Abstract
A three-dimensional reconstruction of Sindbis virus at 7.0 Å resolution presented here provides a detailed view of the virion structure and includes structural evidence for key interactions that occur between the capsid protein (CP) and transmembrane (TM) glycoproteins E1 and E2. Based on crystal structures of component proteins and homology modeling, we constructed a nearly complete, pseudo-atomic model of the virus. Notably, this includes identification of the 33-residue cytoplasmic domain of E2 (cdE2), which follows a path from the E2 TM helix to the CP where it enters and exits the CP hydrophobic pocket and then folds back to contact the viral membrane. Modeling analysis identified three major contact regions between cdE2 and CP, and the roles of specific residues were probed by molecular genetics. This identified R393 and E395 of cdE2 and Y162 and K252 of CP as critical for virus assembly. The N-termini of the CPs form a contiguous network that interconnects 12 pentameric and 30 hexameric CP capsomers. A single glycoprotein spike cross-links three neighboring CP capsomers as might occur during initiation of virus budding.
Keywords: alphavirus, cryo-reconstruction, glycoprotein, virus assembly, cytoplasmic domain
Introduction
Alphaviruses (family Togaviridae) are a group of ~30, small, enveloped viruses that belong to category B bioterrorism agents and have provided excellent models for studying other enveloped viruses. Sindbis virus (SINV) causes epidemic polyarthritis in humans,1 and studies of this otherwise mild, disease-causing virus provide important clues about the structures, posttranslational modifications, and localization of membrane glycoproteins in the more pathogenic alphaviruses. Some alphaviruses produce asymptomatic infections in humans and have proved to be quite useful as gene expression vectors for mammalian and insect cell lines, but many others produce fever, rash, encephalitis, and arthritis.2 Recent extensive as well as alarming outbreaks of Chikungunya highlight the threat of global travel and underscore the need to better understand the mechanisms of alphavirus infection and transmission.3
SINV virions have an overall spherical morphology with an external diameter of ~700 Å and contain three virally encoded proteins arranged with T=4 icosahedral symmetry. The virion contains 240 copies each (four per asymmetric unit) of three structural proteins, which are distributed in two concentric shells.4 The outer shell is solely composed of glycoproteins E1 [439 amino acids (aa); 47 kDa] and E2 (423 aa; 46 kDa), and the inner shell is primarily made of capsid protein (CP) (264 aa; 29 kDa). These concentric shells sandwich a lipid bilayer membrane that is acquired from the host cell during virus budding and penetrated by a transmembrane (TM) helix from each glycoprotein.1 Virions also contain substoichiometric amounts of a small “6K” (55 aa; 6 kDa) protein.5,6 The single-stranded, positive-sense 11. 7-kb viral RNA genome is encapsidated by the inner shell in the nucleocapsid core (NC). Electron cryo-microscopy (cryoEM) and three-dimensional (3D) image reconstructions of several alphaviruses have revealed significant details about the distribution and structures of the E1, E2, and CP proteins and of the viral membrane.4,7–15
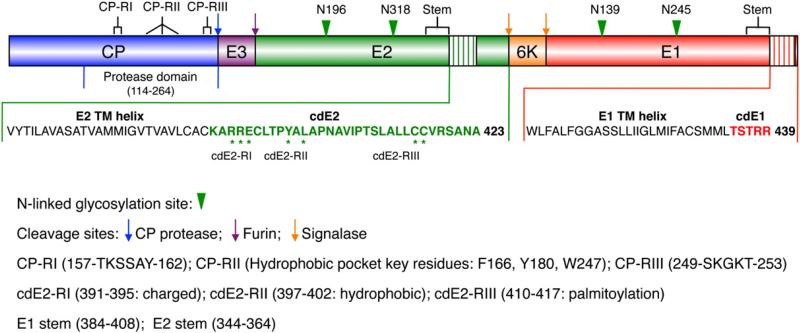
SINV structural proteins (Fig. 1) are translated as a polyprotein (CP–E3–E2–6K–E1) from a subgenomic 26S mRNA and are processed co-translationally.1 CP gets auto-catalytically cleaved from the structural polyprotein, and after a transient interaction with ribosomes, it gets complexed with genomic RNA, resulting in the assembly and accumulation of NC-like particles in the cytoplasm.16 E3 (64 aa; 7 kDa) contains a signal sequence that directs the glycoprotein precursor (E3–E2–6K–E1) into the lumen of the endoplasmic reticulum (ER) where signalase cleaves the precursor at both ends of 6K to generate PE2 (precursor to E2: E3–E2) and E1. PE2 and E1 form heterodimers, and each component is subsequently glycosylated and the heterodimers assemble into trimers of heterodimers.17,18 These trimers are transported through the Golgi to the trans-Golgi network where a furin-like protease cleaves PE2 into E3 and E2, and the trimers then reach the cell surface where they interact with the NC during budding and appear as 80 flower-like spikes in the mature virion.19
Fig. 1.
Organization of the SINV polyprotein. Structural proteins are first translated as a polyprotein, which is then cleaved in a series of distinct processing steps to generate separate CP (blue), E3 (purple), E2 (green), 6K (orange), and E1 (red) proteins. The C-terminal two-thirds of CP adopts a protease domain structure. Each of the E1 and E2 glycoproteins has a large ectodomain containing two N-linked glycosylation sites (green arrowheads), a short stem region, a single TM helix (hatched regions), and a cytoplasmic domain. cdE1 and cdE2 consist of 5 (red characters) and 33 (green characters) residues, respectively. Interactions between cdE2 and CP occur in three regions each in CP (CP-RI, CP-RII, and CP-RIII) and cdE2 (cdE2-RI, cdE2-RII, and cdE2-RIII). Asterisks highlight key residues in the cdE2 that interact with CP as discussed in the text.
These trimeric E1–E2 spikes are the most prominent feature of the alphavirus outer surface.4,11,20,21 The head of each spike adopts a characteristic cloverleaf shape with three petals arranged in a counterclockwise fashion. The stalk of each spike splays tangentially at its base along the outer surface of the viral membrane and forms a tight network of interactions in a thin shell (“skirt”), which is punctuated by small circular openings at the fivefold axes and larger ovoid openings at the twofold axes. Each of the E1 and E2 glycoproteins has a TM helix that anchors the C-terminal ends of the proteins in the lipid bilayer. E1 is primarily noted for its cell fusion activity, and E2 is noted for its role in receptor binding and cell entry.22–24 During infection, exposure of the virion to low pH in the endosome induces dissociation of the E1–E2 heterodimer and E1 then undergoes a series of conformational changes, resulting in the formation of a highly stable E1 homotrimer. In vitro experiments have shown that liposome-stimulated trimerization of E1 represents the fusion-active conformation of the viral spike.25–28 E1 then associates with the target membrane via the putative fusion peptide, and the dual interaction of E1 with the target and virus membranes drives the fusion reaction.29
In SINV, the cytoplasmic domains of E1 and E2 (cdE1 and cdE2) contain 5 and 33 residues, respectively.9,30 Previous mutational analyses had suggested that cdE1 had no direct role in virus budding.31 cdE2 has three conserved Cys residues that are modified by palmitoylation and a unique Tyr residue that is phosphorylated transiently.30,32,33 The detailed structures of these cytoplasmic domains remain undetermined.
Alphavirus CP interacts with genomic RNA to form the NC. The first 80 residues in CP are involved in nonspecific binding and charge neutralization of the genome; residues 81 to 113 bind to the encapsidation signal on the viral RNA; and residues 99 to 113 bind to the large ribosomal subunit.34–37 The crystal structures of several alphavirus CPs have been determined, and although the 113 N-terminal residues are disordered, residues 114–264 adopt a chymotrypsin-like fold.38–40 cdE2 has been shown to interact specifically with CP and forms a link between the outer glycoprotein and inner protein shells.41 Mutagenesis studies coupled with modeling experiments were used to predict that residues Y400 and L402 in cdE2 bind to the hydrophobic pocket in CP as a critical step in the virus budding process.42,43 Many studies have explored interactions between cdE2 and CP to determine their influence on virus assembly, infection, and fusion,44–46 but none of them have provided direct structural evidence about the details of these interactions.
Here, we have employed cryoEM, 3D image reconstruction, and site-directed mutagenesis to explore the detailed interactions between cdE2 and CP. Prior to this, we had generated a pseudo-atomic model for the Sindbis virion based on a 9-Å cryo-reconstruction,9 coupled with information gleaned from the crystal structures of component viral proteins such as the protease domain of the SINV CP47 and the ectodomain of the Semliki Forest virus E1 protein.48 Until very recently, the structure of E2 remained a major missing piece of the puzzle. The report of crystal structures for the low-pH form of the SINV E1–E2 heterotrimer49 and that for neutral-pH forms of the precursor p62–E1 heterodimer and of the mature E3–E2–E1 complex for Chikungunya virus (CHIKV)50 provide the first detailed views of the E2 ectodomain. A recent cryoEM structure of Barmah Forest virus showed density for the E2 endodomain and suggested contact between the end of the TM helix of E2 and CP.15 We now report a cryo-reconstruction of SINV at 7.0 Å resolution, and armed with crystal structure data for CP, E1, and E2 and the results of several structure-guided muta-genesis experiments, we have built a complete model of the E1–E2 glycoproteins. This model includes a detailed structure of cdE2 as it exits from the membrane and then enters the CP hydrophobic pocket where it makes extensive interactions with CP. Our mutagenesis experiments confirm that these interactions are important in virus assembly, and the new SINV model provides insights about intersubunit interactions among the SINV structural proteins that drive virus assembly and budding. Of note, after the submission of this report, advance online publication of a separate cryoEM and modeling study of Venezuelan equine encephalitis virus (VEEV)51 showed that VEEV and SINV, which are representative members of the New and Old World alphavirus lineages, have remarkably similar structures, and the two independently derived pseudo-atomic models are quite compatible. The only significant difference between VEEV and SINV is that VEEV was shown to contain substoichiometric amounts of E351 whereas mature SINV contains no E3.52
Results
SINV virion cryo-reconstruction at 7 Å resolution
The map of SINV E2-N318Q, computed from ~24,000 images of unstained, vitrified virions at a resolution of 7 Å (Materials and Methods), reveals improved structural details (e.g., for cdE2) when compared to our previous 9-Å map.9 The virion has distinct surface protrusions and well-separated density layers within the virion (Fig. 2). The E1–E2 glycoproteins form the outer spike and skirt structures (blue and magenta, respectively, in Fig. 2). The host-derived lipid bilayer lies just below the skirt and is penetrated by the E1–E2 TM helices (both bilayer and helices are in cyan in Fig. 2). The C-terminal protease domains of the 240 CPs (green in Fig. 2b) are arranged inside the inner leaflet of the bilayer and form a highly ordered arrangement of pentameric and hexameric capsomers in the nucleocapsid. These in turn surround a contiguous layer of density (yellow in Fig. 2b) that is mainly composed of the flexible CP N-terminal region (amino acids 1–113). The remaining density at the center of the particle (red in Fig. 2b) exhibits no sign of icosahedral order and is attributed primarily to the single-stranded RNA genome but may include some portion of the highly basic CP N-termini.
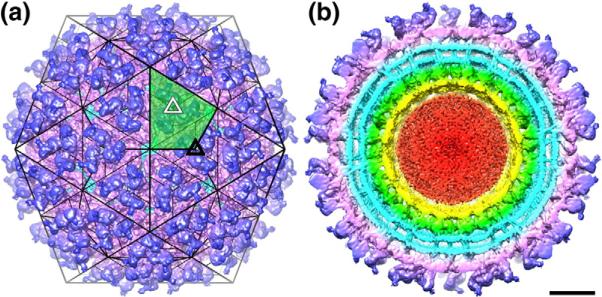
Fig. 2.
Overview of the SINV structure. (a) Shaded-surface representation of SINV cryo-reconstruction, color cued radially to emphasize the layered nature of the alphavirus structure (see below). The 80 prominent, trimeric spikes are positioned at threefold (black triangle) and quasi-threefold (white triangle) points in a T=4, icosahedral lattice (black lines). One of the 60 asymmetric units is highlighted by green shading, and thin black lines approximate the boundaries of individual subunits. (b) Same as (a) but with the front half of the reconstructed density map removed to reveal internal features of the SINV structure. The outer protein shell, composed solely of the E1 and E2 glycoproteins, is divided morphologically into spikes (blue) and the underlying skirt (magenta). The outer and inner leaflets of the viral membrane appear as two concentric, roughly spherical shells of density (cyan), crossed by distinct E1 and E2 TM helices (also in cyan). The ordered, protease domains of the CP (green) lie beneath the membrane and surround a layer of density (yellow) that is attributed primarily to the N-terminal segments of the CP. The remaining disorganized density (red) at the core of the virion is attributed primarily to the RNA genome. The scale bar represents 100 Å.
A complete model of the SINV glycoprotein structure in virions
Density for each glycoprotein was clearly defined in the 7-Å SINV cryo-reconstruction. The low-pH crystal structure of the ectodomain portion of the trimeric SINV E1–E2 heterodimer49 docked nicely into the cryoEM map, although it was apparent that E2 and E1 are more closely associated in virions compared to those seen in crystals. Consequently, we fit the E1 and E2 atomic models as separate rigid bodies (Materials and Methods), with E1 retained in its initial position and portions of E2 moved up to 5 Å closer to E1 as compared to the crystal structure (Fig. 3a). The tight association we observed between E1 and E2 in SINV is the same as that found in the neutral-pH crystal structure of the CHIKV glycoproteins.50 The tight association between E1 and E2 is lost at low pH,49 which is consistent with a previous report that showed that the E1–E2 heterodimer dissociated upon low-pH treatment.25
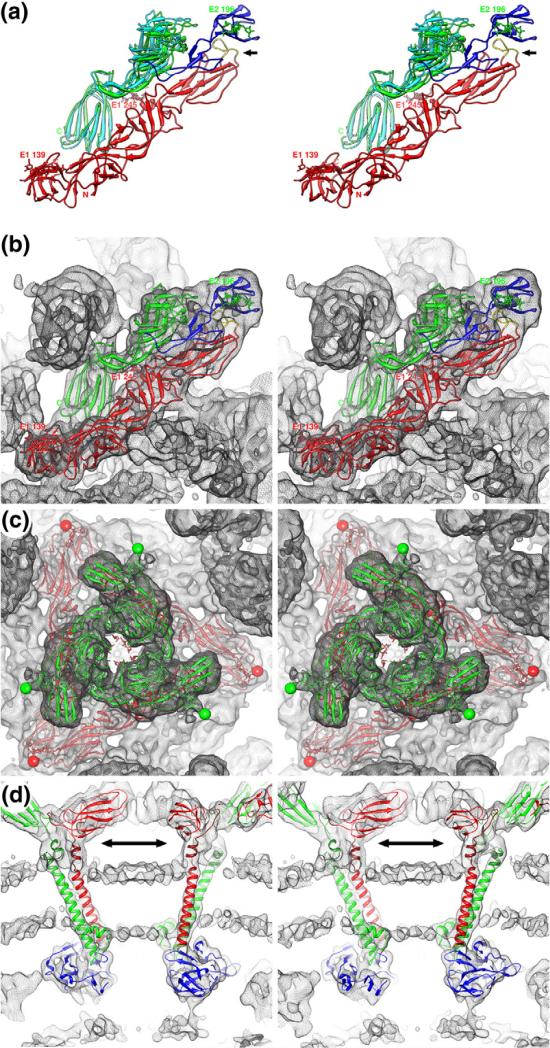
Fig. 3.
Atomic modeling of SINV cryo-reconstruction. (a) Stereo ribbon diagram representation of the E1–E2 heterodimer. Highlighted in the E1 molecule (red) are the two glycosylation sites (N139 and N245) and the fusion peptide (yellow; black arrow). Two models of the E2 structure (A and C domains) are shown: one found in crystals (cyan) and the other obtained after fitting it into the SINV cryoEM density map (green). The E2 B domain (blue), a homology model derived from the CHIKV E2 crystal structure (see Materials and Methods), is shown in its fitted position, where it lies directly above and covers the E1 fusion peptide. The single E2 glycosylation site (N196) in the E2-N318Q mutant is labeled. (b) Same as (a) for the final E1–E2 heterodimer model fitted into the SINV cryoEM density map (grey mesh). (c) Same as (b) but with a complete (E1–E2)3 model fitted into the density map and viewed from above a threefold symmetric spike. The N196 glycosylation sites appear as clearly defined protuberances at the top of the spike (green spheres), the E1-N139 sites are at the far corners of the skirt (red sphere), and density at the E1-N245 sites (center of the spike) is not as well defined as for the other two sites. (d) Pseudo-atomic models of the E1 (red) and E2 (green) stem and TM helix regions, and the CP protease domain (blue) fitted into the SINV density map. At the chosen density threshold, the membrane bilayer shows up as a pair of thin layers that pucker inwards towards the virion center (bottom of panel). The stem regions (double arrow) are depicted in darker colors to distinguish them from the TM regions of the models.
The structures of only two (A and C) of the three immunoglobulin-like domains of the SINV E2 ectodomain were resolved in the crystal structure.49 However, the crystal structure of all domains of the homologous E2 glycoprotein ectodomain from CHIKV50 provided a template from which a homology model of the SINV E2 B domain was built and fitted to the SINV virion density map. As expected and as was shown in the CHIKV crystal structure,50 the E2 B domain drapes over the E1 fusion peptide (yellow backbone trace in Fig. 3a) in the mature virion and prevents premature virus fusion. The SINV E2-N318Q mutant used in this study lacks one of the four glycosylation sites present in wild-type (WT) virions. The three remaining sites showed extra densities at the appropriate locations in the cryoEM map. N139 in E1 lies at the top of the interface between E1 domains I and III, and N245 lies in the middle of E1 domain II (Fig. 3a). N196 in E2 is at the tip of the E2 B domain and appears in the density map as a small surface protrusion on the top of the spike (Fig. 3b and c).
The TM regions of the two glycoproteins are more clearly defined in the new SINV cryoEM reconstruction compared to that seen in the previous 9-Å reconstruction.9 The eight TM helices in the virion asymmetric unit (four each for E1 and E2) appear as distinct columns of density that traverse the entire viral membrane in E1–E2 pairs (Fig. 3d). The E1 and E2 helices in each pair are closely juxtaposed and aligned nearly parallel with one another and make multiple contacts. The existence of such contacts is consistent with a previous report showing that interactions between the E1 and E2 TM helices play a role in virus budding and fusion.53 Our SINV cryo-reconstruction lacks sufficient resolution to build an accurate, full atomic model of the TM helices, but knowledge of the sequence coupled with the observed density features suggests that they belong to the Cluster 4 group of TM helix–helix interactions (parallel helices with an ~20° crossing angle).54 As was indicated for other TM helices surveyed by Moore et al., the pairing interactions of TM helices in SINV might facilitate the formation of glycoprotein heterodimers in the ER.54 The presence of 240 pairs of TM helices in alphavirus virions perturbs the otherwise spherical membrane bilayer (~500 Å diameter) as evidenced by significant inward puckering of the bilayer near the fivefold axes where five TM pairs cluster in a circle above each CP pentamer (Fig. 3d).
Each alphavirus glycoprotein has a stem region that connects the ectodomain to the TM helix (Fig. 1). The E1 stem facilitates the formation of the E1 homotrimers during membrane fusion.55 The recently solved crystal structures of the SINV and CHIKV glycoproteins lack these stem regions,49,50 but our SINV cryo-reconstruction shows density for the entire length of both glycoproteins, including the stems. Our fit of the SINV E2 ectodomain crystal structure into the cryoEM map positioned the last residue of the model (S343) close to the outer leaflet of the membrane. The cryoEM map included a clear path of unassigned density that proceeds from S343 to the start of the E2 TM helix at V365. This density nicely accommodated a model of the 21-residue stem region (amino acids 344–365) (Fig. 3d). The larger, 25-residue E1 stem (amino acids 384–408) was modeled in two separate segments (Materials and Methods) that together formed a connection between the last residue of the E1 ectodomain and the first residue of the E1 TM helix (Fig. 3d).
Organization of the NC
The 240 glycoprotein heterodimers in the outer shell and 240 CP monomers in the NC core of all alphaviruses are arranged in T = 4 icosahedral lattices, yet these similar arrangements yield distinctly different morphological units. Indeed, the glycoproteins form 80 trimeric spikes, whereas the CPs cluster as 12 pentameric and 30 hexameric capsomers. The crystal structure of the protease domain of the CP (amino acids 114 to 264)47 docked quite well as a rigid body into the cryoEM map between radii of ~172 and 212 Å for each of the four CPs in the asymmetric unit (Fig. 4a). The fit of the CP subunit positioned it with its hydrophobic pocket facing the inner leaflet of the lipid bilayer, and this orientation readily allows interactions with cdE2 as it protrudes radially inwards from the membrane towards the CP. Subunit interactions within cap-somers are more extensive and presumably stronger than the interactions between capsomers. Indeed, the intracapsomer interactions in both types of capsomer appear as thick bridges of density, and the pseudo-atomic model of each capsomer includes a putative salt bridge between R174 in one subunit and E236 of a neighboring subunit (blue and red spheres, respectively; Fig. 4a). Weaker intercapsomer interactions are revealed by thin extensions of density that project from the edge of one capsomer towards its neighbor (Fig. 4a). The likely contributors for these weak interactions are two charged residues (E121 and K153) from two neighboring subunits. At lower radii (b170 Å), an extensive and thick layer of density (yellow, Fig. 2b) lies in stark contrast to the limited set of intercapsomer interactions that occur between the CP protease domains. Close inspection of the density map at higher thresholds revealed that this layer has an extensive network of contacts tethering the capsomers together (Fig. 4b). Given the position of this layer, it is likely made up of the first 113 disordered residues of the CP. In fact, a short, contiguous region of density that protrudes radially inwards from the first ordered residue (R114) was modeled as a stretch of eight residues from D113 to M106. A similar region in VEEV was modeled as an α-helix51 (Fig. S1), but there is no evidence from our reconstructed density map or on the basis of secondary-structure prediction to suggest that this region is α-helical in SINV. Beyond this 8-residue stretch, the remaining 105 residues and some of the genome are left to form the network that interconnects the capsomers (Fig. 4c).
Fig. 4.
SINV pentamer model. (a) Five copies of the CP protease atomic model fitted into the SINV density corresponding to the pentameric capsomer as viewed from the center of the virion. The ribbon model is color coded (blue– red rainbow) starting at CP residue 114. Strong intracapsomer subunit–subunit interactions are observed and include a potential salt bridge between R174 (blue sphere) and E236 (red sphere). (b) Same as (a) but including the layer of unmodeled density that lies immediately below the ordered protease domain layer and is ascribed primarily to the disordered, 113-residue, N-terminal region of CP. This layer appears as a network of density that supports both intra- and intercapsomer interactions. (c) Side view of the SINV map with density attributed to the CP protease domains subtracted away, to reveal a stretch of density leading from residue 114 (blue spheres) to the network below and modeled as eight residues ending at residue 106 (yellow sphere). The additional difference density at low radii in this view was interpreted as including most of the remaining residues of the CP N-terminus (amino acids 1–105). Left pair of arrows highlights the upper and lower bounds of the region of the map shown in (a), whereas the right pair highlights the region shown in (b).
Identification and modeling of cdE1 and cdE2
Clearly defined regions of unassigned density were present in the cryoEM map that filled some of the gap between the high radii surface of the CP and the inner leaflet ends of the TM helices. In agreement with the previous studies that had suggested the existence of interactions between cdE2 and CP,41,45 we modeled the bulk of this unassigned density (all 33 residues of cdE2) as a helix–loop–helix structure (dark green; Fig. 5). The cdE2 backbone follows a path from the end of the E2 TM helix towards the CP and into its hydrophobic pocket and then reemerges where it folds back and binds the inner surface of the lipid bilayer. The shape and volume of the density assigned to cdE2 are consistent with the predicted secondary structure for cdE2. The five residues of cdE1 were modeled to fit the unassigned density, extending from the end of the E1 TM helix onto the top of the CP (Fig. 5). Hence, we were able to model all residues of cdE1 and cdE2 to the unassigned densities in this region of the virion map and to follow the paths of these cytoplasmic domains from the respective ends of the TM helices at the inner leaflet of the viral membrane and down to contact sites on the CP surface. In contrast to the recently reported structure of Barmah Forest virus,15 our structure determination of SINV as well as that of VEEV51 clearly reveals the entire path of cdE2 and shows that it penetrates the hydrophobic pocket of CP and makes extensive contact with CP. For SINV, these three attributes were confirmed by the structure-guided mutagenesis studies described below. The Barmah Forest virus report also suggested that an interaction occurs between the endodomain of E2 and the TM portion of E1 at the lipid bilayer. Our structure identifies cdE1, which extends beyond the TM helix and makes contact with the CP. Consistent with the Barmah Forest virus structure, cdE2 in SINV lies in close proximity to E1 from a neighboring subunit (Fig. 5); however, our interpretation of the role of this interaction differs (see Discussion).
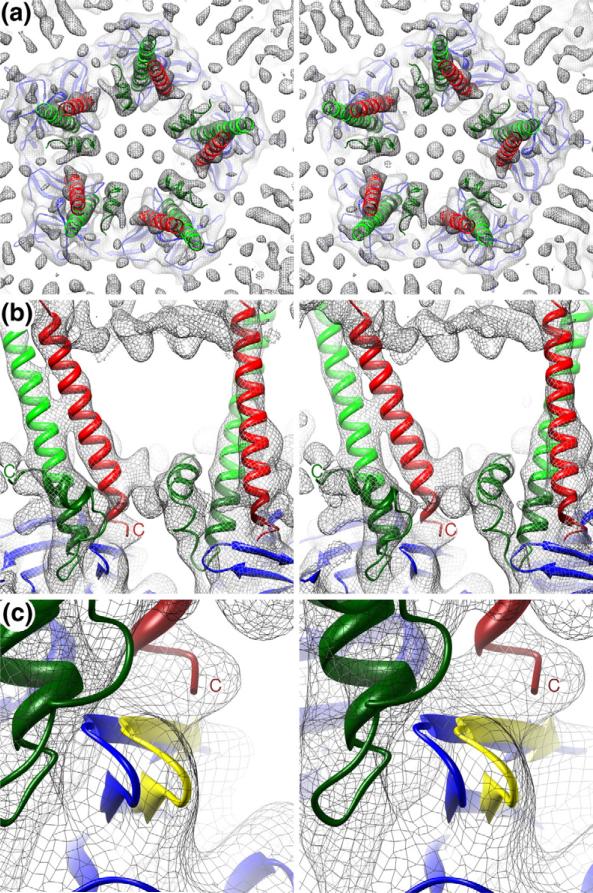
Fig. 5.
TM helices and cdE1 and cdE2. (a) Stereo view of a slab of the SINV density map (gray mesh) that, for clarity, includes just the inner half of the membrane and the protease domain of the CP pentamer and viewed towards the center of the virion. The fitted E1 (red) and E2 (green) TM helix models, the cdE2 model (dark green), and CP (blue) are shown. (b) Close-up, side view in stereo of a small volume of the density map showing two pairs of TM helices, the cdE1 and cdE2 models, and the top portion of CP. Unassigned density at the top and center of the panel is attributed to the phospholipid head groups in the outer and inner leaflets of the bilayer, respectively. (c) Same as (b) showing a highly magnified view of a loop in CP (amino acids 248–252) that moves several angstroms from its position in the crystal structure (blue) to one that fits the density map (yellow). This movement opens the CP pocket and allows cdE2 to bind to it.
Our modeling analyses also revealed that conformational changes occur in CP, most noticeably in the proximity of the hydrophobic pocket. The SINV map clearly showed that the F2–G2 loop (248-NSKGK-252) at the edge of the hydro-phobic pocket in CP47 shifts ~3.5 Å away from the position it adopts in the crystal structure and thus away from the pocket (from blue to yellow in Fig. 5c). Movement of the F2–G2 loop may expand access to the pocket and thereby accommodate cdE2 insertion and binding. This region of CP is known to be flexible as it adopts slightly different conformations in different alphavirus CP crystal structures.42
Contact regions in cdE2 and CP
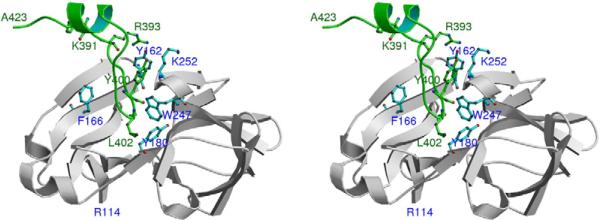
The bulky aromatic residue, W247, divides the CP hydrophobic pocket into two similar-sized cavities, with F166 in the upper half and Y180 in the lower half (Fig. 6). The pocket contains several conserved residues (Table 1) that form hydrophobic interactions with residues of cdE2.41,43 In the original model proposed by Lee et al.,41 Y400 of cdE2 was believed to interact with Y180 in the lower half of the pocket.41 However, the new SINV map showed clearly defined cdE2 density that followed an unambiguous path into the CP hydrophobic pocket, and this enabled us to model residues Y400 and L402 as contacting the upper and lower halves of the pocket, respectively (Fig. 6). Close inspection of the cdE2–CP interactions revealed three contact regions each in cdE2 and CP (Fig. 1), which we designate with Roman numeral labels. In cdE2, these include cdE2-RI (391–395), cdE2-RII (397–402), and cdE2-RIII (410–417). CP contains CP-RI (157–162), CP-RII (W247, F166, and Y180), and CP-RIII (249–253). CPRI and CP-RIII comprise part of two exposed loops on the surface of the protein just outside the hydrophobic pocket.
Fig. 6.
Interactions between cdE2 and CP. Stereo view shows the full-length cdE2 model (from K391 to A423) and the path it follows into and back out of the CP hydrophobic pocket. Residue W247 divides the pocket into upper and lower compartments, which contain F166 and Y180, respectively. The loop in cdE2 starting at R393 dives down into the pocket with Y400 and L402 making contacts with CP residues inside the upper and lower compartments of the pocket. Upon exiting the pocket, cdE2 forms a short helix that approaches the inner membrane and ends at C-terminal residue A423. The N-terminal residue in the CP crystal structure is R114.
Table 1.
Multiple sequence alignment of alphaviruses shows conservation in the CP and cdE2 contact regions
| Antigenic complexa | Virusa | CP–RI (157–162)b | CP–RIII (249–253)b | cdE2–RI (391–395)b |
|---|---|---|---|---|
| BF | BFV | KKSSKY | KDMVT | VARTK |
| EEEV | KKASIY | QKGVT | RTRNL | |
| MIDV | KKSSKY | KDMVT | SARNK | |
| SF | CHIKV | KRSSKY | KDIVT | CARRR |
| MAYV | KKSSKY | KDMVT | VARNK | |
| ONNV | KRSSKY | KDIVT | CARRR | |
| RRV | KKSSKY | KDMVT | TARRK | |
| SFV | KKSSKY | KDMVT | AARSK | |
| VEE | VEEV | KKASKY | EKGVT | KSRAS |
| WEE | AURA | TKSSSY | NKKGA | KARRD |
| SINV | TKSSAY | SKGKT | KARRE | |
| WHAV | TKSSAY | AKGKT | KARRD | |
| WEEV recombinant | WEEV | KKASMY | QKGVT | KARRD |
| HJV | KKSSKY | QKGVT | KARRD | |
| FMV | KKASIY | QKGVT | RARRD | |
| Unclassified | SPDV | LEAAAY | KKMKA | CSRVR |
Standard abbreviations of the virus names and classification according to its antigenic cross-reactivity.56
SINV numbering. Sequence alignments for the contact regions in alphavirus CP and cdE2 with positively and negatively charged residues highlighted in blue and red text, respectively. The Y162 of CP-RI and R393 of cdE2 are conserved in all alphaviruses. Multiple sequence alignments of cdE2 and full-length CP were carried out with Clustal X57 (Figs. S2 and S3).
Density attributed to cdE2 exits from the inner membrane starting with residue K391. The rest of the highly charged cdE2-RI lies near the outer surface of the CP pocket and contacts the two surface-exposed loops that contain CP-RI and CPRIII. RII in cdE2 forms hydrophobic interactions with Y400 and L402 in the two halves of the CP pocket. Next, a stretch of six cdE2 residues (404-PNAVIP-409) emerges from the pocket and the backbone path changes direction twice, possibly influenced by the prolines. The next portion of cdE2 (cdE2-RIII), which contains the signal sequence for translocation of 6K, initially inserts into the ER membrane and later gets reoriented back to the cytoplasmic leaflet of the lipid bilayer.58 The two palmitoylated cysteines in cdE2-RIII tether the C-terminus of cdE2 to the cytoplasmic side of the membrane, as is consistent with earlier studies about its proximity to the membrane.32,47
Analysis of the pseudo-atomic model that we derived as described above led to the identification of several potential residues that contribute to interactions in the contact regions. To confirm and further explore the roles of how these specific residues might affect virus growth, replication, translation, processing, assembly, and release, we designed and performed extensive mutational analyses of cdE2 and CP as described in the following four sections.
Mutations in the contact regions affect virus growth
We generated mutations in all three regions in cdE2 and CP, and this led to several distinguishable plaque phenotypes (Table 2). WT SINV forms large plaques 48 h post-infection whereas mutants form medium, small, very small, or no plaques depending on the severity of the defect (Table 2). Those viruses that gave rise to the very small plaques grew extremely poorly and could not be recovered. Our analysis of the different mutant phenotypes and these altered growth characteristics allowed us to identify key residues in the cdE2 and CP contact regions. For example, cdE2-RI contains a contiguous string of three charged residues (R393, R394, and E395), with R393 being completely conserved in all alphaviruses (Table 1) and our SINV pseudo-atomic model suggested that these residues interact with CP-RI and CP-RIII. Most of the R393 mutants exhibited altered virus production (Table 2). Defects caused by substitutions at R394 and E395 were tolerated, but deletion of either R393 or E395 did significantly affect virus production. Of note, a K393 revertant appeared with the R393E mutant, thereby restoring the positive charge at position 393. Similarly, multiple alanine substitutions in cdE2-RII affected virus production, which is consistent with reports demonstrating the importance of residues that bind to the CP hydrophobic pocket.43,59
Table 2.
Summary of the phenotypic characteristics of CP and cdE2 mutants
| CP |
cdE2 |
||||||
|---|---|---|---|---|---|---|---|
| CP–RI (157–162) |
CP–RII (Hydrophobic pocket) |
CP–RIII (249–253) |
cdE2–RI (391–395) |
||||
| Virusa | Plaque sizeb | Virusa | Plaque sizeb | Virusa | Plaque sizeb | Virusa | Plaque sizeb |
| TKSSAY | lp | SKGKT | lp | KARRE | lp | ||
| AAAA-- | mp | Y180A, S182A | sp | -ΔΔΔ- | np | --AAA | vsp |
| -ΔΔΔΔΔ | np | M132A | sp | A---A | mp | --AA- | vsp |
| --AA-- | mp | E133A | mp | -A-A- | sp | ---AA | sp |
| --RR-- | sp | K135A | sp | -E-E- | vsp | --A-- | vsp |
| --ΔΔ-- | np | -D-D- | vsp | --E-- | vsp | ||
| -E---- | mp | -A--- | mp | --EE- | vsp | ||
| --R--- | sp | -G--- | mp | ----A | sp | ||
| ---R-- | mp | -M--- | mp | ----K | mp | ||
| -----F | lp | -E--- | mp | 391^A | sp | ||
| -----A | sp | --A-- | sp | 392^A | sp | ||
| -----K | vsp | ---A- | sp | 394^A | vsp | ||
| -Δ---- | sp | ---E- | vsp | 395^A | vsp | ||
| -----Δ | vsp | ---D- | vsp | -Δ--- | vsp | ||
| ---R- | lp | --Δ-- | vsp | ||||
| ---Δ- | sp | ----Δ | vsp | ||||
^, insertion; Δ, deletion.
Plaque sizes relative to the WT are as follows: lp, large plaque (3.5–4 mm); mp, medium plaque (2–3 mm); sp, small plaque (1–2 mm); vsp, very small plaque (<1 mm); np, no plaques. Plaque sizes corresponding to the diameter of plaques are measured subsequent to diethylaminoethyl-dextran transfection of the in vitro transcribed SINV RNA into BHK-15 cells after 48 h at 37 °C. Very small plaques are pinpoint in size and cannot be propagated.
Most of the hydrophobic pocket residues (CP-RII) and their interactions with cdE2-RII have been studied extensively.41,43 Residues in cdE2-RIII comprise the signal sequence that acts to translocate 6K during polyprotein processing.1,60 Our experiments showed that multiple alanine substitution of cdE2-RIII produced a lethal phenotype. We therefore focused our attention on identifying and characterizing interactions that occur in cdE2 and CP regions I and III. Mutation of Y162 in CP-RI had a pronounced effect on plaque phenotype. For example, Tyr substitution (Y162F) yielded the large-plaque phenotype whereas Ala (Y162A) and Lys (Y162K) substitutions produced small-plaque and lethal phenotypes, respectively (Table 2). These results suggest that an aromatic residue plays a critical role at this position. The double-alanine mutant with substitutions at positions 250 and 252 in CP-RIII (K250A, K252A) produced small plaques, but no virus could be recovered from the reverse-charge, double-glutamic-acid mutant K250E, K252E. Notably though, as a consequence of a single-nucleotide change (from GAG in E252 to AAG in K252), this double mutant readily converted (in 12 h) to E250, K252.
On the basis of all of the single-site alanine substitutions that we examined, three critical residues were identified. These included Y162 in CP-RI, K252 in CP-RIII, and R393 in cdE2-RI. The intersubunit interaction seen in the SINV CP crystal structure, involving the N-terminal arm of one molecule and the pocket of another molecule, suggested that the conserved E111 in the N-terminal arm forms a salt bridge with K252.41 This is analogous to the interaction that cdE2 makes with the hydrophobic pocket of CP in the virus and we propose that E395 of cdE2-RI forms a salt bridge with K252 of CP-RIII. The coupled pair of charged residues (cdE2 E395 and CP K252 in SINV) exhibits co-variance among the other alphaviruses. That is, depending on the nature of the residue and charge in the critical cdE2 position, there is an oppositely charged residue in the critical CP-RIII position. Hence, this salt bridge appears to be a highly conserved feature and thus a critical component of all alphaviruses. Furthermore, it is noteworthy that R393 of cdE2-RI and Y162 of CP-RI are invariant in all alphaviruses (Table 1), and our model places the Cα positions of these residues in close proximity, which suggests that they may form another key interaction.
Mutations of three critical CP–cdE2 contact residues affect virus replication
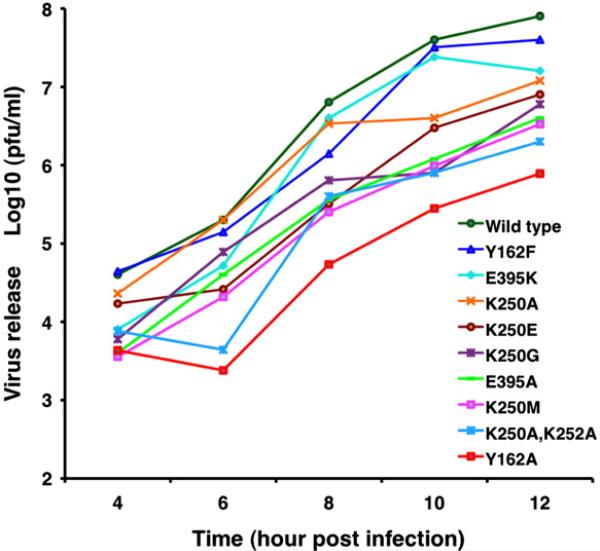
One-step growth curve analysis was used to examine the properties of all the recovered viable virus mutants (Fig. 7). Notably, growth of the G251A mutant was 30-fold less than that of WT, suggesting that flexibility of the F2–G2 loop may be important for virus assembly. Growth of the K250A, K252A double-alanine mutant was down fourfold compared with that of the WT virus, and the single-alanine mutant Y162A was reduced ~70-fold. The growth kinetics of potentially important residues is shown in Fig. 7 only for a small subset of those actually measured. Based on all our mutagenesis experiments, it is quite clear that the observed defects depend on the amino acid substitutions. With the exception of E395K, we did not recover any significant amount of virus from any of the cdE2-RI mutants (Table 2). The R393E (CGC/GAA) mutant rapidly reverted to E393K (GAA/AAA), which restored the positive charge at that position.
Fig. 7.
One-step growth analysis of a subset of CP and cdE2 mutants. BHK-15 cells were infected at a multiplicity of infection of 2 with WT or mutant virus, and the supernatant was collected at hourly intervals. Each experiment was conducted in triplicate, and the virus released in the supernatants at 4, 6, 8, 10, and 12 h post-infection was determined by plaque assay. The mean value of each virus titer at each time point is plotted in the growth curves. For clarity, the labels for the growth curves are arranged from top to bottom to correlate with the amount of virus release at the 12-h post-infection time point.
CP and cdE2 mutants exhibit translation, processing, and assembly defects
Western immunoblot analysis was used to detect defects in protein production in the CP and cdE2 mutants (Fig. S4). For CP-RI and CP-RIII mutants, reversal of charge and residue deletions resulted in adverse defects in folding and processing of the polyprotein. One of the catalytic triad, single-site mutants (D163A) was included in the study as a negative control to allow us to compare the defects in protease activity of the mutants. Except for the region III mutants, most of the cdE2 mutants exhibited no defects in polyprotein processing. CP mutants that showed defects in virus growth but produced detectable levels of CP compared with WT virus were used in the core accumulation assay (Fig. S5). Alanine mutants Y162A and K250A, K252A accumulated cores at near-WT levels. Charge reversal of CP region III did cause defects in NC formation. The Western blot and core accumulation experiments demonstrated that the effects of mutations at critical residues R393 of cdE2 and Y162 and K252 of CP were not caused by defects in poly-protein processing or NC assembly. However, the double-Asp substitution mutant K250D, K252D showed slight defects in core assembly (Fig. S3).
The membrane localization and surface expression of E2 in all the defective CP and cdE2 mutants were analyzed by immunofluorescence assay (IFA) and confocal microscopy (Fig. S6). Most of the cdE2 mutations in regions I and II maintained the expression of E2 on the plasma membrane, whereas mutations in region III negatively affected the surface expression of E2. IFA and confocal microscopy experiments did confirm that severely affected CP mutants such as K250E, K252E do express CP in the cytoplasm, and E2 does traffic normally to the plasma membrane yet the mutants failed to produce infectious particles. As such, these mutants were classified as having defects in NC formation and interaction between NC and cdE2.
Release of virus mutants is greatly reduced
Virus released into the media from transfected cells, as measured by quantitative reverse transcriptase (qRT)-PCR, showed that the budding mutants were significantly reduced in virus yield when compared with the WT virus. Cycle threshold values determined by qRT-PCR (see Materials and Methods) were converted into the number of RNA molecules by using a standard curve based on known amounts of in vitro transcribed, full-length RNA. The number of RNA molecules released into the media in the CP mutant K250D, K252D was comparable to that in the negative control (D163A), which was significantly reduced relative to the positive WT control. The results of qRT experiments with cdE2 and CP mutants show an average ~104- fold reduction in virus release at 12 h for all non-budding mutants, when compared with the number obtained (4.4×107) for the WT virus (Fig. S7). The background level of RNA molecules (~103) released into the media for the mutants could be due to residual RNA from the electroporation, or to cell lysis, or to very limited virus budding.
Discussion
Our new cryoEM reconstruction of SINV and the recent determinations of the E1 and E2 ectodomain crystal structures have allowed us to build essentially complete models for both glycoproteins in the virion. On the inner side of the membrane, the identification of the E1 and E2 cytoplasmic domains and their interactions with the CP reveal new insights into alphavirus structure and the link between glycoproteins and CP. The mutagenesis studies permitted an interrogation of the structural model, and the resulting data provided a means to improve our understanding of virus assembly.
Construction of the E1–E2 model
The crystal structures of the E1 and E2 ectodo-mains fit well after placement within the cryoEM density, suggesting high fidelity and reliability of the virus cryoEM structure. Minor adjustments were made to bring E1 and E2 closer together in the virion, indicating that they interact closely after assembly. The apparent increased contact between the two glycoproteins at neutral pH as opposed to low pH suggests that tighter interactions are required for virion stability. The paths of the glycoprotein stem regions were delineated by following density features in the cryo-reconstruction that connected the ends of the ectodomains to the beginning of each clearly defined TM helix. After fitting the CP X-ray structure in the nucleocapsid density, we unambiguously interpreted the extra densities emerging from the TM helices as cdE2 and cdE1. Both of these cytoplasmic domains were seen to interact with CP, demonstrating a multivalent mode of binding of glycoproteins to CP.53 These contacts are likely important during virus assembly and budding and provide interactions that stabilize the mature virion by tethering the outer and inner protein shells. All of the above conclusions concerning SINV are also consistent with the recent study of VEEV.51
Mutants of cdE2 and CP confirm subunit interactions predicted from the SINV model
We employed extensive structure-guided mutagenesis to interrogate the roles of individual amino acids in cdE2 and CP in various stages of virus assembly (Table 2). Mutations in cdE2-RII were already known to affect virus budding.41,43 Our analyses confirmed that hydrophobic residues in cdE2-RII and CP-RII are critical for efficient virus budding. Substitution of these residues by alanine produced glycoproteins that were still processed and transported to the plasma membrane, but there is a major defect in virus release presumably as a consequence of weaker or even loss of interactions between cdE2 and CP (Table 2).
Charged residues in cdE261,62 and the distance spanned between the lipid bilayer and hydrophobic pocket are both important for normal virus budding.63 Our mutational analyses of cdE2-RI confirmed that the charged residues of cdE2 (R393, R394, and E395) and their locations are critical for interaction with CP. Indeed, virus production was significantly reduced as a result of charge reversals at these residues (Fig. S7). Alanine substitution and reversal of charge for the conserved R393 residue led to revertants. The severely affected phenotype that accompanies insertions or deletions in region I of cdE2 suggests that the spatial orientation of charged residues is critical for proper virus budding. Mutation of the conserved residues in cdE2-RIII (Fig. 1) mainly affects processing and transport of E2, as these residues are part of the signal sequence for 6K translocation.60
The highly conserved Y162 of CP-RI forms a critical interaction with cdE2. Though the Y162F mutant grew like WT, Y162A formed small plaques and Y162K had a severely affected phenotype. This suggests that an aromatic side chain at position 162 is required to facilitate stacking interactions with R393 in cdE2-RI. Given that the CP-RI region is adjacent to D163, a component of the catalytic triad,64,65 some of the deletions and charge reversals introduced into CP-RI affected protease activity. This led to processing defects that were detected by Western analysis and IFA (Figs. S2 and S4). Characterization of defects caused by various mutations revealed a subset of mutants that exhibited normal NC formation, glycoprotein processing, and transport to the plasma membrane but resulted in release of very few particles. Presumably, in these mutants, effective interactions between CP and cdE2 are not produced. The number of viral RNA molecules released into the media for these mutants, measured by qRT-PCR, showed a significant reduction comparable to that of a negative control (D163A).
Nucleocapsid core
The NC structure is well resolved in the cryoEM density map. The crystal structure of the SINV CP protease domain47 fits nicely inside the SINV cryoEM density map as one rigid body. CP subunits display strong intracapsomer interactions but weaker intercapsomer interactions (Fig. 4). Two surface loops of CP appear to change conformation owing to interactions they make with surrounding proteins in the virion. The first loop (F2–G2) expands to adapt to cdE2 binding inside the hydrophobic pocket. The second (A1–B1) forms subunit interactions between neighboring capsomers.
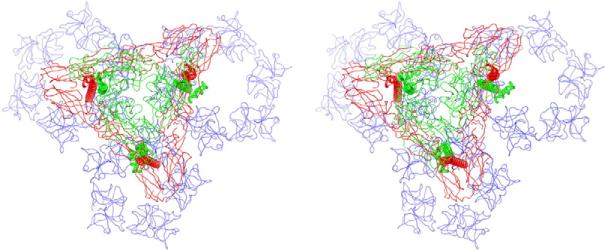
CP residues 1–113 are located below the fitted C-terminal protease domain (residues 114–264) and form a protein network that interconnects the capsomers. Despite the presence of this stabilizing layer, free cores are fragile structures compared with cores in virions.66,67 Our pseudo-atomic model clearly shows that each glycoprotein spike extends three pairs of cytoplasmic domains to three neighboring capsomers, and these further stabilize the core (Fig. 8). This arrangement also promotes spike clustering and induces membrane curvature necessary for virus budding. The membrane bilayer adopts an overall spherical shape, but the membrane curvature changes dramatically around the symmetry axes (Fig. 3d). This membrane puckering might arise from the core reorganization mediated by glycoprotein interactions, which is believed to be a prerequisite for alphavirus budding.68
Fig. 8.
Glycoprotein trimers cross-link groups of three capsomers in the NC. A stereo view down the pseudo-3-fold axis of the SINV virion shows how the three “legs” of an (E1–E2)3 trimer (E1 in red and E2 in green) interconnect a pentamer and two hexamers (all in blue) in the underlying NC. Each leg consists of an E1–E2 TM pair and the corresponding cytoplasmic domains (cdE1 and cdE2).
Model of alphavirus virus assembly
Interaction between cdE2 and the nucleocapsid has been proposed to be the primary driving force for alphavirus budding.19 The SINV cryoEM reconstruction and pseudo-atomic model presented here, coupled with comprehensive structure-based molecular-genetic analyses of CP–cdE2 interactions, refine our understanding of the mechanisms involved in the three components of alphavirus assembly. These include (1) NC formation in the cytoplasm, (2) formation of the E1–E2 spike followed by transport to the plasma membrane, and (3) the budding of NC through the spike-enriched host membrane to form the mature virion.
In the first step of native virus assembly, the NC is formed in the cytoplasm through specific interactions between the RNA genome and the flexible, highly basic, N-terminal region of CP (amino acids 1–113), which results in the encapsidation of the genome.36 Intersubunit interactions between the protease domains of multiple CPs (amino acids 114–264) promote the organization of pentameric and hexameric capsomers and are supported by the underlying network of CP N-terminal domains. Spike-mediated NC assembly has been reported for Semliki Forest virus CP mutants, which yield virions without requiring NC formation to occur in the cytoplasm prior to budding.69,70 Presumably, the NC assembly defect in these mutants was rescued by dominant spike–capsid interactions that form at the plasma membrane.
The E1 and E2 glycoproteins form heterodimers in the ER through multiple contacts, which includes pairing interactions between the E1 and E2 TM helices.49,50,53 The E1–E2 heterodimers then associate as trimers and are transported via type II cytopathic vacuoles to the plasma membrane where they associate as trimeric spikes in hexagonal arrays.71 The budding process is initiated when the NC attaches to one or more cdE2. Charged residues in cdE2-RI facilitate an initial recognition event that directs the cdE2-RII loop towards the CP pocket, where strong hydrophobic interactions form between residues in the cdE2 loop and the CP pocket.41 Additional stabilizing interactions with CP may be contributed by cdE1. Each pair of E1–E2 cytoplasmic domains interacts with a single CP subunit, and the three pairs of glycoproteins within a spike interact with CPs in separate capsomers (Fig. 8).4 This interaction cross-links the capsomers within the NC and stabilizes it for virus release. The natural curvature of the preassembled NC, combined with regularly spaced, strong interactions between it and the glycoprotein domains, allows the membrane and embedded spikes to encircle the NC.72 This proceeds until all sites are saturated and budding is completed. The close association of glycoproteins induced by budding is solidified by strong lateral interactions that comprise the skirt portion of the virion.73,74 The rigidity of the glycoprotein contacts in the outer shell further constrain the capsomers and enhance NC organization and stability.66 Contrary to the hypothesis proposed for Barmah Forest virus,15 we suggest that the lateral interaction between cdE2 and cdE1 is not a driving force for budding. Rather, it is merely a consequence of the CP–cdE2 interactions that induce spikes to cluster around the curved surface of the core.
Currently, there are no antiviral drugs available for managing alphavirus infections. Dioxane-based agents that inhibit SINV replication by binding to the CP hydrophobic pocket have been previously described.75 These compounds were designed based on knowledge gleaned from the crystal structure of CP in which the hydrophobic pocket was occupied by dioxane molecules derived from the crystallization solvent.76 Pseudo-atomic models such as the one derived in the current study and also the one on VEEV51 should help provide additional clues about critical subunit–subunit interactions that can be targeted with drugs and block the final step of assembly and subsequent budding and release.
Materials and Methods
Viruses and cells
Viruses were propagated in BHK-15 cells in Eagle's minimal essential medium supplemented with 10% fetal bovine serum at 37 °C in the presence of 5% CO2 unless otherwise indicated. For the cryoEM analysis, single-site mutant viruses (E2-N318Q) were prepared according to the previously published procedure.20
CryoEM and 3D image reconstruction
A previous cryoEM study of E2-N318Q particles, which led to a virion reconstruction at 9 Å resolution,9 included data from 31 micrographs recorded in a Philips CM200 microscope at 200 keV and a nominal magnification of 38,000×. For the present study, we recorded additional micrographs in a Philips CM300 microscope at 300 kV, at a nominal magnification of 45,000×, and with the objective lens defocused between 1.1 and 3.4 μm underfocus. Micrographs that exhibited astigmatism or specimen drift were eliminated from the data set, and the remaining 46 were digitized on a Zeiss SCAI scanner at 7-μm intervals. These were then interpolated to match the pixel size (equivalent to 1.78 Å at the specimen) of the previous data set.9 Individual virus particle images (5112) were boxed from the new set of micrographs using the program RobEM‡ and, when added to the previous set of images, doubled the number of boxed particles to ~24,000. The defocus level of each micrograph was estimated using RobEM. The previous 9-Å SINV E2-N318Q cryo-reconstruction was used as a starting model to initiate determination and refinement of the centers and orientations of all particles using the AUTO3DEM suite of programs.77 We employed the same procedures used to reconstruct three double-stranded RNA mycoviruses at subnanometer resolutions78,79 to reach a final 7.0-Å resolution with SINV E2-N318Q virions. Resolution was estimated on the basis of standard Fourier shell correlation criteria, using a 0.5 threshold.80 An empirical inverse temperature factor of 1/150 Å2 was employed during the particle parameter refinement and the calculation of the final reconstructed map.
Model building and structure interpretation
The crystal structure of the C-terminal protease domain of CP39 was used to model the corresponding CP density in the SINV cryo-reconstruction. Optimal fitting of the CP model was achieved by rigid-body refinement with the Fit in Map module of Chimera.81 The small F2–G2 loop (amino acids 248–252) that surrounds the hydrophobic pocket of CP was manually adjusted to fit the EM density. The crystal structure of the trimer of the SINV E1–E2 ectodomain49 was fitted into the SINV cryo-reconstruction using Chimera.81 The E1 and E2 models were then treated as separate rigid bodies to further optimize the fit of each glycoprotein. E1 required only minor adjustment from the initial fit but E2 had to be rotated a few degrees and translated closer to E1. Though the B domain of E2 was not seen in the SINV E1–E2 crystal structure, the homologous domain in the CHIKV E2 crystal structure50 was used as a template to build a model through MODELLER.82 An ensemble of the 25 highest-scoring homology models for the B domain of the SINV E2 glycoprotein was generated, and each of these was manually fit into the SINV density map. Based on visual inspection, an optimal B domain model was selected, which was then combined with the A and C domains from the SINV E2 crystal structure to construct an entire pseudo-atomic model of the SINV E2 ectodomain. Coot83 was used to assure that the residues connecting the B and A domains followed allowed bond lengths and angles.
No crystal structure data were available to directly model the complete TM, stem, or cytoplasmic domain regions of the glycoproteins, but the virion cryo-reconstruction contained density that could be reliably ascribed to each of these components. The TM regions were modeled as standard α-helices and fitted into the SINV cryo-reconstruction with Chimera. The stem region in E2 was modeled to closely follow the density feature that connects the last residue of the ectodomain (S343) with the first residue in the cognate TM helix (V365). Modeling of the 25-residue E1 stem region was subdivided into two steps. First, a homology model for the 10-residue, N-terminal part of the E1 stem (A384–K393) was generated based on the CHIKV E1 crystal structure.50 Then, the remaining C-terminal part of the E1 stem (N394–S408) was constructed in the same manner as the E2 stem, to connect it to the E1 TM helix at W409. This part of the modeling (for E2 as well) was also guided by secondary-structure predictions based on the sequence of the stem.
The five-residue cdE1 was modeled by adding residues to the end of the E1 TM helix and fitting them inside the unmodeled density in this region of the cryoEM map. The 33-residue cdE2 was modeled as a helix–turn–helix structure as determined by the morphology of the density features in the appropriate region of the SINV map and according to secondary-structure predictions given by Coot.83 Mutagenesis studies were used to help fine-tune and refine the cdE2 model and its interactions with CP.
Construction and characterization of CP and cdE2 mutant viruses
CP and cdE2 clones were constructed from pToto64, a full-length cDNA clone of SINV that has been previously described84 by overlap PCRs. The cDNA clones were linearized and in vitro transcribed with SP6 RNA polymerase, transfected into BHK-15 cells as previously described.85,86 Plaque phenotypes were determined 48 h posttransfection by comparing the mutant with WT Toto64 plaques, and virus titers were determined after plaque purification. The presence of the mutation in each virus was confirmed by sequencing the RT-PCR products corresponding to the CP and E2 coding region from RNA purified from cytoplasmic extracts of infected BHK-15 cells. One-step growth analyses were performed as described previously36,87 to measure growth kinetics of the virus. The expression of E2 and CP and polyprotein processing in lysates of BHK-15 cells transfected with mutant RNA were analyzed by Western immunoblots using polyclonal anti-CP and anti-E2 antibodies. BHK-15 cells grown on glass coverslips were examined by an IFA as explained elsewhere.86 Fluorescence images were obtained at room temperature using a confocal microscope (MRC1024; Bio-Rad Laboratories) (Nikon 60×, 1.4 NA lens) and LaserSharp software (Bio-Rad Laboratories). NC accumulation assays were carried out as previously described84 with certain modifications. Briefly, cytoplasmic extracts of the mutants were prepared at 12 h post-electroporation and NCs were purified by ultracentrifugation from the cytoplasmic extracts through a 0–54% Optiprep (Sigma) linear density gradient. The gradients were fractionated into 1-ml fractions, and 20- μl aliquots were assayed for the presence of CP by Western analysis using anti-CP antibody. Determination of virus release by qRT-PCR was performed as previously described.36 BHK-15 cells were electroporated using ~40 μg of in vitro transcribed RNA from WT and mutant cDNA clones, and the cells (~1×106) were plated onto a six-well plate. Virus supernatant was collected after 12 h, and viral RNA was purified from 140 μl of each supernatant using a QIAamp Viral RNA Mini Kit (Qiagen) according to the manufacturer's instructions. The number of molecules of viral RNA was determined using the standard curve of the cycle threshold values determined by qRT-PCR versus the number of molecules of in vitro transcribed genomic RNA.
One-step growth analysis of CP mutants
One-step growth analyses were performed as described previously.36,87 The cells were treated at a multiplicity of infection of 2. Following adsorption of viruses to cells for 1 h at room temperature, the cells were incubated at 37 °C, and the medium was replaced every 30 min for the first 2 h and every hour thereafter until the 12-h time point. The supernatant was collected at defined time points, and the titer of the virus released was determined by plaque assays of BHK-15 cell monolayers.
Polyprotein processing and transport of E2
The expression of E2 and CP and polyprotein processing in lysates of BHK-15 cells transfected with mutant RNA were analyzed by Western immunoblots using polyclonal anti-CP and anti-E2 antibodies. Samples were run under denaturing conditions in 10% or 13% 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol acrylamide gels. BHK-15 cells grown on glass coverslips were examined by an IFA.86 After staining with secondary antibody, we washed the cells three times with phosphate-buffered saline and then placed coverslips on microscope slides, cell side down, in the presence of FluorSave reagent (Gibco). Fluorescence images were obtained at room temperature using a confocal microscope (MRC1024; Bio-Rad Laboratories) (Nikon 60×, 1.4 NA lens) and LaserSharp software (Bio-Rad Laboratories).
Determination of virus release by qRT-PCR
qRT-PCR was performed using the SuperScript III Platinum SYBR Green One-Step qRT-PCR Kit (Invitrogen) with primers 5′-TTCCCTgTgTgCACgTACAT-3′ and 5′-TgAgCCCAACCAgAAgTTTT-3′, which bind to nucleotides 1044–1063 and nucleotides 1130–1149 of the SINV genome, respectively.36 Amplification reactions were carried out in 25-μl sample volumes that contained a 5-μl aliquot of purified viral RNA. All reactions were performed in triplicate. Cycling conditions were 20 min at 50 °C and 5 min at 95 °C, followed by 40 cycles of 5 s at 95 °C and 1 min at 60 °C.
Accession numbers
The density map of the 3D reconstruction of SINV virion has been deposited in the Electron Microscopy Data Bank at the European Bioinformatics Institute with accession code EMD-5251. The coordinates of the fitted CP, E1, and E2 proteins have been deposited with the Protein Data Bank (accession code: 3J0F).
Supplementary Material
Acknowledgements
We thank R. Ashmore, Y. Ji, D. C. Marinescu, and R. S. Sinkovits for providing software and advice. This work was supported in part by National Institutes of Health (NIH) grants P01 AI-55672 to R.J.K. and T.S.B., NIH grant GM56279 to R.J.K., NIH grant R37 GM-033050 to T.S.B., a grant from the W. M. Keck Foundation to the Purdue Structural Biology Group, and National Science Foundation shared instrument grant BIR-9112921, the University of California, San Diego, and a gift from the Agouron foundation (all to T.S.B.).
Abbreviations used
- aa
amino acids
- cdE1
cytoplasmic domain of E1
- cdE2
cytoplasmic domain of E2
- CHIKV
Chikungunya virus
- CP
capsid protein
- cryoEM
electron cryo-microscopy
- ER
endoplasmic reticulum
- IFA
immunofluorescence assay
- NC
nucleocapsid core
- qRT
quantitative reverse transcriptase
- SINV
Sindbis virus
- TM
transmembrane
- VEEV
Venezuelan equine encephalitis virus
- WT
wild type
- 3D
three-dimensional
Footnotes
http://cryoem.ucsd.edu/programDocs/runRobem.txt
Supplementary Data
Supplementary data to this article can be found online at doi:10.1016/j.jmb.2011.09.045
References
- 1.Strauss JH, Strauss EG. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Toivanen A. Alphaviruses: an emerging cause of arthritis? Curr. Opin. Rheumatol. 2008;20:486–490. doi: 10.1097/BOR.0b013e328303220b. [DOI] [PubMed] [Google Scholar]
- 3.Chevillon C, Briant L, Renaud F, Devaux C. The Chikungunya threat: an ecological and evolutionary perspective. Trends Microbiol. 2008;16:80–88. doi: 10.1016/j.tim.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 4.Cheng RH, Kuhn RJ, Olson NH, Rossmann MG, Choi HK, Smith TJ, Baker TS. Nucleocapsid and glycoprotein organization in an enveloped virus. Cell. 1995;80:621–630. doi: 10.1016/0092-8674(95)90516-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaedigk-Nitschko K, Schlesinger MJ. The Sindbis virus 6K protein can be detected in virions and is acylated with fatty acids. Virology. 1990;175:274–281. doi: 10.1016/0042-6822(90)90209-a. [DOI] [PubMed] [Google Scholar]
- 6.Lusa S, Garoff H, Liljestrom P. Fate of the 6K membrane protein of Semliki Forest virus during virus assembly. Virology. 1991;185:843–846. doi: 10.1016/0042-6822(91)90556-q. [DOI] [PubMed] [Google Scholar]
- 7.Zhang W, Mukhopadhyay S, Pletnev SV, Baker TS, Kuhn RJ, Rossmann MG. Placement of the structural proteins in Sindbis virus. J. Virol. 2002;76:11645–11658. doi: 10.1128/JVI.76.22.11645-11658.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang W, Heil M, Kuhn RJ, Baker TS. Heparin binding sites on Ross River virus revealed by electron cryo-microscopy. Virology. 2005;332:511–518. doi: 10.1016/j.virol.2004.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mukhopadhyay S, Zhang W, Gabler S, Chipman PR, Strauss EG, Strauss JH, et al. Mapping the structure and function of the E1 and E2 glycoproteins in alphaviruses. Structure. 2006;14:63–73. doi: 10.1016/j.str.2005.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mancini EJ, Clarke M, Gowen BE, Rutten T, Fuller SD. Cryo-electron microscopy reveals the functional organization of an enveloped virus, Semliki Forest virus. Mol. Cell. 2000;5:255–266. doi: 10.1016/s1097-2765(00)80421-9. [DOI] [PubMed] [Google Scholar]
- 11.Paredes AM, Brown DT, Rothnagel R, Chiu W, Schoepp RJ, Johnston RE, Prasad BV. Three-dimensional structure of a membrane-containing virus. Proc. Natl Acad. Sci. USA. 1993;90:9095–9099. doi: 10.1073/pnas.90.19.9095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang W, Fisher BR, Olson NH, Strauss JH, Kuhn RJ, Baker TS. Aura virus structure suggests that the T = 4 organization is a fundamental property of viral structural proteins. J. Virol. 2002;76:7239–7246. doi: 10.1128/JVI.76.14.7239-7246.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuller SD. The T = 4 envelope of Sindbis virus is organized by interactions with a complementary T = 3 capsid. Cell. 1987;48:923–934. doi: 10.1016/0092-8674(87)90701-x. [DOI] [PubMed] [Google Scholar]
- 14.Paredes A, Alwell-Warda K, Weaver SC, Chiu W, Watowich SJ. Venezuelan equine encephalomyelitis virus structure and its divergence from old world alphaviruses. J. Virol. 2001;75:9532–9537. doi: 10.1128/JVI.75.19.9532-9537.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kostyuchenko VA, Jakana J, Liu X, Haddow AD, Aung M, Weaver SC, et al. The structure of barmah forest virus as revealed by cryo-electron microscopy at a 6-angstrom resolution has detailed transmembrane protein architecture and interactions. J. Virol. 2011;85:9327–9333. doi: 10.1128/JVI.05015-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suomalainen M, Liljestrom P, Garoff H. Spike protein–nucleocapsid interactions drive the budding of alphaviruses. J. Virol. 1992;66:4737–4747. doi: 10.1128/jvi.66.8.4737-4747.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mulvey M, Brown DT. Assembly of the Sindbis virus spike protein complex. Virology. 1996;219:125–132. doi: 10.1006/viro.1996.0229. [DOI] [PubMed] [Google Scholar]
- 18.Carleton M, Brown DT. The formation of intramolecular disulfide bridges is required for induction of the Sindbis virus mutant ts23 phenotype. J. Virol. 1997;71:7696–7703. doi: 10.1128/jvi.71.10.7696-7703.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strauss JH, Strauss EG, Kuhn RJ. Budding of alphaviruses. Trends Microbiol. 1995;3:346–350. doi: 10.1016/s0966-842x(00)88973-8. [DOI] [PubMed] [Google Scholar]
- 20.Pletnev SV, Zhang W, Mukhopadhyay S, Fisher BR, Hernandez R, Brown DT, et al. Locations of carbohydrate sites on alphavirus glyco-proteins show that E1 forms an icosahedral scaffold. Cell. 2001;105:127–136. doi: 10.1016/s0092-8674(01)00302-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.von Bonsdorff CH, Harrison SC. Hexagonal glycoprotein arrays from Sindbis virus membranes. J. Virol. 1978;28:578–583. doi: 10.1128/jvi.28.2.578-583.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klimstra WB, Ryman KD, Johnston RE. Adaptation of Sindbis virus to BHK cells selects for use of heparan sulfate as an attachment receptor. J. Virol. 1998;72:7357–7366. doi: 10.1128/jvi.72.9.7357-7366.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gardner JP, Frolov I, Perri S, Ji Y, MacKichan ML, zur Megede J, et al. Infection of human dendritic cells by a sindbis virus replicon vector is determined by a single amino acid substitution in the E2 glycoprotein. J. Virol. 2000;74:11849–11857. doi: 10.1128/jvi.74.24.11849-11857.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Omar A, Koblet H. Semliki Forest virus particles containing only the E1 envelope glycoprotein are infectious and can induce cell–cell fusion. Virology. 1988;166:17–23. doi: 10.1016/0042-6822(88)90141-9. [DOI] [PubMed] [Google Scholar]
- 25.Waarts BL, Smit JM, Aneke OJ, McInerney GM, Liljestrom P, Bittman R, Wilschut J. Reversible acid-induced inactivation of the membrane fusion protein of Semliki Forest virus. J. Virol. 2005;79:7942–7948. doi: 10.1128/JVI.79.12.7942-7948.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smit JM, Bittman R, Wilschut J. Low-pH-dependent fusion of Sindbis virus with receptor-free cholesterol- and sphingolipid-containing liposomes. J. Virol. 1999;73:8476–8484. doi: 10.1128/jvi.73.10.8476-8484.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gibbons DL, Vaney MC, Roussel A, Vigouroux A, Reilly B, Lepault J, et al. Conformational change and protein–protein interactions of the fusion protein of Semliki Forest virus. Nature. 2004;427:320–325. doi: 10.1038/nature02239. [DOI] [PubMed] [Google Scholar]
- 28.Abell BA, Brown DT. Sindbis virus membrane fusion is mediated by reduction of glyco-protein disulfide bridges at the cell surface. J. Virol. 1993;67:5496–5501. doi: 10.1128/jvi.67.9.5496-5501.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gibbons DL, Kielian M. Molecular dissection of the Semliki Forest virus homotrimer reveals two functionally distinct regions of the fusion protein. J. Virol. 2002;76:1194–1205. doi: 10.1128/JVI.76.3.1194-1205.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu N, Brown DT. Phosphorylation and dephosphorylation events play critical roles in Sindbis virus maturation. Virology. 1993;196:703–711. doi: 10.1006/viro.1993.1527. [DOI] [PubMed] [Google Scholar]
- 31.Barth BU, Suomalainen M, Liljestrom P, Garoff H. Alphavirus assembly and entry: role of the cytoplasmic tail of the E1 spike subunit. J. Virol. 1992;66:7560–7564. doi: 10.1128/jvi.66.12.7560-7564.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ivanova L, Schlesinger MJ. Site-directed mutations in the Sindbis virus E2 glycoprotein identify palmitoylation sites and affect virus budding. J. Virol. 1993;67:2546–2551. doi: 10.1128/jvi.67.5.2546-2551.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gaedigk-Nitschko K, Schlesinger MJ. Site-directed mutations in Sindbis virus E2 glycoprotein's cytoplasmic domain and the 6K protein lead to similar defects in virus assembly and budding. Virology. 1991;183:206–214. doi: 10.1016/0042-6822(91)90133-v. [DOI] [PubMed] [Google Scholar]
- 34.Perera R, Owen KE, Tellinghuisen TL, Gorbalenya AE, Kuhn RJ. Alphavirus nucleocapsid protein contains a putative coiled coil alpha-helix important for core assembly. J. Virol. 2001;75:1–10. doi: 10.1128/JVI.75.1.1-10.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wengler G, Wurkner D. Identification of a sequence element in the alphavirus core protein which mediates interaction of cores with ribosomes and the disassembly of cores. Virology. 1992;191:880–888. doi: 10.1016/0042-6822(92)90263-o. [DOI] [PubMed] [Google Scholar]
- 36.Warrier R, Linger BR, Golden BL, Kuhn RJ. Role of sindbis virus capsid protein region II in nucleocapsid core assembly and encapsidation of genomic RNA. J. Virol. 2008;82:4461–4470. doi: 10.1128/JVI.01936-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weiss B, Geigenmuller-Gnirke U, Schlesinger S. Interactions between Sindbis virus RNAs and a 68 amino acid derivative of the viral capsid protein further defines the capsid binding site. Nucleic Acids Res. 1994;22:780–786. doi: 10.1093/nar/22.5.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tong L, Choi HK, Minor W, Rossmann MG. The structure determination of Sindbis virus core protein using isomorphous replacement and molecular replacement averaging between two crystal forms. Acta Crystallogr., Sect. A: Found. Crystallogr. 1992;48:430–442. doi: 10.1107/s0108767391014630. [DOI] [PubMed] [Google Scholar]
- 39.Choi HK, Tong L, Minor W, Dumas P, Boege U, Rossmann MG, Wengler G. Structure of Sindbis virus core protein reveals a chymotrypsin-like serine proteinase and the organization of the virion. Nature. 1991;354:37–43. doi: 10.1038/354037a0. [DOI] [PubMed] [Google Scholar]
- 40.Choi HK, Lu G, Lee S, Wengler G, Rossmann MG. Structure of Semliki Forest virus core protein. Proteins. 1997;27:345–359. doi: 10.1002/(sici)1097-0134(199703)27:3<345::aid-prot3>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 41.Lee S, Owen KE, Choi HK, Lee H, Lu G, Wengler G, et al. Identification of a protein binding site on the surface of the alphavirus nucleocapsid and its implication in virus assembly. Structure. 1996;4:531–541. doi: 10.1016/s0969-2126(96)00059-7. [DOI] [PubMed] [Google Scholar]
- 42.Choi HK, Lee S, Zhang YP, McKinney BR, Wengler G, Rossmann MG, Kuhn RJ. Structural analysis of Sindbis virus capsid mutants involving assembly and catalysis. J. Mol. Biol. 1996;262:151–167. doi: 10.1006/jmbi.1996.0505. [DOI] [PubMed] [Google Scholar]
- 43.Owen KE, Kuhn RJ. Alphavirus budding is dependent on the interaction between the nucleocapsid and hydrophobic amino acids on the cytoplasmic domain of the E2 envelope glycoprotein. Virology. 1997;230:187–196. doi: 10.1006/viro.1997.8480. [DOI] [PubMed] [Google Scholar]
- 44.Hernandez R, Sinodis C, Horton M, Ferreira D, Yang C, Brown DT. Deletions in the transmembrane domain of a sindbis virus glycoprotein alter virus infectivity, stability, and host range. J. Virol. 2003;77:12710–12719. doi: 10.1128/JVI.77.23.12710-12719.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.West J, Hernandez R, Ferreira D, Brown DT. Mutations in the endodomain of Sindbis virus glycoprotein E2 define sequences critical for virus assembly. J. Virol. 2006;80:4458–4468. doi: 10.1128/JVI.80.9.4458-4468.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whitehurst CB, Willis JH, Sinodis CN, Hernandez R, Brown DT. Single and multiple deletions in the transmembrane domain of the Sindbis virus E2 glycoprotein identify a region critical for normal virus growth. Virology. 2006;347:199–207. doi: 10.1016/j.virol.2005.11.029. [DOI] [PubMed] [Google Scholar]
- 47.Tong L, Wengler G, Rossmann MG. Refined structure of Sindbis virus core protein and comparison with other chymotrypsin-like serine proteinase structures. J. Mol. Biol. 1993;230:228–247. doi: 10.1006/jmbi.1993.1139. [DOI] [PubMed] [Google Scholar]
- 48.Lescar J, Roussel A, Wien MW, Navaza J, Fuller SD, Wengler G, et al. The Fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell. 2001;105:137–148. doi: 10.1016/s0092-8674(01)00303-8. [DOI] [PubMed] [Google Scholar]
- 49.Li L, Jose J, Xiang Y, Kuhn RJ, Rossmann MG. Structural changes of envelope proteins during alphavirus fusion. Nature. 2010;468:705–708. doi: 10.1038/nature09546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Voss JE, Vaney MC, Duquerroy S, Vonrhein C, Girard-Blanc C, Crublet E, et al. Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature. 2010;468:709–712. doi: 10.1038/nature09555. [DOI] [PubMed] [Google Scholar]
- 51.Zhang R, Hryc CF, Cong Y, Liu X, Jakana J, Gorchakov R, et al. 4.4 Å cryo-EM structure of an enveloped alphavirus Venezuelan equine encephalitis virus. EMBO J. 2011;30:3854–3863. doi: 10.1038/emboj.2011.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paredes AM, Heidner H, Thuman-Commike P, Prasad BV, Johnston RE, Chiu W. Structural localization of the E3 glycoprotein in attenuated Sindbis virus mutants. J. Virol. 1998;72:1534–1541. doi: 10.1128/jvi.72.2.1534-1541.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sjoberg M, Garoff H. Interactions between the transmembrane segments of the alphavirus E1 and E2 proteins play a role in virus budding and fusion. J. Virol. 2003;77:3441–3450. doi: 10.1128/JVI.77.6.3441-3450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moore DT, Berger BW, DeGrado WF. Protein–protein interactions in the membrane: sequence, structural, and biological motifs. Structure. 2008;16:991–1001. doi: 10.1016/j.str.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liao M, Kielian M. Site-directed antibodies against the stem region reveal low pH-induced conformational changes of the Semliki Forest virus fusion protein. J. Virol. 2006;80:9599–9607. doi: 10.1128/JVI.01054-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Powers AM, Brault AC, Shirako Y, Strauss EG, Kang W, Strauss JH, Weaver SC. Evolutionary relationships and systematics of the alphaviruses. J. Virol. 2001;75:10118–10131. doi: 10.1128/JVI.75.21.10118-10131.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 58.Liu LN, Lee H, Hernandez R, Brown DT. Mutations in the endo domain of Sindbis virus glycoprotein E2 block phosphorylation, reorientation of the endo domain, and nucleocapsid binding. Virology. 1996;222:236–246. doi: 10.1006/viro.1996.0414. [DOI] [PubMed] [Google Scholar]
- 59.Zhao H, Lindqvist B, Garoff H, von Bonsdorff CH, Liljestrom P. A tyrosine-based motif in the cytoplasmic domain of the alphavirus envelope protein is essential for budding. EMBO J. 1994;13:4204–4211. doi: 10.1002/j.1460-2075.1994.tb06740.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liljestrom P, Garoff H. Internally located cleavable signal sequences direct the formation of Semliki Forest virus membrane proteins from a polyprotein precursor. J. Virol. 1991;65:147–154. doi: 10.1128/jvi.65.1.147-154.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lopez S, Yao JS, Kuhn RJ, Strauss EG, Strauss JH. Nucleocapsid–glycoprotein interactions required for assembly of alphaviruses. J. Virol. 1994;68:1316–1323. doi: 10.1128/jvi.68.3.1316-1323.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hernandez R, Lee H, Nelson C, Brown DT. A single deletion in the membrane-proximal region of the Sindbis virus glycoprotein E2 endodomain blocks virus assembly. J. Virol. 2000;74:4220–4228. doi: 10.1128/jvi.74.9.4220-4228.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hernandez R, Ferreira D, Sinodis C, Litton K, Brown DT. Single amino acid insertions at the junction of the sindbis virus E2 transmembrane domain and endodomain disrupt virus envelopment and alter infectivity. J. Virol. 2005;79:7682–7697. doi: 10.1128/JVI.79.12.7682-7697.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gorbalenya AE, Donchenko AP, Koonin EV, Blinov VM. N-terminal domains of putative helicases of flavi- and pestiviruses may be serine proteases. Nucleic Acids Res. 1989;17:3889–3897. doi: 10.1093/nar/17.10.3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hahn CS, Strauss JH. Site-directed mutagenesis of the proposed catalytic amino acids of the Sindbis virus capsid protein autoprotease. J. Virol. 1990;64:3069–3073. doi: 10.1128/jvi.64.6.3069-3073.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mukhopadhyay S, Chipman PR, Hong EM, Kuhn RJ, Rossmann MG. In vitro-assembled alphavirus core-like particles maintain a structure similar to that of nucleocapsid cores in mature virus. J. Virol. 2002;76:11128–11132. doi: 10.1128/JVI.76.21.11128-11132.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Paredes A, Alwell-Warda K, Weaver SC, Chiu W, Watowich SJ. Structure of isolated nucleocapsids from Venezuelan equine encephalitis virus and implications for assembly and disassembly of enveloped virus. J. Virol. 2003;77:659–664. doi: 10.1128/JVI.77.1.659-664.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Garoff H, Sjoberg M, Cheng RH. Budding of alphaviruses. Virus Res. 2004;106:103–116. doi: 10.1016/j.virusres.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 69.Skoging-Nyberg U, Liljestrom P. M-X-I motif of Semliki Forest virus capsid protein affects nucleocapsid assembly. J. Virol. 2001;75:4625–4632. doi: 10.1128/JVI.75.10.4625-4632.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Forsell K, Griffiths G, Garoff H. Preformed cytoplasmic nucleocapsids are not necessary for alphavirus budding. EMBO J. 1996;15:6495–6505. doi: 10.1002/j.1460-2075.1996.tb01040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Soonsawad P, Xing L, Milla E, Espinoza JM, Kawano M, Marko M, et al. Structural evidence of glycoprotein assembly in cellular membrane compartments prior to Alphavirus budding. J. Virol. 2010;84:11145–11151. doi: 10.1128/JVI.00036-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Skoging U, Vihinen M, Nilsson L, Liljestrom P. Aromatic interactions define the binding of the alphavirus spike to its nucleocapsid. Structure. 1996;4:519–529. doi: 10.1016/s0969-2126(96)00058-5. [DOI] [PubMed] [Google Scholar]
- 73.Ekstrom M, Liljestrom P, Garoff H. Membrane protein lateral interactions control Semliki Forest virus budding. EMBO J. 1994;13:1058–1064. doi: 10.1002/j.1460-2075.1994.tb06354.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fuller SD, Berriman JA, Butcher SJ, Gowen BE. Low pH induces swiveling of the glycoprotein heterodimers in the Semliki Forest virus spike complex. Cell. 1995;81:715–725. doi: 10.1016/0092-8674(95)90533-2. [DOI] [PubMed] [Google Scholar]
- 75.Kim HY, Kuhn RJ, Patkar C, Warrier R, Cushman M. Synthesis of dioxane-based antiviral agents and evaluation of their biological activities as inhibitors of Sindbis virus replication. Bioorg. Med. Chem. 2007;15:2667–2679. doi: 10.1016/j.bmc.2007.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kim HY, Patkar C, Warrier R, Kuhn R, Cushman M. Design, synthesis, and evaluation of dioxane-based antiviral agents targeted against the Sindbis virus capsid protein. Bioorg. Med. Chem. Lett. 2005;15:3207–3211. doi: 10.1016/j.bmcl.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 77.Yan X, Sinkovits RS, Baker TS. AUTO3DEM—an automated and high throughput program for image reconstruction of icosahedral particles. J. Struct. Biol. 2007;157:73–82. doi: 10.1016/j.jsb.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tang J, Pan J, Havens WM, Ochoa WF, Guu TS, Ghabrial SA, et al. Backbone trace of partitivirus capsid protein from electron cryomicroscopy and homology modeling. Biophys. J. 2010;99:685–694. doi: 10.1016/j.bpj.2010.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tang J, Ochoa WF, Li H, Havens WM, Nibert ML, Ghabrial SA, Baker TS. Structure of Fusarium poae virus 1 shows conserved and variable elements of partitivirus capsids and evolutionary relationships to picobirnavirus. J. Struct. Biol. 2010;172:363–371. doi: 10.1016/j.jsb.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Van Heel M, Harauz G. Resolution criteria for three dimensional reconstruction. Optik. 1986;73:119–122. [Google Scholar]
- 81.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 82.Marti-Renom MA, Stuart AC, Fiser A, Sanchez R, Melo F, Sali A. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 83.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Owen KE, Kuhn RJ. Identification of a region in the Sindbis virus nucleocapsid protein that is involved in specificity of RNA encapsidation. J. Virol. 1996;70:2757–2763. doi: 10.1128/jvi.70.5.2757-2763.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kuhn RJ, Niesters HG, Hong Z, Strauss JH. Infectious RNA transcripts from Ross River virus cDNA clones and the construction and characterization of defined chimeras with Sindbis virus. Virology. 1991;182:430–441. doi: 10.1016/0042-6822(91)90584-x. [DOI] [PubMed] [Google Scholar]
- 86.Navaratnarajah CK, Kuhn RJ. Functional characterization of the Sindbis virus E2 glycoprotein by transposon linker-insertion mutagenesis. Virology. 2007;363:134–147. doi: 10.1016/j.virol.2007.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kuhn RJ, Hong Z, Strauss JH. Mutagenesis of the 3′ nontranslated region of Sindbis virus RNA. J. Virol. 1990;64:1465–1476. doi: 10.1128/jvi.64.4.1465-1476.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.