Abstract
Signaling networks in living systems are coordinated through subcellular compartmentalization and precise timing of activation. These spatiotemporal aspects ensure the fidelity of signaling while contributing to the diversity and of downstream events. This is studied through development of molecular tools that generate localized and precisely timed protein activity in living systems. To study the molecular events responsible for cytoskeletal changes in real time, we generated versions of Rho family GTPases whose interactions with downstream effectors is controlled by light. GTPases were grafted to the phototropin LOV (light, oxygen, or voltage) domain (Huala, E., Oeller, P. W., Liscum, E., Han, I., Larsen, E., and Briggs, W. R. (1997). Arabidopsis NPH1: A protein kinase with a putative redox-sensing domain. Science 278, 2120–2123.) via an alpha helix on the LOV C-terminus (Wu, Y. I., Frey, D., Lungu, O. I., Jaehrig, A., Schlichting, I., Kuhlman, B., and Hahn, K. M. (2009). A genetically encoded photoactivatable Rac controls the motility of living cells. Nature 461, 104–108.). The LOV domain sterically blocked the GTPase active site until it was irradiated. Exposure to 400–500 nm light caused unwinding of the helix linking the LOV domain to the GTPase, relieving steric inhibition. The change was reversible and repeatable, and the protein could be returned to its inactive state simply by turning off the light. The LOV domain incorporates a flavin as the active chromophore. This naturally occurring molecule is incorporated simply upon expression of the LOV fusion in cells or animals, permitting ready control of GTPase function in different systems. In cultured single cells, light-activated Rac leads to membrane ruffling, protrusion, and migration. In collectively migrating border cells in the Drosophila ovary, focal activation of photoactivatable Rac (PA-Rac) in a single cell is sufficient to redirect the entire group. PA-Rac in a single cell also rescues the phenotype caused by loss of endogenous guidance receptor signaling in the whole group. These findings demonstrate that cells within the border cell cluster communicate and are guided collectively. Here, we describe optimization and application of PA-Rac using detailed examples that we hope will help others apply the approach to different proteins and in a variety of different cells, tissues, and organisms.
1. Introduction
In living cells, transient protein interactions, conformational changes, and posttranslational modifications are controlled with precise kinetics and localization. To better understand how location and timing impact signaling, and to harness local signaling to produce cell behaviors, approaches have been developed to modulate protein activity in living cells and animals. Modulating activity with light was initially accomplished through covalent attachment of photolabile protecting groups to specific residues of proteins, blocking activity until irradiation broke the covalent bond and freed the “caged” residue. These techniques require high-energy light to break covalent bonds and lead to irreversible protein activation. They are technically challenging in that specific residues must be modified, and the resulting covalent protein adduct must be loaded into cells via microinjection or other techniques that perturb the cell membrane. In contrast, the methods described here and elsewhere in this volume enable reversible photoactivation of modified proteins that are entirely genetically encoded, using less harmful wavelengths and naturally occurring cofactors that permit ready application in living animals. Here, we use photo-activatable Rac (PA-Rac) as an example and provide detailed procedures for generating and applying a PA protein.
2. The LOV Domain as a Tool for Protein Caging
The LOV (light, oxygen, or voltage) domain denotes a subgroup of the Per-ARNT-Sim (PAS) domains, named for homologous domains that occur within signaling proteins initially identified in the Drosophila period circadian rhythm (Per) and single-minded (Sim) proteins and in the vertebrate aryl hydrocarbon receptor nuclear transporter (ARNT). The LOV domain has a conserved central antiparallel β sheet with five strands, and several α helices, within which resides a flavin analogue, typically a flavin mononucleotide (FMN) (Möglich et al., 2009). Upon 400–500 nm illumination, excitation of the flavin molecule leads to the formation of a covalent linkage between the C4(a) atom in the flavin and a thiol from a conserved Cys residue in the LOV domain. This reaction is reversible and undergoes recovery in the dark over seconds to minutes depending on the protein (Swartz et al., 2001). Attached to the LOV domain in phototropin is a helix (named Jα) that is docked on the β-sheet of the LOV domain (Harper et al., 2003). Upon illumination, conformational changes occur throughout the LOV domain, leading to dissociation and unwinding of the Jα helix.
As shown in Fig. 16.1A, this light-induced helix unwinding can be the basis of a generalizable approach to protein caging. When the helix unwinds, the LOV domain is no longer held tightly against the GTPase, but is now on the end of a long tether. This relieves steric inhibition and “activates” the protein. Advantages of the LOV domain include its small size (~15 kDa), well-characterized light-induced conformational change, and the prevalence of the flavin cofactor, which occurs in cultured cells and in vivo. For some applications, the ability to reverse activation simply by turning off the light will be valuable, although the current time constants for dark recovery are on the order of seconds to minutes.
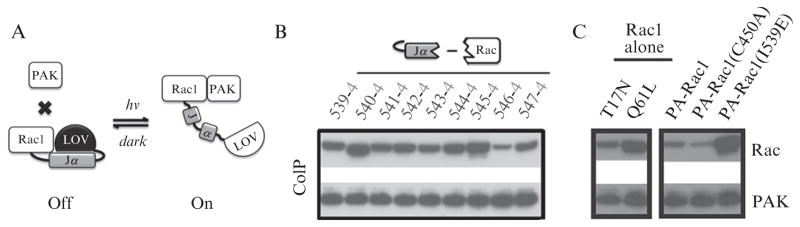
Figure 16.1.
Design and characterization of photoactivatable Rac (PA-Rac). (A) cartoon diagram of PA-Rac light-induced conformational change and the resultant binding of PA-Rac to effector PAK. (B) pull down results of different linkages between Jα and Rac using PAK as bait. (C) light control of the binding of PA-Rac to PAK exceeded the difference in binding between inactive (T17N) and active (Q61L) Rac mutants (comparing PA-Rac in the dark, the dark state mutant C450A, and the lit state mutant I539E)
3. Design and Structure Optimization of PA-Rac
We were interested in grafting the LOV-Ja domain onto Rac such that activity would be inhibited in the dark, but restored upon illumination. The interconversion of Rho GTPases between active (GTP-bound) and inactive (GDP-bound) states is catalyzed by guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs). We wanted control of Rac activity to be solely a function of our cell irradiation, and not subject to normal upstream regulatory pathways. We therefore locked PA-Rac in its GTP-loaded state by using a hydrolysis-deficient mutant (Q61L in Rac1), which also blocks its interaction with GEFs. We introduced mutations at the interface between GTPases and GAP proteins (N91H/E92H in Rac) based on known crystal structures, leading to decreased binding of p50GAP.
Because the C-terminus of Jα was well packed against the LOV domain in the dark, we reasoned that the globular LOV domain could establish a steric block, occluding effector binding, if the C-terminus of Jα were tethered near Rac’s effector binding interface. Irradiation and unwinding of the helix should relieve this block. The steric block was achieved by attaching the LOV-Jα to the N-terminus of Rac. We sampled different linkages by varying the junctional residues at the C-terminus of Jα and the N-terminus of Rac, leading to a working linkage (L546-I4) that significantly decreased the interaction of active Rac with downstream effectors. To optimize GTPase caging, we developed a simple pull down assay taking advantage of the efficient FLAG epitope-M2 monoclonal antibody system (Sigma). Different linkers and Rac/Lov truncations were tested to find those best able to inhibit Rac interactions with effectors in the dark, and to restore them in the light (Fig. 16.1B). HEK293 cells were transfected with both FLAG-tagged p21-activated kinase (PAK), a Rac effector, and different LOV domain-Rac fusions. Pull down of Rac by Pak was used to gauge the effect of structural modification.
To avoid unintended Rac activation by light, the cells were maintained in an incubator in a dark room and all steps were performed under red light. The cells were lysed on ice in prechilled lysis buffer (50 mM Tris, pH 7.5, 150 mM NaCl, and 1% Triton X-100 with an EDTA-free protease inhibitor cocktail). EDTA was avoided because GTP binding to Rac requires Mg2+ ions, which are chelated by EDTA. Two wells (φ = 2.2 cm) from a 6-well plate with cells at 50–75% confluency, lysed in 400 μL lysis buffer, provided enough protein at an appropriate concentration for this assay. Incubation on ice for 20 min was sufficient for lysis. The cell lysates were carefully collected and cleared of insoluble debris by centrifuging at 10,000×g for 5 min. Small portions of the supernatants (30–40 μL) were sampled at this time for western blot analysis of protein expression. Forty microliters (50% slurry) of anti-FLAG antibody-conjugated agarose (Sigma) was mixed with the rest of the samples and incubated for 1 h in the dark at 4 °C, followed by three washes with 500 μL of lysis buffer. Handee spin columns (Pierce) made sample handling in the dark less challenging and also minimized the variation between experiments. The bound proteins were eluted with 40 μL of 200 μg/mL 3× FLAG peptide after 5 min of incubation. The pull down samples were subjected to western blot analysis using antibodies against fluorescent protein JL-8 (Clontech) and PAK N-20 (Santa Cruz) or rabbit polyclonal anti-FLAG antibody (Sigma).
To examine enhancement of Pak interaction with light, we first attempted to carry out these assays in room light, but this produced inconsistent results, potentially due to the rapid dark recovery of the LOV domain during sample preparation. Instead, we compared point mutations that mimic the closed/dark (C450A) and open/lit (I539E) conformations of the LOV domain (Fig. 16.1C). These screens led to the optimized PA-Rac described in the original publication (Wu et al., 2009).
4. Activation of PA-Rac in Living Cells
4.1. Cell handling
Constructs derived from the pTriEx vector (Novagen) carry a CMV promoter. These were used, typically at 0.2–0.3 μg DNA per well of a 6-well plate (φ = ~ 2.2 cm) for transient transfection of mouse embryo fibroblasts with Fugene 6 reagents. The cells were imaged between 12–24 h after transfection. Alternatively, lower but uniform expression of PA-Rac can be achieved in retrovirus-infected MEF cells. We chose a retroviral vector that carries a TetCMV promoter. The expression of PA-Rac was suppressed until the time of imaging using retroviral constructs in which the expression of PA-Rac was driven by a TetCMV promoter. Using this approach, the expression level could be controlled through titration of doxycycline. We used the fluorescent proteins attached to the PA-Rac to gauge expression level, determining optimum expression for light-induced Rac activation with minimal effects from low-level PA-Rac activity in the dark.
Because of the light sensitivity of the LOV domain, cells expressing PA-Rac should not be exposed to light within the LOV action spectrum (<500 nm) immediately before imaging experiments. It is safe to prepare live samples under yellow or red light, available in most dark rooms. If DIC or phase-contrast images are needed, caution must be taken to avoid photoactivation from the transmitted light source. A red filter, such as a Schott RG610 Glass filter (610 nm long pass), can be placed in the transmitted light path (most commercial microscopes have a transmitted light filter holder). Even light emitted from bright computer monitors can be of concern, but this is less consequential at a distance. We transferred the cells immediately after transfection into incubators located in isolated dark rooms to avoid unintended light exposure. In some experiments, extensive manipulations were required prior to imaging. When this was difficult in a dark room, cells could be incubated in the dark for an hour to reduce the effect of prior light exposure and restore responsiveness to photoactivation. It can be useful to place a black paper cone over the objective and live cell chamber of the microscope to eliminate small amounts of ambient light in some rooms.
We constructed PA-Rac constructs with various fluorescent protein tags including mCerulean, mVenus, and mCherry, to facilitate the identification of cells expressing PA-Rac. Based on the excitation wavelengths of these fluorescent proteins, mCherry (590 nm max) can be monitored without concern for excitation of the LOV domain. The common excitation filters for mVenus or YFP do fall in the 480–500 nm range that can activate the LOV domain. We recommend using excitation filters that cut off below 515–520 nm, together with minimal intensity and exposure time, to avoid unintended photoconversion of the LOV domain. Because the excitation wavelengths of mCerulean or CFP (433 nm max) overlap with the photo-activation spectrum, fluorescence from these proteins is best used for confirmation of expression at the end of experiments.
Our preparation of cells for live cell imaging has been previously described (Hodgson et al., 2010). Briefly, cells were seeded onto φ25 mm coverslips coated with 1–10 μg/mL fibronectin overnight. The imaging medium was Ham’s F-12K medium without phenol red, containing 2% fetal bovine serum. Coverslips were mounted in an Attofluor live cell chamber (Invitrogen) and placed in a microscope stage with a heated stage adaptor (Warner).
4.2. Irradiation in living cells—Light sources, dosage, and spatial control
The action spectrum of plant phototropin is in the UV-A and blue light range (360–500 nm). We tested several common laser lines for their ability to induce membrane ruffles in MEF cells expressing PA-Rac. The wavelengths 405, 458, 473, and 488 nm all proved to be effective. The power dosage of the PA-Rac to 458 nm line was measured in stable MEF cell lines, where expression levels could be well controlled and the areas of induced protrusions readily measured. A light dose of 6.2 μJ over a 10 μm spot at 458 nm induced a cellular response with a single exposure. This was the lowest power setting (0.1% of total power on the mW scale) of our Fluoview 1000 confocal microscope at very fast scan rate (10 μs/pixel). Following this exact photoactivation regime but with increasing laser power or scan duration, we determined that the cellular response (protrusion area) stopped increasing when we reached 1000-fold higher dose. A 100 W Mercury arc lamp used for fluorescence imaging was also an effective source for photo-activation. We can qualitatively state that we found no difficulties in activating PA-Rac through global cell irradiation using a 100 W mercury source filtered through a ND 2.0 (1.0% transmission) filter and a CFP excitation filter (ET430/24 nm). This was sufficient to induce membrane ruffles with a 500 ms exposure. One of the main peaks of emission from the mercury light source falls at these wavelengths, and these observations were made with cells expressing low levels of PA-Rac (empirically optimized for optimal light response without induction of a Rac phenotype in the dark).
Spatial control is perhaps the most valuable feature of light-mediated protein activation. The ability to target a defined cell area enables study of localized signaling milieus. With Rac, localized irradiation can be used to control cell motility. Most conventional wide-field microscopes can be modified to incorporate a field stop/diaphragm or pinhole in a conjugate image plane. The light source used for fluorescence excitation can be used to illuminate a small region of the cell. By modulating illumination intensity, low intensity can be used to target the protein uncaging light, followed by high intensity radiation to activate protein. Alternatively, various scan modes on laser scanning confocal microscopes can be used. Commercial vendors have devised a variety of solutions for laser irradiation of small portions of the field of view, usually for FRAP studies. A laser beam can be coupled into the light path and focused on the focal plane to a diffraction-limited spot or dilated to bigger areas through z offset. These spots can also be mobilized to scan across different shapes either manually or using galvanometer-driven mirrors. Complex patterns can be achieved using digital micromirror-based devices such as the commercially available Mosaic digital illumination system (Levskaya et al., 2009), or more advanced methods based on liquid-crystal spatial light modulators, realized in applications involving laser tweezers (Curtis et al., 2002) and adaptive optics (Girkin et al., 2009).
4.3. Detection and quantitation of Rac activation in cells
For any given protein, it will be important to devise an assay that can define successful activation in living cells. For Rac, overexpression in many cell types induces membrane ruffles and lamellipodial protrusions, both phenotypes readily identifiable with DIC or phase-contrast imaging. These phenotypes were scored after global illumination to examine the effects of different mutations on PA-Rac. Induction was much clearer when cells were serum-starved to minimize background activity prior to irradiation. Effects were easier to observe in MEF, HEK293, and HeLa cells, but less apparent in COS-7 cells. This was perhaps due to differences in basal Rac activity and/or the abundance of downstream effectors. Similar differences were seen with dominant-negative mutants of PA-Rac that inhibited endogenous Rac activity through sequestration of Rac GEFs. The production of protrusions was quantified from line scans of the cell border in time-lapse images, or by monitoring protrusion area.
To analyze Rac initiation of downstream signaling, we also tracked translocation of downstream effectors. For wide-field microscopy, artifacts of changing cell volume were avoided using ratiometric imaging. The fluorescence intensity of the tagged target protein was divided by the fluorescence intensity of a separately expressed volume indicator, that is, YFP or mCherry. As demonstrated previously (Wu et al., 2009), PA-Rac redistributed slowly out of sites of illumination with a diffusion coefficient of 0.55 μm2s−1. This led to detectable accumulation of downstream effectors where Rac was activated.
Phosphorylation of endogenous PAK was also traced using immunofluorescence of fixed cells after local activation of PA-Rac. MEF cells stably expressing PA-Rac were plated onto coverslips with etched grids (Bellco), used to locate the cells that had been irradiated. Cells were irradiated at 473 nm through a 20× phase-contrast objective. Immediately after protrusions were induced, the cells were fixed in 3.7% formalin (Sigma), permeabilized in 0.2% Triton X-100, incubated with anti-phospho-PAK antibody (Cell Signaling), and finally incubated with Alexa Fluor 594-conjugated secondary antibody (Molecular Probes). Imaging was performed as described above.
5. Application of PA-Rac in Drosophila Ovarian Border Cell Migration
5.1. What is border cell migration?
To test whether PA-Rac would be useful in an intact three-dimensional tissue and in a different organism, we chose to study border cell migration in the Drosophila ovary, a well-characterized example of collective cell movement. Border cells are a group of 6–8 cells that arise from a monolayer of ~650 epithelial follicle cells that surround 15 nurse cells and one oocyte in a structure called an egg chamber (Fig. 16.2). Border cells migrate ~175 μm in between the nurse cells, as an interconnected group of two distinct cell types: 4–8 migratory cells surrounding two central polar cells. Polar cells cannot migrate but secrete a cytokine that activates the JAK/STAT pathway rendering the outer cells motile. The outer cells carry the polar cells and lose the ability to move in the absence of continuous JAK/STAT activation. Thus each cell type requires the other. Border cells also require steroid hormone, receptor tyrosine kinase, Notch, and other signaling cascades. Thus border cells experience a rich and complex signaling environment, as do most cells in vivo.
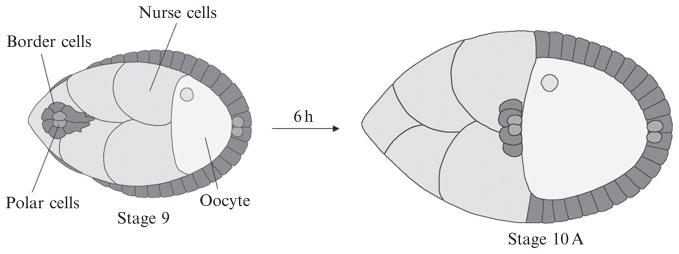
Figure 16.2.
Schematic drawing of border cell migration. A pair of polar cells secretes a cytokine that activates the JAK/STAT pathway in immediately adjacent cells to specify the border cells at early stage 9. Then, border cells detach from the epithelium and migrate in between the germ-line cells during stage 9 and arrive at the boundary between nurse cells and oocyte at stage 10.
There are several prominent differences between the migration of single cultured cells and border cells. Firstly, single cells in vitro typically migrate on stiff 2-dimensional surfaces coated with fibronectin or collagen substrates, while border cells migrate in a more pliable environment surrounded on all sides by nurse cells. Secondly, border cells experience a complex signaling environment that includes many nondirectional signals that could potentially activate small GTPases uniformly, in addition to directional signals, which could possibly create a high background and interfere with the effect of light-induced Rac activation. Thirdly, single cells in culture have unlimited time and space in which to move, whereas border cells are normally limited to a straight path between the nurse cells, starting from the anterior tip and ending at the oocyte and only during developmental stage 9. Lastly and most importantly, border cells migrate as a coherent cluster. Therefore, it was unclear whether activation of Rac in one cell would have an effect on the other cells.
Previous studies had shown that constitutive activation or inhibition of Rac led to complete inhibition of migration. However, one could only speculate as to why activation or inactivation produced the same effect. The ability to control Rac activity both temporally and spatially with PA-Rac allowed us to address these critical open questions.
5.2. Genetics
Considering the different excitation wavelengths of mCerulean, mVenus, and mCherry and PA-Rac activation, we chose to insert N-terminal-mCherry-tagged PA-RacQ61L, PA-RacT17N, and the light-insensitive control C450M-PA-RacQ61L into pUASt Drosophila expression vector, using the Invitrogen Gateway recombination system. Transgenic flies were generated by Bestgene Inc., and confirmed by the detection of mCherry fluorescence. We used the mammalian Rac rather than substituting Drosophila Rac for two reasons. Drosophila and mammalian Rac proteins share more than 90% similarity and are known to function similarly (Luo et al. 1994). We were concerned, however, that the 6 amino-acid difference between the carboxy-terminal tails of Drosophila and mammalian Rac might alter the physical interaction with the Lov-Jα domain. We are currently testing PA-Drosophila Rac and comparing it with the mammalian protein.
Initially, all fly stocks and crosses were maintained at room temperature in normal light. We used the Gal4/UAS system (Brand and Perrimon, 1993) to drive cell-type specific expression of PA-Rac. Specifically, we used transgenic flies expressing Gal4 under the control of the slow border cells (slbo) gene, to drive expression of pUASt-PA-Rac. During stage 9 of oogenesis, Slbo-Gal4 drives expression of PA-Rac and other UAS transgenes primarily in border cells. Expression of PA-RacQ61L or PA-RacT17N driven by Slbo-Gal4 had no detectable deleterious effect on border cell migration in the absence of laser illumination. Another Gal4 commonly used to drive border cell expression of UAS transgenes, c306-Gal4, was lethal in combination with UAS-PA-RacQ61L. c306 is not as specific as slbo-Gal4 and the lethality was probably due to expression of the transgene earlier in development and in appropriate activation of Rac due to ambient light. UAS-PA-RacT17N driven by C306-Gal4 caused a moderate delay in detachment of border cells from the epithelium in early stage 9. C306-Gal4 drives UAS transgene expression earlier and to higher levels than slbo-Gal4, which starts weakly in a few anterior and posterior follicle cells around stage 7 and gradually becomes stronger during stages 8–10, when border cells complete their migration. Thus, the restricted spatial and temporal expression of Slbo-Gal4 has important advantages in alleviating side effects of “leaky” Rac activity. Newer PA-Rac mutations currently being characterized have greatly reduced residual Rac activity in the dark. To further prevent the effects of residual Rac activity, we maintained progeny flies carrying both Gal4 and UAS-PA-Rac constructs in a culture incubator with lower temperature (18 °C), which decreases Gal4-mediated gene expression due to temperature sensitivity of the Gal4 transcription factor. Prior to dissection, flies were transferred to foil-wrapped vials and incubated at 29 °C overnight to induce UAS transgene expression. This incubation had no negative effect on border cell migration in the absence of laser illumination.
5.3. Illumination of border cells—In vitro culture, live imaging, and photomanipulation
Drosophila egg chambers were dissected in Schneider’s insect medium supplemented with 20% fetal bovine serum and 0.10 mg/mL insulin, and then mounted in the same medium on a 50 mm Greiner Lumox culture hydrophilic dish (also known as a petriperm plate) and coved with a 22 mm coverslip, as described (Prasad et al. 2007). Figure 16.3 shows a schematic drawing of the culture chamber mounting. The following points are critical for successful live imaging of border cells: The final culture medium pH was adjusted to 6.85–6.95, which is essential for normal border cell migration (Prasad et al. 2007). Secondly, the surface of the petriperm plate was kept clean to allow O2/CO2 exchange. Thirdly, the volume of culture medium between the petriperm plate and coverslip was adjusted carefully. Too little medium can crush egg chambers and prevent normal border cell migration. Excess medium allows the egg chamber to float and change position, making subsequent manipulations impossible. This problem is particularly noticeable when the mounted petriperm plate needs to be inverted (as shown in Fig. 16.3). The mounting volume is appropriate when the stages 9 and 10 egg chambers have normal morphology (not too flat) and their position remains stable even when one lightly taps the edge of the petriperm plate with a finger. The dissection and mounting process should be complete within 30 min to prevent general activation of PA-Rac, since these steps are carried out in ambient light.
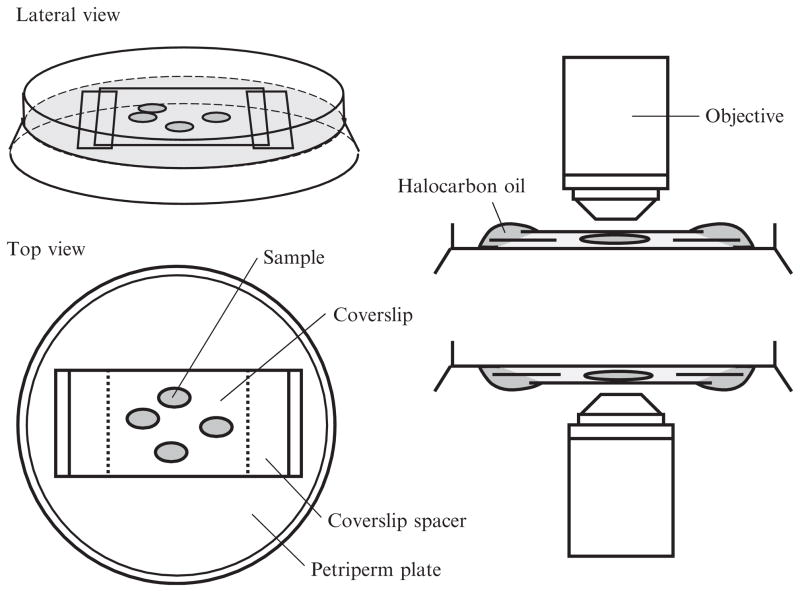
Figure 16.3.
Schematic drawing of mounting of the cultured chamber. The final setup after mounting the cultured egg chamber as viewed from either lateral or top view in left panel. The coverslip spacers are put upon the membrane of petriperm plate to prevent crushing egg chambers. Halocarbon oil is used to prevent evaporation of cultured medium. The mounting process is basically the same whether using an upright or inverted microscope.
Photoactivation, time-lapse-imaging, and 3-dimensional morphological reconstruction were carried out using a Zeiss 510-Meta or 710-Meta confocal microscope using a 63×, 1.4 numerical aperture oil lens with 2× zoom. To photoactivate PA-Rac in border cells, a 458 nm laser was set at 10% power for 0.1 ms scan per pixel in a 7 μm spot, and the photoactivation scan took ~25 s at a scan speed of 2× and scan number of 8. Laser scanning with 477 and 488 nm wavelengths produced a much weaker effect. Lower scan energy, less scan time, or smaller scan regions also produced weaker effects. Optimal photomanipulation conditions will likely depend on both the specific tissue under study and the particular microscope and laser setup. Approximately 20–30 s after photoactivation, border cells were imaged by capturing the mCherry-tagged PA-Rac signal using a 568 nm wavelength scan at the minimally detectable laser power, lasting 6–7 s at scan speed of 8× and scan number of 8. This series of photoactivation and live imaging steps was repeated for a few minutes or up to 5 h depending on the experiment. Egg chambers survived this repeated light exposure for up to 5 h before showing signs of phototoxicity. However, the effect of light-induced Rac activation diminished after 2.5–3.5 h of repeated treatment. The 3-dimensional reconstruction of border cell morphology was captured before and after repeated phototreatment of border cells, using 20 focal planes 1.5 μm apart in the z-axis, using 568 nm light. The laser energy at 568 nm was adjusted to higher levels to show the detailed filopodia.
5.4. Detection and quantification of effects of Rac activation on border cells
Analysis of the effect of light-induced Rac activation in border cells requires careful quantification and comparison to controls to distinguish induced effects from endogenous cell behavior. As mentioned above, border cells experience a rich cocktail of endogenous signals that lead to substantial Rac activation, protrusion formation, and directional migration. So, for example, when membrane ruffles or lamellipodial protrusions are observed following photo-activation of Rac, how do we distinguish whether PA-Rac caused that effect or whether the cell might have done the same thing without light-activated Rac due to endogenous signals? To clearly identify effects of light-induced Rac activity, we measured multiple parameters including migration speed, protrusion area, number, and density, as well as a directionality index (DI). We compared these features in wild-type and various genetic backgrounds in the presence or absence of light. The light-insensitive versions of PA-Rac proteins were also essential to control for nonspecific effects of the phototreatment itself. Substantial variation in border cell responses were observed, which might be caused by slight variations in the condition of the egg chambers, PA-Rac expression levels, starting position of the cells along the travel path, etc. Statistical analyses were therefore essential.
Migration speed is the simplest parameter to measure. The distance of the center of the border cell cluster between the first and final time points in a time-lapse series was measured using Imaris software. This distance divided by the elapsed time gave the speed. Imaris software can also measure speeds during shorter intervals within the overall experiment. We typically measured the migration speed beginning ~1 h after initiating the light treatment, and we observed border cell migration speeds ranging from 0.1 to 0.5 μm per minute in response to light-induced Rac.
Protrusion area was measured as follows: a confocal image of the cluster edge was captured before light treatment as shown in Fig. 16.4 (left panel). After repeated light treatments, another image was captured, as shown in Fig. 16.4 (middle panel). The difference in position of the cell boundary between the two images, as shown in Fig. 16.4 (right panel), was used to estimate the protrusion area using ImageJ software.
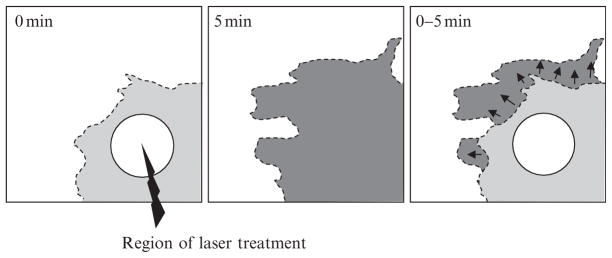
Figure 16.4.
Schematic drawing of change in protrusion area in response to light-induced Rac. The light or dark gray region shows the protrusion region at the beginning or end of laser treatment. Black arrows represent the direction of expansion of the protruding region in response to light-induced Rac.
Cell protrusions were defined and counted automatically using MatLab as follows: a circle corresponding to the average cluster diameter was drawn, and major protrusions were defined as extensions at least 2 μm beyond the circle and broader than 2 μm. The number of major protrusions at any given time varied between 2 and 5 in different genetic backgrounds. The DI represents the fraction of forward-directed protrusions relative to the total number of protrusions. And DI was calculated using the following equation:
where N is the total number of major protrusions, is the ith protrusion vector, and d⃗ is the unit vector of migration direction. The protrusion vector is calculated by fitting the major protrusion by a parabola whose peak together with the cluster center gives the vector’s direction and length. Protrusion density was generated by dividing the number of all the recognizable membrane protrusions by the measured cell perimeter in micron.
5.5. What do we learn from PA-Rac application to border cell migration?
The studies in cultured cells showed that PA-Rac could induce local membrane ruffling and protrusion and even cell movement. The effects were stronger in some cell types than others and were most pronounced in low-serum or serum-free conditions. It was not obvious therefore how effective the treatment would be in vivo, where cells are exposed to myriad endogenous signals both directional and nondirectional. Our results indicated that despite the intense and complex signaling environment, local activation of Rac is sufficient to cause local ruffling and protrusion and to set the direction of movement for cells, forward, backward, or sideways, in vivo. Secondly, we found that there were regions of the egg chamber into which the cells could not move. Thus PA-Rac could be used to “map” the permissive and nonpermissive regions of the tissue. Thirdly, border cells migrate collectively as an interconnected group, as do a variety of other cell types including vertebrate neural crest cells and invading carcinomas (Gaggioli et al. 2007; Theveneau et al. 2010). We found that local activation of Rac in a single cell was sufficient to redirect the entire cluster. Most surprisingly, light-induced Rac activity in one border cell altered the morphology of the other cells. Photo-inactivation of Rac in the leading cell “confused” all of the cells. These findings demonstrate that a group of cells can sense direction collectively according to which cell has the highest level of Rac activity, a finding that would have been difficult or impossible to make with traditional genetic methods. Moreover, these studies demonstrate that PA proteins should be generally useful in a variety of cell types, tissues, and organisms.
References
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118(2):401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Curtis JE, Koss BA, Grier DG. Dynamic holographic optical tweezers. Opt Commun. 2002;207:169–175. [Google Scholar]
- Gaggioli C, Hooper S, Hidalgo-Carcedo C, Grosse R, Marshall JF, Harrington K, Sahai E. Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nat Cell Biol. 2007;9(12):1392–1400. doi: 10.1038/ncb1658. [DOI] [PubMed] [Google Scholar]
- Girkin JM, Poland S, Wright AJ. Adaptive optics for deeper imaging of biological samples. Curr Opin Biotechnol. 2009;20:106–110. doi: 10.1016/j.copbio.2009.02.009. [DOI] [PubMed] [Google Scholar]
- Harper SM, Neil LC, Gardner KH. Structural basis of a phototropin light switch. Science. 2003;301:1541–1544. doi: 10.1126/science.1086810. [DOI] [PubMed] [Google Scholar]
- Hodgson L, Shen F, Hahn K. Biosensors for characterizing the dynamics of rho family GTPases in living cells. Curr Protoc Cell Biol. 2010;Chapter 14(Unit 14.11):1–26. doi: 10.1002/0471143030.cb1411s46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huala E, Oeller PW, Liscum E, Han I, Larsen E, Briggs WR. Arabidopsis NPH1: A protein kinase with a putative redox-sensing domain. Science. 1997;278:2120–2123. doi: 10.1126/science.278.5346.2120. [DOI] [PubMed] [Google Scholar]
- Levskaya A, Weiner OD, Lim WA, Voigt CA. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature. 2009;461:997–1001. doi: 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Liao YJ, Jan LY, Jan YN. Distinct morphogenetic functions of similar small GTPases: Drosophila Drac1 is involved in axonal outgrowth and myoblast fusion. Genes Dev. 1994;8(15):1787–1802. doi: 10.1101/gad.8.15.1787. [DOI] [PubMed] [Google Scholar]
- Möglich A, Ayers RA, Moffat K. Structure and signaling mechanism of Per-ARNT-Sim domains. Structure. 2009;17:1282–1294. doi: 10.1016/j.str.2009.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad M, Jang AC, Starz-Gaiano M, Melani M, Montell DJ. A protocol for culturing Drosophila melanogaster stage 9 egg chambers for live imaging. Nat Protoc. 2007;2(10):2467–2473. doi: 10.1038/nprot.2007.363. [DOI] [PubMed] [Google Scholar]
- Swartz TE, Corchnoy SB, Christie JM, Lewis JW, Szundi I, Briggs WR, Bogomolni RA. The photocycle of a flavin-binding domain of the blue light photoreceptor phototropin. J Biol Chem. 2001;276:36493–36500. doi: 10.1074/jbc.M103114200. [DOI] [PubMed] [Google Scholar]
- Theveneau E, Marchant L, Kuriyama S, Gull M, Moepps B, Parsons M, Mayor R. Collective chemotaxis requires contact-dependent cell polarity. Dev Cell. 2010;19(1):39–53. doi: 10.1016/j.devcel.2010.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YI, Frey D, Lungu OI, Jaehrig A, Schlichting I, Kuhlman B, Hahn KM. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature. 2009;461:104–108. doi: 10.1038/nature08241. [DOI] [PMC free article] [PubMed] [Google Scholar]