SPG15 | AR | ZFYVE26 | 14q 24.1 |
| SPG16 | XL | ? | Xq11.2 |
| SPG17 | AD | BSCL2 | 11q 12.3 |
| SPG18 | AR | ? | 8p12-p11.21 |
| SPG19 | AD | ? | 9q33-q34 |
| SPG20 | AR | SPG20 | 13q 13.3 |
| SPG21 | AR | SPG21 | 15q 22.31 |
| SPG22 | XL | SLC16A2 | Xq 13.2 |
| SPG23 | AR | ? | 1q24- q32 |
| SPG24 | AR | ? | 13q 14.3 |
| SPG25 | AR | ? | 6q23.3-q24.1 |
| SPG26 | AR | ? | 12p11.1-q15 |
| SPG27 | AR | ? | 10q 22 1-q24 1 |
| SPG28 | AR | ? | 14q 21.3-q22.3 |
| SPG29 | AD | ? | 1p31.1-p21.1 |
| SPG30 | AR | ? | 2q37 3 |
| SPG31 | AD | REEP1 | 2p11.2 |
| SPG32 | AR | ? | 14q12-q21 |
| SPG33 | AD | ZFYVE27 | 10q 24.2 |
| SPG34 | XL | ? | Xq24-q25 |
| SPG35 | AR | ? | 16q 21-q 23.1 |
| SPG36 | AD | ? | 12q 23-q 24 |
| SPG37 | AD | ? | 8p21.1-q13.3 |
| SPG38 | AD | ? | 4p16-p15 |
| SPG39 | AR | PN PLA6 | 19p13 2 |
| SPG40 | AD | ATL1 | 14q 22.1 |
| SPG41 | AD | ? | 11p14.1-p11.2 |
| SPG42 | AD | SLC33A1 | 3q25.31 |
| SPG44 | AR | GJC2 | 1q42 13 |
| SPG45 | AR | ? | 10q24.3-q25.1 |
| ? | AR | AIMP1 | 4q24 |
| Optic atrophy( OA) | |||
| OPA1 | AD | OPA1 | 3q29 |
| OPA2 | XL | ? | Xp11.4-p11.21 |
| OPA3 | AD | OPA3 | 19q 13 32 |
| OPA4 | AD | ? | 18q12.2-q123 |
| OPA5 | ADAR | ? | 22q12.1-q13.1 |
| OPA6 | AR | ? | 8q21 13-q22 1 |
| OPA7 | AR | TM EM126 | A 11q 14.1 |
| LHON | M | ND genes | mtDNA |
| Parkinson disease ( PD)) | |||
| PARK1/4 | AD | SN CA | 4q22.1 |
| PARK2 | AR | PARK2 | 6q26 |
| PARK3 | AD | ? | 2p13 |
| PARK5 | AD | UCHL1 | 4p13 |
| PARK6 | AR | PIN K1 | 1p36 12 |
| PARK7 | AR | PARK7/Park | in 1p36 23 |
| PARK8 | AD | LRRK2 | 12q 12 |
| PARK9 | AR | ATP13A2 | 1p36 13 |
| PARK10 | ? | ? | 1p32 |
| PARK11 | AD | GIGYF2 | 2q37 1 |
| PARK12 | ? | ? | Xq21-q25 |
| PARK13 | ? | HTRA2 | 2p13 1 |
| PARK14 | ? | PLA2G6 | 22q 13 1 |
| PARK15 | AR | FBXO7 | 22q 12 3 |
| PARK16 | ? | ? | 1q32 |
| ? | AR | SPG11 | 15q21.1 |
| ? | AR | NDUFV2 | 18p11 22 |
| Spinocerebellar ataxia ( SCA) | |||
| SCA1 | AD | ATXN1 | 6p22.3 |
| SCA2 | AD | ATXN2 | 12q 24.12 |
| SCA3 | AD | ATXN3 | 14q 32.12 |
| SCA4 | AD | PLEKHG4 | 16q 22.1 |
| SCA5 | AD | SPTBN2 | 11q 13.2 |
| SCA6 | AD | CACNA1A | 19p13.2 |
| SCA7 | AD | ATXN7 | 3p14.1 |
| SCA8 | AD | ATXN8 | 13q 21.33 |
| SCA8 | AD | ATXN 8OS | 13q 21.33 |
| SCA9 | AD | ? | ? |
| SCA10 | AD | ATXN10 | 22q 13.31 |
| SCA11 | AD | TTBK2 | 15q15.2 |
| SCA12 | AD | PPP2R2B | 5q32 |
| SCA13 | AD | KCNC3 | 19q 13.33 |
| SCA14 | AD | PR KCG | 19q 13.42 |
| SCA15 | AD | ITPR1 | 3p26.1 |
| SCA16 | AD | CN TN4 | 3p26.2 |
| SCA17 | AD | TBP | 6q27 |
| SCA18 | AD | ? | 7q22-q23 |
| SCA19 | AD | ? | 1p21-q21 |
| SCA20 | AD | ? | 11p13-q 11 |
| SCA21 | AD | ? | 7p21.3-p15.1 |
| SCA22 | AD | ? | 1p21-q23 |
| SCA23 | AD | ? | 20p13-p12.3 |
| SCA24 | AR | ? | 1p36 |
| SCA25 | AD | ? | 2p21-p13 |
| SCA26 | AD | ? | 19p13.3 |
| SCA27 | AD | FGF14 | 13q 33.1 |
| SCA28 | AR | AFG3L2 | 18p11.21 |
| SCA29 | AD | ? | 3p26 |
| SCA30 | AD | ? | 4q34.3-q35.1 |
| SCA31 | AD | BEAN -TK2 | 16q 21 |
| SCAN1 | AR | TDP1 | 14q 32.1 |
| SCAR1 | AR | SETX | 9q34.13 |
| SCAR2 | AR | ? | 9q34-qter |
| SCAR3 | AR | ? | 6p23-p21 |
| SCAR4 | AR | ? | 1p36 |
| SCAR5 | AR | ? | 15q25.3 |
| SCAR6 | AR | ? | 20q 11-q 13 |
| SCAR7 | AR | ? | 11p15 |
| SCAR8 | AR | SYNE1 | 6q25.2 |
| SCAR9 | AR | ADCK3 | 1q42 13 |
| SCAX1 | XL | ? | Xp11 21-q21 3 |
| DR PLA | AD | ATN1 | 12p13 31 |
| FR DA1 | AR | FXN | 9q21 11 |
| FR DA2 | AR | ? | 9p23-p11 |
| IOSCA | AR | C10orl2 | 10q 24 31 |
| MIRAS | AR | POLG | 15q26.1 |
| ? | AR | AN O10 | 3p22 1 |
| ? | AD | SCN8A | 12q 13.3 |
Disease classification as listed in OMIM. Inh., Inheritance. AD, autosomal dominant; AR, autosomal recessive; M, mitochondrial; XL, X-linked.
SINCE WE ALL THINK FIRST ABOUT BIOENERGETICS, LET’S TALK ABOUT IT
Mitochondria are organelles present in all cells of the body (erythrocytes excluded), ranging from a few hundred to many thousands, depending on cell type. Maternally inherited, they are the locus for many of the body’s “housekeeping” functions, including the biosynthesis of amino acids and steroids and the beta-oxidation of fatty acids; they also play a central role in apoptosis. However, the function that sets this organelle apart, and which is responsible for the cliché that mitochondria are the “powerhouses of the cell,” is the production of adenosine triphosphate (ATP), via the combined efforts of the tricarboxylic acid cycle and the respiratory chain/oxidative phosphorylation system (OxPhos). The respiratory chain is a set of biochemically linked multi-subunit complexes (complexes I, II, III, and IV), and two electron carriers (ubiquinone/coenzyme Q and cytochrome c). It uses the energy stored in food to generate a proton gradient across the mitochondrial inner membrane, while at the same time transferring electrons to oxygen, producing water. The energy of the proton gradient drives ATP synthesis via ATP synthase (complex V); the ATP is then distributed throughout the cell.
The central importance of mitochondria for cellular energy production is underscored by the discovery in the last 20 years of numerous syndromes resulting from OxPhos defects (DiMauro and Schon, 2003). The mitochondrial respiratory chain is the product of a joint effort between the mitochondrial and nuclear genomes. Mitochondria harbor their own DNA (mtDNA) which is a 16.6-kb double-stranded circular DNA that encodes for 13 of the ~92 polypeptides of the OxPhos system (DiMauro and Schon, 2003), while the nuclear DNA (nDNA) specifies ~79 OxPhos structural polypeptides and more than 100 other proteins required for the proper incorporation of cofactors (e.g. iron-sulfur proteins, hemes, copper) and for the assembly of the five respiratory chain complexes into an integrated system (Fernandez-Vizarra et al., 2009).
Patients with OxPhos dysfunction who carry mutations in either mtDNA or nDNA present with a host of clinical features, many of which are neurological, such as seizures, myoclonus, ataxia, progressive muscle weakness, stroke-like episodes, and cognitive impairment (DiMauro and Schon, 2003). However, these manifestations do not typically overlap either with the clinical or the neuropathological hallmarks of any of our selected adult-onset neurodegenerative disorders (Table 1). Furthermore, to a remarkable degree, mutations in both mtDNA and nDNA that affect the integrity or functioning of the OxPhos complexes typically do not strike in adulthood, but rather in infancy (e.g. Leigh syndrome, which is a fatal, necrotizing encephalopathy). Yet, some patients with OxPhos dysfunction do succumb later, in their twenties or thirties (e.g. Kearns-Sayre syndrome; which is a sporadically-occurring, fatal, multisystem disorder featuring paralysis of the extraocular muscles, retinal degeneration, and heart block), but it is atypical for mitochondrial patients to survive much longer, and it is exceptional for any individual to experience an onset of an OxPhos disease beyond the age of 40. However, the age at onset and the severity of the disorder correlate well with the degree of ATP deficit caused by the mutation. Thus, “mild” mutations could theoretically give rise to a slowly progressive, late-onset neurodegenerative disease, such as AD or PD. Such mild mutations typically arise in one of two ways: either because the mutation per se does not cause a severe OxPhos impairment (e.g. mutations in complex I subunits causing Leber’s hereditary optic neuropathy, or LHON (Sadun et al., 2010), a maternally-inherited form of blindness), or because the proportion of mutated mtDNAs coexisting with normal mtDNAs (i.e. heteroplasmy) within affected neurons is relatively low, such that the deficit in ATP production is only partial, as is typically the case in oligosymptomatic mothers of affected children (DiMauro and Schon, 2003). Still, even if mtDNA mutations have the potential to provoke neuronal death, the fact remains that there are now more than 200 documented mutations in the 37 mtDNA-encoded genes, and an equal number in almost 100 nDNA-encoded OxPhos-related genes (Smits et al., 2010), yet only a handful are associated with adult-onset neurodegenerative disease. Among these, only two well-documented mtDNA mutations are associated with adult-onset neurodegeneration - one with Parkinsonism (De Coo et al., 1999) and one with SCA (Silvestri et al., 2000) – but, as far as we can tell, none with AD, ALS, CMT, HD, or HSP. A number of mtDNA polymorphisms have also been associated with some of these disorders, but their pathogenicity remains to be established, and except for a few isolated reports (Swerdlow et al., 1998), there is little evidence of maternal inheritance of neurodegenerative disease. Furthermore, mutations in proteins required for mtDNA replication, such as those in mitochondrial DNA polymerase γ and in the helicase Twinkle, cause rare forms of cerebellar degeneration (Hakonen et al., 2008). Also rare are mutations in frataxin, which is required for the synthesis of mitochondrial iron-sulfur proteins that are components of respiratory complexes, causing Friedreich’s ataxia (Schmucker and Puccio, 2010), and mutations in ADCK3/CABC1 that affect the synthesis of coenzyme Q of the respiratory chain, causing a recessive form of SCA (Gerards et al., 2010).
The above discussion emphasizes that neurodegenerative disorders, especially those of late-onset, cannot be classified neatly as canonical “primary mitochondrial cytopathies.” And yet, it is possible that much is to be gained by viewing neurodegeneration through the prism of primary mitochondrial cytopathies, because if we do not, we may fail to recognize a bioenergetic component in the disease process. Take PD as an example. A meta-analysis of genome-wide gene expression microarray studies revealed the strongest association between PD and genes encoding for OxPhos subunits and for enzymes involved in glucose metabolism, all of which are regulated by PGC-1α (Zheng et al., 2010), a transcriptional co-activator of mitochondrial biogenesis (Puigserver et al., 1998). Relevant to this observation is the identification of PARIS (Parkin-interacting substrate), a partner of the PD-related protein Parkin (see below), that represses PGC-1α expression (Shin et al., 2011). These authors propose that inactivation of Parkin, either by mutation or by environmental stress, leads to the accumulation of PARIS and the ensuing inhibition of PGC-1α transcription, which in turn may reduce mitochondrial biogenesis and cause OxPhos deficiency. Thus, if PD is any guide, bioenergetic defects could indeed play a role in common neurodegenerative disorders, not so much as the initiating factor of the neurodegenerative cascade but more as a pathogenically meaningful consequence of some other perturbation (e.g. loss of Parkin activity).
SO WHAT OTHER MITOCHONDRIAL PROBLEMS CAN CAUSE NEURODEGENERATION?
The textbook image of mitochondria as bean-shaped organelles that populate the cytoplasm in apparently random fashion belies a far more dramatic reality (Braschi and McBride, 2010). Mitochondria are constantly on the go. They fuse and divide, branch and fragment, swell and extend, exist in clusters and as individual entities. Importantly, they travel throughout the cell, from the cell body outwards (anterograde movement) and “homeward-bound” in the opposite direction (retrograde movement). When not moving, they periodically anchor themselves on - and then disengage from - other organelles, such as the ER, endocytic vesicles, and the plasma membrane. In short, mitochondria are dynamic organelles that move from the cell body to regions of the cell to deliver ATP and other metabolites where they are most required, and then return. This is seen most strikingly in highly elongated cells such as neurons: mitochondria are enriched at presynaptic terminals at the ends of axons and at postsynaptic terminals at the ends of dendrites, where bioenergetic demand is particularly high. In addition, while this constant motion helps the cell redirect and recycle mitochondria in an efficient manner, “worn-out” mitochondria are ultimately disposed of (and their component parts recycled) via autophagy (“mitophagy”) or via extrusion of “mitochondria-derived vesicles” (Braschi et al., 2010). The inability of mitochondria to execute these functions would be expected to disrupt cellular physiology and viability, and the degree of impairment likely corresponds to that cell’s requirements for having well-functioning mitochondria positioned in the right place at the right time. For these reasons, there is growing enthusiasm for the notion that defects in mitochondrial dynamics might play a pivotal role in the pathogenesis of neurodegenerative disorders.
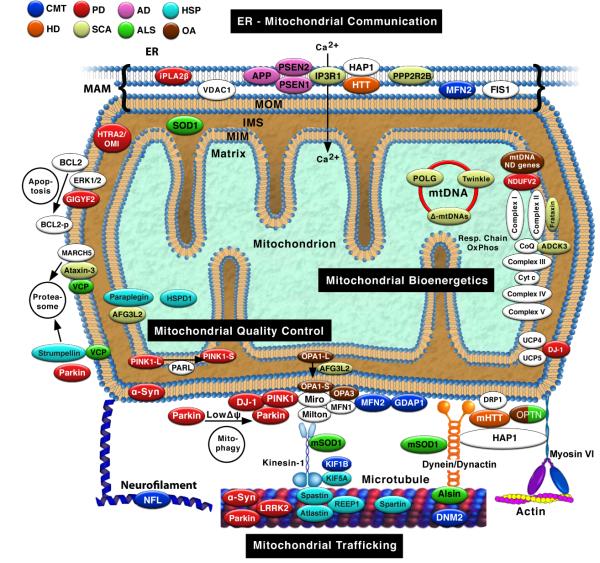
We will focus here on three ways that altered “mitodynamics” could contribute to adult-onset neurodegeneration (Chen and Chan, 2009): aberrant mitochondrial trafficking, altered interorganellar communication, and impaired mitochondrial quality control (Fig. 1).
Fig. 1. Mitochondrial interactions in neurodegenerative diseases.
Proteins associated with mutations causing neurodegenerative disorders are in colored ovals (colored according to the key), and are associated with four broad mitochondrial functions (black rectangles). White ovals indicate selected relevant mitochondrial proteins not currently associated with neurodegenerative disease. In general, only pathogenic proteins discussed in the text are shown; note that some proteins not discussed here are also associated with mitochondria, either directly or indirectly (Table 3); HTRA2/OMI and HSPD1, associated with PD and HSP, respectively, are included in the figure for completeness, but were not discussed in the text. Proteins that “touch” each other indicate a physical or genetic interaction. For simplicity, the figure does not show all interactions, or shows some interactions that occur only in some tissues or at specific times, or both; conversely, some interactions are speculative, based on extrapolations from the literature. See text for details. IMS, intermembrane space; MAM, mitochondria-associated ER membranes; MIM, mitochondrial inner membrane; MOM, mitochondrial outer membrane; m, mutant; Δ-mtDNAs, large-scale partial deletions of mtDNA.
Neurodegenerative disorders and mitochondrial trafficking
Organelles such as lysosomes, peroxisomes, and mitochondria are not positioned statically within cells. Rather, they are transported on cytoskeletal elements, that is, microtubules and actin cables, often in association with intermediate filaments (Jung et al., 2004). Short-range movement on actin cables requires myosin motors, whereas long-range movement on microtubules requires two other types of motors: dynein/dynactin for retrograde transport and kinesins for anterograde transport (Hollenbeck, 1996). Dynein-mediated retrograde movement appears to be promiscuous, with no specific adaptor for mitochondria. Kinesins, on the other hand, comprise a large superfamily, among which is a subset that has been reported to associate specifically with mitochondria (Zinsmaier et al., 2009).
Given the critical role of mitochondria in maintaining cell viability, it stands to reason that defects in mitochondrial trafficking could underlie neurodegenerative processes. Is there evidence to support this view? We have approached this question in two ways. First, we asked if there were evidence for perturbed mitochondrial trafficking in any of our selected set of neurodegenerative diseases. Conversely, we examined situations where mitochondrial trafficking is known to be perturbed, and asked whether the ensuing phenotypes were reminiscent of any of our selected diseases.
Direct evidence that mitochondrial trafficking is altered in human neurodegenerative disease patients is actually quite limited. This paucity of data is not surprising, given both the logistical hurdles in obtaining human samples and the difficulty in analyzing mitochondrial transport in autoptic material. Nevertheless, of the disorders on our list, such evidence has been reported in autoptic samples from patients with sporadic AD: defects in axonal trafficking of molecular motor proteins and organelles, including mitochondria, were inferred from the observation of axonal swellings containing vesicles, vacuoles, multilamellar bodies, and especially mitochondria, in the nucleus basalis of Meynert; the formation of these vesicles was apparently mediated by the expression of kinesin-1, a microtubule motor (Stokin et al., 2005).
On the other hand, ample data for trafficking defects exist in experimental models - mainly genetically-engineered mice - of a number of adult-onset neurodegenerative disorders. Both anterograde (De Vos et al., 2007) and retrograde (Shi et al., 2010) mitochondrial transport were reduced in motor neurons from ALS mice expressing mutant superoxide dismutase-1 (SOD1). Even more remarkable, misfolded wild-type SOD1 immunopurified from a subset of patients with sporadic ALS and perfused into isolated squid axoplasm inhibited fast axonal transport (Bosco et al., 2010). This latter observation is particularly noteworthy, as it reveals a remarkable potential connection between the sporadic and familial forms of the disease.
Altered mitochondrial trafficking and integrity have also been observed upon overexpression of at least two other proteins whose mutations cause familial forms of ALS. First, increased expression of the wild-type guanine-nucleotide exchange factor alsin in monkey COS7 cells was associated with disorganization of the microtubule network and with organellar abnormalities, including perinuclear clustering of mitochondria (Millecamps et al., 2005). Second, transgenic mice expressing human TAR DNA binding protein 43 (TDP-43) showed mitochondrial maldistribution, with an excess of mitochondria in the cell body of motor neurons and a paucity of mitochondria in distal motor axon terminals (Shan et al., 2010).
Thus far, our discussion of experimental models has revolved mainly around pathologies of the motor neurons, in which mitochondria must travel exceedingly long distances. But what about trafficking in disorders in which neurons with much shorter axons are the primary target of the disease? In fact, Alzheimer’s disease, a disorder primarily of “short” neurons in the cortex and hippocampus, displays features of aberrant axonal trafficking of cargo (Stokin et al., 2005), and especially of altered mitochondrial trafficking (Wang et al., 2009a) and dynamics (Wang et al., 2008; Wang et al., 2009b). Moreover, published data suggest that Huntington’s disease, an adult-onset fatal chorea involving relatively “short” striatopallidal neurons, may also be a disorder of mitochondrial trafficking. HD is caused by mutations - specifically expansions of a polyglutamine stretch - in huntingtin (HTT), a protein of unknown function. In transfected primary rat cortical neurons, mutant, but not wild-type, HTT blocked mitochondrial movement (Chang et al., 2006). Expression of mutant HTT in transgenic mice impaired trafficking of vesicles and mitochondria, and mutant HTT preferentially redistributed kinesin- and dynein-related proteins in extracts from human HD brain (Trushina et al., 2004). These effects on mitochondria and on trafficking were likely due specifically to the polyglutamine expansion located within the N-terminal region of HTT, as truncated fragments containing the N-terminal region associated preferentially with mitochondria in HTT knock-in mice, and these mutant HTT fragments affected mitochondrial trafficking in both the anterograde and retrograde directions (Orr et al., 2008).
Other aspects of HTT function also point to mitochondrial trafficking (Sack, 2010). The HTT binding partner huntingtin-associated protein (HAP1) associates with membranous organelles, including mitochondria (Gutekunst et al., 1998), and interacts with both kinesin and dynein/dynactin to regulate the transport of cargo on microtubules (Bossy-Wetzel et al., 2008). Interestingly, Milton, one of two mitochondrial microtubule adaptor proteins (the other is Miro; see below), is a HAP1 homolog, and it too binds HTT and dynactin (Stowers et al., 2002). Taken together, these data support the possibility that altered mitochondrial trafficking contributes to neurodegeneration in HD.
More speculative, but still worth mentioning, is the potential link between proteins known to cause familial PD and defects in microtubule-mediated trafficking. The mitochondrial kinase PINK1 (PTEN-induced putative kinase-1) may play a role in mitochondrial transport, as it was shown to form a multiprotein complex with Milton and Miro (Weihofen et al., 2009), but the effects of PD-linked mutations on this relationship are currently unknown. Upon overexpression of wild-type α-synuclein in differentiated SH-SY5Y neuroblastoma cells (which mimics the multiplications of the normal gene found in some PD patients), aggregates of the protein disrupted the microtubule network and microtubule-dependent trafficking of cargoes (Lee et al., 2006). On the other hand, both the PD-linked protein LRRK2 (leucine-rich repeat kinase-2) and Parkin were found to alter the balance between polymerized and depolymerized tubulin (Gillardon, 2009; Yang et al., 2005), with downstream effects on trafficking of cargo that still remain to be demonstrated.
To make matter even more complicated, just because a neurodegenerative disease gene is associated with the trafficking machinery for intracellular cargo does not necessarily mean that trafficking is the main problem. For example, in transfection experiments, the HSP-related protein spartin was localized to microtubules and mitochondria via determinants located in the N- and C-terminal regions of the protein, respectively (Lu et al., 2006). However, proteomic analysis implied that spartin plays a different role, in protein folding and turnover, both in mitochondria and ER (Milewska et al., 2009), and may also be in involved in lipid droplet formation (Hooper et al., 2010). A similar dilemma surrounds another HSP-related protein, REEP1 (receptor expression-enhancing protein 1). One group localized REEP1 to mitochondria (Zuchner et al., 2006), while another group found that REEP1 interacted with atlastin-1, another HSP-related protein, within tubular ER membranes, thereby coordinating ER shaping with microtubule dynamics (Bian et al., 2011; Park et al., 2010). However, despite the potential connection of both spartin and REEP1 to microtubules and mitochondria, there is no evidence that either one plays any role in mitodynamics, even though mutations in both cause neurodegeneration.
These examples illustrate the challenge in relating pathology to specific problems in mitochondrial dynamics. Perhaps a more fruitful approach might be to start from situations where mitochondrial trafficking is known to be perturbed, and then see whether they produce phenotypes mimicking aspects of neurodegenerative disease. From the outset, it should be noted that there are hardly any mutations in the structural components of actin, dynein, or kinesin known to cause neurodegenerative disease. In our survey, we found only three: mutations in kinesin heavy chain isoform 1Bβ cause CMT (Zhao et al., 2001) and in isoform 5A cause HSP (Ebbing et al., 2008), while mutations in the p150Glued subunit of the dynein-associated protein dynactin increase the risk of developing ALS (Munch et al., 2004). This state of affairs probably reflects the essentiality of these motor molecules to life. Nonetheless, a number of disorders are caused by mutations in proteins that are associated with actin, dynein, and kinesin in a secondary manner, among which five appear to affect mitochondrial behavior.
Actin cables, like microtubules, have a polarity, with myosin motors typically moving towards the “barbed” (+) end of actin filaments and away from the “pointed” (-) end (Wells et al., 1999). One exception to this rule is myosin VI, which moves in the opposite direction (Wells et al., 1999). It apears to play a role in asymmetric partitioning of organelles and cytoskeletal components during cell division, at least in worms, as deletion of myosin VI in C. elegans resulted in a failure to deliver mitochondria to budding spermatids (Kelleher et al., 2000). This “unconventional” myosin has an indirect connection to at least two neurodegenerative diseases, ALS and HD, via one of its binding partners, the cargo adaptor protein optineurin (Sahlender et al., 2005). Mutations in optineurin, which have already been reported to cause primary open-angle glaucoma (Fuse, 2010), cause ALS (Maruyama et al., 2010). Optineurin also binds HTT, and plays a role in cellular signaling, membrane trafficking, and cellular morphogenesis (Anborgh et al., 2005; Hattula and Peränen, 2000), providing further support that altered mitochondrial trafficking plays a role in HD.
With respect to microtubule function, mutations in spastin, a microtubule-severing protease causing HSP, resulted in abnormal perinuclear clustering of mitochondria and peroxisomes in transfected HEK293 cells (McDermott et al., 2003) and in axonal transport defects and mitochondrial clustering on microtubules in spastin-mutated mice (Kasher et al., 2009).
Regarding intermediate filaments, mutations in the neurofilament light chain (NFL) cause CMT (Brownlees et al., 2002; Pérez-Ollé et al., 2005). Expression of mutant NFL in explanted embryonic mouse motor neurons disrupted the neurofilament network, but notably, rounding of mitochondria and reduction in axonal diameter occurred prior to this event, implying that mitochondrial dysfunction contributes to the pathogenesis of the disease (Tradewell et al., 2009). Moreover, expression of heat shock protein B1 in neurons expressing some CMT-mutant forms of NFL abrogated the mitochondrial and trafficking phenotypes. This result is not only consistent with the role of this chaperone in neurofilament assembly, but also helps explain why mutations in this heat shock protein also cause CMT (Tradewell et al., 2009).
The strategy of examining defects in mitochondria-related proteins has yielded a more compelling connection with adult-onset neurodegenerative disorders, but this relationship is not particularly obvious when viewing in toto all eight of the neurodegenerative disorders that we have selected. In fact, as can be seen from the above discussion, mitochondrial connections are prevalent in only two specific disorders, HSP and CMT, both of which are axonopathies often associated with myelin pathology (Table 1).
Neurodegenerative disorders and mitochondrial-ER communication
Mitochondria do not exist, or operate, in isolation, but associate with many other subcellular organelles. Aside from connections to cytoskeletal elements, mitochondria interact with, for example, peroxisomes, lysosomes, Golgi, and ER. Among these, the most intriguing is the connection between mitochondria and ER. These two orgenelles are linked, both biochemically and physically (Csordas et al., 2006), via mitochondria-associated ER membranes (ER-MAM, or MAM) (Rusinol et al., 1994). Located mainly in the perinuclear region of cells (Area-Gomez et al., 2009; Schon and Area-Gomez, 2010), MAM has been reported to be enriched in more than 75 proteins, including those involved in calcium homeostasis (e.g. inositol-1,4,5-triphosphate [IP3] receptors [IP3Rs] and ryanodine receptors), in lipid metabolism (e.g. phosphatidylethenolamine N-methyltransferase), in intermediate metabolism (e.g. glucose-6-phosphatase), in cholesterol metabolism (e.g. acyl-coenzyme A:cholesterol acyltransferase 1 [ACAT1]), in the transfer of lipids between the ER and mitochondria (e.g. fatty acid transfer proteins 1 and 4), and in ER stress (e.g. glucose-regulated proteins 75 and 78) (Hayashi et al., 2009b). Contacts between the two organelles are maintained by MAM-associated proteins, such as phosphofurin acidic cluster sorting protein-2 (Simmen et al., 2005) and mitofusin-2 (MFN2), which is also required for mitochondrial fusion (de Brito and Scorrano, 2008). Interestingly, fission-1 (FIS1), a protein required for mitochondrial fission, has recently also been localized to the MAM (Iwasawa et al., 2011).
The relationship between MAM and calcium trafficking (Csordas et al., 2010) is worthy of some elaboration. As alluded to above, two cargo adaptor proteins discovered initially in Drosophila - Miro and Milton - are implicated in the specific linkage of mitochondria to kinesin-1 in neurons. Miro is anchored to the mitochondrial outer membrane (Guo et al., 2005), and binds to the mitochondrial-specific adaptor protein Milton, which is linked to the kinesin-1 heavy chain (Brickley et al., 2005; Glater et al., 2006; Koutsopoulos et al., 2010). Miro is a calcium-binding protein (Fransson et al., 2003), and thus has the potential for being a regulator of mitochondrial motility in neurons, in essence operating as a sensor of local [Ca2+] and ATP. It has been proposed that in the Ca2+-unbound state, Miro binds Milton and mitochondria are attached to microtubules, whereas in the Ca2+-bound state, Miro cannot bind Milton and mitochondria are uncoupled from microtubules (Rice and Gelfand, 2006). This model is consistent with the “saltatory movement” model proposed by Hajnóczky (Liu and Hajnóczky, 2009; Yi et al., 2004), in which mitochondria move only when local [Ca2+] is low, and stop when the local [Ca2+] is high. Notably, only Ca2+ mobilized via IP3Rs (or, in muscle, via the related ryanodine receptors) could generate this result. We note, however, that very few of the experiments supporting this model have been conducted in mammalian neurons.
The tethering of mitochondria to ER via MAM is a dynamic process, as organelles must disengage from the ER in order to engage, and then travel on, microtubules. Any defect that alters this equilibrium could conceivably result in a mismatch between the number of mitochondria required in specific regions of a neuron and the demand for mitochondrial cargo in those regions (Schon and Area-Gomez, 2010). Given the dynamic nature of MAM, and the role of IP3Rs in maintaining the proper equilibrium between ER and mitochondrial [Ca2+], one can easily imagine that neurodegenerative disorders in which calcium homeostasis is disrupted could arise from altered ER-mitochondrial communication, or conversely, that alterations in calcium homeostasis from some other cause could affect this communication indirectly. Among our selected adult-onset neurodegenerative diseases, two candidates are HD, in which both HTT and HAP1 interact with IP3R1 (Tang et al., 2003), and a form of SCA associated with loss of IP3R1 function (van de Leemput et al., 2007).
However, the most compelling case for a role for MAM in pathogenesis is familial AD due to mutations in presenilin-1 and -2, which are components of the γ-secretase complex that cleaves the amyloid precursor protein (APP) to produce amyloid-β, a constituent of the extracellular neuritic “plaques” that accumulate in the brains of AD patients (Schon and Area-Gomez, 2010). Apart from the accumulation of hyperphosphorylated forms of the microtubule-associated protein tau in intraneuronal “tangles” (the other prominent aspect of AD pathology), both the familial and sporadic forms of the disease are characterized by a number of other features that have received less attention. These include altered lipid, cholesterol, and glucose metabolism (Schon and Area-Gomez, 2010), aberrant calcium homeostasis (Supnet and Bezprozvanny, 2010), ER stress and the unfolded protein response (Hoozemans et al., 2005), aberrant mitochondrial dynamics (e.g. fragmented and perinuclear mitochondria, associated with, for example, altered levels (Wang et al., 2009a) or post-translational modifications (Cho et al., 2009) of the mitochondrial fission protein DRP1), and defects in energy metabolism (Ferreira et al., 2010), but it remains to be determined to what degree these phenomena are causally linked. It is in this context that a recent report that presenilin-1 and -2, and γ-secretase activity itself, are highly enriched in the MAM (Area-Gomez et al., 2009) is so interesting, because the functions noted above that are perturbed in AD are in fact the very functions associated with MAM. Moreover, even the generation of the plaques might be explained by altered MAM function, as MAM-localized ACAT1, which is required to convert intracellular cholesterol to cholesteryl esters that are deposited in lipid droplets, is apparently a modulator of APP processing and amyloid-β production (Puglielli et al., 2001), for currently unknown reasons. Thus, pathogenic mutations in the presenilins could alter ER-mitochondrial communication (Zampese et al., 2011), leading to the features of the disease (Schon and Area-Gomez, 2010).
One other neurodegenerative disease that may be associated with MAM dysfunction is CMT, which can be caused by mutations both in MFN2 (Chen and Chan, 2009) and in ganglioside-induced differentiation-associated protein 1 (GDAP1) (Pedrola et al., 2005), which interacts with MFN2 (Niemann et al., 2005). MFN2, like mitofusin-1 (MFN1), is required for mitochondrial fusion (Chen and Chan, 2009). However, a portion of MFN2 is enriched in the MAM, where it is required for the tethering of ER to mitochondria (de Brito and Scorrano, 2008). We note that CMT-mutant MFN2 expressed in cultured dorsal root ganglion neurons induced abnormal clustering of fragmented mitochondria, as well as impaired axonal transport of mitochondria (Baloh et al., 2007). Perhaps these abnormalities resulted from an underlying defect in ER-mitochondrial communication.
The study of MAM is a nascent field that has just begun to be recognized as a contributor to neurodegeneration, and likely will expand beyond the diseases cited above. For example, there may be a “MAM connection” in at least two other diseases in which the relevant proteins - both involved in phospholipid metabolism - appear to be enriched in the MAM. These are SCA due to mutations in PPP2R2B, a regulatory subunit of protein phosphatase 2A (Giorgi et al., 2010) that promotes mitochondrial fission (Dagda et al., 2008a), presumably via MAM-localized FIS1 (Iwasawa et al., 2011), and PD due to mutations in subunit β of the calcium-independent phospholipase A2 (iPLA2β; gene PLA2G6), which plays a key role in ER-mitochondrial cross-talk during ER stress-induced apoptosis (Lei et al., 2008). It would thus be fascinating to see if future studies on PPP2R2B and iPLA2β provide insight into a potential link between MAM and neurodegeneration in SCA, in PD, and perhaps even beyond.
Neurodegenerative disorders and mitochondrial quality control
Besides alterations in trafficking and in ER-mitochondrial communication, mitochondria can also fail to reach their destinations due to dysregulation of quality control systems. The cell has surveillance mechanisms to eliminate mutated, unfolded, and otherwise unwanted proteins, via autophagic and ubiquitin-proteasome systems located in the cytosol. In a similar manner, “unwanted” mitochondria can be disposed of, and their contents recycled, by mitophagy. Although there is currently no evidence that mitochondria contain proteasomes, they do have mechanisms to eliminate misfolded or unneeded polypeptides, via, for example, the AAA (ATPase associated with diverse cellular activities) protease paraplegin/SPG7 and the paraplegin-related protease AFG3L2, and their regulators, the prohibitins PHB and PHB2 (Osman et al., 2009). In addition, mitochondrial proteins, especially those in the outer membrane, can be retrotranslocated into the cytosol by, for example, mitochondrially-targeted valosin-containing protein (VCP/p97, also a AAA protease) (Xu et al., 2011), for subsequent clearance by the proteasome. This process constitutes, as it were, a mitochondrial version of ER-associated degradation (Heo et al., 2010). Interestingly, mutations in VCP were recently found to cause familial ALS (Johnson et al., 2010).
Thus, mutations in mitochondrial quality control genes could prevent the efficient elimination of damaged mitochondria and the degradation of superfluous and potentially deleterious polypeptides, hence leading to neuronal dysfunction and perhaps ultimately to cell death. In order for quality control to operate at the level of the mitochondrion, cells must be able to distinguish between “good” and “bad” organelles, and in fact, such discrimination does occur. Mitochondria apparently are deemed to be “good” if they have a high membrane potential (Δψ), and perhaps low levels of reactive oxygen species (ROS) as well, both presumably indicative of a well-functioning respiratory chain. Conversely, they are deemed “bad” if they have a low Δψ and elevated ROS, indicative of defective OxPhos; these are the organelles that are eliminated via selective mitophagy (Twig and Shirihai, 2011). Mitophagy of damaged organelles, however, is a last resort, as cells initially try to prevent the accumulation of “bad” mitochondria via maintenance of a dynamic equilibrium between mitochondrial fission and fusion, which “homogenizes” organellar contents. This mixing of a few bad mitochondria within a larger pool of good ones allows for complementation of genes and gene products to take place after mitochondria have exchanged contents (Gilkerson et al., 2008), thereby blunting, or even eliminating, the deleterious effects of misfolded proteins and randomly mutated mtDNAs (Twig and Shirihai, 2011).
Thus, from a quality control standpoint, one might predict that mutations in genes encoding proteins required for mitochondrial dynamics, and especially organellar fission and fusion, would result in compromised organellar “mixing,” leading to an excess accumulation of bad mitochondria, perhaps causing disease, and this is indeed the case. Gene products in this category include four associated with fusion (although interestingly, none with fission): MFN2 and GDAP1, both causing CMT, and optic atrophy proteins OPA1 and OPA3, both causing optic atrophy (OA). Even though OPA1 and OPA3 (Huizing et al., 2010; Ryu et al., 2010) and GDAP1 (Niemann et al., 2005) interact with mitofusins to regulate the mitochondrial network, it is again worth noting that the four genes are associated with two totally different clinical presentations. Mitochondrial dynamics are also altered in HD (Bossy-Wetzel et al., 2008; Kim et al., 2010; Oliveira, 2010), as the expression of mitochondrial fission-related proteins, such as FIS1 and dynamin-related protein-1 (DRP1) (Costa et al., 2010), which happens to interact with HTT (Song et al., 2011), are increased in striatum and frontal cortex of patients, whereas that of fusion-related proteins, such as MFN1, MFN2, and OPA1 are decreased (Shirendeb et al., 2011), likely explaining the fragmented mitochondria and altered mitochondrial dynamics seen in the disease (Pandey et al., 2010; Shirendeb et al., 2011).
Among diseases in this category, PD stands out, as it is becoming apparent that some genetic forms of the disease may be in essence disorders of mitochondrial quality control. Paradoxically, the history of PD, at least from a genetic/biochemical perspective, pointed away from such a conclusion, as the earliest observations regarding pathogenesis implied a deficiency of complex I of the respiratory chain as the key culprit. That conclusion was based on the findings that (1) MPTP (1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine), a complex I inhibitor similar to rotenone, caused PD-like symptoms, (2) complex I activity was reported to be reduced in PD post-mortem tissues, (3) mutations in complex I subunits, such as nDNA-encoded NDUFV2, were associated with PD (Nishioka et al., 2010), and (4) accumulations of large-scale deletions of mtDNA were found specifically in the substantia nigra of sporadic PD patients (Bender et al., 2006; Kraytsberg et al., 2006), the signature target region of the brain in this disease (Dauer and Przedborski, 2003). However, a notable challenge to this concept was the failure to find clear evidence of mutations in mtDNA that cause PD (Simon et al., 2010). Moreover, the identification in the last decade of at least a dozen genetic loci associated with familial PD (Table 2) has changed our perspective dramatically, as many of these gene products are associated with mitochondria but have no obvious or direct connection to OxPhos, and many of those proteins appear to be involved in quality control.
Two of those PD-related proteins may be involved in quality control in an indirect manner. PLA2G6 (phospholipase A2, group VI) (Seleznev et al., 2006) is a mitochondrial lipase that deacetylates cardiolipin and is involved in ER stress and ER-mitochondrial crosstalk via ceramide (Lei et al., 2008). GIGYF2 (GRB10 interacting GYF protein-2) enhances the activation of mitochondrially-localized (Deng et al., 2000; Galli et al., 2009) extracellular signal-regulated kinases ERK1 and ERK2 (Deng et al., 2000; Higashi et al., 2010), both of which are involved in mitophagy (Dagda et al., 2008b) and apoptosis (Deng et al., 2000; Higashi et al., 2010).
A much stronger case for a role in mitochondrial quality control can be made for Parkin, a cytosolic E3 ubiquitin ligase. However, in this role, Parkin does not act alone, as mounting evidence implicates the necessary interaction with another PD-related and mitochondrially-localized protein, PINK1. PINK1 is a kinase of unknown specificity that displays a possible dual location in the organelle, i.e. it has been found in both the outer (Zhou et al., 2008) and inner (Jin et al., 2010; Silvestri et al., 2005) membranes. Mitochondrial PINK1 is present in two forms, “long” and “short,” with the long form (PINK1-L, ~64 kDa) cleaved to the short form (PINK1-S, ~52 kDa) within the mitochondrial inner membrane by PARL (presenilin-associated rhomboid-like protein), an intramembrane protease (Deas et al., 2011; Jin et al., 2010).
As an aside, like PINK1, OPA1 also has long and short forms, with OPA1-L cleaved, not by PARL, but by AFG3L2, to produce OPA1-S (Duvezin-Caubet et al., 2007). The long forms of both OPA1 and PINK1 are targeted to the inner membrane, where cleavage occurs, essentially “releasing” the short forms of both proteins to perform their functions. Whereas the function of OPA1-S is clearly fusion of the mitochondrial outer membrane, the precise role of PINK1-S remains to be determined. One possibility is that PINK1 (Weihofen et al., 2009), which, like MFN2 (Misko et al., 2010), interacts with the mitochondria-kinesin adaptors Miro and Milton, and with mitofilin (Weihofen et al., 2009), another mitochondrial morphology-related protein (John et al., 2005), assists in the “offloading” of mitochondria from microtubules in order to allow them to fragment and become autophagized (Gomes et al., 2011; Hailey et al., 2010).
However, the lion’s share of attention to PINK1 is devoted to its relationship with Parkin in cooperating in a signaling pathway (Clark et al., 2006) to maintain mitochondrial integrity, presumably by eliminating “bad” mitochondria via mitophagy (Vives-Bauza and Przedborski, 2011). As such, we deem the elucidation of the biology of Parkin and PINK1 to be far more important in illustrating the “mitochondrial connection” to neurodegenerative disease than the few number of patients harboring mutations in these proteins might warrant.
In the current view, upon loss of Δψin damaged mitochondria, PINK1 residing in the outer membrane triggers, in some unknown fashion, the recruitment of cytosolic Parkin to the mitochondria (Jin et al., 2010; Narendra et al., 2010b; Vives-Bauza et al., 2010). Mitochondrial proteins located in the outer membrane, such as the voltage-dependent anion channel 1 (VDAC1; also called porin) are then ubiquitinated in a Parkin-dependent manner (Geisler et al., 2010). The ubiquitination of outer membrane proteins recruits the autophagy molecule LC3 (microtubule-associated protein-1 light chain-3) to build the autophagosome around the damaged mitochondrion (Vives-Bauza and Przedborski, 2011), apparently mediated by the adaptor proteins HDAC6 (Okatsu et al., 2010) and p62 (Geisler et al., 2010; Narendra et al., 2010a; Okatsu et al., 2010). Upon membrane depolarization, according to some studies, Parkin also induces ubiquitination of mitofusins (Gegg et al., 2010; Ziviani et al., 2010), which are then degraded by the proteasome via VCP (Tanaka et al., 2010), although others found that it is DRP1 and not MNF1/2 or FIS1 that is degraded by the proteasome in a Parkin-dependent manner (Wang et al., 2011). This discrepancy needs to be resolved, as the exact fusion/fission factors that are regulated by Parkin would indicate whether Parkin, either alone or in conjunction with PINK1, promotes fragmentation or elongation of mitochondria. Perhaps α-synuclein may begin to provide some hints into this outstanding issue, as overexpression of PD-mutant and wild-type α-synuclein (which, as noted above, mimics the gene multiplications found in some PD patients) were reported to promote fragmentation of mitochondria (Kamp et al., 2010; Nakamura et al., 2011). Conversely, downregulation of wild-type α-synuclein in C. elegans resulted in elongated mitochondria (Kamp et al., 2010). Although changes in the fusion/fission balance have not yet been demonstrated in PD samples, on the surface, one would predict that mutations in α-synuclein would enhance, rather than hamper, mitochondrial turnover, because fragmentation, and not elongation, of mitochondria into “bite-sized” pieces facilitates mitophagy (Twig et al., 2008). Alternatively, rather than altering mitophagy, perhaps α-synuclein influences quality control through its effect on the fusion/fission balance, by affecting the ability of good mitochondria to complement bad ones.
The PD-related protein DJ-1 may also have a relationship to quality control, as it has a number of proposed disparate connections to mitochondria. In addition to possibly binding to the NDUFA4 and ND1 subunits of complex I (Hayashi et al., 2009a), DJ-1 has been reported to interact with both PINK1 and Parkin (Moore et al., 2005) and to modulate mitochondrial fission/fusion in a ROS-dependent manner (Irrcher et al., 2010). This latter effect is consistent with its proposed function as an atypical peroxiredoxin-like peroxidase that scavenges mitochondrial H2O2 (Andres-Mateos et al., 2007). Moreover, DJ-1 seems to regulate the expression of the mitochondrial uncoupling (UCP) proteins, as its ablation in mice is associated with reduced expression of UCP4 and UCP5 in brain (Guzman et al., 2010). While these two UPCs are among the least well-characterized members of this family, it is tantalizing to suggest that changes in their expression in brain could alter mitochondrial Δψ, which, if confirmed, would be an important clue as to how DJ-1 participates in mitochondrial quality control. Indeed, if as suggested from the PINK1/Parkin story, a loss of Δψ is a prerequisite for the disposal of bad mitochondria, the loss-of-function mutations in DJ-1 that cause PD may impair mitochondrial quality control by distorting the relationships among mitochondrial damage, Δψ, and mitophagy. In this scenario, DJ-1 would operate upstream of PINK1/Parkin within the mitophagy pathway, an idea consistent with the demonstration that silencing DJ-1 in human cell lines does not affect PINK1-dependent recruitment of Parkin and ensuing mitophagy in response to Δψ collapse by protonophores (Vives-Bauza et al., 2010). Clearly, further work is needed to determine how the function of PD-related proteins intersect with mitodynamics to contribute to the pathogenesis of the disease.
The mitophagy model for pathogenesis in PD is appealing, as it explains many of the features of the disease that have been ascribed to mitochondrial dysfunction noted above. However, there are aspects to this developing story that suggest caution in accepting such a scenario uncritically. First, deletion of PINK1, Parkin, or DJ-1 in mice, either alone or in combination, had little perceptible effect on neuronal function (Kitada et al., 2009), calling the role of mitophagy in the pathogenesis of the disease into question. Equally important is the relative artificiality of some of the experimental manipulations upon which the role of these proteins has been based. Because both PINK1 and Parkin are present at low levels, most conclusions are derived from overexpression experiments. Furthermore, the lack of good antibodies has required the use of epitope tags to detect these proteins. Finally, the complete disruption of mitochondrial Δψ using ionophores such as CCCP (carbonyl cyanide m-chlorophenyl hydrazone) does not mimic the much lower degree of disruption of Δψ that likely occurs in patients; even cells that lack mtDNA and OxPhos function entirely can maintain about 50% of the wild-type Δψ. Thus, while the concept of mitochondrial quality control as a pathogenic principle in PD remains appealing, some aspects of the current model may require modification.
We would be remiss if we failed to mention that quality control has more than a janitorial function, as it is also required to maintain normal cellular and organellar processes. For example, the major mitochondrial matrix AAA protease, besides degrading misfolded proteins (Thomas Langer, personal communication), regulates mitochondrial ribosome biogenesis, by processing the mitochondrial ribosomal protein MRPL32 for proper incorporation into, and functioning of, mitochondrial ribosomes. Consistent with this function, the loss of either SPG7 or AFG3L2 (Nolden et al., 2005), the two subunits that compose the matrix AAA protease, compromise mitochondrial translation, resulting in bioenergetic impairment (Atorino et al., 2003; Nolden et al., 2005). Together with the fact that mutations in SPG7 cause HSP (Casari et al., 1998) and mutations in AFG3L2 cause SCA (Di Bella et al., 2010), the aforementioned findings suggest that defects in mitochondrial ribosomal biogenesis via defects in quality control can provoke neurodegeneration.
CONCLUDING REMARKS
For years, defects in OxPhos and oxidative stress have been two of the most popular hypotheses put forward to explain pathogenesis of almost all neurodegenerative disorders. It is clear that “classical” mitochondrial diseases, many of which are myopathies and encephalopathies in children and young adults, are unquestionably provoked by bioenergetic defects. However, when we look at the data critically, the role of impaired bioenergetics as the primary cause of late-onset neurodegenerative diseases is far less compelling, even in the case of disorders when there are known mutations in OxPhos genes, such as in LHON. If so, how can we reconcile a role for mitochondria in these disorders, given the large literature implicating energy metabolism? Perhaps the recent shift in emphasis regarding the role of mitochondria in neurodegenerative disorders reflects a better appreciation of the relationship between cause and effect in these diseases, namely, that impaired OxPhos is not the cause of neurodegeneration, but is one result of other underlying mitochondrial problems. Furthermore, we recognize that while changes in mitochondrial function do not necessarily have to affect bioenergetic output, the fact remains that if, for example, mitodynamics are perturbed, the absolute production and local delivery of ATP will be reduced, and that at some point in the disease process bioenergetic failure will occur, probably delivering the coup de grâce.
We have entered a new era of mitochondrial biology, one in which the focus is no longer solely on bioenergetics per se but on mitochondria as an integrated subcellular system (Fig. 1). Under this rubric, a central theme that has emerged is one of altered mitochondrial dynamics. While important advances have been made in this area in a relatively short period of time, some key outstanding questions still remain to be addressed. For example, if diseases such as AD, ALS, and PD are due to errors in mitochondrial quality control overseen by a suite of ubiquitous “housekeeping” proteins, why do these diseases display a predilection for specific subpopulations of neurons? To almost belabor the obvious, the simple answer is that some specific neurons may be more vulnerable to the pathological process than others; clearly, such a differential neuronal susceptibility will only reveal itself if the defect in question is mild, as would be expected for an adult-onset neurodegenerative disorder.
Based on this premise, let us use PD to illustrate a putative pathogenic scenario, by comparing two subpopulations of dopaminergic neurons from the ventral midbrain that are affected differentially in the disease, namely those in the substantia nigra (severely affected) and those in the ventral tegmental area (mildly effected) (Dauer and Przedborski, 2003). One compelling difference between these two groups of neurons is that recruitment of L-type calcium channels during normal autonomous pacemaking is associated with a high ROS signal in dopaminergic neurons of the substantia nigra but not of the ventral tegmental area (Guzman et al., 2010). Thus, one could speculate that the former region accumulates a much higher burden of ROS-related mtDNA mutations than the latter, a view that is, in fact, supported by the observation that dopaminergic neurons in the substantia nigra of both aged normal subjects (Kraytsberg et al., 2006) and PD patients (Bender et al., 2006) contain more mtDNA deletions than do those from controls. Thus, if the “mitochondrial quality control” hypothesis for the pathogenesis of neurodegenerative disorders is correct, one would predict that unless properly eliminated by mitophagy, the number of functionally deficient mitochondria would slowly increase over time to a much greater extent in those neurons generating more mutated mtDNAs (i.e. substantia nigra), eventually causing functional perturbations and neuronal death. Extending this idea further, if mitophagy is important in other adult-onset neurodegenerative disorders, many of which are sporadic, one might also expect that other risk factors, both genetic and environmental, would affect mitophagy and thereby induce the pathology. These risk factors, if they exist, remain to be uncovered.
We have divided our discussion of mitodynamics into three areas - trafficking, organelle interconnectivity, and quality control - mainly for convenience, but we consider all three to be intertwined aspects of a larger whole. In keeping with this view, we note that the analysis of pathogenic mechanisms in essentially all of our selected disorders encompassed more than one of these areas, underscoring the integrative nature of mitodynamics, in which a problem in one area can readily have consequences in another one, including bioenergetics.
Finally, while we have focused in this review almost exclusively on mitochondria, we do not want to leave the impression that mitochondrial defects are the sine qua non of neurodegenerative disease. Far from it: of the 104 genes that were mentioned at the outset, we have discussed fewer than half; the remainder have no obvious connection to mitochondria, and yet they cause neurodegeneration. Moreover, we wish to reiterate that we have focused on familial forms of common neurodegenerative disease as one way to provide a window onto pathogenesis of their sporadic counterparts. This assumption, of course, remains to be validated.
Can any of the above discussion inform ideas about therapeutic strategies for neurodegenerative disorders? Based on the insights into mitochondrial behavior in these disorders, one can begin to envision pharmacological approaches to treatment. For example, regarding mutations in mtDNA, one strategy could be to eliminate mutated mtDNAs while leaving wt-mtDNAs intact, in order to reduce the load of mutated mtDNAs below a critical threshold. One such way to “shift heteroplasmy” is to force cells harboring high levels of partially-deleted mtDNAs to eliminate “bad” mitochondria that contain predominantly mutated mtDNAs while, at the same time, sparing “good” ones that contain predominantly normal mtDNAs, by growing them in ketogenic media that selects for well-functioning mitochondria (Santra et al., 2004). A shifting approach might work particularly well in diseases like PD, in which substantia nigra is known to contain relatively high levels of mtDNA deletions (Bender et al., 2006). The role of the PD-related proteins PINK1 and Parkin in mitophagy points to another way to eliminate “bad” mitochondria selectively, namely, by upregulating the autophagic pathway. An obvious pro-autophagic candidate drug would be rapamycin, which has already been shown to protect against neuronal death in mouse models of PD (Malagelada et al., 2010). For mutations in other genes associated with mitochondrial function, and especially those that impair function only partially, a third promising approach might be to increase energy production in patients by upregulating PGC-1α expression, using compounds such as bezafibrate, a PPAR pan-agonist (Santra et al., 2004) or AICAR (5-aminoimidazole-4-carboxamide ribonucleoside), which acts as an AMP agonist by mimicking AMP (Viscomi et al., 2011). Finally, it may be possible to alter mitodynamics directly by, for example, shifting the relationship between fission and fusion pharmacologically, using the quinazolinone mdivi1 (mitochondrial division inhibitor 1), which enhances mitochondrial fusion in yeast by inhibiting the mitochondrial dynamin Dnm1 that is required for organelle fission (Cassidy-Stone et al., 2008).
Taken together, we may view the role of mitochondria in the pathogenesis of neurodegenerative disorders, and the ways in which we have begun to think about therpaeutics, as multifaceted, and going well beyond the “mere” synthesis and distribution of ATP throughout cells. Mitochondria encompass numerous functions, include many important ones that have not even been discussed here (e.g. amino acid metabolism; steroid metabolism; apoptosis; xenobiotic detoxification; immunological defense), all of which could play a role in neurodegenerative disorders. To the cliché that mitochondria are the “powerhouses” of the cell, let us add one more: what has been uncovered in the last 10 years regarding the role of mitochondria in neurodegenerative disorders is merely the “tip of the iceberg.” Far more exciting findings lay ahead.
Table 3.1.
| Gene | Su btype | Protein | Fu nction/comment | 1Mito? |
|---|---|---|---|---|
| Proteins associated with neurotransmission (12) | ||||
| AN O10 | None | Anoctamin 10; Ca 2+-activated Cl - channel | Chloride transport | N |
| CACNA1A | SC A6 | Ca2+ channel, α-1A subunit | Calcium transport | N |
| EGR2 | CM T1D/4E | Eary growth response 2 protein | Regulates myelin transcription | N |
| GJB1 | CM TX1 | Gap juncton protein β 1 (connexn-32) | Role in myelination | N |
| GJC2 | SPG44 | Gap junction protein γ2 (GJA12) (connexin-47) | Role in myelination | N |
| ITPR1 | SC A15 | IP3 receptor 1 | Calcium transport (enriched in MAM) | N |
| KC NC3 | SC A13 | K + channel | Potassium transport | N |
| MPZ | CM T1B/2I/2J | Myelin protein P0 | Myelin protein | N |
| NDRG1 | CM T4D | N-myc downstream regulated 1 | Myelin maintenance protein, putative | N |
| PM P22 | CM T1A | Peripheral myelin protein 22 | Myelin protein | N |
| PR X | CM T4F | Per iaxin | Myelin protein | N |
| SC N8A | None | Na+ channel, type VIII, α subunit | Sodium transport | N |
| Proteins associated with the cytoskeleton (25) | ||||
| ATL1 | SPG3A /40 | Atlastin 1 GTPase (also called SPG3) | ER-modeling dynamin; interacts with spastin and REEP1 | N |
| ATN1 | DR PLA | Atrophin-1 | Mayinteract with spartin via AIP4 | N |
| DNM2 | CMT2M/D IB | Dynamin-2 | Microtubule-associated force-producing protein | N |
| FGD4 | CM T4H | Frabn FYVE/RhoGEF/PH doman containing 4 | Binds, regulates actin | N |
| HSPB1 | CM T2F | Heat-shock proten 27 (HSP27) | Actin organzation; birds microtubules | N |
| HSPB8 | CM T2L | Heat-shock protein 22 (HSP22) | Chaperone; associated with autophag y | N |
| KIF1B | CM T2A1 | Kinesin 1B (CMT mutation in non-mito KIF1B β isoform) | KIF 1b α isoform transports mitochondria, myelin mRNAs | Y |
| KIF5A | SPG10 | Kinesin 5A | Microtubule motor protein; binds Milton | Y |
| L1CAM | SPG1 | L1 cell adhesion molecule | Axonal glycoprotein | N |
| MTMR2 | CM T4B1 | Myotubularin-related protein 2 | Phosphoinositol-related phosphatase; interacts with SBF2 | N |
| NEFL | CM T1F/2E | Neurofilament, light chain (NFL) | Intracellular transport to axons and dendrites | Y |
| NIPA1 | SPG6 | Non-imprinted in Prader-Willi/Angelman syndromes | Mg2+ transporter; interacts wATL1 | N |
| OPTN | ALS12 | Optineurin | Function unclear; binds ubiquitin; also causes glaucoma | Y |
| PLEKHG4 | SC A4 | Puratrophin-1 (Puknje cell atrophy associated) | Actin dynamics; has a spectrin repeat domain | N |
| REEP1 | SPG31 | Receptor expression-enhancing protein 1 | Binds spastin and atlastin; associates with microtubules | Y |
| SBF2 | CM T4B2 | Myotubularin-related protein 13 (MTMR13) | Pseudophosphatase; interacts with MTMR 2 | N |
| SH 3TC2 | CM T4C | SH 3 domain and tetratricopeptde repeats 2 | Endosomal recycling with Rab11 | N |
| SPAST | SPG4 | Spastin | Severs microtubules; axonal branching | Y |
| SPG20 | SPG20 | Spartin | Binds microtubules; protein folding and turnover? | Y |
| SPG21 | SPG21 | Maspardin (ACP33 acidic cluster protein) | Axonal branching | N |
| SPTBN2 | SC A5 | Spectri n, β-III | Cytoskeletal protein | N |
| SYNE1 | SC AR8 | Synaptic nuclear envelope protein (nesprin-1) | Links organelles to the actin cytoskeleton; has spectrin repeats | N |
| TTBK2 | SC A11 | Tau tubulin kinase 2 | Phosphorylates tau and tubulin | N |
| VAPB | ALS8 | VAMP-associated protein B | Associates with microtubules; membrane transport | N |
| ZF YVE27 | SPG33 | Zinc finger FYVE domain containing 27 (protrudin) | Interacts w spastin; may not be pathogenic (Martignoni et al, 2008) | N |
| Mitochon dria-localized proteins (24) | ||||
| ADCK3 | SC AR9 | Ubiquinone synthesis regulatory kinase (CABC1) | CoQ synthesis | Y |
| AFG3L2 | SC A28 | Paraplegin-like AAA protease | Mitochondrial protein degradation | Y |
| ATXN 3 | SC A3 | Ataxin-3 deubiquitinase | DN A repar; binds mtochondrial E3 ubquitin lig ase MARCH 5 | Y |
| C10orf2 | IOSCA | Twinkle DNA/RNA helicase (PEO1) | mtDN A replication | Y |
| FXN | FR DA1 | Frataxin | Mitochondrial iron metabolism | Y |
| GD AP1 | CM T2K/4A | Gang loside-induced differentiation-associated protein 1 | Interacts wi th mi tofusins | Y |
| HSPD1 | SPG13 | Heat-shock proten 60 (HSP6C) | Mi tochondrial chaperone | Y |
| HTRA2 | PARK13 | HtrA serine peptidase 2 (OMI) | Apoptosis | Y |
| HTT | HD | Hunti ng tin | Mutant HTT is mtochond"ia; interacts wth microtubules | Y |
| MFN2 | CM T2A2 | Mitofusin 2 | Mitochondrial fusion; MAM integrity | Y |
| mtDN A | LHON | Complex I subunits (mtD NA-encoded) | Respiratory chain function | Y |
| NDUFV2 | PD | Complex I subunit (nD NA-encoded) | Respiratory chain function | Y |
| OPA1 | OPA1 | Dynamin-related GTPase | Mitochondrial fusion | Y |
| OPA3 | OPA3 | Dynamin-related GTPase | Mitochondrial fusion? | Y |
| PARK7 | PARK7 | DJ-1 | Atypca peroxidase; mtochondria protein quaity control? | Y |
| PARKIN | PARK2 | Parkin | Mi tophagy | Y |
| PINK1 | PARK6 | PTEN -induced putative kinase 1 | Mi tophagy | Y |
| POLG | MIRAS | Mitochondral DNA poymerase y | mtDN A replication | Y |
| SN CA | PARK1/4 | α-synuclein | Function unclear | Y |
| SOD1 | ALS1 | Superoxide dismutase, Cu,Zn-containing | Redox regulation | Y |
| SPG7 | SPG7 | Paraplegin AAA protease | Mitochondrial protein quality control | Y |
| TD P1 | SC AN1 | Tyrosyl-DN A phosphodiesterase 1 | Topoisomerase I; DN A repair | Y |
| TM EM126A | OPA7 | Transmembrane protein 126A | Function inknown | Y |
| VC P | ALS | Valosin-containing protein/p97 | Retrotranslocation of proteins for proteasome (cyto->mto) | Y |
| Potential relationship with mitochondria and/or mitochondria-associated ER membranes (MAM) (17) | ||||
| ALS2 | ALS2 | Alsin | Guanine-nucleotide exchange factor for RAB5 | N |
| APP | AD 1 | Amyloid precursor protein | Presenilin substrate (present in MAM) | N |
| ATXN 1 | SC A1 | Ataxin-1 | RN A metabolism | N |
| ATXN 10 | SC A10 | Ataxin-10 | Binds GNB2 (mitochonsrial), which binds MF N1 (mitochondrial) | N |
| BEAN -TK2 | SC A31 | BEAN -thymidine kinase 2 overlap region | TK2 is mitochondrial; BEAN is not | N |
| EGR2 | CM T1D | Early growth response 2 protein | Transcription factor | N |
| GARS | CM T2D | Glycyl-tRN A synthetase | Protein synthesis (mitochondrial/cytosolic isoforms) | Y |
| GIGYF2 | PARK11 | GR B10 interacting GYF protein 2 | Enhances activaton of ERK1/2 which is mtochondrial | Y |
| KIAA0196 | SPG8 | Strumpellin; AAA protease | Binds VCP; degrades MOM proteins | Y |
| LMNA | CM T2B1 | Lamin A/C | Nuclear membrane | N |
| LRRK2 | PARK8 | Leucine-rich repeat Ser/Thr-protein kinase 2 | Function not clear | N |
| PLA2G6 | PARK14 | Phospholipase A2, group VI (iPLA2 β) | ER-mitochondria crosstakvia ceramide (probaby in MAM) | Y |
| PPP2R2B | SC A12 | PP2A regulatory subunit 2B β | Signaling (probably in M AM) | Y |
| PSEN 1 | AD 3 | Preseni lin- 1 | Aspartyl protease; in MAM | N |
| PSEN 2 | AD 4 | Preseni lin- 2 | Aspartyl protease; in MAM | N |
| TARDBP | ALS10 | TAR DNA bnding protein 43 (TDP43) | DN A/RNA-binding protein, regulates transcription/splicing | N |
| TBP | SC A17 | TATA box-binding protein | Transcription factor | N |
Table 3.2.
| Gene | Subtype | Protein | Fu nction/comment | 1Mito? |
|---|---|---|---|---|
| No obvious relationship to mitochondria (26) | ||||
| AARS | CM T2N | A anyl-tRNA synthetase | Protein synthesis (cytoplasmic) | N |
| ANG | ALS9 | Angiogenin; RNAse A (RNASE4) | tRNA-specific RNAse; binds actin on endothelial cells | N |
| ATP13A2 | PARK9 | AT Pase, P-type | Cation transporter | N |
| ATXN2 | ALS13/SCA2 | Ataxn-2 | Function unknown | N |
| ATXN7 | SCA7 | Ataxn-7 | Transcriptional regulation | N |
| ATXN8 | SCA8 | Ataxn-8 | Affects RNA-bndng protein MBNL1 | N |
| ATXN8OS | SCA8 | Ataxn-8 opposite strand | Function unknown | N |
| BSCL2 | SPG17 | Se p n | Lipid droplet morphology; in ER | N |
| CYP7B1 | SPG5A | 25-HydroxychOesterol 7- α-hycroxylase | Cholesterol catabolism in ER, 1st step | N |
| FBXO7 | PARK15 | F-box only protein 7 | Ubiquitination | N |
| FGF14 | SCA27 | Fibroblast growth factor 14 | Signalling | N |
| FIG4 | ALS11/CMT4J | Polyphosphoinositide phosphatase (SAC3) | Synthesisof phosphatidylinositol-3,5-bisphosphate | N |
| FUS | ALS6 | Fused in sarcoma/translocated in liposarcoma (FUSTLS) | hnRN P protein | N |
| LITAF | CM T1C | Lipopoysaccharide-induced TNF- α factor | Stimulates monocytes/macrophages | N |
| MED25 | CM T2B2 | Mediator compex subunit 25 | Transcriptional coactivator | N |
| PNPLA6 | SPG39 | Neuropathy targ et esterase | Deacetylates intraceluar phosphatdylchoine (in ER) | N |
| PRKCG | SCA14 | Protein kinase c, γ type | Signaling; activated by Ca2+ and DAG | N |
| RAB7A | CM T2B | Ras-reated GTPase RAB7A | Endosomal; vesicle transport; phagosome maturation | N |
| SETX | ALS4, SCAR1 | Senataxin | Putative DNA/RNA helicase | N |
| SLC16A2 | SPG22 | Monocarboxylate transporter 8 (MCT8) | Thyroid hormone transporter | N |
| SLC33A1 | SPG42 | Acetyl-CoA transporter (AT-1; ACATN1) | ER -Golgi sialylation | N |
| SPG11 | SPG11, PD | Spatacsi n | Function unknown | N |
| TRPV4 | CM T2C | Transient receptor potential cation channel | Osmoregulation | N |
| UCHL1 | PARK5 | Ubiquitin C-termina hydrolase | Protei n deg radation | N |
| YARS | CM TDIC | Tyrosyl-tRNA synthetase | Protein synthesis (cytoplasmic) | N |
| ZFYVE26 | SPG15 | Zinc finger FYVE domain containing 26 | Spastizin (FYVE-CENT); centrosomal proten | N |
indicates whether the listed protein is known to be targeted directly to, or interacts indirectly with, mitochondria (Y, yes; N, no).
Acknowledgments
We thank Drs. William Dauer, Salvatore DiMauro, Michio Hirano, Peter Hollenbeck, Orian Shirihai, and Jean Paul Vonsattel for critical comments, and Robert Lee and Arnaud Jacquier for their expert assistance with the figure. This work was supported by grants from the National Institutes of Health (HD32062 to EAS; NS042269, NS064191, NS38370, NS070276, and NS072182 to SP), the U.S. Department of Defense (W81XWH-08-1-0522, W81XWH-08-1-0465, and W81XWH-09-1-0245 to SP), the Parkinson Disease Foundation, the Thomas Hartman Foundation For Parkinson’s Research, Project A.L.S, the Muscular Dystrophy Association, the Ellison Medical Foundation, the Alzheimer Drug Discovery Foundation, and the Marriott Mitochondrial Disorder Clinical Research Fund (MMDCRF).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anborgh PH, Godin C, Pampillo M, Dhami GK, Dale LB, Cregan SP, Truant R, Ferguson SS. Inhibition of metabotropic glutamate receptor signaling by the huntingtin-binding protein optineurin. J. Biol. Chem. 2005;280:34840–34848. doi: 10.1074/jbc.M504508200. [DOI] [PubMed] [Google Scholar]
- Andres-Mateos E, Perier C, Zhang L, Blanchard-Fillion B, Greco TM, Thomas B, Ko HS, Sasaki M, Ischiropoulos H, Przedborski S, et al. DJ-1 gene deletion reveals that DJ-1 is an atypical peroxiredoxin-like peroxidase. Proc. Natl. Acad. Sci. USA. 2007;104:14807–14812. doi: 10.1073/pnas.0703219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Area-Gomez E, de Groof AJ, Boldogh I, Bird TD, Gibson GE, Koehler CM, Yu WH, Duff KE, Yaffe MP, Pon LA, Schon EA. Presenilins are enriched in endoplasmic reticulum membranes associated with mitochondria. Am. J. Pathol. 2009;175:1810–1816. doi: 10.2353/ajpath.2009.090219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atorino L, Silvestri L, Koppen M, Cassina L, Ballabio A, Marconi R, Langer T, Casari G. Loss of m-AAA protease in mitochondria causes complex I deficiency and increased sensitivity to oxidative stress in hereditary spastic paraplegia. J. Cell Biol. 2003;163:777–787. doi: 10.1083/jcb.200304112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baloh RH, Schmidt RE, Pestronk A, Milbrandt J. Altered axonal mitochondrial transport in the pathogenesis of Charcot-Marie-Tooth disease from mitofusin 2 mutations. J. Neurosci. 2007;27:422–430. doi: 10.1523/JNEUROSCI.4798-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender A, Krishnan KJ, Morris CM, Taylor GA, Reeve AK, Perry RH, Jaros E, Hersheson JS, Betts J, Klopstock T, et al. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat. Genet. 2006;38:515–517. doi: 10.1038/ng1769. [DOI] [PubMed] [Google Scholar]
- Bian X, Klemm RW, Liu TY, Zhang M, Sun S, Sui X, Liu X, Rapoport TA, Hu J. Structures of the atlastin GTPase provide insight into homotypic fusion of endoplasmic reticulum membranes. Proc. Natl. Acad. Sci. USA. 2011;108:3976–3981. doi: 10.1073/pnas.1101643108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco DA, Morfini G, Karabacak NM, Song Y, Gros-Louis F, Pasinelli P, Goolsby H, Fontaine BA, Lemay N, McKenna-Yasek D, et al. Wild-type and mutant SOD1 share an aberrant conformation and a common pathogenic pathway in ALS. Nat. Neurosci. 2010;13:1396–1403. doi: 10.1038/nn.2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossy-Wetzel E, Petrilli A, Knott AB. Mutant huntingtin and mitochondrial dysfunction. Trends Neurosci. 2008;31:609–616. doi: 10.1016/j.tins.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braschi E, Goyon V, Zunino R, Mohanty A, Xu L, McBride HM. Vps35 mediates vesicle transport between the mitochondria and peroxisomes. Curr. Biol. 2010;20:1310–1315. doi: 10.1016/j.cub.2010.05.066. [DOI] [PubMed] [Google Scholar]
- Braschi E, McBride HM. Mitochondria and the culture of the Borg: understanding the integration of mitochondrial function within the reticulum, the cell, and the organism. Bioessays. 2010;32:985–966. doi: 10.1002/bies.201000073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickley K, Smith MJ, Beck M, Stephenson FA. GRIF-1 and OIP106, members of a novel gene family of coiled-coil domain proteins: association in vivo and in vitro with kinesin. J. Biol. Chem. 2005;280:14723–14732. doi: 10.1074/jbc.M409095200. [DOI] [PubMed] [Google Scholar]
- Brownlees J, Ackerley S, Grierson AJ, Jacobsen NJ, Shea K, Anderton BH, Leigh PN, Shaw CE, Miller CC. Charcot-Marie-Tooth disease neurofilament mutations disrupt neurofilament assembly and axonal transport. Hum. Mol. Genet. 2002;11:2837–2844. doi: 10.1093/hmg/11.23.2837. [DOI] [PubMed] [Google Scholar]
- Casari G, De Fusco M, Ciarmatori S, Zeviani M, Mora M, Fernandez P, De Michele G, Filla A, Cocozza S, Marconi R, et al. Spastic paraplegia and OXPHOS impairment caused by mutations in paraplegin, a nuclear-encoded mitochondrial metalloprotease. Cell. 1998;93:973–983. doi: 10.1016/s0092-8674(00)81203-9. [DOI] [PubMed] [Google Scholar]
- Cassidy-Stone A, Chipuk JE, Ingerman E, Song C, Yoo C, Kuwana T, Kurth MJ, Shaw JT, Hinshaw JE, Green DR, Nunnari J. Chemical inhibition of the mitochondrial division dynamin reveals its role in Bax/Bak-dependent mitochondrial outer membrane permeabilization. Dev. Cell. 2008;14:193–204. doi: 10.1016/j.devcel.2007.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang DT, Rintoul GL, Pandipati S, Reynolds IJ. Mutant huntingtin aggregates impair mitochondrial movement and trafficking in cortical neurons. Neurobiol. Dis. 2006;22:388–400. doi: 10.1016/j.nbd.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Chen H, Chan DC. Mitochondrial dynamics - fusion, fission, movement, and mitophagy -in neurodegenerative diseases. Hum. Mol. Genet. 2009;18:R169–R176. doi: 10.1093/hmg/ddp326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YZ, Bennett CL, Huynh HM, Blair IP, Puls I, Irobi J, Dierick I, Abel A, Kennerson ML, Rabin BA, et al. DNA/RNA helicase gene mutations in a form of juvenile amyotrophic lateral sclerosis (ALS4) Am. J. Hum. Genet. 2004;74:1128–1135. doi: 10.1086/421054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho DH, Nakamura T, Fang J, Cieplak P, Godzik A, Gu Z, Lipton SA. S-nitrosylation of Drp1 mediates β-amyloid-related mitochondrial fission and neuronal injury. Science. 2009;324:102–105. doi: 10.1126/science.1171091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark IE, Dodson MW, Jiang C, Cao JH, Huh JR, Seol JH, Yoo SJ, Hay BA, Guo M. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162–1166. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- Costa V, Giacomello M, Hudec R, Lopreiato R, Ermak G, Lim D, Malorni W, Davies KJ, Carafoli E, Scorrano L. Mitochondrial fission and cristae disruption increase the response of cell models of Huntington’s disease to apoptotic stimuli. EMBO Mol. Med. 2010;2:490–503. doi: 10.1002/emmm.201000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csordas G, Renken C, Varnai P, Walter L, Weaver D, Buttle KF, Balla T, Mannella CA, Hajnoczky G. Structural and functional features and significance of the physical linkage between ER and mitochondria. J. Cell Biol. 2006;174:915–921. doi: 10.1083/jcb.200604016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csordas G, Varnai P, Golenar T, Roy S, Purkins G, Schneider TG, Balla T, Hajnoczky G. Imaging interorganelle contacts and local calcium dynamics at the ER-mitochondrial interface. Mol. Cell. 2010;39:121–132. doi: 10.1016/j.molcel.2010.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagda RK, Merrill RA, Cribbs JT, Chen Y, Hell JW, Usachev YM, Strack S. The spinocerebellar ataxia 12 gene product and protein phosphatase 2A regulatory subunit Bβ2 antagonizes neuronal survival by promoting mitochondrial fission. J. Biol. Chem. 2008a;283:36241–36248. doi: 10.1074/jbc.M800989200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagda RK, Zhu J, Kulich SM, Chu CT. Mitochondrially localized ERK2 regulates mitophagy and autophagic cell stress: implications for Parkinson’s disease. Autophagy. 2008b;4:770–782. doi: 10.4161/auto.6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauer W, Przedborski S. Parkinson’s disease: mechanisms and models. Neuron. 2003;39:889–909. doi: 10.1016/s0896-6273(03)00568-3. [DOI] [PubMed] [Google Scholar]
- de Brito OM, Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 2008;456:605–610. doi: 10.1038/nature07534. [DOI] [PubMed] [Google Scholar]
- De Coo IF, Renier WO, Ruitenbeek W, Ter Laak HJ, Bakker M, Schägger H, Van Oost BA, Smeets HJ. A 4-base pair deletion in the mitochondrial cytochrome b gene associated with parkinsonism/MELAS overlap syndrom. Ann. Neurol. 1999;45:130–133. doi: 10.1002/1531-8249(199901)45:1<130::aid-art21>3.3.co;2-q. [DOI] [PubMed] [Google Scholar]
- De Vos KJ, Chapman AL, Tennant ME, Manser C, Tudor EL, Lau KF, Brownlees J, Ackerley S, Shaw PJ, McLoughlin DM, et al. Familial amyotrophic lateral sclerosis-linked SOD1 mutants perturb fast axonal transport to reduce axonal mitochondria content. Hum. Mol. Genet. 2007;16:2720–2728. doi: 10.1093/hmg/ddm226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deas E, Plun-Favreau H, Gandhi S, Desmond H, Kjaer S, Loh SH, Renton AE, Harvey RJ, Whitworth AJ, Martins LM, et al. PINK1 cleavage at position A103 by the mitochondrial protease PARL. Hum. Mol. Genet. 2011;20:867–879. doi: 10.1093/hmg/ddq526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Ruvolo P, Carr B, May WS., Jr. Survival function of ERK1/2 as IL-3-activated, staurosporine-resistant Bcl2 kinases. Proc. Natl. Acad. Sci. USA. 2000;97:1578–1583. doi: 10.1073/pnas.97.4.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Bella D, Lazzaro F, Brusco A, Plumari M, Battaglia G, Pastore A, Finardi A, Cagnoli C, Tempia F, Frontali M, et al. Mutations in the mitochondrial protease gene AFG3L2 cause dominant hereditary ataxia SCA28. Nat. Genet. 2010;42:313–321. doi: 10.1038/ng.544. [DOI] [PubMed] [Google Scholar]
- DiMauro S, Schon EA. Mitochondrial respiratory-chain diseases. New Engl. J. Med. 2003;348:2656–2668. doi: 10.1056/NEJMra022567. [DOI] [PubMed] [Google Scholar]
- Duvezin-Caubet S, Koppen M, Wagener J, Zick M, Israel L, Bernacchia A, Jagasia R, Rugarli EI, Imhof A, Neupert W, et al. OPA1 processing reconstituted in yeast depends on the subunit composition of the m-AAA protease in mitochondria. Mol. Biol. Cell. 2007;18:3582–3590. doi: 10.1091/mbc.E07-02-0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbing B, Mann K, Starosta A, Jaud J, Schols L, Schule R, Woehlke G. Effect of spastic paraplegia mutations in KIF5A kinesin on transport activity. Hum. Mol. Genet. 2008;17:1245–1252. doi: 10.1093/hmg/ddn014. [DOI] [PubMed] [Google Scholar]
- Elden AC, Kim HJ, Hart MP, Chen-Plotkin AS, Johnson BS, Fang X, Armakola M, Geser F, Greene R, Lu MM, et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature. 2010;466:1069–1075. doi: 10.1038/nature09320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Vizarra E, Tiranti V, Zeviani M. Assembly of the oxidative phosphorylation system in humans: what we have learned by studying its defects. Biochim. Biophys. Acta. 2009;1793:200–211. doi: 10.1016/j.bbamcr.2008.05.028. [DOI] [PubMed] [Google Scholar]
- Ferreira IL, Resende R, Ferreiro E, Rego AC, Pereira CF. Multiple defects in energy metabolism in Alzheimer’s disease. Curr. Drug Targets. 2010;11:1193–1206. doi: 10.2174/1389450111007011193. [DOI] [PubMed] [Google Scholar]
- Fransson A, Ruusala A, Aspenstrom P. Atypical Rho GTPases have roles in mitochondrial homeostasis and apoptosis. J. Biol. Chem. 2003;278:6495–6502. doi: 10.1074/jbc.M208609200. [DOI] [PubMed] [Google Scholar]
- Fuse N. Genetic bases for glaucoma. Tohoku J. Exp. Med. 2010;221:1–10. doi: 10.1620/tjem.221.1. [DOI] [PubMed] [Google Scholar]
- Galli S, Jahn O, Hitt R, Hesse D, Opitz L, Plessmann U, Urlaub H, Poderoso JJ, Jares-Erijman EA, Jovin TM. A new paradigm for MAPK: structural interactions of hERK1 with mitochondria in HeLa cells. PLoS One. 2009;4:e7541. doi: 10.1371/journal.pone.0007541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gegg ME, Cooper JM, Chau KY, Rojo M, Schapira AH, Taanman JW. Mitofusin 1 and mitofusin 2 are ubiquitinated in a PINK1/parkin-dependent manner upon induction of mitophagy. Hum. Mol. genet. 2010;19:4861–4870. doi: 10.1093/hmg/ddq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, Holmström KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- Gerards M, van den Bosch B, Calis C, Schoonderwoerd K, van Engelen K, Tijssen M, de Coo R, van der Kooi A, Smeets H. Nonsense mutations in CABC1/ADCK3 cause progressive cerebellar ataxia and atrophy. Mitochondrion. 2010;10:510–515. doi: 10.1016/j.mito.2010.05.008. [DOI] [PubMed] [Google Scholar]
- Gilkerson RW, Schon EA, Hernandez E, Davidson MM. Mitochondrial nucleoids maintain genetic autonomy but allow for functional complementation. J. Cell Biol. 2008;181:1117–1128. doi: 10.1083/jcb.200712101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillardon F. Leucine-rich repeat kinase 2 phosphorylates brain tubulin-beta isoforms and modulates microtubule stability - a point of convergence in parkinsonian neurodegeneration? J. Neurochem. 2009;110:1514–1522. doi: 10.1111/j.1471-4159.2009.06235.x. [DOI] [PubMed] [Google Scholar]
- Giorgi C, Ito K, Lin HK, Santangelo C, Wieckowski MR, Lebiedzinska M, Bononi A, Bonora M, Duszynski J, Bernardi R, et al. PML regulates apoptosis at endoplasmic reticulum by modulating calcium release. Science. 2010;330:1247–1251. doi: 10.1126/science.1189157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glater EE, Megeath LJ, Stowers RS, Schwarz TL. Axonal transport of mitochondria requires milton to recruit kinesin heavy chain and is light chain independent. J. Cell Biol. 2006;173:545–557. doi: 10.1083/jcb.200601067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes LC, Benedetto GD, Scorrano L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat. Cell Biol. 2011;13:589–598. doi: 10.1038/ncb2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Macleod GT, Wellington A, Hu F, Panchumarthi S, Schoenfield M, Marin L, Charlton MP, Atwood HL, Zinsmaier KE. The GTPase dMiro is required for axonal transport of mitochondria to Drosophila synapses. Neuron. 2005;47:379–393. doi: 10.1016/j.neuron.2005.06.027. [DOI] [PubMed] [Google Scholar]
- Gutekunst CA, Li SH, Yi H, Ferrante RJ, Li XJ, Hersch SM. The cellular and subcellular localization of huntingtin-associated protein 1 (HAP1): comparison with huntingtin in rat and human. J, Neurosci. 1998;18:7674–7686. doi: 10.1523/JNEUROSCI.18-19-07674.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman JN, Sanchez-Padilla J, Wokosin D, Kondapalli J, Ilijic E, Schumacker PT, Surmeier DJ. Oxidant stress evoked by pacemaking in dopaminergic neurons is attenuated by DJ-1. Nature. 2010;468:696–700. doi: 10.1038/nature09536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, Lippincott-Schwartz J. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakonen AH, Goffart S, Marjavaara S, Paetau A, Cooper H, Mattila K, Lampinen M, Sajantila A, Lönnqvist T, Spelbrink JN, Suomalainen A. Infantile-onset spinocerebellar ataxia and mitochondrial recessive ataxia syndrome are associated with neuronal complex I defect and mtDNA depletion. Hum. Mol. Genet. 2008;17:3822–3835. doi: 10.1093/hmg/ddn280. [DOI] [PubMed] [Google Scholar]
- Hattula K, Peränen J. FIP-2, a coiled-coil protein, links Huntingtin to Rab8 and modulates cellular morphogenesis. Curr. Biol. 2000;10:1603–1606. doi: 10.1016/s0960-9822(00)00864-2. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Ishimori C, Takahashi-Niki K, Taira T, Kim YC, Maita H, Maita C, Ariga H, Iguchi-Ariga SM. DJ-1 binds to mitochondrial complex I and maintains its activity. Biochem. Biophys. Res. Commun. 2009a;390:667–672. doi: 10.1016/j.bbrc.2009.10.025. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Rizzuto R, Hajnoczky G, Su TP. MAM: more than just a housekeeper. Trends Cell Biol. 2009b;19:81–88. doi: 10.1016/j.tcb.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo JM, Livnat-Levanon N, Taylor EB, Jones KT, Dephoure N, Ring J, Xie J, Brodsky JL, Madeo F, Gygi SP, et al. A stress-responsive system for mitochondrial protein degradation. Mol. Cell. 2010;40:465–480. doi: 10.1016/j.molcel.2010.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi S, Iseki E, Minegishi M, Togo T, Kabuta T, Wada K. GIGYF2 is present in endosomal compartments in the mammalian brains and enhances IGF-1-induced ERK1/2 activation. J. Neurochem. 2010;115:423–437. doi: 10.1111/j.1471-4159.2010.06930.x. [DOI] [PubMed] [Google Scholar]
- Hollenbeck PJ. The pattern and mechanism of mitochondrial transport in axons. Front. Biosci. 1996;1:d91–d102. doi: 10.2741/a118. [DOI] [PubMed] [Google Scholar]
- Hooper C, Puttamadappa SS, Loring Z, Shekhtman A, Bakowska JC. Spartin activates atrophin-1-interacting protein 4 (AIP4) E3 ubiquitin ligase and promotes ubiquitination of adipophilin on lipid droplets. BMC Biol. 2010;8:72. doi: 10.1186/1741-7007-8-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoozemans JJ, Veerhuis R, Van Haastert ES, Rozemuller JM, Baas F, Eikelenboom P, Scheper W. The unfolded protein response is activated in Alzheimer’s disease. Acta Neuropathol. 2005;110:165–172. doi: 10.1007/s00401-005-1038-0. [DOI] [PubMed] [Google Scholar]
- Huizing M, Dorward H, Ly L, Klootwijk E, Kleta R, Skovby F, Pei W, Feldman B, Gahl WA, Anikster Y. OPA3, mutated in 3-methylglutaconic aciduria type III, encodes two transcripts targeted primarily to mitochondria. Mol. Genet. Metab. 2010;100:149–154. doi: 10.1016/j.ymgme.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irrcher I, Aleyasin H, Seifert EL, Hewitt SJ, Chhabra S, Phillips M, Lutz AK, Rousseaux MW, Bevilacqua L, Jahani-Asl A, et al. Loss of the Parkinson’s disease-linked gene DJ-1 perturbs mitochondrial dynamics. Hum. Mol. Genet. 2010;19:3734–3746. doi: 10.1093/hmg/ddq288. [DOI] [PubMed] [Google Scholar]
- Iwasawa R, Mahul-Mellier AL, Datler C, Pazarentzos E, Grimm S. Fis1 and Bap31 bridge the mitochondria-ER interface to establish a platform for apoptosis induction. EMBO J. 2011;30:556–568. doi: 10.1038/emboj.2010.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SM, Lazarou M, Wang C, Kane LA, Narendra DP, Youle RJ. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J. Cell Biol. 2010;191:933–942. doi: 10.1083/jcb.201008084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John GB, Shang Y, Li L, Renken C, Mannella CA, Selker JM, Rangell L, Bennett MJ, Zha J. The mitochondrial inner membrane protein mitofilin controls cristae morphology. Mol. Biol. Cell. 2005;16:1543–1554. doi: 10.1091/mbc.E04-08-0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JO, Mandrioli J, Benatar M, Abramzon Y, Van Deerlin VM, Trojanowski JQ, Gibbs JR, Brunetti M, Gronka S, Wuu J, et al. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010;68:857–864. doi: 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung C, Chylinski TM, Pimenta A, Ortiz D, Shea TB. Neurofilament transport is dependent on actin and myosin. J. Neurosci. 2004;24:9486–9496. doi: 10.1523/JNEUROSCI.1665-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamp F, Exner N, Lutz AK, Wender N, Hegermann J, Brunner B, Nuscher B, Bartels T, Giese A, Beyer K, et al. Inhibition of mitochondrial fusion by alpha-synuclein is rescued by PINK1, Parkin and DJ-1. EMBO J. 2010;29:3571–3589. doi: 10.1038/emboj.2010.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasher PR, De Vos KJ, Wharton SB, Manser C, Bennett EJ, Bingley M, Wood JD, Milner R, McDermott CJ, Miller CC, et al. Direct evidence for axonal transport defects in a novel mouse model of mutant spastin-induced hereditary spastic paraplegia (HSP) and human HSP patients. J. Neurochem. 2009;110:34–44. doi: 10.1111/j.1471-4159.2009.06104.x. [DOI] [PubMed] [Google Scholar]
- Kelleher JF, Mandell MA, Moulder G, Hill KL, L’Hernault SW, Barstead R, Titus MA. Myosin VI is required for asymmetric segregation of cellular components during C. elegans spermatogenesis. Curr. Biol. 2000;10:1489–1496. doi: 10.1016/s0960-9822(00)00828-9. [DOI] [PubMed] [Google Scholar]
- Kim J, Moody JP, Edgerly CK, Bordiuk OL, Cormier K, Smith K, Beal MF, Ferrante RJ. Mitochondrial loss, dysfunction and altered dynamics in Huntington’s disease. Hum. Mol. Genet. 2010;19:3919–3935. doi: 10.1093/hmg/ddq306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada T, Tong Y, Gautier CA, Shen J. Absence of nigral degeneration in aged parkin/DJ-1/PINK1 triple knockout mice. J. Neurochem. 2009;111:696–702. doi: 10.1111/j.1471-4159.2009.06350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutsopoulos OS, Laine D, Osellame L, Chudakov DM, Parton RG, Frazier AE, Ryan MT. Human Miltons associate with mitochondria and induce microtubule-dependent remodeling of mitochondrial networks. Biochim. Biophys. Acta. 2010;1803:564–574. doi: 10.1016/j.bbamcr.2010.03.006. [DOI] [PubMed] [Google Scholar]
- Kraytsberg Y, Kudryavtseva E, McKee AC, Geula C, Kowall NW, Khrapko K. Mitochondrial DNA deletions are abundant and cause functional impairment in aged human substantia nigra neurons. Nat. Genet. 2006;38:518–520. doi: 10.1038/ng1778. [DOI] [PubMed] [Google Scholar]
- Lee HJ, Khoshaghideh F, Lee S, Lee SJ. Impairment of microtubule-dependent trafficking by overexpression of α-synuclein. Eur. J. Neurosci. 2006;24:3153–3162. doi: 10.1111/j.1460-9568.2006.05210.x. [DOI] [PubMed] [Google Scholar]
- Lei X, Zhang S, Bohrer A, Ramanadham S. Calcium-independent phospholipase A2 (iPLA2β)-mediated ceramide generation plays a key role in the cross-talk between the endoplasmic reticulum (ER) and mitochondria during ER stress-induced insulin-secreting cell apoptosis. J. Biol. Chem. 2008;283:34819–34832. doi: 10.1074/jbc.M807409200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Hajnóczky G. Ca2+-dependent regulation of mitochondrial dynamics by the Miro-Milton complex. Int. J. Biochem. Cell Biol. 2009;41:1972–1976. doi: 10.1016/j.biocel.2009.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Rashid F, Byrne PC. The hereditary spastic paraplegia protein spartin localises to mitochondria. J. Neurochem. 2006;98:1908–1919. doi: 10.1111/j.1471-4159.2006.04008.x. [DOI] [PubMed] [Google Scholar]
- Malagelada C, Jin ZH, Jackson-Lewis V, Przedborski S, Greene LA. Rapamycin protects against neuron death in in vitro and in vivo models of Parkinson’s disease. J. Neurosci. 2010;30:1166–1175. doi: 10.1523/JNEUROSCI.3944-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama H, Morino H, Ito H, Izumi Y, Kato H, Watanabe Y, Kinoshita Y, Kamada M, Nodera H, Suzuki H, et al. Mutations of optineurin in amyotrophic lateral sclerosis. Nature. 2010;465:223–226. doi: 10.1038/nature08971. [DOI] [PubMed] [Google Scholar]
- McDermott CJ, Grierson AJ, Wood JD, Bingley M, Wharton SB, Bushby KM, Shaw PJ. Hereditary spastic paraparesis: disrupted intracellular transport associated with spastin mutation. Ann. Neurol. 2003;54:748–759. doi: 10.1002/ana.10757. [DOI] [PubMed] [Google Scholar]
- Milewska M, McRedmond J, Byrne PC. Identification of novel spartin-interactors shows spartin is a multifunctional protein. J. Neurochem. 2009;111:1022–1030. doi: 10.1111/j.1471-4159.2009.06382.x. [DOI] [PubMed] [Google Scholar]
- Millecamps S, Gentil BJ, Gros-Louis F, Rouleau G, Julien JP. Alsin is partially associated with centrosome in human cells. Biochim. Biophys. Acta. 2005;1745:84–100. doi: 10.1016/j.bbamcr.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Misko A, Jiang S, Wegorzewska I, Milbrandt J, Baloh RH. Mitofusin 2 is necessary for transport of axonal mitochondria and interacts with the Miro/Milton complex. J. Neurosci. 2010;30:4232–4240. doi: 10.1523/JNEUROSCI.6248-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore DJ, Zhang L, Troncoso J, Lee MK, Hattori N, Mizuno Y, Dawson TM, Dawson VL. Association of DJ-1 and parkin mediated by pathogenic DJ-1 mutations and oxidative stress. Hum. Mol. Genet. 2005;14:71–84. doi: 10.1093/hmg/ddi007. [DOI] [PubMed] [Google Scholar]
- Moreira MC, Klur S, Watanabe M, Németh AH, Le Ber I, Moniz JC, Tranchant C, Aubourg P, Tazir M, Schöls L, et al. Senataxin, the ortholog of a yeast RNA helicase, is mutant in ataxia-ocular apraxia 2. Nat. Genet. 2004;36:225–227. doi: 10.1038/ng1303. [DOI] [PubMed] [Google Scholar]
- Munch C, Sedlmeier R, Meyer T, Homberg V, Sperfeld AD, Kurt A, Prudlo J, Peraus G, Hanemann CO, Stumm G, Ludolph AC. Point mutations of the p150 subunit of dynactin (DCTN1) gene in ALS. Neurology. 2004;63:724–726. doi: 10.1212/01.wnl.0000134608.83927.b1. [DOI] [PubMed] [Google Scholar]
- Nakamura K, Nemani VM, Azarbal F, Skibinski G, Levy JM, Egami K, Munishkina L, Zhang J, Gardner B, Wakabayashi J, et al. Direct membrane association drives mitochondrial fission by the Parkinson Disease-associated protein α-synuclein. J. Biol. Chem. 2011 doi: 10.1074/jbc.M110.213538. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D, Kane LA, Hauser DN, Fearnley IM, Youle RJ. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy. 2010a;6:1090–1106. doi: 10.4161/auto.6.8.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010b;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemann A, Ruegg M, La Padula V, Schenone A, Suter U. Ganglioside-induced differentiation associated protein 1 is a regulator of the mitochondrial network: new implications for Charcot-Marie-Tooth disease. J. Cell Biol. 2005;170:1067–1078. doi: 10.1083/jcb.200507087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka K, Vilariño-Güell C, Cobb SA, Kachergus JM, Ross OA, Hentati E, Hentati F, Farrer MJ. Genetic variation of the mitochondrial complex I subunit NDUFV2 and Parkinson’s disease. Parkinsonism Relat. Disord. 2010;16:686–687. doi: 10.1016/j.parkreldis.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolden M, Ehses S, Koppen M, Bernacchia A, Rugarli EI, Langer T. The m-AAA protease defective in hereditary spastic paraplegia controls ribosome assembly in mitochondria. Cell. 2005;123:277–289. doi: 10.1016/j.cell.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Okatsu K, Saisho K, Shimanuki M, Nakada K, Shitara H, Sou YS, Kimura M, Sato S, Hattori N, Komatsu M, et al. p62/SQSTM1 cooperates with Parkin for perinuclear clustering of depolarized mitochondria. Genes Cells. 2010;15:887–900. doi: 10.1111/j.1365-2443.2010.01426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira JM. Mitochondrial bioenergetics and dynamics in Huntington’s disease: tripartite synapses and selective striatal degeneration. J. Bioenerg. Biomembr. 2010;42:227–234. doi: 10.1007/s10863-010-9287-6. [DOI] [PubMed] [Google Scholar]
- Orr AL, Li S, Wang CE, Li H, Wang J, Rong J, Xu X, Mastroberardino PG, Greenamyre JT, Li XJ. N-terminal mutant huntingtin associates with mitochondria and impairs mitochondrial trafficking. J. Neurosci. 2008;28:2783–2792. doi: 10.1523/JNEUROSCI.0106-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman C, Merkwirth C, Langer T. Prohibitins and the functional compartmentalization of mitochondrial membranes. J. Cell Sci. 2009;122:3823–3830. doi: 10.1242/jcs.037655. [DOI] [PubMed] [Google Scholar]
- Pandey M, Mohanakumar KP, Usha R. Mitochondrial functional alterations in relation to pathophysiology of Huntington’s disease. J. Bioenerg. Biomembr. 2010;42:217–226. doi: 10.1007/s10863-010-9288-5. [DOI] [PubMed] [Google Scholar]
- Park SH, Zhu PP, Parker RL, Blackstone C. Hereditary spastic paraplegia proteins REEP1, spastin, and atlastin-1 coordinate microtubule interactions with the tubular ER network. J. Clin. Invest. 2010;120:1097–1110. doi: 10.1172/JCI40979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrola L, Espert A, Wu X, Claramunt R, Shy ME, Palau F. GDAP1, the protein causing Charcot-Marie-Tooth disease type 4A, is expressed in neurons and is associated with mitochondria. Hum. Mol. Genet. 2005;14:1087–1094. doi: 10.1093/hmg/ddi121. [DOI] [PubMed] [Google Scholar]
- Pérez-Ollé R, López-Toledano MA, Goryunov D, Cabrera-Poch N, Stefanis L, Brown K, Liem RK. Mutations in the neurofilament light gene linked to Charcot-Marie-Tooth disease cause defects in transport. J. Neurochem. 2005;93:861–874. doi: 10.1111/j.1471-4159.2005.03095.x. [DOI] [PubMed] [Google Scholar]
- Puglielli L, Konopka G, Pack-Chung E, Ingano LA, Berezovska O, Hyman BT, Chang TY, Tanzi RE, Kovacs DM. Acyl-coenzyme A:cholesterol acyltransferase modulates the generation of the amyloid β-peptide. Nat. Cell Biol. 2001;3:905–912. doi: 10.1038/ncb1001-905. [DOI] [PubMed] [Google Scholar]
- Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- Pulst SM, Nechiporuk A, Nechiporuk T, Gispert S, Chen XN, Lopes-Cendes I, Pearlman S, Starkman S, Orozco-Diaz G, Lunkes A, et al. Moderate expansion of a normally biallelic trinucleotide repeat in spinocerebellar ataxia type 2. Nat. Genet. 1996;14:269–276. doi: 10.1038/ng1196-269. [DOI] [PubMed] [Google Scholar]
- Rice SE, Gelfand VI. Paradigm lost: milton connects kinesin heavy chain to miro on mitochondria. J. Cell Biol. 2006;173:459–461. doi: 10.1083/jcb.200604071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusinol AE, Cui Z, Chen MH, Vance JE. A unique mitochondria-associated membrane fraction from rat liver has a high capacity for lipid synthesis and contains pre-Golgi secretory proteins including nascent lipoproteins. J. Biol. Chem. 1994;269:27494–27502. [PubMed] [Google Scholar]
- Ryu SW, Jeong HJ, Choi M, Karbowski M, Choi C. Optic atrophy 3 as a protein of the mitochondrial outer membrane induces mitochondrial fragmentation. Cell. Mol. Life Sci. 2010;67:2839–2850. doi: 10.1007/s00018-010-0365-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sack GH., Jr. Mitochondrial matters in Huntington disease. J. Bioenerg. Biomembr. 2010;42:189–191. doi: 10.1007/s10863-010-9291-x. [DOI] [PubMed] [Google Scholar]
- Sadun AA, Morgia CL, Carelli V. Leber’s Hereditary Optic Neuropathy. Curr Treat Options Neurol. 2010;13:109–117. doi: 10.1007/s11940-010-0100-y. [DOI] [PubMed] [Google Scholar]
- Sahlender DA, Roberts RC, Arden SD, Spudich G, Taylor MJ, Luzio JP, Kendrick-Jones J, Buss F. Optineurin links myosin VI to the Golgi complex and is involved in Golgi organization and exocytosis. J. Cell Biol. 2005;169:285–295. doi: 10.1083/jcb.200501162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santra S, Gilkerson RW, Davidson M, Schon EA. Ketogenic treatment reduces deleted mitochondrial DNAs in cultured human cells. Ann. Neurol. 2004;56:662–669. doi: 10.1002/ana.20240. [DOI] [PubMed] [Google Scholar]
- Schmucker S, Puccio H. Understanding the molecular mechanisms of Friedreich’s ataxia to develop therapeutic approaches. Hum. Mol. Genet. 2010;19:R103–R110. doi: 10.1093/hmg/ddq165. [DOI] [PubMed] [Google Scholar]
- Schon EA, Area-Gomez E. Is Alzheimer’s disease a disorder of mitochondria-associated membranes? J. Alzheimers Dis. 2010;20:S281–S292. doi: 10.3233/JAD-2010-100495. [DOI] [PubMed] [Google Scholar]
- Seleznev K, Zhao C, Zhang XH, Song K, Ma ZA. Calcium-independent phospholipase A2 localizes in and protects mitochondria during apoptotic induction by staurosporine. J. Biol. Chem. 2006;281:22275–22288. doi: 10.1074/jbc.M604330200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan X, Chiang PM, Price DL, Wong PC. Altered distributions of Gemini of coiled bodies and mitochondria in motor neurons of TDP-43 transgenic mice. Proc. Natl. Acad. Sci. USA. 2010;107:16325–16330. doi: 10.1073/pnas.1003459107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi P, Strom AL, Gal J, Zhu H. Effects of ALS-related SOD1 mutants on dynein- and KIF5-mediated retrograde and anterograde axonal transport. Biochim. Biophys. Acta. 2010;1802:707–716. doi: 10.1016/j.bbadis.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JH, Ko HS, Kang H, Lee Y, Lee YI, Pletinkova O, Troconso JC, Dawson VL, Dawson TM. PARIS (ZNF746) repression of PGC-1α contributes to neurodegeneration in Parkinson’s disease. Cell. 2011;144:689–702. doi: 10.1016/j.cell.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirendeb U, Reddy AP, Manczak M, Calkins MJ, Mao P, Tagle DA, Reddy PH. Abnormal mitochondrial dynamics, mitochondrial loss and mutant huntingtin oligomers in Huntington’s disease: implications for selective neuronal damage. Hum. Mol. Genet. 2011;20:1438–1455. doi: 10.1093/hmg/ddr024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvestri G, Mongini T, Odoardi F, Modoni A, deRosa G, Doriguzzi C, Palmucci L, Tonali P, Servidei S. A new mtDNA mutation associated with a progressive encephalopathy and cytochrome c oxidase deficiency. Neurology. 2000;54:1693–1696. doi: 10.1212/wnl.54.8.1693. [DOI] [PubMed] [Google Scholar]
- Silvestri L, Caputo V, Bellacchio E, Atorino L, Dallapiccola B, Valente EM, Casari G. Mitochondrial import and enzymatic activity of PINK1 mutants associated to recessive parkinsonism. Hum. Mol. Genet. 2005;14:3477–3492. doi: 10.1093/hmg/ddi377. [DOI] [PubMed] [Google Scholar]
- Simmen T, Aslan JE, Blagoveshchenskaya AD, Thomas L, Wan L, Xiang Y, Feliciangeli SF, Hung CH, Crump CM, Thomas G. PACS-2 controls endoplasmic reticulum-mitochondria communication and Bid-mediated apoptosis. EMBO J. 2005;24:717–729. doi: 10.1038/sj.emboj.7600559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon DK, Pankratz N, Kissell DK, Pauciulo MW, Halter CA, Rudolph A, Pfeiffer RF, Nichols WC, Foroud T, Parkinson Study Group, P.I. Maternal inheritance and mitochondrial DNA variants in familial Parkinson’s disease. BMC Med. Genet. 2010;11:53. doi: 10.1186/1471-2350-11-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits P, Smeitink J, van den Heuvel L. Mitochondrial translation and beyond: processes implicated in combined oxidative phosphorylation deficiencies. J. Biomed. Biotech. 2010;2010:737385. doi: 10.1155/2010/737385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Chen J, Petrilli A, Liot G, Klinglmayr E, Zhou Y, Poquiz P, Tjong J, Pouladi MA, Hayden MR, et al. Mutant huntingtin binds the mitochondrial fission GTPase dynamin-related protein-1 and increases its enzymatic activity. Nat. Med. 2011;17:377–382. doi: 10.1038/nm.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokin GB, Lillo C, Falzone TL, Brusch RG, Rockenstein E, Mount SL, Raman R, Davies P, Masliah E, Williams DS, Goldstein LS. Axonopathy and transport deficits early in the pathogenesis of Alzheimer’s disease. Science. 2005;307:1282–1288. doi: 10.1126/science.1105681. [DOI] [PubMed] [Google Scholar]
- Stowers RS, Megeath LJ, Gorska-Andrzejak J, Meinertzhagen IA, Schwarz TL. Axonal transport of mitochondria to synapses depends on milton, a novel Drosophila protein. Neuron. 2002;36:1063–1077. doi: 10.1016/s0896-6273(02)01094-2. [DOI] [PubMed] [Google Scholar]
- Supnet C, Bezprozvanny I. Neuronal calcium signaling, mitochondrial dysfunction, and Alzheimer’s disease. J. Alzheimers Dis. 2010;20:S487–S498. doi: 10.3233/JAD-2010-100306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swerdlow RH, Parks JK, Davis JN, 2nd, Cassarino DS, Trimmer PA, Currie LJ, Dougherty J, Bridges WS, Bennett JP, Jr., Wooten GF, Parker WD. Matrilineal inheritance of complex I dysfunction in a multigenerational Parkinson’s disease family. Ann. Neurol. 1998;44:873–881. doi: 10.1002/ana.410440605. [DOI] [PubMed] [Google Scholar]
- Tanaka A, Cleland MM, Xu S, Narendra DP, Suen DF, Karbowski M, Youle RJ. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J. Cell Biol. 2010;191:1367–1380. doi: 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang TS, Tu H, Chan EY, Maximov A, Wang Z, Wellington CL, Hayden MR, Bezprozvanny I. Huntingtin and huntingtin-associated protein 1 influence neuronal calcium signaling mediated by inositol-(1,4,5) triphosphate receptor type 1. Neuron. 2003;39:227–239. doi: 10.1016/s0896-6273(03)00366-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tradewell ML, Durham HD, Mushynski WE, Gentil BJ. Mitochondrial and axonal abnormalities precede disruption of the neurofilament network in a model of Charcot-Marie-Tooth disease type 2E and are prevented by heat shock proteins in a mutant-specific fashion. J. Neuropathol. Exp. Neurol. 2009;68:642–652. doi: 10.1097/NEN.0b013e3181a5deeb. [DOI] [PubMed] [Google Scholar]
- Trushina E, Dyer RB, Badger JD, 2nd, Ure D, Eide L, Tran DD, Vrieze BT, Legendre-Guillemin V, McPherson PS, Mandavilli BS, et al. Mutant huntingtin impairs axonal trafficking in mammalian neurons in vivo and in vitro. Mol. Cell. Biol. 2004;24:8195–8209. doi: 10.1128/MCB.24.18.8195-8209.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twig G, Shirihai OS. The interplay between mitochondrial dynamics and mitophagy. Antioxid. Redox Signal. 2011;14:1939–1951. doi: 10.1089/ars.2010.3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Leemput J, Chandran J, Knight MA, Holtzclaw LA, Scholz S, Cookson MR, Houlden H, Gwinn-Hardy K, Fung HC, Lin X, et al. Deletion at ITPR1 underlies ataxia in mice and spinocerebellar ataxia 15 in humans. PLoS Genet. 2007;3:e108. doi: 10.1371/journal.pgen.0030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viscomi C, Bottani E, Civiletto G, Cerutti R, Moggio M, Fagiolari G, Schon EA, Lamperti C, Zeviani M. In vivo correction of COX deficiency by activation of the AMPK/PGC1α axis. Cell Metab. 2011 doi: 10.1016/j.cmet.2011.04.011. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vives-Bauza C, Przedborski S. Mitophagy: the latest problem for Parkinson’s disease. Trends Mol. Med. 2011;17:158–165. doi: 10.1016/j.molmed.2010.11.002. [DOI] [PubMed] [Google Scholar]
- Vives-Bauza C, Zhou C, Huang Y, Cui M, de Vries RL, Kim J, May J, Tocilescu MA, Liu W, Ko HS, et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc. Natl. Acad. Sci. USA. 2010;107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Song P, Du L, Tian W, Yue W, Liu M, Li D, Wang B, Zhu Y, Cao C, et al. Parkin ubiquitinates Drp1 for proteasome-dependent degradation: implication of dysregulated mitochondrial dynamics in parkinson disease. J. Biol. Chem. 2011;286:11649–11658. doi: 10.1074/jbc.M110.144238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Su B, Fujioka H, Zhu X. Dynamin-like protein 1 reduction underlies mitochondrial morphology and distribution abnormalities in fibroblasts from sporadic Alzheimer’s disease patients. Am. J. Pathol. 2008;173:470–482. doi: 10.2353/ajpath.2008.071208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Su B, Lee HG, Li X, Perry G, Smith MA, Zhu X. Impaired balance of mitochondrial fission and fusion in Alzheimer’s disease. J. Neurosci. 2009b;29:9090–9103. doi: 10.1523/JNEUROSCI.1357-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Su B, Zheng L, Perry G, Smith MA, Zhu X. The role of abnormal mitochondrial dynamics in the pathogenesis of Alzheimer’s disease. J. Neurochem. 2009a;109:153–159. doi: 10.1111/j.1471-4159.2009.05867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weihofen A, Thomas KJ, Ostaszewski BL, Cookson MR, Selkoe DJ. Pink1 forms a multiprotein complex with Miro and Milton, linking Pink1 function to mitochondrial trafficking. Biochemistry. 2009;48:2045–2052. doi: 10.1021/bi8019178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells AL, Lin AW, Chen LQ, Safer D, Cain SM, Hasson T, Carragher BO, Milligan RA, Sweeney HL. Myosin VI is an actin-based motor that moves backwards. Nature. 1999;401:505–508. doi: 10.1038/46835. [DOI] [PubMed] [Google Scholar]
- Xu S, Peng G, Wang Y, Fang S, Karbowski M. The AAA-ATPase p97 is essential for outer mitochondrial membrane protein turnover. Mol. Biol. Cell. 2011;22:291–300. doi: 10.1091/mbc.E10-09-0748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Jiang Q, Zhao J, Ren Y, Sutton MD, Feng J. Parkin stabilizes microtubules through strong binding mediated by three independent domains. J. Biol. Chem. 2005;280:17154–17162. doi: 10.1074/jbc.M500843200. [DOI] [PubMed] [Google Scholar]
- Yi M, Weaver D, Hajnoczky G. Control of mitochondrial motility and distribution by the calcium signal: a homeostatic circuit. J. Cell Biol. 2004;167:661–672. doi: 10.1083/jcb.200406038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zampese E, Fasolato C, Kipanyula MJ, Bortolozzi M, Pozzan T, Pizzo P. Presenilin 2 modulates endoplasmic reticulum (ER)-mitochondria interactions and Ca2+ cross-talk. Proc. Natl. Acad. Sci. USA. 2011;108:2777–2782. doi: 10.1073/pnas.1100735108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Takita J, Tanaka Y, Setou M, Nakagawa T, Takeda S, Yang HW, Terada S, Nakata T, Takei Y, et al. Charcot-Marie-Tooth disease type 2A caused by mutation in a microtubule motor KIF1Bβ. Cell. 2001;105:587–597. doi: 10.1016/s0092-8674(01)00363-4. [DOI] [PubMed] [Google Scholar]
- Zheng B, Liao Z, Locascio JJ, Lesniak KA, Roderick SS, Watt ML, Eklund AC, Zhang-James Y, Kim PD, Hauser MA, et al. PGC-1α, a potential therapeutic target for early intervention in Parkinson’s disease. Sci. Transl. Med. 2010;2:52ra73. doi: 10.1126/scitranslmed.3001059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Huang Y, Shao Y, May J, Prou D, Perier C, Dauer W, Schon EA, Przedborski S. The kinase domain of mitochondrial PINK1 faces the cytoplasm. Proc. Natl. Acad. Sci. USA. 2008;105:12022–12027. doi: 10.1073/pnas.0802814105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinsmaier KE, Babic M, Russo GJ. Mitochondrial transport dynamics in axons and dendrites. Results Probl. Cell Differ. 2009;48:107–139. doi: 10.1007/400_2009_20. [DOI] [PubMed] [Google Scholar]
- Ziviani E, Tao RN, Whitworth AJ. Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin. Proc. Natl. Acad. Sci. USA. 2010;107:5018–5023. doi: 10.1073/pnas.0913485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuchner S, Wang G, Tran-Viet KN, Nance MA, Gaskell PC, Vance JM, Ashley-Koch AE, Pericak-Vance MA. Mutations in the novel mitochondrial protein REEP1 cause hereditary spastic paraplegia type 31. Am. J. Hum. Genet. 2006;79:365–369. doi: 10.1086/505361. [DOI] [PMC free article] [PubMed] [Google Scholar]