Abstract
Salmonellosis caused by Salmonella bacteria is a food-borne disease and worldwide health threat causing millions of infections and thousands of deaths every year. This pathogen infects an usually broad range of host organisms including human and plants. A better understanding of the mechanisms of communication between Salmonella and its hosts requires identifying the interactions between Salmonella and host proteins. Protein-protein interactions (PPIs) are the fundamental building blocks of communication. Here we utilize the prediction platform BIANA to obtain the putative Salmonella-human and Salmonella-Arabidopsis interactomes based on sequence and domain similarity to known PPIs. A gold standard list of Salmonella-host PPIs served to validate the quality of the human model. 24,726 and 10,926 PPIs comprising interactions between 38 and 33 Salmonella effectors and virulence factors with 9,740 human and 4,676 Arabidopsis proteins, respectively, were predicted. Putative hub proteins could be identified and parallels between the two interactomes were discovered. This approach can provide insight into possible biological functions of so far uncharacterized proteins. The predicted interactions are available via a web interface which allows filtering of the database according to parameters provided by the user to narrow down the list of suspected interactions. The interactions are available via a webinterface at http://sbi.imim.es/web/SHIPREC.php
1. Introduction
During infection, Salmonella expresses a variety of virulence factors and effectors that are delivered into the host cell triggering cellular responses through protein-protein interactions (PPIs) with host cell proteins which make the pathogen’s invasion and replication possible. To decipher the molecular details of the communication between host and pathogen, it is necessary to identify Salmonella-host PPIs as well as their biological consequences. Methods to discover and characterize PPIs within an organism (“intraspecies”) or between a host and its pathogen (“interspecies”) have been applied widely and include small scale experiments such as pull-down, co-localization, co-immunoprecipitation assays as well as high-throughput experiments such as yeast-2-hybrid, and mass spectrometry identification of binding partners. Examples of intraspecies interactomes experimentally studied with high-throughput approaches include yeast [1], worm [2], Drosophila [3] and Arabidopsis [4] and a number of bacteria, such as Mycobacterium tuberculosis [5], Escherichia coli [6], Helicobacter pylori [7], Staphylococcus aureus [8] and Campylobacter jejuni [9]. Less high-throughput experimental data exists regarding interspecies interactomes, so far only for Bacillus anthracis-human, Francisella tularensis-human, and Yersinia pestis-human [10]. To fill this gap, numerous computational approaches have been developed to predict pathogen-host interactions, most prominently between HIV and human [11], and other virus-host or bacteria-host interactions [12][13]. Computational methods can also greatly help in interpreting the data with respect to comparing networks and finding general strategies of pathogens [9][14].
Towards identifying Salmonella-host interactions, in a recent survey of the literature and databases, we obtained a small gold standard dataset of 62 Salmonella-host interactions, involving interactions of Salmonella proteins with mostly human host proteins [15]. This gold standard can be used to develop and validate predictions for Salmonella-host interactions. Here we present a computational model to predict PPIs between Salmonella and human and validate the model with the gold standard. We then expanded the model towards predicting PPIs between Salmonella and Arabidopsis as a representative of the plant kingdom to exemplify the most extreme in difference between Salmonella’s hosts. While we include all Salmonella proteins in both models, their in-depth analysis focuses on subnetworks of the interactomes that include known Salmonella effectors and virulence factors and the comparison of the two host systems. The work described here is the first effort to predict Salmonella-Arabidopsis PPIs and compare Salmonella’s interactions with host organisms as extreme as animal and plant kingdoms.
2. Results and discussion
2.1. Salmonella-human interactome and overlap with gold standard
First, we predicted the set of Salmonella-human interactions based on sequence identity or domain assignments using iPfam and 3DID databases and compared the model’s predictions with the set of known Salmonella-host interactions. Since the gold standard dataset contains a small number of non-human host proteins, we retrieved the respective human homologues for these proteins to allow direct comparison. For the recovery analysis 59 interactions of the gold standard dataset were used, excluding the three clearly indirect ones.
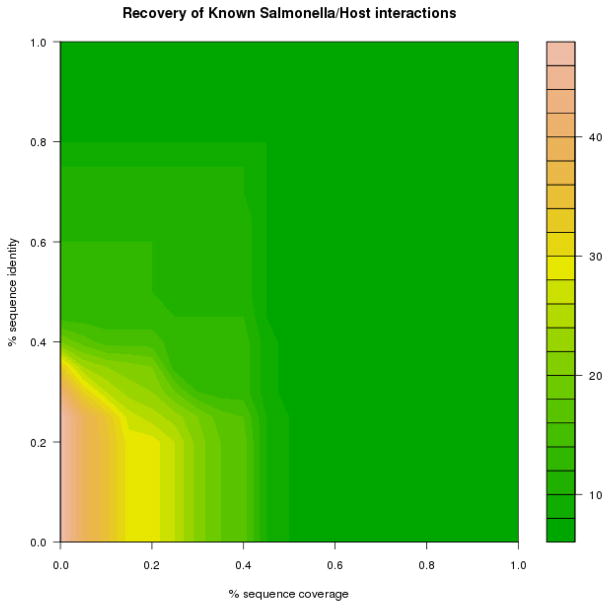
A plot showing the number of gold standard pairs retrieved as a function of sequence coverage and sequence identity is shown in Fig. 1. The maximum retrieval of known interactions was 48 of the 59 gold standard interactions with the lowest sequence identity and coverage requirement. This is because the gold standard contains interactions that are not present in any database yet. If we increase the stringency on the sequence identity and coverage, with a sequence identity cut-off of 60 % and a sequence coverage greater than 70%, six PPIs are predicted. Lowering the sequence identity and coverage both to 21 %, 29 out of 59 gold standard PPIs are retrieved.
Fig. 1.
Recovery of known Salmonella-host interactions using the model based on sequence identity.
Using the domain-based prediction feature, nine of the gold standard interactions are predicted. These nine interactions are also part of the set of 29 PPIs that can be predicted by the model using a sequence-based query. Furthermore, there are six PPIs that are listed in PPI databases and would thus be retrieved by our model as known interactions.
Thus, our model proved to be a valuable source for predicting Salmonella-host PPIs as 49% of the gold standard interactions can be predicted by the model using a sequence and coverage cut-off of 21%.
2.2. Predicted Salmonella-human interactome
The total number of predicted Salmonella-human PPIs based on all interolog evidence [16], i.e. sequence identity (e-value 10−3, sequence identity 60%, sequence coverage 70%) and domain (iPfam and/or 3DID) identity for all Salmonella species and all proteins is ~44,8 million (Table 1). This list of interactions contains a lot of redundancy because it treats each Salmonella species separately. This has an advantage, if one is interested in the interaction specific to a given Salmonella species, strain or serovar. More commonly, the results would be clustered by the sequence of Salmonella proteins so that only one pair of protein is predicted for any Salmonella species. Using a sequence identity of 95%, and sequence coverage of 90% or using the Salmonella gene symbol directly, grouping of the results leads to reduction of the predicted pairs. For simplicity, we here only consider this reduced set of interactions. The results are listed in Table 1. Since we are primarily interested in putative interactions involving known Salmonella effectors, in the following we restrict our analysis to this subnetwork. The predicted number of PPIs for Salmonella effectors is 46,200 when grouping by Salmonella protein sequence and 26,592 when grouping by Salmonella gene symbol. Analysis of these PPIs as described in the experimental part revealed a dataset of 24,726 interactions that were analyzed in detail (Table 2). There are 38 of the 108 known Salmonella effectors (Table S3) in the set of predicted PPIs.
Table 1.
Total numbers of predicted PPIs of all Salmonella proteins with human and Arabidopsis proteins.
| Ungrouped | Grouped by sequence | Grouped by gene symbol | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Union of sequence and PFAM based |
Intersection of sequence and PFAM based |
Sequence based only |
PFAM based only |
Union of sequence and PFAM based |
Intersection of sequence and PFAM based |
Sequence based only |
PFAM based only |
Union of sequence and PFAM based |
Intersection of sequence and PFAM based |
Sequence based only |
PFAM based only |
||
|
| |||||||||||||
| Human | All species (Taxonomy ID: 59201) | 44,794,281 | 190,609 | 4,034,639 | 40950251 | 1,610,538 | 4,606 | 79,833 | 1,535,311 | 988,961 | 3,364 | 77,114 | 91,5211 |
| Human | S. enterica | 43,226,893 | 187,083 | 3,775,390 | 39,638,586 | 1,462,795 | 4,523 | 74,712 | 1,392,606 | 967,901 | 3,347 | 76,869 | 894,379 |
| Human | S. typhi | 727,047 | 3,030 | 56,671 | 673,406 | 694,069 | 3,009 | 56,220 | 640,858 | 552,262 | 3,029 | 53,533 | 501,758 |
| Human | S. typhimurium | 4,158,239 | 18,233 | 362,197 | 3,814,275 | 803,042 | 3,071 | 60,684 | 745,429 | 769,911 | 3,276 | 66,834 | 706,353 |
| Human | S. paratyphi B | 153,607 | 1,487 | 22,313 | 132,781 | 152,939 | 1,487 | 22,313 | 132,113 | 159,830 | 1,519 | 22,466 | 138,883 |
| Human | S. paratyphi A | 1,257,714 | 6,072 | 110,839 | 1,152,947 | 645,535 | 3,049 | 56,868 | 591,716 | 575,351 | 3,100 | 58,321 | 520,130 |
|
| |||||||||||||
| Arabidopsis | All species | 15,932,356 | 7,791 | 702,738 | 15,237,409 | 573,746 | 178 | 14,306 | 559,618 | 342,611 | 268 | 14,582 | 328,297 |
| Arabidopsis | S. enterica | 15,389,512 | 7,298 | 668,228 | 14,728,582 | 505,290 | 165 | 13,130 | 492,325 | 334,545 | 255 | 14,309 | 320,491 |
| Arabidopsis | S. typhi | 256,523 | 119 | 10,111 | 246,531 | 243,677 | 117 | 9,926 | 233,868 | 187,475 | 126 | 9,534 | 178,067 |
| Arabidopsis | S. typhimurium | 1,443,998 | 757 | 64,031 | 1,380,724 | 278,581 | 139 | 10,899 | 267,821 | 269,384 | 215 | 12,705 | 256,894 |
| Arabidopsis | S. paratyphi B | 44,149 | 70 | 3,877 | 40,342 | 43,441 | 70 | 3,877 | 39,634 | 43,684 | 86 | 4,001 | 39,769 |
| Arabidopsis | S. paratyphi A | 445,182 | 240 | 20,499 | 424,923 | 226,649 | 120 | 10,336 | 216,433 | 203,540 | 152 | 10,551 | 193,141 |
Table 2.
Total number of predicted PPIs of Salmonella effectors and virulence factors with human and Arabidopsis proteins.
| ungrouped | Grouped by sequence | Grouped by gene symbol | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Union of sequence and PFAM based |
Intersection of sequence and PFAM based |
Sequence based only |
PFAM based only |
Union of sequence and PFAM based |
Intersection of sequence and PFAM based |
Sequence based only |
PFAM based only |
Union of sequence and PFAM based |
Intersection of sequence and PFAM based |
Sequence based only |
PFAM based only |
||
|
| |||||||||||||
| Human | All species (Taxonomy ID: 59201) | 293,811 | 800 | 3,208 | 291,403 | 46,200 | 118 | 213 | 46,105 | 26,592 | 67 | 161 | 26,498 |
| Human | S. enterica | 269,168 | 684 | 3,006 | 266,846 | 41,223 | 94 | 189 | 41,128 | 26,592 | 67 | 161 | 26,498 |
| Human | S. typhi | 14,609 | 32 | 86 | 14,555 | 14,242 | 32 | 86 | 14,188 | 14,609 | 32 | 32 | 14,555 |
| Human | S. typhimurium | 84,003 | 229 | 486 | 83,746 | 24,765 | 67 | 147 | 24,685 | 26,577 | 67 | 146 | 26,498 |
| Human | S. paratyphi B | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Human | S. paratyphi A | 10,608 | 46 | 100 | 10,554 | 10,608 | 46 | 100 | 10,554 | 10,608 | 46 | 100 | 10,554 |
|
| |||||||||||||
| Arabidopsis | All species | 107,127 | 276 | 1,114 | 106,289 | 18,732 | 37 | 70 | 18,699 | 10,966 | 25 | 52 | 10,939 |
| Arabidopsis | S. enterica | 99,491 | 238 | 1,049 | 98,680 | 18,092 | 37 | 70 | 18,059 | 10,966 | 25 | 52 | 10,939 |
| Arabidopsis | S. typhi | 6,641 | 12 | 33 | 6,620 | 6,303 | 12 | 33 | 6,282 | 6,641 | 12 | 33 | 6,620 |
| Arabidopsis | S. typhimurium | 35,476 | 88 | 186 | 35,378 | 10,903 | 25 | 58 | 10,870 | 10,966 | 25 | 52 | 10,939 |
| Arabidopsis | S. paratyphi B | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Arabidopsis | S. paratyphi A | 4,432 | 12 | 33 | 4,411 | 4,432 | 12 | 33 | 4,411 | 4,432 | 12 | 33 | 4,411 |
The basis for most of the PPIs predictions is the domain similarity. Only less than 1%, namely 155, of the predicted pairs are based on sequence identity. The overlap between the two predictions is low: the number of PPIs predicted by both, sequence (e-value 10−3, sequence identity 60%, sequence coverage 70%) and domain (iPfam and/or 3DID) identity is only 67.
2.3. Predicted Salmonella-Arabidopsis interactome
The total number of predicted PPIs based on all interolog evidence is ~15,9 million for Arabidopsis (Table 1). The total number of predicted PPIs involving Salmonella effectors only in the ungrouped mode is 107,127 which decreases to 10,926 when grouping by Salmonella gene symbol and analyzing as described above which corresponds to ~10.2% of the ungrouped pairs. As with human, the majority of the predictions is based on domain (iPfam and/or 3DID) evidence. The number of PPIs predicted based on sequence alone is 52. The intersection is 25. There are 33 of the 108 known Salmonella effectors (Table S3) in the set of predicted PPIs.
2.4. Comparison of Salmonella effectors and their binding partners
Based on the above considerations, the two predicted interactomes that will be compared in the following comprise 24,726 and 10,926 edges between human and Salmonella proteins and between Arabidopsis and Salmonella proteins, respectively. Within these, 38 Salmonella effectors interact with 9,740 human proteins and 33 Salmonella effectors interact with 4,676 Arabidopsis proteins. For ease of identification, we use gene symbols to represent Salmonella proteins and uniprot entry names for host proteins.
30 Salmonella effectors are common for both networks, while the rest is unique for each predicted interactome (see Table 3). In Table 3, the number of predicted interactions is given for each Salmonella effector based on sequence and/or domain based predictions or the intersection of the two. Despite most predictions being domain-based, the predictions for SipB and SpvC with human proteins are inferred from sequence identity only. Unlike in the Salmonella-human network, within the Salmonella-Arabidopsis interactions there is no Salmonella effector having PPIs predicted based on sequence identity only.
Table 3.
Number of predicted interactions specified for each Salmonella effector involved in the Salmonella-human and Salmonella-Arabidopsis interactome divided into sequence and domain based predictions.
| Salmonella-human | Salmonella-Arabidopsis | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Salmonella effector | Union | Sequence based | Domain based | Intersection | Salmonella effector | Union | Sequence based | Domain based | Intersection |
| BarA | 381 | 19 | 367 | 5 | BarA | 341 | 3 | 338 | - |
|
| |||||||||
| HilA | 104 | - | 104 | - | HilA | 70 | - | 70 | - |
|
| |||||||||
| - | - | - | - | - | HilC | 1 | - | 1 | - |
|
| |||||||||
| - | - | - | - | - | HilD | 1 | - | 1 | - |
|
| |||||||||
| InvB | 2 | - | 2 | - | - | - | - | - | - |
|
| |||||||||
| InvC | 31 | - | 31 | - | InvC | 33 | - | 33 | - |
|
| |||||||||
| InvG | 1,764 (1,741) | - | 1764 | - | - | - | - | - | - |
|
| |||||||||
| Orf408 | 34 (17) | - | 34 (17) | - | Orf408 | 80 (40) | - | 80 (40) | - |
|
| |||||||||
| Orf48 | 1,764 (1,741) | - | 1,764 (1,741) | - | - | - | - | - | - |
|
| |||||||||
| PipB | 1 | - | 1 | - | PipB | 5 | - | 5 | - |
|
| |||||||||
| PipB2 | 1 | - | 1 | - | PipB2 | 5 | - | 5 | - |
|
| |||||||||
| SifA | 631 | 2 | 631 | 2 | SifA | 27 | - | 27 | - |
|
| |||||||||
| SifB | 1,080 | - | 1,080 | - | SifB | 153 | - | 153 | - |
|
| |||||||||
| SipA | 2 | - | 2 | - | - | - | - | - | - |
|
| |||||||||
| SipB | 4 | 4 | - | - | - | - | - | - | - |
|
| |||||||||
| SirA | 186 | 21 | 165 | - | SirA | 257 | 8 | 249 | - |
|
| |||||||||
| - | - | - | - | - | SirC | 1 | - | 1 | - |
|
| |||||||||
| SlrP | 2,852 | - | 2,852 | - | SlrP | 1,673 | - | 1,673 | - |
|
| |||||||||
| SopA | 19 | 7 | 12 | - | SopA | 31 | 6 | 25 | - |
|
| |||||||||
| SopB | 40 | - | 40 | - | SopB | 42 | - | 42 | - |
|
| |||||||||
| SopE | 455 | 14 | 455 | 14 | SopE | 126 | - | 126 | - |
|
| |||||||||
| SopE2 | 455 | 14 | 455 | 14 | SopE2 | 126 | - | 126 | - |
|
| |||||||||
| SpaK | 2 | - | 2 | - | - | - | - | - | - |
|
| |||||||||
| SpaL | 31 | - | 31 | - | SpaL | 33 | - | 33 | - |
|
| |||||||||
| SpiA | 3,528 (1741) | - | 3,528 (1741) | - | - | - | - | - | - |
|
| |||||||||
| SpiR | 367 | - | 367 | - | SpiR | 338 | - | 338 | - |
|
| |||||||||
| SptP | 2,562 (2560) | 11 | 2,562 | 11 | SptP | 1,561 | 12 | 1,561 | 12 |
|
| |||||||||
| SpvB | 637 | 21 | 637 | 21 | SpvB | 168 | 13 | 168 | 13 |
|
| |||||||||
| SpvC | 18 (6) | 18 (6) | - | - | - | - | - | - | - |
|
| |||||||||
| SsaN | 31 | - | 31 | - | SsaN | 33 | - | 33 | - |
|
| |||||||||
| SscB | 1,081 (1,080) | - | 1,081 | - | SscB | 521 | - | 521 | - |
|
| |||||||||
| SseA | 159 | - | 159 | - | SseA | 108 | - | 108 | - |
|
| |||||||||
| SseJ | 1,500 | - | 1,500 | - | SseJ | 497 | - | 497 | - |
|
| |||||||||
| SspA | 133 | 30 | 103 | - | SspA | 167 | 10 | 157 | - |
|
| |||||||||
| SspH1 | 2,852 | - | 2,852 | - | SspH1 | 1,673 | - | 1,673 | - |
|
| |||||||||
| SspH2 | 2,852 | - | 2,852 | - | SspH2 | 1,673 | - | 1,673 | - |
|
| |||||||||
| SsrA | 367 | - | 367 | - | SsrA | 338 | - | 338 | - |
|
| |||||||||
| SsrB | 165 | - | 165 | - | SsrB | 233 | - | 233 | - |
|
| |||||||||
| TtrB | 126 (125) | - | 126 | - | TtrB | 138 | - | 138 | - |
|
| |||||||||
| TtrR | 165 | - | 165 | - | TtrR | 249 | - | 249 | - |
|
| |||||||||
| TtrS | 210 | - | 210 | - | TtrS | 264 | - | 264 | - |
|
| |||||||||
| Total | 26,592 (24,726) | 161 (155) | 26,498 (24,671) | 67 | Total | 10,966 (10,926) | 52 | 10,939 (10,899) | 25 |
Analogous to SipB and SpvC being present only in the sequence-based predictions with one organism, there are many other such examples, when looking at the domain-based predictions. Unique to Arabidopsis are HilC, HilD and SirC. Unique to human are InvB, InvG, Orf48, SipA, SpaK, and SpiA.
2.5. Predicted effector hubs
The effectors of Salmonella with the highest number of edges (hubs) are SspH1, SspH2, SlrP and SptP with more than 2,500 PPIs in the Salmonella-human interactome and more than 1,500 PPIs in the Salmonella-Arabidopsis interactome, respectively. Although not as extreme, there are also several effectors with more than 500 predicted PPIs. These effectors are InvG, Orf48, SipA, SseJ, SifB, SscB, SifA and SpvB for the Salmonella-human network and SscB for the Salmonella-Arabidopsis network. In contrast to these hub proteins, several effectors are predicted to interact with very few proteins, namely InvB, HilC, HilD, SirC, PipB, PipB2, SipA, SipB, SpaK and SpvC, which are all predicted to interact with 6 or less host proteins.
2.6. Predicted central role of SptP
The Salmonella effector that seems to play a central role is SptP, especially when considering the domain-based predictions. On the one hand this effector is predicted to interact with 2,560 and 1,561 unique human and Arabidopsis proteins, respectively. Furthermore, SptP has common binding partners with 23 Salmonella effectors in the Salmonella-human and 15 Salmonella effectors the Salmonella-Arabidopsis network thereby sharing ~25% of its interaction partners in both interactomes (Supplementary Table S1).
2.7. Comparison of human and Arabidopsis proteins that are predicted to interact with the same Salmonella effector
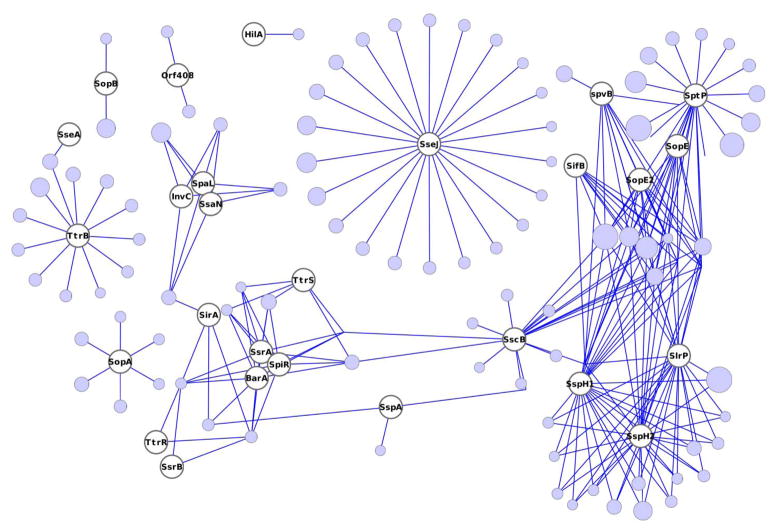
Next, we focused on the homologous proteins shared between Arabidopsis and human hosts and their interaction with the same Salmonella effector(s) by applying a sequence identity and coverage cut-off of 50 %. 2,416 human proteins were similar to 1,507 Arabidopsis proteins. Table 4 summarizes the Salmonella effectors that are involved in both interactomes as well as the numbers of similar human and Arabidopsis proteins. Almost all Salmonella effector proteins share at least one homologue binding partner in human and Arabidopsis. The only effectors that are predicted to interact only with host binding partners that do not reveal any sequence similarity between human and Arabidopsis proteins are PipB, PipB2 and SifA. Fig. 2 visualizes the intersection of the Salmonella-human and the Salmonella-Arabidopsis predicted interactomes. It involves 27 Salmonella effector proteins. Human and Arabidopsis proteins are clustered into the same node according to their sequence similarity. This illustration shows the many indirect connections between the Salmonella proteins. Furthermore, SptP, SspH1, SspH2 and SlrP are hub proteins each with more than 300 interacting host proteins. Finally, SscB is a central protein in the intersected network, predicted to be engaged in interactions with more than 300 host proteins. Examples of human and Arabidopsis proteins that share sequence similarity are given in Table S4.
Table 4.
Similarity between human and Arabidopsis proteins based on sequence identity.
| Salmonella effector | A | B | C | D |
|---|---|---|---|---|
| BarA | 41 | 20 | 340 | 321 |
| HilA | 2 | 1 | 102 | 69 |
| InvC | 16 | 16 | 15 | 17 |
| Orf408 | 4 | 5 | 13 | 35 |
| PipB | 0 | 0 | 1 | 5 |
| PipB2 | 0 | 0 | 1 | 5 |
| SifA | 0 | 0 | 631 | 27 |
| SifB | 190 | 104 | 890 | 49 |
| SirA | 9 | 9 | 177 | 248 |
| SlrP | 222 | 154 | 2,630 | 1,519 |
| SopA | 13 | 14 | 6 | 17 |
| SopB | 10 | 5 | 30 | 37 |
| SopE | 190 | 104 | 265 | 22 |
| SopE2 | 190 | 104 | 265 | 22 |
| SpaL | 16 | 16 | 15 | 17 |
| SpiR | 38 | 19 | 329 | 319 |
| SptP | 312 | 200 | 2,248 | 1361 |
| SpvB | 241 | 136 | 396 | 32 |
| SsaN | 16 | 16 | 15 | 17 |
| SscB | 248 | 148 | 832 | 373 |
| SseA | 6 | 2 | 153 | 106 |
| SseJ | 73 | 51 | 1,427 | 446 |
| SspA | 12 | 11 | 121 | 156 |
| SspH1 | 222 | 154 | 2,630 | 1,519 |
| SspH2 | 222 | 154 | 2,630 | 1,519 |
| SsrA | 38 | 19 | 329 | 319 |
| SsrB | 4 | 3 | 161 | 230 |
| TtrB | 43 | 23 | 82 | 115 |
| TtrR | 4 | 3 | 161 | 246 |
| TtrS | 34 | 16 | 176 | 248 |
Fig. 2.
Intersection network of Salmonella effector proteins interacting with similar human and Arabidopsis proteins.
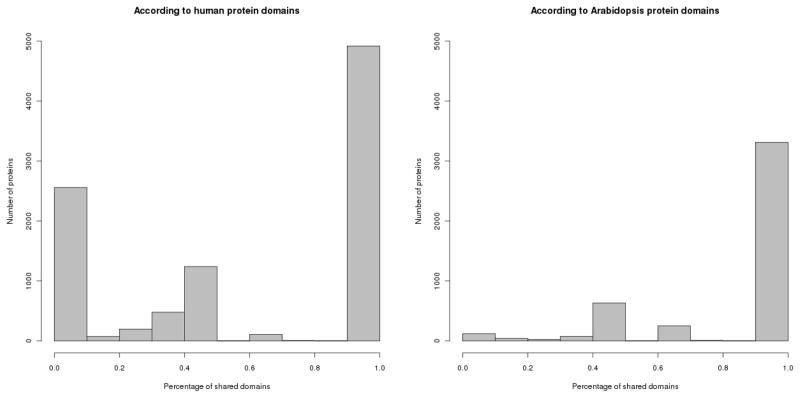
Similar to the sequence-based comparison of the host proteins, we analyzed the human and Arabidopsis proteins that are predicted to interact with the same Salmonella effector by means of domains composition. For each human-Arabidopsis PPI comparison, the percentage of shared Pfam domains was calculated in relation to the total number of domains of the human protein or the Arabidopsis protein, respectively (Fig. 3). 4,919 human proteins predicted to interact with Salmonella effectors share all their domains with Arabidopsis proteins that are predicted to interact with the same Salmonella proteins. There are 3,313 Arabidopsis proteins sharing all their domains with human proteins. Interestingly, 2,559 human proteins did not share any of their domains with Arabidopsis proteins, while only 120 Arabidopsis proteins did not share any of their domains with human proteins. This difference could be explained by the nature of the data: most of the predictions are obtained based on domain interactions reported between domains in high-resolution three dimensional structures. As the Protein Data Bank [17] contains more domain structures related to human and other mammalian proteins than for plant proteins, using this inference method a higher number of predictions is retrieved for human than for Arabidopsis. Furthermore, there are more human-specific domains than there are for Arabidopsis.
Fig. 3.
Similarity between human and Arabidopsis proteins based on domain composition.
Currently, more than 60 % of the Arabidopsis thaliana protein-coding genes are uncharacterized [4]. Thus, the comparison approach utilized here may contribute to elucidating possible functions of Arabidopsis proteins for which direct functional information is lacking.
2.8. Identification of proteins involved in pathogenicity using GUILD
Network biology recently proved its use in identifying candidate genes associated with a disease based on the observation that proteins translated by phenotypically related genes tend to interact, the so called guilt-by-association principle [18]. GUILD [submitted] is a network-based prioritization framework of methods that was used here to unveil genes associated with the infection of hosts by Salmonella. Using GUILD to obtain Salmonella and host proteins that may be important during Salmonella infection and host response is one possibility to filter the predicted subnetworks between Salmonella effectors and host proteins on the one hand and all possible interactions between Salmonella and its host on the other hand to identify interesting and so far undiscovered target candidates in pathogenicity. Four examples of host proteins with high GUILD-rankings that are predicted to interact with Salmonella effector proteins are described below. The top GUILD-ranked Salmonella and host proteins are listed in Table 6 and examples are discussed below in subsections (a)–(d).
Table 6.
High GUILD-ranked Salmonella and host proteins.
| High GUILD-ranked proteins in the Salmonella-human predicted interactome | High GUILD-ranked proteins in the Salmonella-Arabidopsis predicted interactome | ||||||
|---|---|---|---|---|---|---|---|
| Human uniprot entry | score | Salmonella gene name | score | Arabidopsis uniprot entry | score | Salmonella gene name | score |
| EHMT1 | 0.139 | ipgD | 0.589 | PP2A5 | 0.096 | pipC | 0.129 |
|
| |||||||
| AHCYL1 | 0.139 | sigE | 0.390 | PP2A3 | 0.095 | sigE | 0.129 |
|
| |||||||
| PECI | 0.139 | pipC | 0.390 | PP2A4 | 0.095 | sicP | 0.078 |
|
| |||||||
| AHCYL2 | 0.139 | yopH | 0.344 | PP2A1 | 0.095 | ycgB | 0.064 |
|
| |||||||
| ERP29 | 0.139 | stpA | 0.344 | PP2A2 | 0.095 | cheB | 0.058 |
|
| |||||||
| SYTL3 | 0.127 | sicP | 0.229 | PPX2 | 0.090 | modB | 0.044 |
|
| |||||||
| CARD17 | 0.102 | ipaB | 0.174 | PPX1 | 0.090 | corC | 0.034 |
|
| |||||||
| CASP1 | 0.070 | ycgB | 0.077 | RPS27AA | 0.032 | ybeX | 0.034 |
|
| |||||||
| ECI2 | 0.058 | cheB | 0.072 | UBQ12 | 0.032 | sgaB | 0.030 |
|
| |||||||
| NA | 0.056 | modB | 0.054 | UBQ13 | 0.032 | ulaB | 0.030 |
|
| |||||||
| CARD16 | 0.049 | ybeX | 0.042 | At5g20620 | 0.032 | eutM | 0.026 |
|
| |||||||
| COP | 0.047 | corC | 0.042 | UBQ8 | 0.032 | yqiB | 0.025 |
|
| |||||||
| IL18 | 0.043 | sgaB | 0.038 | UBQ9 | 0.032 | hha | 0.024 |
|
| |||||||
| CARD18 | 0.037 | ulaB | 0.038 | F15I1.4 | 0.032 | mfd | 0.021 |
|
| |||||||
| IL1F7 | 0.036 | eutM | 0.034 | RUB1 | 0.032 | diaA | 0.018 |
|
| |||||||
| IL37 | 0.036 | yqiB | 0.032 | RUB2 | 0.032 | yraO | 0.018 |
|
| |||||||
| CASP5 | 0.035 | hha | 0.030 | RPS27AB | 0.032 | rlmH | 0.017 |
|
| |||||||
| ERP29 | 0.035 | yfjD | 0.029 | RPS27AC | 0.032 | ybeA | 0.017 |
|
| |||||||
| AHCYL1 | 0.030 | corB | 0.029 | UBQ13 | 0.030 | fliG | 0.016 |
|
| |||||||
| CARD8 | 0.028 | mfd | 0.028 | SEN3 | 0.030 | mutS | 0.016 |
|
| |||||||
| TYSND1 | 0.027 | yraO | 0.022 | UBQ3 | 0.030 | proC | 0.016 |
|
| |||||||
| JOSD2 | 0.027 | diaA | 0.022 | UBQ4 | 0.030 | rimJ | 0.016 |
|
| |||||||
| NOD2 | 0.027 | rlmH | 0.021 | UBQ13 | 0.030 | serS | 0.016 |
|
| |||||||
| IL18BP | 0.026 | ybeA | 0.021 | At4g05050 | 0.030 | ahpF | 0.015 |
|
| |||||||
| IL1RL2 | 0.026 | mutS | 0.021 | UBQ10 | 0.030 | ptsI | 0.014 |
|
| |||||||
| JOSD2 | 0.026 | rimJ | 0.021 | UBQ14 | 0.030 | udk | 0.014 |
|
| |||||||
| AHCYL2 | 0.025 | serS | 0.020 | UBQ11 | 0.030 | phoB | 0.013 |
|
| |||||||
| PYCARD | 0.022 | ahpF | 0.020 | RPL40A | 0.029 | rplI | 0.013 |
|
| |||||||
| IL1A | 0.017 | proC | 0.020 | RPL40B | 0.029 | pflB | 0.012 |
|
| |||||||
| IL18RAP | 0.017 | fliG | 0.020 | At5g62880 | 0.023 | rpoA | 0.012 |
|
| |||||||
| NRXN1 | 0.017 | uvrY | 0.019 | ARAC7 | 0.022 | pez | 0.012 |
|
| |||||||
| IL1B | 0.017 | pheS | 0.019 | ARAC2 | 0.022 | rpoB | 0.012 |
|
| |||||||
| IL18R1 | 0.017 | pepA | 0.019 | ARAC8 | 0.022 | rpoC | 0.012 |
|
| |||||||
| SYT13 | 0.016 | ptsI | 0.019 | ARAC10 | 0.022 | groL | 0.012 |
|
| |||||||
| Nbla00697 | 0.016 | pyrB | 0.017 | At5g62880 | 0.022 | groEL | 0.012 |
|
| |||||||
| NXPH1 | 0.016 | udk | 0.017 | ARAC9 | 0.022 | dnaK | 0.011 |
|
| |||||||
| NXPH2 | 0.016 | asd | 0.017 | ARAC3 | 0.021 | prsA | 0.011 |
|
| |||||||
| PYDC2 | 0.015 | cmk | 0.017 | ARAC4 | 0.021 | prs | 0.011 |
|
| |||||||
| CARD6 | 0.015 | pfs | 0.017 | At1g20090 | 0.021 | dps | 0.011 |
|
| |||||||
| EHMT1 | 0.015 | mtnN | 0.017 | ARAC5 | 0.021 | lpd | 0.011 |
|
| |||||||
| TIRAP | 0.015 | mtn | 0.017 | ARAC6 | 0.021 | lpdA | 0.011 |
|
| |||||||
| PLA2G4A | 0.014 | eutB | 0.017 | ARAC11 | 0.021 | rpsT | 0.011 |
|
| |||||||
| NLRP3 | 0.014 | gcvA | 0.017 | ARAC1 | 0.021 | rho | 0.011 |
|
| |||||||
| TRAPPC2 | 0.014 | gmd | 0.017 | ACT5 | 0.015 | rplF | 0.011 |
|
| |||||||
| TRAPPC2P1 | 0.014 | srlD | 0.017 | ACT9 | 0.015 | atpD | 0.011 |
|
| |||||||
| TRIM15 | 0.014 | yhbW | 0.017 | ACT2 | 0.015 | rplQ | 0.011 |
|
| |||||||
| IL1R2 | 0.013 | gutD | 0.017 | ACT8 | 0.015 | rplO | 0.011 |
|
| |||||||
| PLA2G5 | 0.013 | hydN | 0.017 | AT3G18780 | 0.015 | rpsI | 0.011 |
|
| |||||||
| C17orf59 | 0.013 | gudD | 0.017 | ACT4 | 0.015 | rpsM | 0.011 |
|
| |||||||
| NOD1 | 0.013 | ygcX | 0.017 | At5g59370 | 0.015 | rplE | 0.011 |
|
| |||||||
| CAST | 0.013 | ygcY | 0.017 | ACT12 | 0.014 | rpsA | 0.011 |
|
| |||||||
| SEPT9 | 0.013 | fdnI | 0.017 | ACT1 | 0.014 | rplL | 0.011 |
|
| |||||||
| MFSD1 | 0.013 | rpsJ | 0.016 | ACT3 | 0.014 | tuf | 0.011 |
|
| |||||||
| RAB27A | 0.013 | phoB | 0.016 | ACT7 | 0.014 | tuf_1 | 0.011 |
|
| |||||||
| CLIC2 | 0.013 | cysD | 0.016 | ACT11 | 0.014 | tuf1 | 0.011 |
|
| |||||||
| HSPA9 | 0.012 | metA | 0.016 | At3g12110/T21B14_108 | 0.014 | tuf2 | 0.011 |
|
| |||||||
| NLRP1 | 0.012 | pflB | 0.016 | F8M21_110 | 0.010 | tufA | 0.011 |
|
| |||||||
| SYTL1 | 0.012 | gntK | 0.016 | RPL27 | 0.010 | tufB | 0.011 |
|
| |||||||
| C20orf196 | 0.012 | galU | 0.016 | At4g02930 | 0.010 | rpsD | 0.011 |
|
| |||||||
| FAM35A | 0.012 | rpoC | 0.016 | TUFA | 0.010 | rpsE | 0.011 |
|
| |||||||
| ZNF644 | 0.012 | rpoB | 0.016 | At5g08670 | 0.010 | rpsB | 0.011 |
|
| |||||||
| ZNF828 | 0.012 | rplA | 0.016 | atpB | 0.010 | rpsC | 0.011 |
|
| |||||||
| TRAPPC6B | 0.012 | metF | 0.016 | At5g08690/T2K12_40 | 0.010 | rplD | 0.011 |
(a) BarA may interact with human Synaptotagmin-like protein 3
One of the predicted interactions that is ranked highly by GUILD is between the Salmonella protein BarA and the human Synaptotagmin-like protein 3. Synaptotagmin-like proteins 1, 2 and 3 (SYTL1-3) have been identified as a specific and direct binding partners of the GTP-bound form of Rab27A in vitro and in vivo [19]. Rab27A has been reported to be essential for exocytosis of granules from polymorphonuclear leukocytes [19]. Rab27A-deficiency leads to diminished secrection of myeloperoxidase in mice and it was proposed that SYTL1 and Rab27A are necessary for release of this enzyme [20]. Myeloperoxidase produces e.g. HOCl, a bactericidal oxidant [21]. Thus, it might be that Salmonella impairs vesicle trafficking and release of cytotoxic components by interacting with SYTL3.
(b) Salmonella dampens immune response by blocking IL18R1
The Secretin_N domain of Salmonella proteins InvG, Orf48 and SipA is predicted to interact with the immunoglobulin-like domain (V-set) of Interleukin-18 receptor 1 (IL18R1). IL18R1 belongs to the Interleukin-1 Receptor/Toll-Like Receptor Superfamily. This receptor has been shown to be expressed on intestinal epithelial cells. Studies with Cryptosporidium parvum, a parasitic protozoan, revealed that expression of antimicrobial peptides due to signaling through this receptor upon response to IL18 may contribute to innate defense against this pathogen [22]. Secondly, IL18 is known to stimulate IFNgamma production in T cells and natural killer cells which contributes to innate and adaptive immune responses. Moreover stimulation of IL18R1 leads to NF-kB activation [23]. Thus, the predicted interaction of the Salmonella proteins InvG, Orf48 and SipA with IL18R1 may block signaling through this receptor, thereby preventing an immune response. This is in line with the observation that Salmonella effector proteins AvrA, SseI, SseL and SspH1 are said to dampen the immune response by inhibiting activation of NF-kB [24–26].
(c) Salmonella invasion of the host cell
Salmonella proteins SopE, SopE2 and SptP are predicted to interact with Arabidopsis Rac-like GTP-binding proteins. This is in line with the findings that the same Salmonella effectors interact with human GTPase Rac1. Interaction of the guanine nucleotide exchange factor (GEF) SopE with human Rac1 leads to activation of this small GTPase, resulting in the stimulation of actin polymerization [27]. This along with other processes contributes to actin modification and membrane ruffling promoting the internalization of the bacteria into the host cell. Once Salmonella has been taken up by the cell, the process of actin remodeling is reversed by SptP. SptP inactivates Rac1 and down-regulates signaling through this GTPase [28]. To our knowledge, it is not known if the activation or down-regulation of Rac-like GTP-binding proteins is important for the response of Arabidopsis or other plants to pathogen infection.
(d) Interaction of SpvB with Arabidopsis actin proteins
It is known that SpvB interacts with mouse G-actin [29]. This interaction leads to the inhibition of actin polymerization based on the ADP-ribosyltransferase activity of SpvB. This is thought to result in reduced vacuole-associated actin polymerizations around the Salmonella-containing vacuole as well as disruption of the host cells’ cytoskeleton and induction of apoptosis [29]. To our knowledge, a similar mechanism of bacteria infecting plants is not known. However, targeting of plant actin by effector proteins of other phytopathogenic bacteria as well as actins playing a role in defense against pathogens is well established. The Pseudomonas syringae effector AvrPphB is believed to target the plant actin cytoskeleton in order to inhibit cellular trafficking processes [30]. The Arabidopsis protein that appears to respond to the effector is the Actin-Depolymerizing Factor (ADF), AtADF4. AtADF4 binds to G-actin and thereby prevents actin polymerization but also binds F-actin promoting depolymerization, believed to be one line of host defense against Pseudomonas syringae [31].
2.9. Putative roles of Salmonella effectors in suppressing host defense response based on predicted interactions
A number of key observations are outlined in sections (a)–(d), below.
(a) SptP may target the JAK/STAT signaling pathway
The model predicts the interaction of SptP with JAK1 (JAK1_HUMAN, Q4LDX3_HUMAN), JAK2 (JAK2_HUMAN, Q506Q0_HUMAN, Q8IXP2_HUMAN) and JAK3 (JAK3_HUMAN, Q8N1E8_HUMAN). These predictions are based on the contact between the Y_phosphatase domain of SptP and the Pkinase_Tyr domain of JAK proteins and additionally the SH2 domain of JAK2 (iPfam and 3DID). Moreover, the interaction of SptP with human STAT proteins (STAT1_HUMAN, STAT2_HUMAN, Q6LD48_HUMAN, STAT3_HUMAN, B5BTZ6_HUMAN, STAT4_HUMAN, E7EWJ5_HUMAN, Q53S87_HUMAN, STA5A_HUMAN, Q8WWS9_HUMAN, STA5B_HUMAN, STAT6_HUMAN) based on the interaction of the Y_phosphatase domain with the SH2 domain is predicted.
JAK proteins associate with cytokine receptors and mediate signal transduction by phosphorylation and thereby activation of STAT proteins which are transcription factors that regulate the transcription of selected genes in the cell nucleus. Rodig et al. demonstrated that JAK1 is essential for mediating biological responses induced by certain cytokine receptors [32]. For example, JAK1 deficient mice do not respond to INFalpha, IFNgamma and IL-10 [33]. This would indicate the possibility that Salmonella may interfere with the ability of the host cell to respond to cytokine signaling. Indeed, this was found to be the case in macrophages [34].
(b) SlrP, SspH1 and SspH2 are predicted to interact with Toll-like receptors (TLRs)
The Salmonella effectors SlrP, SspH1 and SspH2 are predicted to interact with human TLR1 to 10 (TLR1_HUMAN, TLR2_HUMAN, …, TLR10_HUMAN). The prediction is based on the interaction of the LRR_1 domains of both binding partners (iPfam). TLRs are involved in mediating immune responses to bacteria, NFKB activation, cytokine secretion and inflammatory responses. TLRs recognize a variety of microbial components, e.g TLR4 – lipopolysaccharides, TLR5 – flagellin, and thereby trigger antimicrobial responses of immune cells. Several TLR have been shown to be responsible for recognition of Salmonella. Salmonella enterica Choleraesuis is recognized by pig TLR5 and TLR1/2 [35]. TLR5-mediated recognition of Salmonella plays a role in many host species. Recent findings demonstrated that single amino acid exchanges in Salmonella flagellin alter species-specific host response (human, mouse, chicken) [36] as well as the occurrence of SNPs in TLR5 and TLR2 of different pig populations [35]. Beside those receptors TLR4, TLR9 (and/or TLR3) are involved in Salmonella enterica Typhimurium recognition [37]. On the other hand Salmonella requires TLRs for its virulence as bacteria cannot replicate in the absence of TLR2, 4 and 9. There is evidence that TLR-mediated acidification is necessary to induce SPI-2 encoded genes [37]. Flagellin also triggers defense signaling in plants, indicating that these effectors may play a similar role in plants. Domain-based comparison of TLR5_HUMAN with all Arabidopsis proteins that are predicted to interact with the same Salmonella proteins as TLR5 revealed that human TLR5 shares all its domains with 56 Arabidopsis proteins. The shared domains are the TIR-domain (PF01582), the LRR_1-domain (PF00560) and the LRR_4-domain (PF12799) which overlaps with the LRR_1-domain. These Arabidopsis proteins mainly comprise putative or uncharacterized disease resistance proteins (Table 5). Further implications of TLRs are discussed below.
Table 5.
Arabidopsis proteins that share domains with human TLR5.
| Uniprot entry name | Protein name | Gene name |
|---|---|---|
| Q9SZ66_ARATH | Putative disease resistance protein (TMV N-like) | F16J13.80 |
| Q9FKR7_ARATH | Disease resistance protein-like | |
| Q9FKB9_ARATH | Disease resistance protein | |
| Q9FGW1_ARATH | Disease resistance protein-like | |
| Q9SSP0_ARATH | Similar to downy mildew resistance protein RPP5 | F3N23.6 |
| Q9ZVX6_ARATH | Disease resistance protein (TIR-NBS-LRR class), putative | |
| A7LKN2_ARATH | TAO1 | |
| Q9LSV1_ARATH | Disease resistance protein RPP1-WsB | |
| O04264_ARATH | Downy mildew resistance protein RPP5 | RPP5 |
| B7U887_ARATH | Disease resistance protein RPP1-like protein R7 | |
| B7U885_ARATH | Disease resistance protein RPP1-like protein R5 | |
| B7U884_ARATH | Disease resistance protein RPP1-like protein R4 | |
| B7U888_ARATH | Disease resistance protein RPP1-like protein R8 | |
| Q9M285_ARATH | Disease resistence-like protein | T22K7_80 |
| Q9M1N7_ARATH | Disease resistance protein homlog | T18B22.70 |
| O49470_ARATH | Resistance protein RPP5-like | F24J7.80 |
| Q9SCZ3_ARATH | Disease resistance-like protein | F26O13.200 |
| Q9FI14_ARATH | Disease resistance protein-like | TAO1 |
| Q9ZSN4_ARATH | Disease resistance protein RPP1-WsC | |
| Q9ZSN5_ARATH | Disease resistance protein RPP1-WsB | |
| Q9ZSN6_ARATH | Disease resistance protein RPP1-WsA | |
| Q0WQ93_ARATH | Putative uncharacterized protein At1g72840 | |
| Q9FMB7_ARATH | Disease resistance protein-like | |
| A7LKN1_ARATH | TAO1 | |
| Q9FTA6_ARATH | T7N9.23 | |
| Q0WVG8_ARATH | Disease resistance like protein | |
| Q9SUK3_ARATH | Disease resistance RPP5 like protein | dl4500c |
| Q9CAE0_ARATH | Putative disease resistance protein; 17840-13447 | F24D7.6 |
| Q9CAD8_ARATH | Putative disease resistance protein; 27010-23648 | F24D7.8 |
| Q9FKN9_ARATH | Disease resistance protein | |
| O49468_ARATH | Resistence protein-like | F24J7.60 |
| Q9FHF0_ARATH | Disease resistance protein-like | |
| Q9FTA5_ARATH | T7N9.24 | |
| Q8S8G3_ARATH | Disease resistance protein (TIR-NBS-LRR class), putative | |
| Q9SW60_ARATH | Putative uncharacterized protein AT4g08450 | C18G5.30 |
| Q8GUQ4_ARATH | TIR-NBS-LRR | SSI4 |
| Q9FGT2_ARATH | Disease resistance protein-like | |
| Q9FH20_ARATH | Disease resistance protein-like | |
| Q9CAK1_ARATH | Putative disease resistance protein; 24665-28198 | T12P18.10 |
| Q9FNJ2_ARATH | Disease resistance protein-like | |
| Q9CAK0_ARATH | Putative disease resistance protein; 28811-33581 | T12P18.11 |
| Q9FKE2_ARATH | Disease resistance protein RPS4 | |
| Q9FFS5_ARATH | Disease resistance protein-like | |
| B7U882_ARATH | Disease resistance protein RPP1-like protein R2 | |
| B7U883_ARATH | Disease resistance protein RPP1-like protein R3 | |
| B7U881_ARATH | Disease resistance protein RPP1-like protein R1 | |
| Q7FKS0_ARATH | Putative disease resistance protein | At1g63880/T12P18_10 |
| O48573_ARATH | Disease resistance protein-like | T19K24.2 |
| Q0WNV7_ARATH | Resistence protein-like | |
| Q9M1P1_ARATH | Disease resistance protein homolog | T18B22.30 |
| O23536_ARATH | Disease resistance RPP5 like protein | dl4510c |
| C0KJS9_ARATH | Disease resistance protein (TIR-NBS-LRR class) | |
| Q9FN83_ARATH | Disease resistance protein-like | |
| Q56YL9_ARATH | Disease resistance-like protein | At3g44400 |
| Q9FKE5_ARATH | Disease resistance protein RPS4 | |
| Q9M8X8_ARATH | Putative disease resistance protein | T6K12.16 |
(c) SlrP, SspH1, SspH2 and SptP are predicted to interact with the Arabidopsis protein with EFR
The LRR_1 domain of SlrP, SspH1 and SspH2 may interact with the LRR_1 and/or the LRRNT_2 domain (iPfam) of EFR (EFR_ARATH, LRR receptor-like serine/threonine-protein kinase EFR or Elongation factor Tu receptor) whereas the Y_phosphatase domain of SptP is predicted to interact with the Pkinase_Tyr domain of this Arabidopsis protein (iPfam and 3DID). EFR is a plant pathogen recognition receptor (PRR) that binds the PAMP (pathogen associated molecular pattern) elf18 peptide of elongation factor EF-Tu and thereby triggers the host defense [38]. The Pseudomonas syringae effector AvrPto is known to bind EFR which inhibits PAMP-triggered immunity and thereby promotes virulence [39]. It is possible that a similar mechanism is used by Salmonella.
(d) Interaction of Salmonella effectors with Arabidopsis disease resistance proteins
Another PPI that may be based on the contact between two LRR_1 domains is the interaction of SlrP, SspH1 and SspH2 with RPS2 (RPS2_ARATH, Disease resistance protein RPS2 or Resistance to Pseudomonas syringae protein 2). Based on the same domain interaction and additionally on the interaction between LRR_1 (SlrP, SspH1, SspH2) and LRRNT_2 (RPP27) these Salmonella effectors are also predicted to interact with other Arabidopsis disease resistance proteins. These are RPP1 (D9IW02_ARATH, Recognition of Peronospora parasitica 1), RPP4 (Q8S4Q0_ARATH, Disease resistance protein RPP4), RPP5 (O04264_ARATH, Downy mildew resistance protein RPP5) and RPP27 (Q70CT4_ARATH, RPP27 protein). Plant disease resistance proteins specifically recognize pathogenic avirulence proteins (Avr) and share high structural and functional similarity with mammalian TLRs [40]. Arabidopsis RPS2 recognizes Pseudomonas syringae AvrRpt2 and thereby triggers a defense response. A homologue with 58 % identity in the functional domain, AvrRpt2EA, is present in Erwinia amylovora and has been shown to contribute to virulence [41]. RPP1, RPP4, RPP5 and RPP27 are known to contribute to disease resistance against the Peronospora parasitica, the causal agent of downy mildew, and recognize a variety of avirulence proteins (ATR Arabidopsis thaliana recognized proteins) resulting in host resistance (for details see [42]).
2.10. Topological network analysis
The network topology of the different predicted Salmonella-host networks was analyzed by in-depth analysis of its components and clusters. Components refer to sub-networks in which any two nodes are connected to each other by paths. Clusters are groups of nodes in the network having a high connectivity between them. We measured different parameters relating the properties of these bipartite graphs (Table 7). Pathogen-host PPI networks are bipartite graphs because they are composed of two independent sets of proteins (namely from two different species) having edges (predicted interactions) between them. There are no predicted interactions between proteins within the same species, which makes these networks different from intraspecies interactomes. The following parameters are listed in Table 7: 1) number of connected; 2) number of and average clustering coefficient applied to bipartite graphs, split into Salmonella and host proteins; 3) network density coefficients, split also by pathogen and host proteins; 4) scale-free network properties, based on number of predictions for each protein (node degree).
Table 7.
Network topology.
| Network | Is scale- free? P<0.01 |
Number of clusters (I = 1.7) |
Number of clusters with known effectors |
Number of components |
Number of components with known effectors |
Average clustering |
Average host clustering |
Average Salmonella clustering |
Host density |
Salmonella density |
|---|---|---|---|---|---|---|---|---|---|---|
| Human_union | Yes | 372 | 13 | 35 | 2 | 0.29 | 0.21 | 0.3 | 0.006 | 0.006 |
| Human_intersection | Yes | 28 | 4 | 26 | 4 | 0.94 | 0.95 | 0.66 | 0.033 | 0.033 |
| Human_sequence-based | Yes | 292 | 6 | 49 | 3 | 0.38 | 0.38 | 0.33 | 0.003 | 0.003 |
| Human_domain-based | Yes | 311 | 12 | 148 | 1 | 0.4 | 0.4 | 0.38 | 0.007 | 0.007 |
| Arabidopsis_union | Yes | 319 | 9 | 61 | 1 | 0.38 | 0.4 | 0.25 | 0.006 | 0.006 |
| Arabidopsis_intersection | No (P = 0.0116) | 22 | 2 | 17 | 2 | 0.76 | 0.84 | 0.56 | 0.037 | 0.037 |
| Arabidopsis_sequence-based | Yes | 13 | 2 | 3 | 1 | 0.39 | 0.41 | 0.38 | 0.008 | 0.008 |
| Arabidopsis_domain-based | Yes | 342 | 9 | 173 | 2 | 0.45 | 0.47 | 0.38 | 0.007 | 0.007 |
All predicted networks contained few components and clusters containing a large number of proteins and several components and cluster with few proteins. Predictions based only on domain composition produced more unconnected networks (i.e. having more components), while sequence-based predictions produced a more connected network. The number of components containing known Salmonella effectors is low, indicating that effectors are found in a small number of groups. The same pattern was observed when clustering. Clustering coefficients and network densities were very similar when comparing the human and Arabidopsis networks, as well as when comparing sequence and domain based inference methods. In contrast, these parameters change when applied to the intersection network (Table 7), probably due to the smaller size of this network.
PPI network topologies are generally characterized by a low number of highly connected hubs, and a large number of proteins with few connections, referred to as a scale-free network topology [43]. A power-law distribution of the number of PPIs is a characteristic of a scale-free network. This distribution was indeed observed here with statistical significance (P < 0.01), except the prediction based on the intersection of sequence-based and domain-based methods. In this case, a power-law distribution was fit with a value of P < 0.05. Probably this difference is due to the small size of the network.
2.11. Functional enrichment analysis
Interacting proteins are likely to share biological processes or share similar locations compared to non-interacting proteins [44]. The results are shown in Table S5. Three clusters of the Salmonella-human sequence-based predicted network are significantly enriched with GO-terms. Two human proteins of cluster40, SAHH2 and SAHH3, are annotated with the GO-terms “adenosylhomocysteinase activity” and “trialkylsulfonium hydrolase activity”. The GO-term “interleukin-8 binding” is significant for cluster2 and associated with the human proteins CXCR1 and CXCR2. Proteins in cluster0 are annotated with 36 unique GO-terms which allow the proteins to function e.g. in antigen processing and presentation, the MHC complex, translation and protein disassembly (Table S5).
Eight of the 13 Salmonella effector-containing clusters in the Salmonella-human network are significantly enriched with 329 unique GO-terms. When building logical and functional related groups of the most prominent GO-terms enriched within one cluster, the results can be summarized as follows: Cluster0 harbors proteins that play a role in the MHC protein complex, small GTPase mediated signaling and protein kinase activity. Proteins in cluster1 dominantly play a role in processes and molecules related to gene expression and cellular component disassembly. The five human protein of cluster186 for which GO-terms are enriched are UBC, UBB, UB2L3, RL40 and RS27. These proteins function in cell cycle regulation, ubiquitination, antigen processing and presentation, TLR signaling as well as kinase and ligase activity. Many proteins of cluster3 are associated with proteolysis, peptidase and serine hydrolase activity. Cluster38 comprises protein related to transferase activity and several metabolic processes. In cluster5 e.g. the GO-term “actin binding” as well as those pointing at phosphatase activity are enriched. Proteins in cluster7 predominantly are associated with cell adhesion. 665 proteins of cluster8 are integral membrane proteins of which many are annotated to have receptor activity (Table S6).
Finally, we calculated the functional enrichment in the GUILD dataset. To this end, the union networks of sequence- and domain-based predictions, was subjected to GUILD analysis (see above). Those host proteins with the top 100 GUILD-netscores were selected. In the case of the human proteins, the highly ranked GUILD proteins function in cell death and apoptosis as well as immune response, cytokine production and secretion, protein secretion, transport and localization, peptidase activity and kinase cascades (Table S7). Annotations for the top 100 GUILD-scored Arabidopsis proteins are quite different from those for the human ones. When analyzing the over-representation of GO-terms related to biological processes only, only 19 of the top-scored proteins reveal a GO-term annotation. These are “protein tetramerization”, “ATP hydrolysis coupled proton transport”, “energy coupled proton transport, against electrochemical gradient” and “small GTPase mediated signal transduction” (Table S8a). Because of this low number of process terms presumably due to the lesser annotation of Arabiopsis proteins as compared to human proteins, we subsequently included all GO-terms in the analysis. This resulted in enrichment of Arabidopsis proteins with GO function annotations, such as “GTP binding”, “phosphatase activity” and ATPase activity (Table S8b).
3. Conclusions
The present work demonstrates that retrieval of putative interactions based on sequence and domain similarity to known interactions are valuable in predicting host-pathogen interactions. First, the model presented successfully predicted a set of gold standard Salmonella-host PPIs. Furthermore, so far undiscovered interactions between Salmonella effector proteins and host targets were predicted and used successfully to formulate biological hypotheses. These include helping identify conserved or distinguishing mechanisms used by Salmonella when infecting and proliferating in humans and plants. We specifically suggested a number of putative mechanisms by which Salmonella proteins may suppress the immune response elicited by the host, for both, plant and human hosts. Finally, this approach may also be useful to predict the function of so far uncharacterized proteins.
Interolog information has been used previously to predict PPIs, both for intraspecies and interspecies predictions [16][45]. With particular relevance to this work, Krishnadev et al. [13] obtained a list of predicted interactions between human host and Salmonella using a conceptually similar approach. However, there are a number of differences that should be highlighted. Numerous publications show that there is low overlap in public PPI repositories [46]. As a consequence, by using a single database of PPIs chosen by Krishnadev et al. as opposed to a database in which several resources are integrated as is done in the BIANA framework [47] employed here, one would expect to obtain a larger number of predicted pairs. More specifically, in the work of Krishnadev et al. DIP was used as the source for protein pairs and iPfam was used to identify homologues. In contrast, we have integrated interactions from 10 different resources instead of just DIP, and furthermore included domain relationships from 3DiD, where interactions can be structurally modeled. Finally, Krishnadev et al. used only Salmonella enterica Typhimurium, while we applied it to different Salmonella species and two different hosts (human and Arabidopsis). This enables the user with the flexibility of searching for interactions for a specific Salmonella species. Since the approach is general, this work can easily be extended to comparison of other hosts.
While a number of interesting biological hypotheses were derived from the predictions, these have to be seen with caution as they are only based on sequence and domain similarity. Although similar proteins often can interact with the same or similar proteins, there a many examples where it has been shown that very similar proteins do not interact with the same target protein. In the specific case of Salmonella-host interactions, for example, SspH1 is known to bind to PKN1 but as shown by immunoprecipitation, SspH2 and SlrP do not interact with this protein [26]. Vice versa SspH2 binds Filamin A and Profilin-1 whereas these interactions could not be shown for SspH1 and SlrP in a yeast-2-hybrid experiment [48]. One more example is the interaction between PipB2 and KLC which could not be detected for PipB using co-immunoprecipitation [49].
The quality of the putative interactomes could further be improved by combining this method with other computational approaches and by including other biological data sources, e.g. transcriptomic other -omics or localization data, in the predictions. This would reduce the number of false positive predictions. In any case, predicted interactions require experimental validation.
To enable other users to benefit from the models developed and stimulate experimentalists to inspect and validate the predicted interactions, a web interface is available at http://sbi.imim.es/web/SHIPREC.php.
Experimental Part
Prediction of interactions based on homology detection and domain assignment
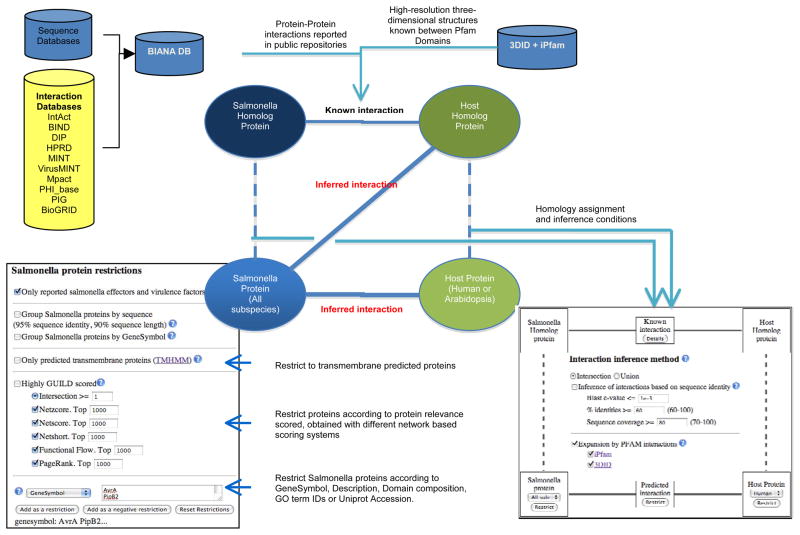
Salmonella-Host (Salmonella-human and Salmonella-Arabidopsis) interacting proteins have been predicted using the interologs approach [16]. The hypothesis of this approach is that two proteins (A and B) interact if it exists a known interaction between two proteins (A′ and B′) such that A is similar to A′ and B similar to B′. Proteins A and B are named target proteins and A′ and B′ template proteins. The basis of the hypothesis is to assume the similar behavior of homolog proteins. However, other approaches have only required the similarity of the residues of interface of the interaction [50], which means that non-homologous proteins can also reproduce the same interaction. Therefore, we have used two different criteria to measure this similarity. The first approach uses sequence similarity between proteins based on the sequence alignment. We align the sequences of two proteins to measure their similarity as a function of the percentage of identical residues and the percentage of their sequence being aligned (i.e. using 60 % identity and 70 % of the total length of the target protein and 90 % of the template). In the second approach we measure the similarity of the target sequences (A and B) with PFAM domains as a function of the e-value calculated with the package HMMER [51]. This results in the assignation of one or several PFAM domains to the target sequences. Then, we use the database of iPfam and 3DiD to check for domain-domain interactions. We hypothesize that A interacts with B if a domain A′ can be assigned to A and a domain B′ to B such that A′ and B′ are interacting domains in iPfam or 3DiD. Furthermore, it has been shown that the specificity of some interactions depends on a set of interacting domains [52]. Therefore, the most restrictive set of predictions will be those for which both criteria of similarity are required, using stringent values of sequence identity, coverage and domain assignation (Fig. 4).
Fig. 4.
Schematic representation of the prediction model and database availability via web interface.
The last step to generate the network of interactions between proteins of Salmonella and human and between Salmonella and Arabidopsis is a clustering of similar pairs. Pairs of interactions can be grouped by gene symbol or by sequence similarity. Grouping by gene symbol is obtained by joining all PPIs containing Salmonella proteins that correspond to the same gene symbol (see the correspondence between gene symbols and Uniprot entry name in Table S2). Grouping by sequence similarity is obtained by joining all pairs of PPIs for which the similarity of their sequences is calculated with an alignment and this shows more than 95 % identical residues and for more than 90 % of the sequence length.
Database sources
Protein sequences of Salmonella, Human and Arabidopsis were extracted from the Uniprot Knowledgebase [53]. In order to avoid missing proteins annotated only in one or few subspecies of Salmonella, we considered all proteins belonging to a taxon inside the Salmonella genus (taxonomy ID 590) to generate a virtual Salmonella proteome. For Human and Arabidopsis proteomes we took all proteins of the taxon 9,606 and 3,702, respectively.
PPIs used as templates for the prediction were extracted using BIANA framework [47] that integrates 10 different databases: DIP [54], HPRD [55], IntAct [56], MINT [57], MPact [58], PHI_base [59], PIG [60], BioGRID [61], BIND [62] and VirusMINT [63]. Using the integration of multiple sources instead of a single source allows a more comprehensive view of interactions and enlarges the set of predictions (as it is known different databases contain a high number of non-overlapping PPIs [46] [http://www.omicsonline.com/Archive/HTMLJuly2008/JPB1.166.html].
Domain-domain interactions used as templates were extracted from the union of the 3DID [64] and iPfam [65] databases. Both databases define interactions between Pfam domains for which high-resolution three-dimensional structures are known. Also, the use of more than a single source allowed a more comprehensive view of known domain-domain interactions.
Gold standard
A dataset of known Salmonella-host protein-protein interactions (PPIs) was obtained by intensive literature and database search screening more than 2,200 journal articles and over 100 databases [15]. This yielded a set of 59 direct and three indirect Salmonella-host PPIs involving 22 Salmonella effectors and 50 host proteins. Of those 62 PPIs 38 have been reviewed before (Haraga et al. 2008 [66], McGhie et al. 2009 [67], Heffron et al. 2011 [25]) but only 16 can be found in databases including only 6 that are listed in the databases DIP, IntAct, PIG and/or BIND whereas the others are found in the descriptions of the uniprot database (www.uniprot.org). This dataset only contains interactions that have been verified by us based of the reliability of the experiments described in the journal article(s). Thus, this dataset of Salmonella-host PPIs represents the most complete Salmonella-host interactome available to date [15].
Parameters used for homology detection and domain assignment
PPI inference between Salmonella and host proteins has been done by sequence similarity with known interacting pairs as follows: Alignments were done using PSI-BLAST with an e-value cutoff threshold of 10−3. 90 % of the template sequence and 20 % of the target were aligned, and the alignment had a minimum of 20 % of identical residues. For assigning Pfam domains we used an E-value cutoff of 10−5 obtained with HMMER 3.0 [51] and the Pfam A database [68].
Selected sub-networks
The full predicted network of interactions of proteins from Salmonella interacting with proteins of the host (human or Arabidopsis) is very large. To ease interpretation of the predictions, we have designed several filters to select specific sub-networks of interest, and these subnetworks are referred to specifically in the text: a) subnetwork containing interactions with known effectors of Salmonella invasion; b) subnetwork containing interactions with transmembrane proteins (likely involved in the pathogen invasion of the host cell); c) subnetwork of interacting pairs sharing similar functions; and d) subnetwork containing interactions with known and predicted effectors and relevant proteins for Salmonella invasion.
Subnetwork of known Salmonella effectors
The most interesting predicted interactions are those in which known Salmonella effectors are involved, as these proteins are known to enhance pathogen virulence and to alter functions in the host. A list of 59 known Salmonella effectors has been used to filter the prediction set.
Subnetwork of transmembrane proteins
The recognition between pathogens and hosts is mostly due to surface structures [69]. Consequently, in order to select interactions that could be involved in the Salmonella-human and Salmonella-Arabidopsis recognition, we applied the TMHMM software [70] to predict transmembrane proteins and to select the subnetwork containing these proteins.
Subnetwork with functional annotation
Predicted Salmonella-host interactions were filtered if the involved proteins shared similar GO terms. A GO term is considered similar if they are equal or if there is a parenthood relation in the GO ontology hierarchy.
Subnetwork with relevant proteins of Salmonella invasion
The GUILD method [submitted] was used to identify genes associated with the infection of hosts by Salmonella. The GUILD framework has been applied to the predicted networks of Human and Salmonella obtained with the union of sequence and domain based prediction methods, in which gold standard Salmonella-host interactions found by literature search and known interactomes of host and Salmonella were added (both reported in source databases described above). The method requires a set of proteins (or genes) known to be associated with a phenotype. We have used the list of known effectors of Salmonella to infer new putative effectors or proteins of the host that are relevant for the invasion based on ranked GUILD scored.
Network topology analysis
To calculate topological parameters of the network, we have used the networkx.bipartite module of Python [71]. To study possible topological modules in the network, we have divided the network in connected components and clusters. Components consist on subnetworks in which every node is connected to the other nodes of the subnetwork by a path (i.e. there does not exist a path between two nodes from different components). Topological clusters consist of groups of nodes of the network being highly connected between them. We have clustered the networks by using the MCL algorithm, using a granularity coefficient of 1.7 [72]. Scale-freeness of the networks has been calculated as described by Khanin et al. [43].
Functional enrichment analysis
Functional relations in the network modules were analyzed by using the functional enrichment algorithm, FuncAssociate 2.0 [73], applied to the clusters containing known Salmonella effectors and the top scoring GUILD proteins.
Browser of the dataset of predicted cross-talk between Salmonella and hosts (human, Arabidopsis)
The predictions are available at http://sbi.imim.es/web/SHIPREC.php. Users can browse them with the ability to filter the data:
Salmonella and host proteins. It is possible to filter transmembrane predicted proteins or specific groups of proteins (identified or excluded by gene symbol or uniprot accession identifiers, having a specific annotated functionality, domain or keyword). For Salmonella proteins, it is also possible to show only known effectors or virulence factors, and to select which Salmonella subspecies to use. For host proteins, the user has to select which host to use (human or Arabidopsis). Also, Salmonella and host proteins can be selected according to top ranked GUILD scores. PPIs can be grouped in sets of proteins of each partner of the interaction with similar sequence (using 95 % sequence identity and 90 % sequence length) or by gene symbol.
Prediction conditions. It is possible to select the prediction method (based on sequence similarity or based on known interacting domains) and the conditions used to obtain them. The user can combine results from both methods by union or intersection.
Predicted interactions. Predicted interaction pairs can be filtered according to GO annotation terms of involved proteins (biological process, cellular component or molecular function).
Output. The result of the applied filter and selection criteria entered by the user, is a table with the details of each prediction, and the details of the PPI template used for inference.
Interactome prediction and analysis
The Salmonella-host interactomes described and analyzed here in detail have been obtained by applying the following parameters using the web interface: union of sequence identity (e-value 10−3, sequence identity 60 %, sequence coverage 70 %) and domain (iPfam and/or 3DID) identity for all Salmonella species; restrict to only Salmonella effectors and virulence factors; group Salmonella proteins by gene symbol. The received PPI dataset has been edited by deleting gene symbol duplicates. This was necessary as some Salmonella effectors had two or more gene symbols which were ordered in different ways depending on the Salmonella serovar. E.g. the three gene symbols of SpvC (SpvC, MkaD, MkfA) were ordered in five different ways which resulted in duplications of PPIs. All these different entries were substituted by SpvC. The final step was the analysis and visualization of the obtained datasets with Cytoscape 2.8. [74].
Supplementary Material
Acknowledgments
This work was funded by the Federal Ministry of Education and Research (BMBF) and the Euroinvestigacion program of MICINN (Spanish Ministry of Science and Innovation), partners of the ERASysBio+ initiative supported under the EU ERA-NET Plus scheme in FP7.
Contributor Information
Judith Klein-Seetharaman, Email: jks33@pitt.edu.
Baldo Oliva, Email: baldo.oliva@upf.edu.
References
- 1.Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. Proc Natl Acad Sci U S A. 2001;98:4569. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]; Tarassov K, Messier V, Landry CR, Radinovic S, Serna Molina MM, Shames I, Malitskaya Y, Vogel J, Bussey H, Michnick SW. Science. 2008;320:1465. doi: 10.1126/science.1153878. [DOI] [PubMed] [Google Scholar]
- 2.Li S, Armstrong CM, Bertin N, Ge H, Milstein S, Boxem M, Vidalain PO, Han JD, Chesneau A, Hao T, Goldberg DS, Li N, Martinez M, Rual JF, Lamesch P, Xu L, Tewari M, Wong SL, Zhang LV, Berriz GF, Jacotot L, Vaglio P, Reboul J, Hirozane-Kishikawa T, Li Q, Gabel HW, Elewa A, Baumgartner B, Rose DJ, Yu H, Bosak S, Sequerra R, Fraser A, Mango SE, Saxton WM, Strome S, Van Den Heuvel S, Piano F, Vandenhaute J, Sardet C, Gerstein M, Doucette-Stamm L, Gunsalus KC, Harper JW, Cusick ME, Roth FP, Hill DE, Vidal M. Science. 2004;303:540. [Google Scholar]
- 3.Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, Vijayadamodar G, Pochart P, Machineni H, Welsh M, Kong Y, Zerhusen B, Malcolm R, Varrone Z, Collis A, Minto M, Burgess S, McDaniel L, Stimpson E, Spriggs F, Williams J, Neurath K, Ioime N, Agee M, Voss E, Furtak K, Renzulli R, Aanensen N, Carrolla S, Bickelhaupt E, Lazovatsky Y, DaSilva A, Zhong J, Stanyon CA, Finley RL, Jr, White KP, Braverman M, Jarvie T, Gold S, Leach M, Knight J, Shimkets RA, McKenna MP, Chant J, Rothberg JM. Science. 2003;302:1727. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]; Formstecher E, Aresta S, Collura V, Hamburger A, Meil A, Trehin A, Reverdy C, Betin V, Maire S, Brun C, Jacq B, Arpin M, Bellaiche Y, Bellusci S, Benaroch P, Bornens M, Chanet R, Chavrier P, Delattre O, Doye V, Fehon R, Faye G, Galli T, Girault JA, Goud B, de Gunzburg J, Johannes L, Junier MP, Mirouse V, Mukherjee A, Papadopoulo D, Perez F, Plessis A, Rosse C, Saule S, Stoppa-Lyonnet D, Vincent A, White M, Legrain P, Wojcik J, Camonis J, Daviet L. Genome Res. 2005;15:376. doi: 10.1101/gr.2659105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braun P, Carvunis AR, Charloteaux B, Dreze M, Ecker JR, Hill DE, Roth FP, Vidal M, Galli M, Balumuri P, Bautista V, Chesnut JD, Kim RC, de los Reyes C, Gilles P, Kim C, Matrubutham U, Mirchandani J, Olivares E, Patnaik S, Quan R, Ramaswamy G, Shinn P, Swamilingiah GM, Wu S, Ecker JR, Dreze M, Byrdsong D, Dricot A, Duarte M, Gebreab F, Gutierrez BJ, MacWilliams A, Monachello D, Mukhtar MS, Poulin MM, Reichert P, Romero V, Tam S, Waaijers S, Weiner EM, Vidal M, Hill DE, Braun P, Galli M, Carvunis AR, Cusick ME, Dreze M, Romero V, Roth FP, Tasan M, Yazaki J, Braun P, Ecker JR, Carvunis AR, Ahn YY, Barabási AL, Charloteaux B, Chen H, Cusick ME, Dangl JL, Dreze M, Ecker R, Fan C, Gai L, Galli M, Ghoshal G, Hao T, Hill DE, Lurin C, Milenkovic T, Moore J, Mukhtar MS, Pevzner SJ, Przulj N, Rabello S, Rietman EA, Rolland T, Roth FP, Santhanam B, Schmitz RJ, Spooner W, Stein J, Tasan M, Vandenhaute J, Ware D, Braun P, Vidal M. Science. 2011;333:601. doi: 10.1126/science.1203877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y, Cui T, Zhang C, Yang M, Huang Y, Li W, Zhang L, Gao C, He Y, Li Y, Huang F, Zeng J, Huang C, Yang Q, Tian Y, Zhao C, Chen H, Zhang H, He ZG. J Proteome Res. 2010;9:6665. doi: 10.1021/pr100808n. [DOI] [PubMed] [Google Scholar]
- 6.Arifuzzaman M, Maeda M, Itoh A, Nishikata K, Takita C, Saito R, Ara T, Nakahigashi K, Huang HC, Hirai A, Tsuzuki K, Nakamura S, Altaf-Ul-Amin M, Oshima T, Baba T, Yamamoto N, Kawamura T, Ioka-Nakamichi T, Kitagawa M, Tomita M, Kanaya S, Wada C, Mori H. Genome Res. 2006;16:686. doi: 10.1101/gr.4527806. [DOI] [PMC free article] [PubMed] [Google Scholar]; Butland G, Peregrin-Alvarez JM, Li J, Yang W, Yang X, Canadien V, Starostine A, Richards D, Beattie B, Krogan N, Davey M, Parkinson J, Greenblatt J, Emili A. Nature. 2005;433:531. doi: 10.1038/nature03239. [DOI] [PubMed] [Google Scholar]
- 7.Rain JC, Selig L, De Reuse H, Battaglia V, Reverdy C, Simon S, Lenzen G, Petel F, Wojcik J, Schachter V, Chemama Y, Labigne A, Legrain P. Nature. 2001;409:211. doi: 10.1038/35051615. [DOI] [PubMed] [Google Scholar]
- 8.Cherkasov A, Hsing M, Zoraghi R, Foster LJ, See RH, Stoynov N, Jiang J, Kaur S, Lian T, Jackson L, Gong H, Swayze R, Amandoron E, Hormozdiari F, Dao P, Sahinalp C, Santos-Filho O, Axerio-Cilies P, Byler K, McMaster WR, Brunham RC, Finlay BB, Reiner NE. J Proteome Res. 2011;10:1139. doi: 10.1021/pr100918u. [DOI] [PubMed] [Google Scholar]
- 9.Parrish JR, Yu J, Liu G, Hines JA, Chan JE, Mangiola BA, Zhang H, Pacifico S, Fotouhi F, DiRita VJ, Ideker T, Andrews P, Finley RL., Jr Genome Biol. 2007;8:R130. doi: 10.1186/gb-2007-8-7-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dyer MD, Neff C, Dufford M, Rivera CG, Shattuck D, Bassaganya-Riera J, Murali TM, Sobral BW. PLoS One. 2010;5:e12089. doi: 10.1371/journal.pone.0012089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tastan O, Qi Y, Carbonell JG, Klein-Seetharaman J. Pac Symp Biocomput. 2009;516 [PMC free article] [PubMed] [Google Scholar]; Nouretdinov I, Gammerman A, Qi Y, Klein-Seetharaman J. Pacific Symposium Biocomputing. 2012 in press. [PMC free article] [PubMed] [Google Scholar]
- 12.Dyer MD, Murali TM, Sobral BW. PLoS Pathog. 2008;4:e32. doi: 10.1371/journal.ppat.0040032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krishnadev O, Srinivasan N. Int J Biol Macromol. 2011;48:613. doi: 10.1016/j.ijbiomac.2011.01.030. [DOI] [PubMed] [Google Scholar]
- 14.Zhao Z, Xia J, Tastan O, Singh I, Kshirsagar M, Carbonell J, Klein-Seetharaman J. Int J Comput Biol Drug Des. 2011;4:83. doi: 10.1504/IJCBDD.2011.038658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schleker S, Sun J, Raghavan B, Srnec M, Mueller N, Koepfinger M, Murthy L, Zhao Z, Klein-Seetharaman J. Proteomics Clin Appl. 2012 doi: 10.1002/prca.201100083. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu H, Luscombe NM, Lu HX, Zhu X, Xia Y, Han JD, Bertin N, Chung S, Vidal M, Gerstein M. Genome Res. 2004;14:1107. doi: 10.1101/gr.1774904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. Nucleic Acids Res. 2000;28:235. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gandhi TK, Zhong J, Mathivanan S, Karthick L, Chandrika KN, Mohan SS, Sharma S, Pinkert S, Nagaraju S, Periaswamy B, Mishra G, Nandakumar K, Shen B, Deshpande N, Nayak R, Sarker M, Boeke JD, Parmigiani G, Schultz J, Bader JS, Pandey A. Nat Genet. 2006;38:285. doi: 10.1038/ng1747. [DOI] [PubMed] [Google Scholar]; Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabasi AL. Proc Natl Acad Sci U S A. 2007;104:8685. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lim J, Hao T, Shaw C, Patel AJ, Szabo G, Rual JF, Fisk CJ, Li N, Smolyar A, Hill DE, Barabasi AL, Vidal M, Zoghbi HY. Cell. 2006;125:801. doi: 10.1016/j.cell.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 19.Kuroda TS, Fukuda M, Ariga H, Mikoshiba K. J Biol Chem. 2002;277:9212. doi: 10.1074/jbc.M112414200. [DOI] [PubMed] [Google Scholar]
- 20.Munafo DB, Johnson JL, Ellis BA, Rutschmann S, Beutler B, Catz SD. Biochem J. 2007;402:229. doi: 10.1042/BJ20060950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hampton MB, Kettle AJ, Winterbourn CC. Blood. 1998;92:3007. [PubMed] [Google Scholar]
- 22.McDonald V, Pollok RC, Dhaliwal W, Naik S, Farthing MJ, Bajaj-Elliott M. Clin Exp Immunol. 2006;145:555. doi: 10.1111/j.1365-2249.2006.03159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dinarello CA. J Allergy Clin Immunol. 1999;103:11. doi: 10.1016/s0091-6749(99)70518-x. [DOI] [PubMed] [Google Scholar]
- 24.Ye Z, Petrof EO, Boone D, Claud EC, Sun J. Am J Pathol. 2007;171:882. doi: 10.2353/ajpath.2007.070220. [DOI] [PMC free article] [PubMed] [Google Scholar]; Le Negrate G, Faustin B, Welsh K, Loeffler M, Krajewska M, Hasegawa P, Mukherjee S, Orth K, Krajewski S, Godzik A, Guiney DG, Reed JC. J Immunol. 2008;180:5045. doi: 10.4049/jimmunol.180.7.5045. [DOI] [PubMed] [Google Scholar]
- 25.Heffron F, Nieman G, Yoon H, Kidwai A, Brown RNE, McDermott JD, Smith R, Adkins JN. In: Salmonella: From Genome to Function. Porwollik S, editor. Caister Academic Press; Norfolk: 2011. p. 187. [Google Scholar]
- 26.Haraga A, Miller SI. Cell Microbiol. 2006;8:837. doi: 10.1111/j.1462-5822.2005.00670.x. [DOI] [PubMed] [Google Scholar]
- 27.Hardt WD, Chen LM, Schuebel KE, Bustelo XR, Galan JE. Cell. 1998;93:815. doi: 10.1016/s0092-8674(00)81442-7. [DOI] [PubMed] [Google Scholar]
- 28.Fu Y, Galan JE. Nature. 1999;401:293. doi: 10.1038/45829. [DOI] [PubMed] [Google Scholar]; Rodriguez-Pachon JM, Martin H, North G, Rotger R, Nombela C, Molina M. J Biol Chem. 2002;277:27094. doi: 10.1074/jbc.M201527200. [DOI] [PubMed] [Google Scholar]
- 29.Tezcan-Merdol D, Nyman T, Lindberg U, Haag F, Koch-Nolte F, Rhen M. Mol Microbiol. 2001;39:606. doi: 10.1046/j.1365-2958.2001.02258.x. [DOI] [PubMed] [Google Scholar]; Margarit SM, Davidson W, Frego L, Stebbins CE. Structure. 2006;14:1219. doi: 10.1016/j.str.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 30.Day B, Graham T. Ann N Y Acad Sci. 2007;1113:123. doi: 10.1196/annals.1391.029. [DOI] [PubMed] [Google Scholar]
- 31.Tian M, Chaudhry F, Ruzicka DR, Meagher RB, Staiger CJ, Day B. Plant Physiol. 2009;150:815. doi: 10.1104/pp.109.137604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodig SJ, Meraz MA, White JM, Lampe PA, Riley JK, Arthur CD, King KL, Sheehan KC, Yin L, Pennica D, Johnson EM, Jr, Schreiber RD. Cell. 1998;93:373. doi: 10.1016/s0092-8674(00)81166-6. [DOI] [PubMed] [Google Scholar]
- 33.Kisseleva T, Bhattacharya S, Braunstein J, Schindler CW. Gene. 2002;285:1. doi: 10.1016/s0378-1119(02)00398-0. [DOI] [PubMed] [Google Scholar]
- 34.Salzman AL, Eaves-Pyles T, Linn SC, Denenberg AG, Szabo C. Gastroenterology. 1998;114:93. doi: 10.1016/s0016-5085(98)70637-7. [DOI] [PubMed] [Google Scholar]
- 35.Shinkai H, Suzuki R, Akiba M, Okumura N, Uenishi H. Mol Immunol. 2011;48:1114. doi: 10.1016/j.molimm.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 36.Keestra AM, de Zoete MR, van Aubel RA, van Putten JP. Mol Immunol. 2008;45:1298. doi: 10.1016/j.molimm.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 37.Arpaia N, Godec J, Lau L, Sivick KE, McLaughlin LM, Jones MB, Dracheva T, Peterson SN, Monack DM, Barton GM. Cell. 2011;144:675. doi: 10.1016/j.cell.2011.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, Felix G. Cell. 2006;125:749. doi: 10.1016/j.cell.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 39.Zong N, Xiang T, Zou Y, Chai J, Zhou JM. Plant Signal Behav. 2008;3:583. doi: 10.4161/psb.3.8.5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ausubel FM. Nat Immunol. 2005;6:973. doi: 10.1038/ni1253. [DOI] [PubMed] [Google Scholar]; Parker JE, Coleman MJ, Szabo V, Frost LN, Schmidt R, van der Biezen EA, Moores T, Dean C, Daniels MJ, Jones JD. Plant Cell. 1997;9:879. doi: 10.1105/tpc.9.6.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao Y, He SY, Sundin GW. Mol Plant Microbe Interact. 2006;19:644. doi: 10.1094/MPMI-19-0644. [DOI] [PubMed] [Google Scholar]
- 42.Slusarenko AJ, Schlaich NL. Mol Plant Pathol. 2003;4:159. doi: 10.1046/j.1364-3703.2003.00166.x. [DOI] [PubMed] [Google Scholar]
- 43.Khanin R, Wit E. J Comput Biol. 2006;13:810. doi: 10.1089/cmb.2006.13.810. [DOI] [PubMed] [Google Scholar]
- 44.Jain S, Bader GD. BMC Bioinformatics. 2010;11:562. doi: 10.1186/1471-2105-11-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tyagi N, Krishnadev O, Srinivasan N. Mol Biosyst. 2009;5:1630. doi: 10.1039/b906543c. [DOI] [PubMed] [Google Scholar]; Wang TY, He F, Hu QW, Zhang Z. Mol Biosyst. 2011;7:2278. doi: 10.1039/c1mb05028a. [DOI] [PubMed] [Google Scholar]; He F, Zhang Y, Chen H, Zhang Z, Peng YL. BMC Genomics. 2008;9:519. doi: 10.1186/1471-2164-9-519. [DOI] [PMC free article] [PubMed] [Google Scholar]; Li ZG, He F, Zhang Z, Peng YL. Amino Acids. 2011 doi: 10.1007/s00726-011-0978-z. [DOI] [PubMed] [Google Scholar]
- 46.Mathivanan S, Periaswamy B, Gandhi TK, Kandasamy K, Suresh S, Mohmood R, Ramachandra YL, Pandey A. BMC Bioinformatics. 2006;7(Suppl 5):S19. doi: 10.1186/1471-2105-7-S5-S19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garcia-Garcia J, Guney E, Aragues R, Planas-Iglesias J, Oliva B. BMC Bioinformatics. 2010;11:56. doi: 10.1186/1471-2105-11-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miao EA, Brittnacher M, Haraga A, Jeng RL, Welch MD, Miller SI. Mol Microbiol. 2003;48:401. doi: 10.1046/j.1365-2958.2003.t01-1-03456.x. [DOI] [PubMed] [Google Scholar]
- 49.Henry T, Couillault C, Rockenfeller P, Boucrot E, Dumont A, Schroeder N, Hermant A, Knodler LA, Lecine P, Steele-Mortimer O, Borg JP, Gorvel JP, Meresse S. Proc Natl Acad Sci U S A. 2006;103:13497. doi: 10.1073/pnas.0605443103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Espadaler J, Romero-Isart O, Jackson RM, Oliva B. Bioinformatics. 2005;21:3360. doi: 10.1093/bioinformatics/bti522. [DOI] [PubMed] [Google Scholar]; Tuncbag N, Gursoy A, Nussinov R, Keskin O. Nat Protoc. 2011;6:1341. doi: 10.1038/nprot.2011.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eddy SR. Genome Inform. 2009;23:205. [PubMed] [Google Scholar]
- 52.Hegyi H, Gerstein M. Genome Res. 2001;11:1632. doi: 10.1101/gr.183801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Apweiler R, Martin M, O’Donovan C, Magrane M, Alam-Faruque Y, Antunes R, Barrell D, Bely B, Bingley M, Binns D, Bower L, Browne P, Chan WM, Dimmer E, Eberhardt R, Fazzini F, Fedotov A, Foulger R, Garavelli J, Castro LG, Huntley R, Jacobsen J, Kleen M, Laiho K, Legge D, Lin Q, Liu W, Luo J, Orchard S, Patient S, Pichler K, Poggioli D, Pontikos N, Pruess M, Rosanoff S, Sawford T, Sehra H, Turner E, Corbett M, Donnelly M, van Rensburg P, Xenarios I, Bougueleret L, Auchincloss A, Argoud-Puy G, Axelsen K, Bairoch A, Baratin D, Blatter MC, Boeckmann B, Bolleman J, Bollondi L, Boutet E, Quintaje SB, Breuza L, Bridge A, deCastro E, Coudert E, Cusin I, Doche M, Dornevil D, Duvaud S, Estreicher A, Famiglietti L, Feuermann M, Gehant S, Ferro S, Gasteiger E, Gateau A, Gerritsen V, Gos A, Gruaz-Gumowski N, Hinz U, Hulo C, Hulo N, James J, Jimenez S, Jungo F, Kappler T, Keller G, Lara V, Lemercier P, Lieberherr D, Martin X, Masson P, Moinat M, Morgat A, Paesano S, Pedruzzi I, Pilbout S, Poux S, Pozzato M, Redaschi N, Rivoire C, Roechert B, Schneider M, Sigrist C, Sonesson K, Staehli S, Stanley E, Stutz A, Sundaram S, Tognolli M, Verbregue L, Veuthey AL, Wu CH, Arighi CN, Arminski L, Barker WC, Chen C, Chen Y, Dubey P, Huang H, Mazumder R, McGarvey P, Natale DA, Natarajan TG, Nchoutmboube J, Roberts NV, Suzek BE, Ugochukwu U, Vinayaka CR, Wang Q, Wang Y, Yeh LS, Zhang J. Nucleic Acids Res. 2011;39:D214. doi: 10.1093/nar/gkq1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Salwinski L, Miller CS, Smith AJ, Pettit FK, Bowie JU, Eisenberg D. Nucleic Acids Res. 2004;32:D449. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peri S, Navarro JD, Kristiansen TZ, Amanchy R, Surendranath V, Muthusamy B, Gandhi TK, Chandrika KN, Deshpande N, Suresh S, Rashmi BP, Shanker K, Padma N, Niranjan V, Harsha HC, Talreja N, Vrushabendra BM, Ramya MA, Yatish AJ, Joy M, Shivashankar HN, Kavitha MP, Menezes M, Choudhury DR, Ghosh N, Saravana R, Chandran S, Mohan S, Jonnalagadda CK, Prasad CK, Kumar-Sinha C, Deshpande KS, Pandey A. Nucleic Acids Res. 2004;32:D497. doi: 10.1093/nar/gkh070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kerrien S, Alam-Faruque Y, Aranda B, Bancarz I, Bridge A, Derow C, Dimmer E, Feuermann M, Friedrichsen A, Huntley R, Kohler C, Khadake J, Leroy C, Liban A, Lieftink C, Montecchi-Palazzi L, Orchard S, Risse J, Robbe K, Roechert B, Thorneycroft D, Zhang Y, Apweiler R, Hermjakob H. Nucleic Acids Res. 2007;35:D561. doi: 10.1093/nar/gkl958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chatr-aryamontri A, Ceol A, Palazzi LM, Nardelli G, Schneider MV, Castagnoli L, Cesareni G. Nucleic Acids Res. 2007;35:D572. doi: 10.1093/nar/gkl950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guldener U, Munsterkotter M, Oesterheld M, Pagel P, Ruepp A, Mewes HW, Stumpflen V. Nucleic Acids Res. 2006;34:D436. doi: 10.1093/nar/gkj003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Winnenburg R, Urban M, Beacham A, Baldwin TK, Holland S, Lindeberg M, Hansen H, Rawlings C, Hammond-Kosack KE, Kohler J. Nucleic Acids Res. 2008;36:D572. doi: 10.1093/nar/gkm858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Driscoll T, Dyer MD, Murali TM, Sobral BW. Nucleic Acids Res. 2009;37:D647. doi: 10.1093/nar/gkn799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stark C, Breitkreutz BJ, Chatr-Aryamontri A, Boucher L, Oughtred R, Livstone MS, Nixon J, Van Auken K, Wang X, Shi X, Reguly T, Rust JM, Winter A, Dolinski K, Tyers M. Nucleic Acids Res. 2011;39:D698. doi: 10.1093/nar/gkq1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Isserlin R, El-Badrawi RA, Bader GD. Database (Oxford) 2011:baq037. doi: 10.1093/database/baq037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chatr-aryamontri A, Ceol A, Peluso D, Nardozza A, Panni S, Sacco F, Tinti M, Smolyar A, Castagnoli L, Vidal M, Cusick ME, Cesareni G. Nucleic Acids Res. 2009;37:D669. doi: 10.1093/nar/gkn739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stein A, Ceol A, Aloy P. Nucleic Acids Res. 2011;39:D718. doi: 10.1093/nar/gkq962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Finn RD, Marshall M, Bateman A. Bioinformatics. 2005;21:410. doi: 10.1093/bioinformatics/bti011. [DOI] [PubMed] [Google Scholar]
- 66.Haraga A, Ohlson MB, Miller SI. Nat Rev Microbiol. 2008;6:53. doi: 10.1038/nrmicro1788. [DOI] [PubMed] [Google Scholar]
- 67.McGhie EJ, Brawn LC, Hume PJ, Humphreys D, Koronakis V. Curr Opin Microbiol. 2009;12:117. doi: 10.1016/j.mib.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, Holm L, Sonnhammer EL, Eddy SR, Bateman A. Nucleic Acids Res. 2010;38:D211. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Quattroni P, Exley RM, Tang CM. Expert Rev Anti Infect Ther. 2011;9:577. doi: 10.1586/eri.11.73. [DOI] [PubMed] [Google Scholar]
- 70.Sonnhammer EL, von Heijne G, Krogh A. Proc Int Conf Intell Syst Mol Biol. 1998;6:175. [PubMed] [Google Scholar]
- 71.Hagberg AA, Schult DA, Swart PJ. In: Varoquaux G, Vaught T, Millman J, editors. Proceedings of the 7th Python in Science Conference; Pasadena, CA, USA. 2008. p. 11. [Google Scholar]
- 72.Brohee S, van Helden J. BMC Bioinformatics. 2006;7:488. doi: 10.1186/1471-2105-7-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Berriz GF, Beaver JE, Cenik C, Tasan M, Roth FP. Bioinformatics. 2009;25:3043. doi: 10.1093/bioinformatics/btp498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Bioinformatics. 2011;27:431. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.