Abstract
Mesenchymal stem cells (MSCs) are used in many of the current stem cell-based clinical trials and their therapeutic efficacy has increasingly been attributed to secretion of paracrine factors. We have previously demonstrated that a therapeutic constituent of this secretion is exosome, a secreted bilipid membrane vesicle of ~50–100 nm with a complex cargo that is readily internalized by H9C2 cardiomyocytes. It reduces infarct size in a mouse model of myocardial ischemia/reperfusion (MI/R) injury. We postulate that this therapeutic efficacy is derived from the synergy of a select permutation of individual exosome components. To identify protein candidates in this permutation, the proteome was profiled and here we identified 20S proteasome as a protein candidate. Mass spectrometry analysis detected all seven α and seven β chains of the 20S proteasome, and also the three beta subunits of “immunoproteasome” with a very high confidence level. We demonstrated that a functional proteasome copurified with MSC exosomes with a density of 1.10–1.18 g/mL, and its presence correlated with a modest but significant reduction in oligomerized protein in a mouse model of myocardial infarction. Circulating proteasomes in human blood also copurified with exosomes. Therefore, 20S proteasome is a candidate exosome protein that could synergize with other constituents to ameliorate tissue damage.
1. Introduction
Mesenchymal stem cells (MSCs) are multipotent fibroblast-like cells that reside in many adult tissues such as bone marrow adipose tissue [1, 2], liver [3], muscle connective tissue [4], amniotic fluid [5], placenta [6, 7], umbilical cord blood [1], and dental pulp [8, 9]. Although their differentiation potentials are primarily osteogenesis, chondrogenesis, and adipogenesis, MSCs have been reported to have the potential to differentiate into an amazing array of cell types that include nearly every major cell types in the adult body [10, 11]. MSCs are currently the most evaluated experimental stem cells with more than 100 clinical trials in 2010 to test their efficacy in treating a myriad of diseases such as cardiovascular diseases (e.g., acute myocardial infarction, endstage ischemic heart disease, or prevention of vascular restenosis), osteogenesis imperfecta (OI) or brittle bone disease, amyotrophic lateral sclerosis (ALS), lysosomal storage diseases (e.g., Hurler syndrome), steroid refractory graft versus host disease (GVHD), periodontitis and bone fractures [12].
The use of MSCs as therapeutics was initially predicated on the hypothesis that transplanted MSCs home and engraft in injured tissues, and then differentiate into cells to replace damaged cells. However, it has been estimated that <1% of transplanted cells actually reached the target tissue with most of the cells being trapped in the liver, spleen, and lung [13], and reported evidence for differentiation of transplanted MSCs at the site of injury often cannot eliminate the possibility of cell fusion [14–16]. It has also been increasingly observed that the therapeutic efficacy of MSC therapy is not dependent on the engraftment of MSC at the site of injury or differentiation capability of the transplanted MSC [17–20], essentially eliminating the need for MSCs to be in the vicinity of their target tissue or to differentiate to exert a therapeutic effect. To reconcile this discrepancy between the therapeutic efficacy of MSC and the lack of MSC engraftment or differentiation at the site of injury, it was proposed that MSCs exert their therapeutic effects through secreted trophic mediators. The general acceptance of this proposal is reflected in the MSC clinical trials of 2010 in which 65 of the 101 clinical trials were rationalized on the trophic secretion of MSCs while only 36 were based on the differentiation potential of MSCs [21]. This paradigm shift in the therapeutic mechanism of MSC from one based on cell engraftment, differentiation and replacement to one based on secretion and paracrine signaling could potentially engender the development of biologic instead of cell-based therapeutics.
In 2008, our group demonstrated that intravenous administration of a single bolus of culture medium conditioned by human embryonic stem cell-derived MSCs (hESC-MSCs) reduced relative infarct size in a pig and mouse model of ischemia/reperfusion injury [22]. By molecular weight fractionation of this conditioned medium (CM), we demonstrated that the active component had a presumptive size of 50–200 nm [22]. Using size exclusion high performance liquid chromatography (HPLC), we purified a population of homogenously sized particles that have the biophysical parameters of exosomes, namely, a hydrodynamic radius of 55–65 nm and a flotation density in sucrose of 1.10–1.18 g/mL. We subsequently demonstrated that this exosome population alone could reduce infarct size by ~40% in a mouse model of myocardial ischemia/reperfusion injury and therefore was the therapeutic agent in the secretion of mesenchymal stem cells [23]. Therapeutic exosomes are also found to be secreted by primary MSC cultures [24] and myc-immortalized hESC-MSCs [25]. The exosomes have exosome-associated proteins such as the tetraspanin proteins, CD9 and CD81, Alix, Tsg101, and RNA that consists primarily of short RNAs of less than 300 nt. Some of these RNAs are microRNAs that are predominantly premicroRNAs [26].
As cells for example, H9C2 cardiomyocytes [26] readily internalized MSC exosomes, possibly by endocytosis, the exosome cargo of protein and RNA could be delivered across plasma membranes into cells. This diverse exosome cargo could, in principle provide a molecular basis for the therapeutic efficacy of MSC secretion in treating a diverse range of diseases (e.g., acute myocardial infarction, endstage ischemic heart disease, or prevention of vascular restenosis), osteogenesis imperfecta (OI) or brittle bone disease, amyotrophic lateral sclerosis (ALS), lysosomal storage diseases (e.g., Hurler syndrome), steroid refractory graft versus host disease (GVHD), periodontitis and bone fractures [12]. The cargo complexity of exosome could also provide a rationale for the efficacy of exosomes in treating complex tissue injuries such as myocardial ischemia/reperfusion injury that involves multiple tissues and targets.
We believe that the individual components in exosome cargo are not equally or sufficiently efficacious in ameliorating tissue injury. We hypothesize that the therapeutic efficacy of MSC exosome against a specific injury is derived from the synergy of a select permutation of individual exosome components. To identify optimal permutation of biochemical activities for effective amelioration of myocardial ischemia/reperfusion injury and other injury, we are now systematically analyzing each of the candidate biochemical activities in MSC exosomes to first assess each activity and its potential to ameliorate injury.
In this paper, we focused on the proteome of exosomes to identify candidate proteins or protein complexes that could drive their efficacy against diverse disease targets, the proteome of these purified exosomes was profiled here by mass spectrometry and antibody array, and found to contain 857 unique gene products (http://www.exocarta.org/). These proteins were distributed over a wide array of biochemical and cellular processes such as communication, structure and mechanics, inflammation, exosome biogenesis, tissue repair and regeneration and metabolism (Lai et al., submitted) consistent with the reported proteome complexity of MSC exosomes.
A predominant feature of MSC exosome proteome was the presence of all seven α and seven β chains of the 20S proteasome, and the three beta subunits of “immunoproteasome” which were identified with very high confidence level by mass spectrometry. These observations were indicative of fully assembled and possibly functional 20S proteasome and immunoproteasome which constitute the catalytic core of the 26S proteasome. The presence of 20S proteasome in exosomes also provides a mechanism for cellular extrusion of the relatively large intact 20S proteasomes as extracellular, circulating proteasome [27–31]. Circulating proteasomes are functionally active and could degrade small peptides. They have been shown to correlate with disease state or progression demonstrating that extracellular proteasomes have important physiological and pathological functions [27–31]. It has been postulated that these extracellular proteasomes are important in degrading soluble peptides in extracellular fluids and modulating potentially pathogenic protein aggregation such as Alzheimer's amyloid plaques [32]. A key to understanding the functions of these extracellular proteasomes will be to determine how and when cells extrude proteasome. As the level of circulating proteasome does not correlate with lactate dehydrogenase it is unlikely that extracellular proteasomes are products of cell lysis [33].
In this paper, we demonstrated that MSC exosomes contained functional 20S proteasomes and its presence correlated with a modest but significant reduction in oligomerized protein in a mouse model of myocardial infarction. Therefore, 20S proteasome is a candidate exosome protein that could synergize with other constituents to ameliorate tissue damage.
2. Materials and Methods
2.1. Preparation of Exosomes
Exosomes were purified from culture medium conditioned by huES9.E1, human ESC-derived mesenchymal stem cells [34] by HPLC as previously described [24, 35]. Briefly, CM collected from MSCs culture was concentrated 50x by tangential flow filtration (TFF) using a membrane with a 100 kDa MWCO (Sartorius, Goettingen, Germany). CM was fractionated by high performance liquid chromatography (HPLC) (TSK Guard column SWXL, 6 × 40 mm and TSK gel G4000 SWXL, 7.8 × 300 mm, Tosoh Corp., Tokyo, Japan). Exosomes were collected from the first peak of the elution and concentrated using 100 kDa MWCO filter (Sartorius). Exosomes were filtered with a 0.22 μm filter and stored in −20°C freezer until use.
2.2. LC MS/MS
Three independent preparations (biological replicates) of HPLC-purified exosomes were analyzed by LC-MS/MS over nine months using three different methods of tryptic digestion. In each preparation, about 2 mL proteins of dialyzed exosomes were used. In the first preparation, the sample was in-solution trypsin digested, that is, reduced, alkylated and tryptic digested as described [36]. After desalting using Sep-Pak C-18 SPE cartridge (Waters, Milford, MA, USA), the tryptic digest was directly analyzed with LC-MS/MS twice (two injections) without further fractionation. In the second preparation, the sample was separated by SDS-PAGE and the gel lane was cut into 8 slices for in gel digestion. Tryptic peptides from each gel slice were analyzed by LC-MS/MS. In the third preparation, the sample was tryptic digested and desalted. To reduce sample complexity, offline peptide fractionation was carried out with a HPLC system (Shimadzu, Japan) through a Polysulfoethyl SCX column (200 mm × 4.6 mm) (PolyLC, USA). Mobile phase A (5 mM KH4PO4 + 30% acetonitrile) and mobile phase B (5 mM KH4PO4 + 30% acetonitrile + 350 mM KCl) at 1 mL/min. Eight fractions were collected, desalted, and dried with a vacuum centrifuge. All three independent replicates were analyzed by a LC-MS/MS system including a Shimadzu micro HPLC system coupled online to a LTQ-FT Ultra linear ion trap mass spectrometer (Thermo Electron, Bremem, Germany) fitted with a nanospray source. Injected peptides were trapped and desalted in a Zorvax 300SB-C18 enrichment column (5 mm × 03 mm, Agilent Technologies, Germany) and eluted into a nano-bored C18 packed column (75 μm × 100 Å, Michrom Bioresources, Auburn, CA USA). A 90 minute gradient at a constant flow rate of 20 μL/min with a splitter to an effective flow rate of 300 nl/min was used to elute the peptides into the mass spectrometer. The LTQ was operated in a data-dependent mode by performing MS/MS scans for 8 of the most intense peaks from each MS scan in the FTMS. For each experiment, all MS/MS (dta) spectra of each replicate were combined into a single mascot generic file by a home-written program. Protein identification was achieved by searching the combined data against the IPI human protein database (version 3.34; 69,164 sequences, 29,064,825 residues) via an in-house Mascot server (Version 2.2.04, Matrix Science, UK). The search parameters were a maximum of 2 missed cleavages using trypsin; fixed modification was carbamidomethylation of cysteine and variable modification was oxidation of methionine. The mass tolerances were set to 10 ppm and 0.8 Da for peptide precursor and fragment ions, respectively. Protein identification was accepted as true positive if two different peptides were found to have scores greater than the homology scores.
2.3. Antibody Array
500 μL of non-conditioned media and exosomes were assayed for the presence of cytokines andother proteins using RayBio Biotin Label-based Human Antibody Array I according to manufacturer's instructions (RayBio, Norcross, GA). The cytokines and other proteins were considered to be present in the exosomes if the signal intensity was 2 fold higher (P < 0.05) than that in non-conditioned medium.
2.4. Western Blot Hybridization
3 μg of conditioned medium or exosomes were separated on 4–12% SDS-polyacrylamide gels and electroblotted onto a nitrocellulose membrane. The membrane was transferred to the membrane holder of SNAP i.d. system (Millipore, Billerica, MA), blocked and incubated with 1 : 200 diluted mouse anti-20S proteasome α1–7. The blot was then incubated with a 1 : 1250 diluted horseradish peroxidase-coupled goat anti-mouse IgG. All antibodies were obtained from Santa Cruz Biotechnology, Santa Cruz, CA. The blot was then incubated with HRP-enhanced chemiluminescent substrate (Thermo Fisher Scientific Inc., Waltham, MA, USA) and then exposed to X-ray film.
2.5. 20S Proteasome Enzymatic Assay
The proteasome activity was measured using a 20S proteasome activity assay kit (Millipore) based on detection of the fluorophore 7-Amino-4 methylcoumarin (AMC) after cleavage from the labeled substrate LLVY-AMC by 20S proteasome in the presence or absence of lactacystin, a specific 20S proteasome inhibitor. Briefly, 4 μg of exosome was incubated with a reaction buffer containing LLVY-AMC in the presence or absence of 25 μM lactacystin. The samples and AMC standards were incubated at 37°C and fluorescence intensity at Ex/Em = 380/460 nm was monitored for 2 hours.
2.6. Animals and Experiments
Male C57Bl6/J (10–12 wks, 25–30 g) mice were obtained from Jackson Laboratory (Bar Harbor, USA). Mice received standard diet and water ad libitum. Myocardial infarction was induced between 8 am and 1 pm by left coronary artery ligation, just below the left atrial appendage. All animal experiments are performed in accordance with the national guidelines on animal care and with prior approval by the Animal Experimentation Committee of Utrecht University, The Netherlands.
2.7. Myocardial Infarction In Vivo
Mice were anesthetized with a mixture of Fentanyl (Jansen-Cilag) 0.05 mg/kg, Dormicum (Roche) 5 mg/kg and medetomidine 0.5 mg/kg through an intraperitoneal injection. Core body temperature was maintained around 37°C during surgery by continuous monitoring with a rectal thermometer connected to an automatic heating blanket. Mice were intubated and ventilated (Harvard Apparatus Inc.) with 100% oxygen. The left coronary artery (LCA) was ligated for 30 minutes using an 8-0 Ethilon (Ethicon) with a section of polyethylene-10 tubing placed over the LCA. Ischemia was confirmed by bleaching of the myocardium and ventricular tachyarrhythmia. Five minutes before reperfusion, mice were intravenously infused with 200 μL saline-diluted exosome containing 0.4 μg protein via the tail vein. Control animals were infused with 200 μL saline. Reperfusion was initiated by releasing the ligature and removing the polyethylene-10 tubing. Reperfusion of the endangered myocardium was characterized by typical hyperemia in the first few minutes. The chest wall was closed and the animals received subcutaneously atipamezole (Antisedan, Pfizer) 2.5 mg/kg, flumazenil (Anexate, Roche) 0.5 mg/kg and buprenorphine (Temgesic, Schering-Plough) 0.1 mg/kg. At 15 min, 1 hour, 24 hours, or 3 days after reperfusion (n = 4/group/time point), animals were sacrificed and the area at risk was excised for protein extraction. LCA ligation in mice results in anterior wall ischemia, while the septal wall is perfused by a septal coronary artery that originates separately from the sinus valsalva. The entire anterior wall of the left ventricle below the level of ligation was excised for area at risk protein extraction.
2.8. Antioligomer Dot Plot
Protein was extracted from area at risk of 30 min LCA ligated mouse heart treated with exosome or saline after 15 minutes, 1 hour, 1 day, and 3 days reperfusion using a cell extraction buffer from Biovision, Mountain View, CA, USA according to manufacturer's instruction. Four animals were used at each time point in the exosome or saline treatment arm. One μg of each sample was applied on a nitrocellulose membrane, air dried, stained with Ponceau S, quantified using Image Lab (Bio-rab Laboratories) and destained. The membrane was then blocked with 10% non-fat dry milk TBST solution, incubated with a 1 : 1000 diluted rabbit anti-oligomer antibody (Invitrogen, Carlsbad, CA) for 2 hours and washed 3 times with TBST. This was followed by one hour incubation with a 1 : 2000 diluted horseradish peroxidase-coupled donkey anti-rabbit IgG (Santa Cruz Biotechnology, Santa Cruz, CA, USA). 3 washes with TBST and incubation with HRP-enhanced chemiluminescent substrate (Thermo Fisher Scientific Inc., Waltham, MA USA). The signal was scanned using ChemiDoc System (Bio-rab Laboratories, Philadelphia, PA) and quantified using Image Lab (Bio-rab Laboratories). The signal was normalized against the intensity of the Ponceu S stain.
2.9. Sucrose Density Gradient Equilibrium Centrifugation
To generate the sucrose density gradient for centrifugation, 14 sucrose solutions with concentrations from 22.8 to 60% were prepared and layered sequentially in an ultracentrifuge tube (Beckman Coulter Inc., CA, USA) starting with the most concentrated solution. Exosome or human plasma was loaded on top before ultracentrifugation for 16.5 h at 200 000 g, 4°C in a SW60Ti rotor (Beckman Coulter Inc.). After centrifugation, 13 fractions of 330 μL each were collected sequentially starting from the top of the gradient. The densities of each fraction were determined by weighing a fixed volume. For pretreatment with detergent-based lysis buffer (Cell Extraction Buffer, Biovision), exosome was incubated with an equal volume of the lysis buffer containing protease inhibitors (Halt Protease Inhibitor Cocktail, Thermo Fisher Scientific) for 30 min on ice before loading on the sucrose density gradient. For subsequent western blot hybridization, 20 μL samples from each fraction were used. The mouse anti-CD63, -CD9, -CD81, and -CD59 antibodies, were obtained from Santa Cruz Biotechnology, Santa Cruz, CA, USA (1 : 60 dilution).
2.10. Liquid Phase Isoelectric Focusing
Liquid phase isoelectric focusing was performed using MicroRotofor, Liquid-Phase IEF Cell, from Bio-Rad (Bio-Rad Laboratories). 80 μg exosome was added to 3 mL of 0.5% Bio-Lyte 3/10 Ampholyte (Bio-Rad Laboratories, diluted with PBS). The sample was loaded into the focusing chamber with 0.1 M phosphoric acid on anode and 0.1 M Sodium Hydroxide on cathode. Isoelectric focusing was performed for 215 min at 1 W constant power. The samples were then collected using vacuum suction into 10 fractions. The pH of each fraction was determined using pH indicator paper (Macherey-Nagel, Bethlehem, PA, USA) before concentrating to 25 μL using 100 kDa MWCO filter (Millipore).
2.11. Cholera Toxin B Precipitation
50 μL of each human plasma fraction from sucrose gradient density equilibrium ultracentrifugation as described above was incubated with 0.005 μg biotinylated-cholera toxin B conjugated to streptavidin-conjugated magnetic beads (Invitrogen Corporation) for 30 min with gentle shaking followed by 30 min incubation The supernatant was then discarded and the magnetic beads were washed thrice with 0.1% BSA/PBS before incubating with 50 μL of detergent-based lysis buffer (Cell Extraction Buffer, Biovision) on ice for 10 min. The lysate was then collected and biotinylated by incubating with 4 μL 2.5 mM EZ-Link Sulfo-NHS-LC-LC-Biotin (Thermo Fisher Scientific) for 30 min with gentle shaking. The remaining reactive biotin was then quenched by adding 100 mM glycine buffer. The biotinylated lysate was then incubated with 5 μL anti-CD81 or -20S α1–7 conjugated magnetic beads (Invitrogen Corporation) for 30 min with gentle shaking. The supernatant was then discarded and the magnetic beads were washed thrice with 0.1% BSA/PBS before incubating with 50 μL of 1 : 5000 diluted HRP-conjugated streptavidin (Biolegend, San Diego, CA, USA) for 15 min at room temperature with gentle shaking. Finally the supernatant was discarded and the magnetic beads were washed thrice with 0.1% BSA/PBS. The amount of bound HRP was determined using Amplex UltraRed Reagent (Invitrogen Corporation) according to manufacturer's instruction.
3. Results
3.1. Proteomic Profiling of Exosome
Proteomic profiling using mass spectrometry and antibody array approaches were performed as previously described [24, 35, 37] on three or one independent preparations of HPLC-purified exosomes, respectively. Mass spectrometric analysis of the three exosome preparations identified 379, 432, and 420 proteins, respectively (please refer to Supplementary File 1 for the LC MS/MS raw data). Of these, 154 (~20%) were present in all three preparations, 157 (~20%) were present in two while 455 (~59%) were present in only one (Figure 1(a)). These analyses were subsequently validated by western blot hybridization or biochemical assays using either the three exosome preparations used in the mass spectrometric analysis or other independently prepared exosomes. Several proteins determined by mass spectrometry to be present in two or one of the three preparations were found to be present in these or other subsequent preparations by either western blot hybridization or biochemical assays. For example, PFKFB3 and PGK were detected in only one and two of the three batches, respectively. However, when tested by western blot hybridization or biochemical assays, both proteins were found to be present in two or more of the three preparations and also subsequent exosome preparations (data not shown). This indicated that the three independent exosome preparations analyzed by LC-MS/MS within nine months interval using three different sample preparation methods were complementary to each other in identifying different sets of low abundant proteins or our selection criteria were too stringent. To ensure a more comprehensive coverage of the exosome proteome, we combined the 3 sets of mass spectrometry data. The analysis of the exosome proteome by antibody array identified 101 proteins of which 10 were also detected by mass spectrometry analysis (Figure 1(b)). By combining the results from mass spectrometry and antibody array, we determined that the exosome proteome has a total of 857 proteins (Table 1) and this dataset has been deposited at http://www.exocarta.org/. In Table 1, the regular font symbols represent the proteins identified by LC MS/MS; the underline symbols represent the proteins identified by antibody arrays; the bold and underline symbols represent the proteins identified by both LC MS/MS and antibody arrays; and lastly the grey shaded symbols represent the proteins that were found to be present in at least 50% of exosomes characterized [38]. Of these 857 proteins, 320 were found in the 739 proteins previously identified in the unfractionated conditioned medium [24, 35] (Figure 1(c)). The remaining 537 proteins in the exosome were detected only when HPLC-purified exosomes were used. Since exosomes constituted about 10% of the total proteins in the conditioned medium [35], we postulated that many of the exosome proteins were masked by the more abundant nonexosome proteins in the conditioned medium.
Figure 1.
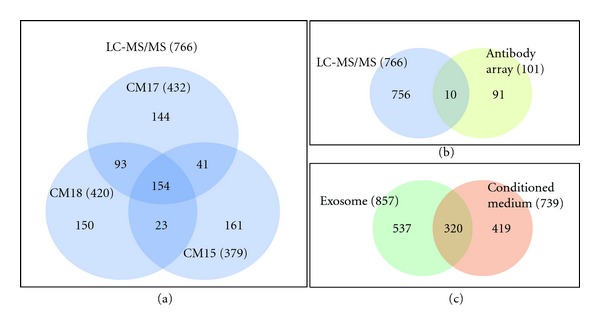
Exosome proteins. (a) Venn diagram of number of proteins detected by LC-MS/MS in 3 independent batches of exosomes, CM15, CM17, and CM18. There are 379, 432, and 420 proteins detected in CM15, CM17 and CM18, respectively. The combined protein number is 766. (b) Venn diagram shows the overlap of proteins detected by LC-MS/MS and antibody array. There are 101 proteins detected by antibody array in CM15. 10 of the 101 proteins detected are overlapping with proteins detected by LC-MS/MS. (c) Intersection of the 739 proteins previously identified in MSC conditioned medium versus the 857 proteins identified in purified exosomes.
Table 1.
Proteomic profile of 3 independently prepared exosomes as determined by LC MS/MS and antibody arrays.
| A2M | C11orf59 | COPS4 | FBXW8 | HNRNPA1 | KRT16 | MMP3 | PRR4 | RPS3 | TGFB2 |
| ABI3BP | C1orf78 | COPS8 | FEN1 | HP | KRT17 | MOS | PRSS23 | RPS4X | TGFBI |
| ACAA2 | C1R | CPS1 | FER1L3 | HPX | KRT18 | MPO | PSMA1 | RPS5 | TGM2 |
| ACAT2 | C1S | CREG1 | FGA | HRSP12 | KRT19 | MPZL1 | PSMA2 | RPSA | TGOLN2 |
| ACLY | C20orf114 | CRIPT | FGB | HSP90AA1 | KRT2 | MRC2 | PSMA3 | RRAS2 | THBS1 |
| ACSL1 | C3 | CRTAP | FGF16 | HSP90AB1 | KRT27 | MSN | PSMA4 | RTN4 | THBS2 |
| ACTA1 | C5orf24 | CSF1 | FGF18 | HSP90B1 | KRT28 | MXRA5 | PSMA5 | RUVBL1 | THY1 |
| ACTA2 | C9orf19 | CSF2 | FGF19 | HSPA1A | KRT3 | MYADM | PSMA6 | S100A11 | TIMP1 |
| ACTB | C9orf91 | CSF3 | FGFRL1 | HSPA1L | KRT4 | MYCBPAP | PSMA7 | S100A13 | TIMP2 |
| ACTG2 | CACNA2D1 | CSPG4 | FGG | HSPA5 | KRT5 | MYH14 | PSMB1 | S100A8 | TIMP3 |
| ACTN1 | CACNA2D4 | CST4 | FLG2 | HSPA6 | KRT6A | MYH9 | PSMB10 | S100P | TKT |
| ACTN2 | CALR | CTA-221G9.4 | FLJ13197 | HSPA8 | KRT6B | MYL6B | PSMB2 | SAA4 | TLN1 |
| ACTN3 | CAND1 | CTBP2 | FLJ22184 | HSPB1 | KRT6C | MYO1C | PSMB3 | SASS6 | TMBIM1 |
| ACTN4 | CAP1 | CTNNA1 | FLJ32784 | HSPD1 | KRT7 | NBL1 | PSMB4 | SCAMP3 | TMED10 |
| ACTR1A | CAPNS1 | CTNNA2 | FLNA | HSPG2 | KRT72 | NEFH | PSMB5 | SCGB2A1 | TMED9 |
| ACTR2 | CAPZA1 | CTNNB1 | FLNB | HTRA1 | KRT73 | NEK10 | PSMB6 | SCYE1 | TMEM16B |
| ACTR3 | CASP14 | CTNND1 | FLNC | HYI | KRT74 | NID1 | PSMB7 | SDC1 | TMEM2 |
| ADAM10 | CAT | CTSG | FLOT1 | ICAM1 | KRT76 | NLRP8 | PSMB8 | SDC2 | TMEM47 |
| ADAM9 | CAV1 | CXCL16 | FLOT2 | ICAM5 | KRT77 | NME1 | PSMB9 | SDC4 | TMEM51 |
| ADAMTS12 | CCDC129 | CXCL2 | FLT1 | IDH3B | KRT78 | NOMO1 | PSMC5 | SDCBP | TNC |
| AEBP1 | CCDC64B | CXorf39 | FN1 | IFITM2 | KRT79 | NRAS | PSMD11 | SEC14L4 | TNFRSF11B |
| AFM | CCL2 | CYBRD1 | FREM3 | IFNG | KRT8 | NRG2 | PSMD14 | SEMA5A | TNFRSF12A |
| AGRN | CCL20 | DBF4B | FST | IFRD1 | KRT80 | NRLN1 | PSMD6 | SEPT2 | TNFRSF1A |
| AHCY | CCL28 | DCD | FTL | IFT140 | KRT84 | NRP1 | PSMD7 | SEPT7 | TNFSF18 |
| AHNAK2 | CCL7 | DCHS2 | FUCA2 | IGF2R | KRT9 | NT5E | PTGFRN | SERINC5 | TNFSF5 |
| AHSG | CCR4 | DCLK2 | GALNT5 | IGFBP3 | LACRT | NTF5 | PTK7 | SERPINA1 | TPBG |
| AKR1B1 | CCR5 | DCN | GANAB | IGFBP4 | LAMA4 | NUSAP1 | PTPRK | SERPINB3 | TPI1 |
| AKR7A2 | CCT5 | DCTN1 | GAPDH | IGFBP6 | LAMB1 | OBFC1 | PTRF | SERPINE1 | TRAP1 |
| ALB | CCT6A | DECR1 | GAPDHS | IGFBP7 | LAMC1 | ODZ3 | PTTG1IP | SERPINE2 | TREM1 |
| ALCAM | CD109 | DEFA1 | GARS | IGHA1 | LAMP1 | OFD1 | PTX3 | SERPINF1 | TREML2P |
| ALDH2 | CD151 | DIP2B | GAS6 | IGHA2 | LAMP2 | OPRM1 | PXDN | SFN | TRIM40 |
| ALDH3A2 | CD248 | DIRAS2 | GDF1 | IGHG1 | LAP3 | OSM | PZP | SFRP1 | TRIM41 |
| ALDH6A1 | CD276 | DKFZp686D0972 | GDF11 | IGHG2 | LCN1 | OTC | QPCTL | SFRP4 | TSN |
| ALDH7A1 | CD44 | DKK1 | GDF3 | IGHG4 | LCN2 | OXNAD1 | QSOX1 | SHANK3 | TSNAX |
| ALDH9A1 | CD47 | DKK3 | GDF5 | IGHM | LDHA | OXTR | RAB10 | SLAIN1 | TSPAN14 |
| ALDOA | CD59 | DMBT1 | GDF8 | IGJ | LDHAL6B | P4HB | RAB11B | SLC16A1 | TSPAN4 |
| ALDOB | CD63 | DNASE1L1 | GDF9 | IGKC | LDHB | PAICS | RAB14 | SLC16A3 | TSPAN6 |
| ALDOC | CD81 | DNPEP | GDI1 | IGKV1-5 | LEPRE1 | PAN3 | RAB15 | SLC1A4 | TSPAN9 |
| ALOX12P2 | CD82 | DPYS | GDI2 | IGL@ | LGALS1 | PAPPA | RAB1A | SLC1A5 | TSTA3 |
| ANG | CD9 | DPYSL2 | GFRA3 | IGLV4-3 | LGALS3 | PARP10 | RAB1B | SLC22A2 | TTLL3 |
| ANGPTL2 | CDC2L5 | DSP | GLDC | IGSF8 | LGALS3BP | PARP16 | RAB2A | SLC25A10 | TTN |
| ANPEP | CDC42 | DULLARD | GLUD1 | IL10 | LGALS8 | PARVG | RAB33B | SLC25A13 | TTYH3 |
| ANXA1 | CDH13 | ECM1 | GNA13 | IL11 | LGR6 | PC | RAB35 | SLC2A1 | TUBA1A |
| ANXA11 | CDIPT | ED1 | GNAI2 | IL13 | LIF | PCOLCE | RAB39B | SLC2A3 | TUBA1B |
| ANXA2 | CDK5R2 | EDG2 | GNAL | IL15RA | LMNA | PDCD6 | RAB5A | SLC38A2 | TUBA1C |
| ANXA2P1 | CEACAM8 | EDIL3 | GNAS | IL17B | LOC124220 | PDCD6IP | RAB5B | SLC38A3 | TUBB |
| ANXA3 | CFB | EEA1 | GNAT3 | IL17R | LOC283523 | PDGFA | RAB5C | SLC39A14 | TUBB2A |
| ANXA4 | CFI | EEF1A1 | GNB1 | IL19 | LOC284297 | PDGFC | RAB6A | SLC3A2 | TUBB2C |
| ANXA5 | CFL1 | EEF1G | GNB2 | IL1F9 | LOC388344 | PDGFRB | RAB7A | SLC44A1 | TUBB3 |
| ANXA6 | CFL2 | EEF2 | GNB4 | IL1RAP | LOC389827 | PDIA3 | RAB8A | SLC44A2 | TUBB6 |
| ANXA7 | CFTR | EFEMP2 | GNG12 | IL1RAPL1 | LOC442497 | PEBP1 | RAB8B | SLC7A10 | UBA52 |
| AP1S1 | CHMP2A | EHD1 | GNPDA1 | IL1RL2 | LOC653269 | PFAS | RAC1 | SLC7A5 | UBB |
| APEH | CHST12 | EHD2 | GOT2 | IL22RA1 | LOC727942 | PFKFB3 | RAC2 | SMARCA4 | UBE1 |
| APOA1 | CITED1 | EHD4 | GPC1 | IL23A | LOC728320 | PFN1 | RAD21 | SMC1A | UBE2N |
| APOE | CLASP2 | EIF4A1 | GPC5 | IL3 | LOC728378 | PFN2 | RALA | SORT1 | UGP2 |
| APP | CLDN1 | EMILIN1 | GPI | IL5 | LOC730013 | PGAM2 | RAN | SPACA1 | UNC13B |
| ARF1 | CLEC11A | ENG | GPR112 | IL6ST | LRP1 | PGD | RAP1A | SPARC | UNC45A |
| ARF4 | CLIC1 | ENO1 | GREM1 | IL7 | LRP6 | PGK1 | RAP1B | SPOCK1 | VAMP3 |
| ARF5 | CLIC6 | ENO2 | GRM2 | IL8 | LRRFIP2 | PGLYRP2 | RAP1B | SPRY4 | VANGL1 |
| ARHGAP18 | CLPX | ENO3 | GRM3 | INHBA | LTBP1 | PIGR | RAP2C | SPTAN1 | VASN |
| ARHGAP23 | CLSTN1 | ENTPD4 | GRM7 | INHBB | LTBP2 | PIP | RARRES1 | SPTBN1 | VAT1 |
| ARHGDIA | CLTA | ENTPD4;LOXL2 | GSN | INSR | LTF | PKM2 | RASA1 | SPTBN4 | VCAN |
| ARHGEF1 | CLTC | EPB41L3 | GSTM1 | IQGAP1 | LYAR | PLAB | RASA4 | SRGN | VCL |
| ARL6IP5 | CLTCL1 | EPHA2 | GSTM2 | ITGA11 | LYZ | PLAU | RB1CC1 | SRI | VCP |
| ARMS2 | CLU | EPO | GSTM5 | ITGA2 | MADH4 | PLEC1 | RCOR2 | SRPX2 | VEGFC |
| ARPC3 | CMIP | ESM1 | GSTO1 | ITGA3 | MAMDC2 | PLEKHG3 | RDH5 | ST6GALNAC6 | VIL1 |
| ARPC4 | CNGB1 | ETFB | GSTP1 | ITGA4 | MAP1A | PLOD1 | RFTN1 | STAT1 | VIL2 |
| ARPC5 | COL12A1 | F2R | GTPBP2 | ITGA5 | MAP2K6 | PLOD2 | RGN | STC1 | VIM |
| ASH1L | COL14A1 | F3 | GYLTL1B | ITGAL | MAP3K1 | PLOD3 | RHOC | STC2 | VTI1A |
| ASL | COL18A1 | F8 | GZMA | ITGAV | MARCKS | PLP2 | RMND5A | STOM | VTN |
| ATP1A1 | COL1A1 | FADD | H2AFV | ITGB1 | MARCKSL1 | PLSCR3 | RNF123 | STOML3 | WDR49 |
| ATP1B3 | COL1A2 | FAH | H2AFX | ITGB5 | MAT1A | PLTP | RNF40 | STX12 | WDR52 |
| ATP2B1 | COL2A1 | FAM108A1 | HBB | ITIH2 | MBD3 | PLUNC | RPL10A | STX2 | WNT5A |
| ATP2B4 | COL3A1 | FAM129B | HBE1 | ITIH4 | MCC | PNO1 | RPL12 | SURF4 | YBX1 |
| ATP5A1 | COL4A1 | FAM29A | HDAC5 | ITPR2 | MCM10 | PODN | RPL15 | SVEP1 | YWHAB |
| ATP5B | COL4A2 | FAM3B | HERC5 | JUP | MDH1 | POLN | RPL18 | SYT1 | YWHAE |
| ATP8B3 | COL4A3 | FAM64A | HGF | KIAA0146 | MDH2 | POSTN | RPL23 | SYT9 | YWHAG |
| ATRN | COL5A1 | FAM71F1 | HGFR | KIAA0256 | ME1 | POTE2 | RPL29 | TAAR2 | YWHAQ |
| ATXN1 | COL5A2 | FAP | HISPPD2A | KIAA0467 | MECP2 | PPIA | RPL35A | TAGLN | YWHAZ |
| AXL | COL6A1 | FASN | HIST1H2AE | KIAA1881 | MFAP4 | PPIB | RPLP0 | TALDO1 | ZBTB4 |
| BASP1 | COL6A2 | FAT | HIST1H2BA | KPNB1 | MFGE8 | PPME1 | RPS10 | TAS2R60 | ZNF134 |
| BDNF | COL6A3 | FAT2 | HIST1H2BL | KRT1 | MFSD2 | PPP1CC | RPS16 | TCN1 | ZNF503 |
| BGN | COL7A1 | FAT4 | HIST1H4H | KRT10 | MIF | PRDM16 | RPS18 | TF | ZNF614 |
| BHMT2 | COMP | FBLN1 | HIST2H2BE | KRT13 | MMP1 | PRDX1 | RPS2 | TFG | |
| BRMS1 | COPB1 | FBN1 | HLAA | KRT14 | MMP10 | PRDX6 | RPS24 | TFRC | |
| BSG | COPS3 | FBN2 | HMGCS2 | KRT15 | MMP2 | PRNP | RPS27A | TGFB1 |
Regular font: identified by LC MS/MS.
Underline: identified by antibody arrays.
Bold and Underline: identified by both LC MS/MS and antibody arrays.
Bold: identified by LC MS/MS and was found to be present in at least 50% of exosomes characterized.
Based on 15 proteomic analyses carried out on exosomes purified from cultured cells and from biological fluids by different groups, Thery et al. had observed that a set of about 17 proteins, namely glyceraldehyde 3-phosphate dehydrogenase (GAPDH), pyruvate kinase (PK), eukaryotic translation elongation factor 1A1 (EEF1A1), milk fat globule EGF factor 8 protein (MFGE8), tetraspanins, 14-3-3 proteins, Gα proteins, clathrin, Alix (PDCD6IP), MHC class1, annexins (ANX), Rab proteins, ezrin(VIL2), radixin(RDX) and moesin (MSN)(ERM), actin, tubulin, HSP70, and HSP90 were found to be present in at least 50% of the exosomes that were characterized [38]. Not unexpectedly, most of these proteins were also found in the proteome of the HPLC-purified MSC exosomes (Table 1). Also consistent with the endosomal origin of exosomes, we detected the presence of endosome-associated proteins such as Alix (PDCD6IP) and Rab (Table 1).
To better understand the biological significance of the proteins in the exosomes, we had previously performed functional clustering of the 857 proteins into biological processes using PANTHER (protein analysis through evolutionary relationships) analytical software [39, 40] (Lai et al.; submitted) The observed frequency of genes from the exosome proteome in each biological process was compared with the reference frequency of 25431 Homo sapiens gene list in the NCBI database for that biological process. The 857 gene products could be clustered into 32 biological processes that were over-represented (P < 0.001) and 3 that were under-represented (P < 0.001) (Lai et al.; submitted). Many of these biological processes are consistent with activities associated with exosome biology for example, communication, cellular motility, inflammation, and exosome biogenesis. One predominant biological process that could not be definitively associated with exosome biology is proteolysis (P = 3.76 × 10−10) and of the unique genes clustered in this process, ~22% (17 out of 78) encode for peptides in the 20S proteasome or immunoproteasome. To ensure the robustness of this observation, examination of the 3 sets of mass spectrometry datasets revealed that with only one exception of PSMB5 in the first exosome preparation, all the 14 peptides of 20S proteasome were detected with very high confidence in all three exosome preparations as demonstrated by the Mascot search results, number of identified peptides with score higher than homology or identity scores, as well as the MS/MS spectra and fragment ions assignment of unique identified peptides (Supplementary data 1–7).
3.2. Presence of 20S Proteasome in MSC Exosome
Mass spectrometry analysis of MSC exosomes not only detected the presence of all seven α- (PSMA1–7) and all seven β-subunits (PSMB1–7) of the 20S core particle with very high confidence (See Supplementary Material available online at doi:10.1155/2012/971907 for the representative spectra, number of peptide detected, and the peptide score of them), but also the three beta subunits of “immunoproteasome,” PSMB8 (β5i or LMP7), PSMB9 (β1i or LMP2), PSMB10 (β2i or LMP10) gene product [41]. To form the 26S proteasome, the 20S core particle must be complemented by other peptides. Although some of these peptides such as PSMC5, PSMD6, PSMD7, PSMD11, and PSMD14 were present, they were not sufficient to form the 26S proteasome. The presence of some of the 20S proteasome peptides was further confirmed by western blot hybridization (Figure 2(a)). As exosomes are known to have a typical density range of 1.10 to 1.18 gmL−1 that could be resolved on sucrose gradients [42], we checked whether these 20S proteasome subunits have the typical exosome flotation density and postulated that the flotation densities of the 20S proteasome subunits would be different before and after release from such vesicles by a detergent-based buffer. Fractionation of exosomes or exosomes pretreated with a detergent-based lysis buffer on a sucrose density gradient by equilibrium ultracentrifugation revealed that 20S proteasome subunits had a similar flotation density as that of exosome-associated proteins, namely, CD9, CD81, CD59, and CD63. All of them had an exosomes-like flotation density of 1.10–1.18 g/mL (Figure 2(b)). Pretreatment with a detergent-based cell lysis buffer decreased the apparent flotation densities of these proteins. Liquid phase isoelectric focusing of exosomes further revealed that 20S proteasome had a similar protein isoelectric point (pI) as that of exosome-associated CD9 (Figure 2(c)). Together the exosome-like flotation density and pI of the 20S proteasome suggested that the 20S proteasome was associated with exosomes and not a contaminant of exosome preparation.
Figure 2.
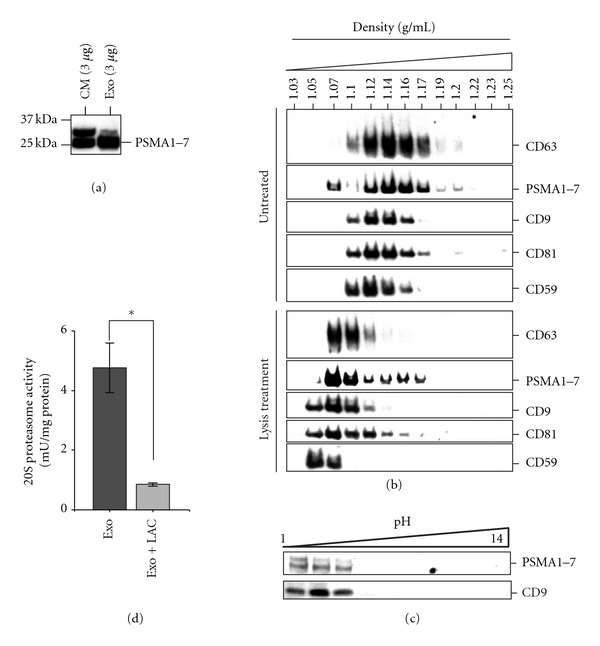
20S proteasome in MSC exosome. (a) Western blot analysis of MSC conditioned medium (CM) and exosome (Exo) using an antibody specific for PMSA 1–7 peptides. (b) Protein analysis of exosome fractionated on a sucrose gradient density. Exosome or exosome pretreated with lysis buffer was loaded on a sucrose density gradient prepared by layering 14 sucrose solutions of concentrations from 22.8 to 60% (w/v) in a SW60Ti centrifuge tube and then ultracentrifuged for 16.5 h at 200 000 g, 4°C, in a SW60Ti rotor. The gradients were removed from the top and the density of each fraction was calculated by weighing a fixed volume of each fraction. The fractions were analyzed by western blot analysis for CD9, PSMA1–7, CD81, CD59, and CD63 in exosome (upper panel) and pretreated exosomes (lower panel). (c) pI of 20S proteasome and CD9 in exosome. Exosome sample was separated by liquid-phase isoelectric focusing into 10 fractions from pH1–14. Each fraction was then concentrated and analyzed by western blot analysis for CD9, PSMA1–7. (d) Proteasome activity in MSC exosome was determined using a commercially available proteasome activity assay kit as described in the Materials and Methods section. Proteasome activity was measured by the rate of degradation of a fluorogenic peptide in the absence or presence of lactacystin, a proteasome inhibitor. One unit (U) enzyme activity is defined as the activity to generate 1 μmole product per minute at 37°C. Each bar represents mean ± SEM of 2 independent assays with each assay performed in triplicate. *P = 0.00023.
3.3. Enzymatic Activity of 20S Proteasome in MSC Exosome
The presence of all seven α- and all seven β-subunits of the 20S core particle indicates that MSC exosomes contain intact 20S proteasome complexes and therefore 20S proteasome enzymatic activity. To test if exosomes contain functional 20S proteasome, purified MSC exosomes were incubated with a fluorophore-labeled peptide, LLVY-AMC (a substrate for the chymotrypsin-like activity; AMC, 7-amido 4-methylcoumarin) [43], that upon cleavage by 20S proteasome released fluorescent AMC. The fluorescence produced by the released AMC would be directly proportional to the proteasome activity. Based on this assay, the 20S proteasome enzymatic activity in MSC exosomes was determined to be 5.00 μU/μg protein where one unit (U) enzyme activity is defined as the activity to generate 1 μmole product per minute at 37°C. This degradation was inhibited by lactacystin, a proteasome-specific inhibitor (Figure 2(d)).
3.4. Exosome Proteasome Reduced Misfolded Proteins In Vivo
To determine if exosome proteasome can reduce misfolded or aberrant proteins in vivo in a disease model and possibly contribute to a therapeutic outcome, we measured the accumulation of misfolded proteins in heart tissue of a mouse model of myocardial ischemia/reperfusion injury with and without exosome treatment.
This model is most appropriate for our aim as accumulation of misfolded proteins has been recognized as an important contributor to tissue damage during myocardial ischemia/reperfusion injury [44–48] (reviewed [49]) and we have previously shown that exosome treatment resulted in extensive cardioprotection [23]. Furthermore, in our hands, the area at risk in this mouse model is also highly reproducible [23, 24, 50–52].
To determine if exosome could reduce the amount of misfolded proteins, we used a conformation-dependent antibody [53] to quantify the amount of misfolded proteins in the area at risk of ischemic mouse heart after 15 min, one hour, one day and three days reperfusion. The level of denatured or misfolded protein was assayed using an antibody against oligomers. Denatured or partially unfolded proteins tend to self-associate to form high molecular aggregates such as oligomers and these oligomers have a common structure that is independent of amino acid sequence [54]. An antibody known as A11 that was raised against synthetic Aβ oligomers was found to specifically recognize this common structure present in misfolded protein oligomers but did not recognize monomers or mature fibers of proteins or peptides [54]. It will react with soluble Aβ40 oligomers and does not react with soluble low molecular weight Aβ40 or Aβ40 fibrils.
Using this antibody, we found that those mice treated with exosome had a modest but significantly lower level of misfolded proteins (P = 0.037) (n = 4 per time point) 1 day after reperfusion compared with those saline treated controls (n = 4 per time point) (Figure 3). This demonstrated that MSC exosome has the potential to reduce misfolded proteins in vivo and may contribute to the exosome-mediated cardioprotection.
Figure 3.
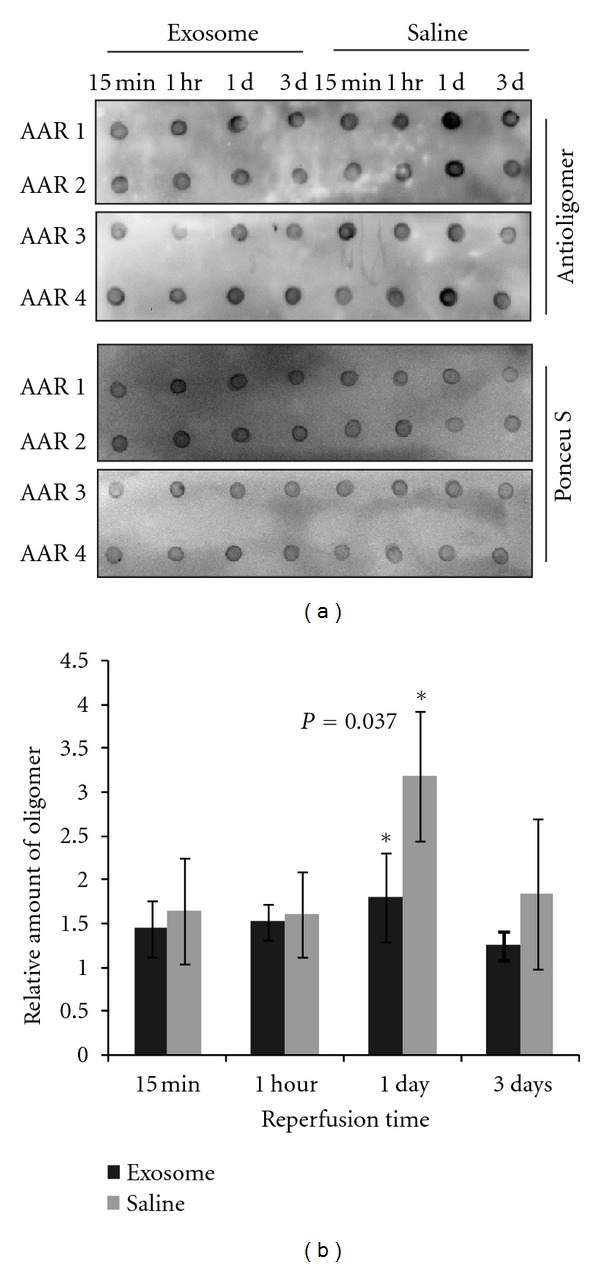
Exosome reduced oxidized protein in vivo. Myocardial ischemia/reperfusion injury was induced by ligation of the left coronary artery (LCA) for 30 min and subsequent reperfusion by releasing the ligation. Five minutes before reperfusion, mice (n = 4/group/time point) were intravenously infused with exosome or saline. At 15 min, 1 hour, 1 day, or 3 days after reperfusion, the animal was scarified and the area at risk (AAR) was excised and extracted for protein. One μg of each sample was applied onto a nitrocellulose membrane, stained with Ponceau S, destained and probed with rabbit antioligomer antibody and a HRP reporting system as described in material and method. The intensity of the antibody binding was quantified using Image Lab and normalized against the intensity of Ponceau S staining. (a) Representative dot plots. (b) Each bar represented mean ± SEM of 4 animal.
3.5. Association of Plasma 20S Proteasome with Exosome
Although our experimental observations strongly supported the presence of functional 20S proteasome in MSC exosomes, the association of 20S proteasome with exosomes could be a culture artifact contaminant. To test this possibility, we determined if circulating 20S proteasome in human plasma is associated with exosomes. 20S proteasomes are widely reported to be present in plasma [27–31] and we confirmed this by immunoblotting plasma for the presence of proteasome subunits (Figure 4(a)). Fractionation of plasma on a sucrose density gradient revealed that the plasma 20S proteasome fractionated at the flotation density of exosome that is, 1.10–1.18 g/mL (Figure 4(a)). CD81, a tetraspanin protein commonly found on exosome membrane also floated at this density. Each of the fractions was then extracted with cholera toxin B chain (CTB) which binds GM1 ganglioside, a glycolipid found to be enriched in the membrane of exosomes [55] and also MSC exosomes (unpublished data). CTB together with cholera toxin A chain forms the cholera toxin. Cholera toxin enters the cytosol by binding GM1 ganglioside through CTB [56]. This cellular entry of cholera toxin is a well characterized highly specific receptor mediated pathway [57]. The CTB extract was then assayed by ELISA for 20S proteasome subunits and CD81. 20S proteasome subunits and CD81 were both found to be present in the CTB extraction (Figure 4(b)). These observations demonstrated that plasma 20S proteasome colocalized with CD81 in GM1 ganglioside-enriched complex that floats at 1.10–1.18 g/mL and that at least some of the circulating proteasomes in the plasma are associated with exosomes. The presence of 20S proteasome in MSC exosomes and plasma exosomes together implicated exosome as a normal physiological conduit for cellular extrusion of 20S proteasome.
Figure 4.
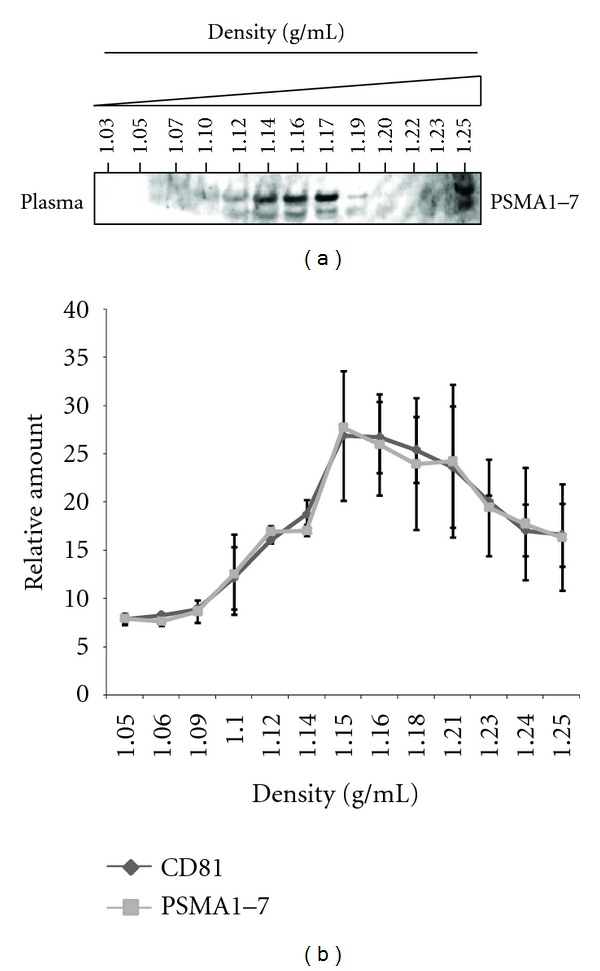
Human plasma contained exosome-associated 20S proteasome. (a) Human plasma was loaded on a sucrose density gradient prepared by layering 14 sucrose solutions of concentrations from 22.8 to 60% (w/v) in a SW60Ti centrifuge tube and then ultracentrifuged for 16.5 h at 200 000 g, 4°C, in a SW60Ti rotor. 330 μL fractions were removed sequentially from the top and the density of each fraction was calculated by weighing a fixed volume of each fraction. The fractions were analyzed by Western blot analysis for PSMA1–7. (b) Relative amount of CD81 and PSMA1–7 in plasma exosome. Exosomes which are known to have membrane enriched in GM1 gangliosides were isolated from the sucrose density gradient by the high affinity of GM1 gangliosides for cholera toxin B (CTB) chains conjugated to magnetic beads. The CTB extract was assayed by ELISA for the amount of PSMA1–7 and CD81. Each point represents mean ± SEM (n = 2).
4. Discussion
In this paper, we investigated the proteome of MSC exosome to identify candidate proteins or protein complexes that could contribute to their therapeutic efficacy in ameliorating myocardial ischemia/reperfusion injury or other pathological conditions. We hypothesize that this therapeutic efficacy against a complex injury is likely to be derived from the synergy of a select permutation of individual exosome components rather than a single component. To elucidate this permutation which may include proteins, RNA and other molecules, we focused first on the proteins by elucidating the exosome proteome to identify candidate proteins or protein complexes that have the biochemical potential to elicit a therapeutic response.
The proteome of 3 independently prepared, HPLC-purified ESC-derived MSC exosome using mass spectrometry and cytokine array were analyzed and 857 proteins were identified. We have previously reported that 739 proteins were profiled in hESC-MSC conditioned medium using LTQ-FT mass spectrometer. These proteins included many proteins commonly found in other exosomes. The proteome of MSC exosomes contained a diverse array of proteins. Clustering of these proteins according to their functions suggested that the exosome has the potential to drive many biological processes (Lai et al., submitted). This is consistent with the reported efficacy of MSCs in treating a myriad of diseases such as cardiovascular diseases (e.g., acute myocardial infarction, endstage ischemic heart disease, or prevention of vascular restenosis), osteogenesis imperfecta (OI) or brittle bone disease, amyotrophic lateral sclerosis (ALS), lysosomal storage diseases (e.g., Hurler syndrome), steroid refractory Graft versus Host Disease (GVHD), and periodontitis and bone fractures [12].
A significant fraction of proteins in MSC exosomes are involved in the highly regulated and complex intracellular membrane trafficking and sorting through the biosynthetic and endocytotic pathways [58], and these processes probably reflect the biogenesis of exosomes. In fact, many of the proteins in these processes constitute a common set of proteins found in exosomes from different cell sources [59]. However, the biological significance of the other proteins remains to be uncovered. Exosomes have been implicated in an increasing number of important physiological and pathological processes such as disposal of unwanted protein [60], antigen presentation [42], genetic exchange [61], immune responses [38, 62] and tumor metastasis [38, 62–66]. Whether the proteome of MSC exosomes could sustain such processes remains to be determined.
Proteasome subunits have been previously reported to be present in exosomes [61, 67–69] but this is the first time that mass spectrometry analysis of exosome proteome had detected the entire protein complement of a 20S proteasome with very high confidence. In addition, we also demonstrated that exosome had the lactacystin-sensitive proteolytic activity of 20S proteasome. Since decreased proteasome activity resulting in accumulation of misfolded proteins has been recognized as an important contributor to tissue damage during myocardial ischemia/reperfusion injury [44–48] (reviewed [49]) and MSC exosomes reproducibly reduce IS/AAR by ~40% in a mouse model of myocardial ischemia/reperfusion injury [23, 24, 52] we tested if cardioprotective MSC exosomes exert some of their therapeutic effects through proteolytic degradation of misfolded proteins. The level of denatured or misfolded protein was assayed using an antibody against oligomers. Using this antibody, we demonstrated that accumulation of misfolded proteins or oligomers was reduced in the heart tissues of a mouse model of myocardial ischemia/reperfusion injury that had been treated with MSC exosomes. This positive correlation between exosome-mediated decreased accumulation of misfolded proteins and exosome-mediated reduction in IS/AAR supports a therapeutic role the 20S proteasome in MSC exosome.
20S proteasome is responsible for the degradation of about 90% of all intracellular oxidatively damaged proteins [70] and reduced proteasomal activity has been postulated to be a contributing factor in the pathogenesis of aging-related neurodegenerative diseases such as Alzheimer's disease and Parkinson's disease [71, 72] or cardiovascular disease [73–75]. The presence of all seven α and β subunits of the 20S proteasome, the three beta subunits of “immunoproteasome” and validation of 20S proteasome enzymatic activity in the exosomes in vitro and in vivo suggested that the cardioprotective activity of MSC exosomes [35] could be partly attributed to the presence of 20S proteasome. This further implied that MSC exosomes have the potential to therapeutically correct for proteasome insufficiency. Many debilitating neurodegenerative diseases such as Alzheimer's, Parkinson's or prion disease that are caused by accumulation of denatured or misfolded proteins could thus be alleviated by MSC exosomes carrying functional proteasomes. The use of exosomes as a delivery vehicle for proteasome has the added advantage of being able to transcend the blood-brain-barrier. This barrier is the main cause in the intractability of neurodegenerative diseases to medical intervention. Unlike most conventional drugs, exosomes could cross the blood-brain-barrier as demonstrated by the recent knockdown of the gene target in the brain by exosomes loaded with exogenous siRNA [76].
The presence of 20S proteasome in MSC exosomes also suggested that cells extruded 20S proteasome through exosomes. Consistent with this, we observed that plasma proteasomes had the flotation density (1.10–1.19 g/mL) and pI of exosomes, and were associated with GM1 gangliosides, a glycolipid found to be enriched in the membrane of exosome [77]. This finding supported our hypothesis that cellular extrusion of 20S proteasomes via exosomes is a normal physiological process.
Together this study demonstrated that MSC and plasma exosomes contain functional 20S proteasome, and cells secrete proteasome via the exosome pathway. MSC exosomes could reduce misfolded proteins in the heart after ischemia/reperfusion injury suggesting that they could also be used to alleviate diseases precipitated by accumulation of denatured proteins such as Alzheimer's disease, Parkinson's disease or prion disease.
In conclusion, MSC exosome has functional 20S proteamsomes and its presence correlated with a modest but significant reduction in oligomerized protein in a mouse model of myocardial infarction. Therefore, 20S proteasome is a candidate exosome protein that could synergize with other constituents in the exosome to ameliorate tissue damage.
Supplementary Material
Supplementary material provides the raw data of the detected 20S proteasome in 3 independent batches of exosomes. Supplementary data 1 provides a summary of number of detected peptides in 3 independent batches of exosomes; Supplementary data 2–4 provide the raw data of detected 20S proteasome peptides in 3 independent batches of exosomes respectively; Supplementary data 5–7 provide the Mascot peptide view of unique 20S proteasome peptides in 3 independent batches of exosomes respectively.
Conflicts of Interest
The authors declare that they have no conflict of interests.
Acknowledgments
The authors gratefully acknowledge Kong Meng Hoi and Eddy Tan at the Bioprocessing Technology Institute (BTI) for helping in the purification of the exosomes and Jayanthi Padmanabhan and Jeremy Lee (BTI) for the preparation and concentration of the conditioned medium.
References
- 1.Kern S, Eichler H, Stoeve J, Klüter H, Bieback K. Comparative analysis of mesenchymal stem cells from bone marrow, umbilical cord blood, or adipose tissue. Stem Cells. 2006;24(5):1294–1301. doi: 10.1634/stemcells.2005-0342. [DOI] [PubMed] [Google Scholar]
- 2.Banas A, Teratani T, Yamamoto Y, et al. Adipose tissue-derived mesenchymal stem cells as a source of human hepatocytes. Hepatology. 2007;46(1):219–228. doi: 10.1002/hep.21704. [DOI] [PubMed] [Google Scholar]
- 3.In ’t Anker PS, Noort WA, Scherjon SA, et al. Mesenchymal stem cells in human second-trimester bone marrow, liver, lung, and spleen exhibit a similar immunophenotype but a heterogeneous multilineage differentiation potential. Haematologica. 2003;88(8):845–852. [PubMed] [Google Scholar]
- 4.Young HE, Steele TA, Bray RA, et al. Human reserve pluripotent mesenchymal stem cells are present in the connective tissues of skeletal muscle and dermis derived from fetal, adult, and geriatric donors. Anatomical Record. 2001;264(1):51–62. doi: 10.1002/ar.1128. [DOI] [PubMed] [Google Scholar]
- 5.Roubelakis MG, Pappa KI, Bitsika V, et al. Molecular and proteomic characterization of human mesenchymal stem cells derived from amniotic fluid: comparison to bone marrow mesenchymal stem cells. Stem Cells and Development. 2007;16(6):931–952. doi: 10.1089/scd.2007.0036. [DOI] [PubMed] [Google Scholar]
- 6.Fukuchi Y, Nakajima H, Sugiyama D, Hirose I, Kitamura T, Tsuji K. Human placenta-derived cells have mesenchymal stem/progenitor cell potential. Stem Cells. 2004;22(5):649–658. doi: 10.1634/stemcells.22-5-649. [DOI] [PubMed] [Google Scholar]
- 7.Miao Z, Jin J, Chen L, et al. Isolation of mesenchymal stem cells from human placenta: comparison with human bone marrow mesenchymal stem cells. Cell Biology International. 2006;30(9):681–687. doi: 10.1016/j.cellbi.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 8.Jo YY, Lee HJ, Kook SY, et al. Isolation and characterization of postnatal stem cells from human dental tissues. Tissue Engineering. 2007;13(4):767–773. doi: 10.1089/ten.2006.0192. [DOI] [PubMed] [Google Scholar]
- 9.Huang GTJ, Gronthos S, Shi S. Critical reviews in oral biology & medicine: mesenchymal stem cells derived from dental tissues vs. those from other sources: their biology and role in regenerative medicine. Journal of Dental Research. 2009;88(9):792–806. doi: 10.1177/0022034509340867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friedenstein AJ, Petrakova KV, Kurolesova AI, Frolova GP. Heterotopic of bone marrow. Analysis of precursor cells for osteogenic and hematopoietic tissues. Transplantation. 1968;6(2):230–247. [PubMed] [Google Scholar]
- 11.Brooke G, Cook M, Blair C, et al. Therapeutic applications of mesenchymal stromal cells. Seminars in Cell & Developmental Biology. 2007;18(6):846–858. doi: 10.1016/j.semcdb.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 12.Giordano A, Galderisi U, Marino IR. From the laboratory bench to the patient’s bedside: an update on clinical trials with mesenchymal stem cells. Journal of Cellular Physiology. 2007;211(1):27–35. doi: 10.1002/jcp.20959. [DOI] [PubMed] [Google Scholar]
- 13.Phinney DG, Prockop DJ. Concise review: mesenchymal stem/multipotent stromal cells: the state of transdifferentiation and modes of tissue repair—current views. Stem Cells. 2007;25(11):2896–2902. doi: 10.1634/stemcells.2007-0637. [DOI] [PubMed] [Google Scholar]
- 14.Ferrand J, Noel D, Lehours P, et al. Human bone marrow-derived stem cells acquire epithelial characteristics through fusion with gastrointestinal epithelial cells. PLoS ONE. 2011;6(5) doi: 10.1371/journal.pone.0019569.e19569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spees JL, Olson SD, Ylostalo J, et al. Differentiation, cell fusion, and nuclear fusion during ex vivo repair of epithelium by human adult stem cells from bone marrow stroma. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(5):2397–2402. doi: 10.1073/pnas.0437997100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vassilopoulos G, Wang PR, Russell DW. Transplanted bone marrow regenerates liver by cell fusion. Nature. 2003;422(6934):901–904. doi: 10.1038/nature01539. [DOI] [PubMed] [Google Scholar]
- 17.Iso Y, Spees JL, Serrano C, et al. Multipotent human stromal cells improve cardiac function after myocardial infarction in mice without long-term engraftment. Biochemical and Biophysical Research Communications. 2007;354(3):700–706. doi: 10.1016/j.bbrc.2007.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meirelles LDS, Caplan AI, Nardi NB. In search of the in vivo identity of mesenchymal stem cells. Stem Cells. 2008;26(9):2287–2299. doi: 10.1634/stemcells.2007-1122. [DOI] [PubMed] [Google Scholar]
- 19.Dai W, Hale SL, Martin BJ, et al. Allogeneic mesenchymal stem cell transplantation in postinfarcted rat myocardium: short- and long-term effects. Circulation. 2005;112(2):214–223. doi: 10.1161/CIRCULATIONAHA.104.527937. [DOI] [PubMed] [Google Scholar]
- 20.Noiseux N, Gnecchi M, Lopez-Ilasaca M, et al. Mesenchymal stem cells overexpressing Akt dramatically repair infarcted myocardium and improve cardiac function despite infrequent cellular fusion or differentiation. Molecular Therapy. 2006;14(6):840–850. doi: 10.1016/j.ymthe.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 21.Ankrum J, Karp JM. Mesenchymal stem cell therapy: two steps forward, one step back. Trends in Molecular Medicine. 2010;16(5):203–209. doi: 10.1016/j.molmed.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Timmers L, Lim S-K, Arslan F, et al. Reduction of myocardial infarct size by human mesenchymal stem cell conditioned medium. Stem Cell Research. 2008;1(2):129–137. doi: 10.1016/j.scr.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 23.Lai RC, Arslan F, Lee MM, et al. Exosome secreted by MSC reduces myocardial ischemia/reperfusion injury. Stem Cell Research. 2010;4(3):214–222. doi: 10.1016/j.scr.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 24.Lai RC, Arslan F, Tan SS, et al. Derivation and characterization of human fetal MSCs: an alternative cell source for large-scale production of cardioprotective microparticles. Journal of Molecular and Cellular Cardiology. 2010;48(6):1215–1224. doi: 10.1016/j.yjmcc.2009.12.021. [DOI] [PubMed] [Google Scholar]
- 25.Chen TS, Arslan F, Yin Y, et al. Enabling a robust scalable manufacturing process for therapeutic exosomes through oncogenic immortalization of human ESC-derived MSCs. Journal of Translational Medicine. 2011;9, article 47 doi: 10.1186/1479-5876-9-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen TS, Lai RC, Lee MM, Choo AB, Lee CN, Lim SK. Mesenchymal stem cell secretes microparticles enriched in pre-microRNAs. Nucleic Acids Research. 2010;38(1):215–224. doi: 10.1093/nar/gkp857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zoeger A, Blau M, Egerer K, Feist E, Dahlmann B. Circulating proteasomes are functional and have a subtype pattern distinct from 20S proteasomes in major blood cells. Clinical Chemistry. 2006;52(11):2079–2086. doi: 10.1373/clinchem.2006.072496. [DOI] [PubMed] [Google Scholar]
- 28.Jakob C, Egerer K, Liebisch P, et al. Circulating proteasome levels are an independent prognostic factor for survival in multiple myeloma. Blood. 2007;109(5):2100–2105. doi: 10.1182/blood-2006-04-016360. [DOI] [PubMed] [Google Scholar]
- 29.Sixt SU, Peters J. Extracellular alveolar proteasome: possible role in lung injury and repair. Proceedings of the American Thoracic Society. 2010;7(1):91–96. doi: 10.1513/pats.200906-035JS. [DOI] [PubMed] [Google Scholar]
- 30.Sixt SU, Adamzik M, Spyrka D, et al. Alveolar extracellular 20s proteasome in patients with acute respiratory distress syndrome. American Journal of Respiratory and Critical Care Medicine. 2009;179(12):1098–1106. doi: 10.1164/rccm.200802-199OC. [DOI] [PubMed] [Google Scholar]
- 31.Ma W, Kantarjian H, Bekele B, et al. Proteasome enzymatic activities in plasma as risk stratification of patients with acute myeloid leukemia and advanced-stage myelodysplastic syndrome. Clinical Cancer Research. 2009;15(11):3820–3826. doi: 10.1158/1078-0432.CCR-08-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haass C, Selkoe DJ. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer’s amyloid β-peptide. Nature Reviews Molecular Cell Biology. 2007;8(2):101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- 33.Sixt SU, Beiderlinden M, Jennissen HP, Peters J. Extracellular proteasome in the human alveolar space: a new housekeeping enzyme? American Journal of Physiology. 2007;292(5):L1280–L1288. doi: 10.1152/ajplung.00140.2006. [DOI] [PubMed] [Google Scholar]
- 34.Lian Q, Lye E, Yeo KS, et al. Derivation of clinically compliant MSCs from CD105+, CD24- differentiated human ESCs. Stem Cells. 2007;25(2):425–436. doi: 10.1634/stemcells.2006-0420. [DOI] [PubMed] [Google Scholar]
- 35.Lai RC, Arslan F, Lee MM, et al. Exosome secreted by MSC reduces myocardial ischemia/reperfusion injury. Stem Cell Research. 2010;4(3):214–222. doi: 10.1016/j.scr.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 36.Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nature Biotechnology. 2001;19(3):242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 37.Sze SK, de Kleijn DP, Lai RC, et al. Elucidating the secretion proteome of human embryonic stem cell-derived mesenchymal stem cells. Molecular and Cellular Proteomics. 2007;6(10):1680–1689. doi: 10.1074/mcp.M600393-MCP200. [DOI] [PubMed] [Google Scholar]
- 38.Thery C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nature Reviews Immunology. 2009;9(8):581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 39.Thomas PD, Campbell MJ, Kejariwal A, et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Research. 2003;13(9):2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thomas PD, Kejariwal A, Guo N, et al. Applications for protein sequence-function evolution data: mRNA/protein expression analysis and coding SNP scoring tools. Nucleic Acids Research. 2006;34:W645–W650. doi: 10.1093/nar/gkl229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanaka K. The proteasome: overview of structure and functions. Proceedings of the Japan Academy B. 2009;85:12–36. doi: 10.2183/pjab.85.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raposo G, Nijman HW, Stoorvogel W, et al. B lymphocytes secrete antigen-presenting vesicles. Journal of Experimental Medicine. 1996;183(3):1161–1172. doi: 10.1084/jem.183.3.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bogyo M, Shin S, McMaster JS, Ploegh HL. Substrate binding and sequence preference of the proteasome revealed by active-site-directed affinity probes. Chemistry and Biology. 1998;5(6):307–320. doi: 10.1016/s1074-5521(98)90169-7. [DOI] [PubMed] [Google Scholar]
- 44.Bulteau A, Lundberg K, Humphries K, et al. Oxidative modification and inactivation of the proteasome during coronary occlusion/reperfusion. Journal of Biological Chemistry. 2001;276(32):30057–30063. doi: 10.1074/jbc.M100142200. [DOI] [PubMed] [Google Scholar]
- 45.Powell SR, Wang P, Katzeff H, et al. Oxidized and ubiquitinated proteins may predict recovery of postischemic cardiac function: essential role of the proteasome. Antioxidants and Redox Signaling. 2005;7(5-6):538–546. doi: 10.1089/ars.2005.7.538. [DOI] [PubMed] [Google Scholar]
- 46.Powell SR, Davies KJ, Divald A. Optimal determination of heart tissue 26S-proteasome activity requires maximal stimulating ATP concentrations. Journal of Molecular and Cellular Cardiology. 2007;42(1):265–269. doi: 10.1016/j.yjmcc.2006.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gurusamy N, Goswami S, Malik G, Das DK. Oxidative injury induces selective rather than global inhibition of proteasomal activity. Journal of Molecular and Cellular Cardiology. 2008;44(2):419–428. doi: 10.1016/j.yjmcc.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 48.Wang X, Li J, Zheng H, Su H, Powell SR. Proteasome functional insufficiency in cardiac pathogenesis- American Journal of Physiology. 2011;301:H2207–H2219. doi: 10.1152/ajpheart.00714.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Powell SR, Divald A. The ubiquitin-proteasome system in myocardial ischaemia and preconditioning. Cardiovascular Research. 2010;85(2):303–311. doi: 10.1093/cvr/cvp321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arslan F, Smeets MB, O’Neill LAJ, et al. Myocardial ischemia/reperfusion injury is mediated by leukocytic toll-like receptor-2 and reduced by systemic administration of a novel anti-toll-like receptor-2 antibody. Circulation. 2010;121(1):80–90. doi: 10.1161/CIRCULATIONAHA.109.880187. [DOI] [PubMed] [Google Scholar]
- 51.Timmers L, Lim SK, Arslan F, et al. Reduction of myocardial infarct size by human mesenchymal stem cell conditioned medium. Stem Cell Research. 2008;1(2):129–137. doi: 10.1016/j.scr.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 52.Chen TS, Arslan F, Yin Y, et al. Enabling a robust scalable manufacturing process for therapeutic exosomes through oncogenic immortalization of human ESC-derived MSCs. Journal of Translational Medicine. 2011;9, article 47 doi: 10.1186/1479-5876-9-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Glabe CG. Conformation-dependent antibodies target diseases of protein misfolding. Trends in Biochemical Sciences. 2004;29(10):542–547. doi: 10.1016/j.tibs.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 54.Kayed R, Head E, Thompson JL, et al. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300(5618):486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 55.de Gassart A, Geminard C, Fevrier B, Raposo G, Vidal M. Lipid raft-associated protein sorting in exosomes. Blood. 2003;102(13):4336–4344. doi: 10.1182/blood-2003-03-0871. [DOI] [PubMed] [Google Scholar]
- 56.Critchley DR, Streuli CH, Kellie S, Ansell S, Patel B. Characterization of the cholera toxin receptor on Balb/c 3T3 cells as a ganglioside similar to, or identical with, ganglioside GM1. No evidence for galactoproteins with receptor activity. Biochemical Journal. 1982;204(1):209–219. doi: 10.1042/bj2040209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Spangler BD. Structure and function of cholera toxin and the related echerichia coli heat-labile enterotoxin. Microbiological Reviews. 1992;56(4):622–647. doi: 10.1128/mr.56.4.622-647.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Simons M, Raposo G. Exosomes—vesicular carriers for intercellular communication. Current Opinion in Cell Biology. 2009;21(4):575–581. doi: 10.1016/j.ceb.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 59.Simpson RJ, Lim JW, Moritz RL, Mathivanan S. Exosomes: proteomic insights and diagnostic potential. Expert Review of Proteomics. 2009;6(3):267–283. doi: 10.1586/epr.09.17. [DOI] [PubMed] [Google Scholar]
- 60.Pan BT, Johnstone RM. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor. Cell. 1983;33(3):967–978. doi: 10.1016/0092-8674(83)90040-5. [DOI] [PubMed] [Google Scholar]
- 61.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature Cell Biology. 2007;9(6):654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 62.Schorey JS, Bhatnagar S. Exosome function: from tumor immunology to pathogen biology. Traffic. 2008;9(6):871–881. doi: 10.1111/j.1600-0854.2008.00734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huber V, Filipazzi P, Iero M, Fais S, Rivoltini L. More insights into the immunosuppressive potential of tumor exosomes. Journal of Translational Medicine. 2008;6, article 63 doi: 10.1186/1479-5876-6-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Muntasell A, Berger AC, Roche PA. T cell-induced secretion of MHC class II-peptide complexes on B cell exosomes. EMBO Journal. 2007;26(19):4263–4272. doi: 10.1038/sj.emboj.7601842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zeelenberg IS, Ostrowski M, Krumeich S, et al. Targeting tumor antigens to secreted membrane vesicles in vivo induces efficient antitumor immune responses. Cancer Research. 2008;68(4):1228–1235. doi: 10.1158/0008-5472.CAN-07-3163. [DOI] [PubMed] [Google Scholar]
- 66.Zöller M. Tetraspanins: push and pull in suppressing and promoting metastasis. Nature Reviews Cancer. 2009;9(1):40–55. doi: 10.1038/nrc2543. [DOI] [PubMed] [Google Scholar]
- 67.Buschow SI, van Balkom BW, Aalberts M, Heck AJ, Wauben M, Stoorvogel W. MHC class II-associated proteins in B-cell exosomes and potential functional implications for exosome biogenesis. Immunology and Cell Biology. 2010;88(8):851–856. doi: 10.1038/icb.2010.64. [DOI] [PubMed] [Google Scholar]
- 68.Gonzales PA, Pisitkun T, Hoffert JD, et al. Large-scale proteomics and phosphoproteomics of urinary exosomes. Journal of the American Society of Nephrology. 2009;20(2):363–379. doi: 10.1681/ASN.2008040406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Carayon K, Chaoui K, Ronzier E, et al. Proteolipidic composition of exosomes changes during reticulocyte maturation. The Journal of Biological Chemistry. 2011;286:34426–34439. doi: 10.1074/jbc.M111.257444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jung T, Grune T. The proteasome and its role in the degradation of oxidized proteins. IUBMB Life. 2008;60(11):743–752. doi: 10.1002/iub.114. [DOI] [PubMed] [Google Scholar]
- 71.Stadtman ER, Levine RL. Protein oxidation. Annals of the New York Academy of Sciences. 2000;899:191–208. doi: 10.1111/j.1749-6632.2000.tb06187.x. [DOI] [PubMed] [Google Scholar]
- 72.Dunlop RA, Brunk UT, Rodgers KJ. Oxidized proteins: mechanisms of removal and consequences of accumulation. IUBMB Life. 2009;61(5):522–527. doi: 10.1002/iub.189. [DOI] [PubMed] [Google Scholar]
- 73.Sohns W, van Veen TA, van der Heyden MA. Regulatory roles of the ubiquitin-proteasome system in cardiomyocyte apoptosis. Current Molecular Medicine. 2010;10(1):1–13. doi: 10.2174/156652410791065426. [DOI] [PubMed] [Google Scholar]
- 74.Willis MS, Townley-Tilson WH, Kang EY, Homeister JW, Patterson C. Sent to destroy: the ubiquitin proteasome system regulates cell signaling and protein quality control in cardiovascular development and disease. Circulation Research. 2010;106(3):463–478. doi: 10.1161/CIRCRESAHA.109.208801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tsukamoto O, Minamino T, Kitakaze M. Functional alterations of cardiac proteasomes under physiological and pathological conditions. Cardiovascular Research. 2010;85(2):339–346. doi: 10.1093/cvr/cvp282. [DOI] [PubMed] [Google Scholar]
- 76.Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nature Biotechnology. 2011;29(4):341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 77.Rabesandratana H, Toutant JP, Reggio H, Vidal M. Decay-accelerating factor (CD55) and membrane inhibitor of reactive lysis (CD59) are released within exosomes during in vitro maturation of reticulocytes. Blood. 1998;91(7):2573–2580. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material provides the raw data of the detected 20S proteasome in 3 independent batches of exosomes. Supplementary data 1 provides a summary of number of detected peptides in 3 independent batches of exosomes; Supplementary data 2–4 provide the raw data of detected 20S proteasome peptides in 3 independent batches of exosomes respectively; Supplementary data 5–7 provide the Mascot peptide view of unique 20S proteasome peptides in 3 independent batches of exosomes respectively.


