Abstract
Viruses depend on their hosts at every stage of their life cycles and must therefore communicate with them via Protein-Protein Interactions (PPIs). To investigate the mechanisms of communication by different viruses, we overlay reported pairwise human-virus PPIs on human signalling pathways. Of 671 pathways obtained from NCI and Reactome databases, 355 are potentially targeted by at least one virus. The majority of pathways are linked to more than one virus. We find evidence supporting the hypothesis that viruses often interact with different proteins depending on the targeted pathway. Pathway analysis indicates overrepresentation of some pathways targeted by viruses. The merged network of the most statistically significant pathways shows several centrally located proteins, which are also hub proteins. Generally, hub proteins are targeted more frequently by viruses. Numerous proteins in virus-targeted pathways are known drug targets, suggesting that these might be exploited as potential new approaches to treatments against multiple viruses.
Keywords: systems biology, protein-protein interaction networks, host-pathogen interactions, protein function, signal transduction, human immunodeficiency virus-1, human papillomavirus, bovine papillomavirus, Epstein-Barr virus, human herpesvirus
1 Introduction
Infectious diseases cause millions of deaths every year. New infectious diseases appear regularly in diverse parts of the globe, even as we fail to eradicate established diseases such as Acquired Immune Deficiency Syndrome (AIDS), malaria and tuberculosis. Virulent strains of known pathogens increasingly appear, as in the recent swine flu outbreak. Current treatments and vaccinations are focused on the pathogen. However, pathogens are able to adapt and rapidly evolve mechanisms to evade anti-bacterial and anti-viral drugs and vaccines. Moreover, once the pathogen has evaded the immune system, the infection may become chronic. Recent efforts in vaccine development therefore include targeting the host by combining pathogen-derived antigens with adjuvants, for example to boost the host immune system (Lederman, 1995). New avenues for drug discovery may arise from study of the interactions between the pathogens and their hosts. When a pathogen invades a cell, it depends on the host’s resources at all stages of its life cycle. To subvert the host cellular machinery for its purposes, the pathogen must communicate with the host, via chains of interactions referred to as signal transduction pathways. These allow cells to respond to the environment, and pathogens to take over the host.
Recently, more than 2500 interactions involving ~1000 human and 17 Human Immunodeficiency Virus (HIV) proteins have been catalogued (Fu et al., 2009). While not all of these interactions may be based on physical contacts, this large number provides a glimpse into the extent of the communication between HIV and its human host, which is particularly remarkable considering the small number of HIV proteins. Small genome size is general for other viral genomes also, which typically encode less than 20 proteins. Increasingly, the interactions involving viruses other than HIV are also being catalogued, e.g., by the Pathogen Interaction Gateway (PIG) (Driscoll et al., 2009) and VirusMINT (Chatr-Aryamontri et al., 2009) efforts. These databases store information on PPIs between multiple hosts and pathogens. The VirusMINT database (Chatr-Aryamontri et al., 2009) integrates interactions involving proteins from 149 virus strains and their human binding partners, obtained from sources such as MINT (Zanzoni et al., 2002), IntAct (Kerrien et al., 2007) and the HIV-1, Human Protein Interaction Database (Fu et al., 2009). PIG, in addition to interactions with viruses, also includes bacteria (Driscoll et al., 2009). These efforts now allow a global view of the PPI networks between pathogens and their hosts. Earlier analysis of proteins targeted by pathogens including bacteria has revealed some common features that appear to be shared amongst several host–pathogen systems (Dyer et al., 2008). The topological properties of proteins in the human PPI network indicate that pathogens target hub proteins, i.e., proteins that have many interacting partners, or bottlenecks, i.e., proteins that have many ‘shortest paths’ going through them (Dyer et al., 2008). A common set of host proteins was identified that are frequently targeted by multiple pathogens, suggesting that at least some of the proteins targeted in humans are conserved across pathogens.
In this study, we strive to understand the functional significance of the interactions between pathogens and their hosts in the context of signal transduction pathways that may be disrupted or altered by PPIs involving the pathogens. The focus here is on viruses. A comparison of the potential effects of various pathogenic viruses on different signal transduction pathways in the host cell will allow addressing questions such as which viruses target the same pathways, which viruses target pathways that are less or not at all targeted by other viruses and which proteins are common targets for many viruses given the current interaction data. Addressing the above-mentioned questions is expected to advance our fundamental understanding of virus-host communication mechanisms and may help antiviral drug discovery.
2 Materials and methods
2.1 Pathway data and path identification
Pathway data was collected from the NCI and Reactome protein pathway databases (Matthews et al., 2009; Schaefer et al., 2009) in November 2008 and processed as described previously (Balakrishnan et al., 2009). Only pathways under “Biomolecular interactions and cellular processes assembled into authoritative human signaling pathways” were downloaded from the PID website, http://pid.nci.nih.gov/. Thus, 671 pathways were analysed, representing 102 and 569 pathways from the NCI and Reactome databases, respectively.
2.2 Interactions data
Information on the interactions involving multiple viruses with human proteins was obtained from the VirusMINT database, which includes manually curated data as well as interactions downloaded from other databases (Chatr-Aryamontri et al., 2009). The VirusMINT database contains PPI data for 557 proteins and 149 different viral strains. This corresponds to 2007 unique interactions supported by 5483 experimental evidences (Chatr-Aryamontri et al., 2009). The interactions were downloaded on 4 August 2009. Data in VirusMINT that was imported from MINT and IntAct contained full experimental details while the HIV-1 Human Protein Interactions Database (Schaefer et al., 2009) does not provide a description of the experimental evidences. Therefore, VirusMINT reports only the subset of data from the HIV-1 Human Protein Interactions Database, which represents enzymatic reactions, physical associations and co-localisation (Chatr-Aryamontri et al., 2009).
Some of the viruses reported in the VirusMINT database were very close relatives. We, therefore, grouped strains of the same virus as one category for each virus. The membership of different strains in their respective group is listed in Table 2, along with the number of interactions that has been reported in the VirusMINT database for each group.
Table 2.
Taxon grouping and summary of data. The table details the grouping of different strains of viruses and lists statistics on their interactions and the pathways they are linked to. Column 1: Taxon. The abbreviation indicated in brackets is used throughout the text. Column 2: Strains grouped together as the taxon indicated in column 1. For abbreviation comma or parenthesis is used; i.e., “Bacteriophage PBS2, lamda” reads “Bacteriophage lambda” and “Bacteriophage PBS2”, whereas Vaccinia virus (Copenhagen) indicates Vaccinia virus and Vaccinia virus Copenhagen. Column 3: Number of interactions in VirusMINT with human proteins for each strain in the grouped taxons. Column 4: Total number of interactions for the taxon group and the number of human proteins involved in the interactions, given in brackets. Column 5: Number of pathways each virus taxon group is linked to by way of PPI. Column 6: Percentage of the human proteins present in pathways that are targeted by the virus in at least one pathway
| Taxon | Grouped taxons | Virus PPI |
Total PPI |
Pathways | % proteins |
|---|---|---|---|---|---|
| Autographa californica nuclear polyhedrosis virus (AcMNPV) | Autographa californica nuclear polyhedrosis virus | 1 | 1(1) | 14 | 100 |
| Avian sarcoma virus (ASV) | Avian sarcoma virus strain PR2257 | 2 | 2(1) | 18 | 50 |
| Bacteriophage (PBS2) | Bacteriophage PBS2, lambda | 1, 1 | 2(1) | 1 | 50 |
| Bovine papillomavirus (BPV) | Bovine papillomavirus type 1, 2 | 28, 1 | 29(1) | 68 | 59 |
| Cercopithecine herpesvirus (CeHV) | Cercopithecine herpesvirus 12 | 4 | 4(1) | 14 | 75 |
| Cottontail rabbit papillomavirus (CRPV) | Cottontail rabbit papillomavirus, Kansas | 2, 3 | 5(4) | 0 | 0 |
| Ebola virus (EBOV) | Ebola virus strain Zaire Mayinga | 1 | 1(1) | 2 | 100 |
| Epstein–Barr virus (EBV) | Epstein-Barr virus strain GD1, B95–8 | 121, 24 | 145(13) | 121 | 57 |
| Fowl adenovirus (FAdV) | Fowl adenovirus 1 strain CELO/Phelps | 2 | 2(1) | 10 | 50 |
| Hepatitis B virus (HBV) | Hepatitis B virus | 8 | 8(3) | 10 | 50 |
| Hepatitis C virus (HCV) | Hepatitis C virus genotype 1a isolate 1, 1b isolate BK, 1a isolate H, 1b isolate Taiwan, 1b isolate Con1, subtype 1a and 1b | 22, 4, 1, 4, 1, 3, 2, 1 | 38(12) | 101 | 59 |
| Hepatitis E virus (HEV) | Hepatitis E virus strain Burma, Mexico | 2, 1 | 3(2) | 0 | 0 |
| Hepatitis delta virus (HDV) | Hepatitis delta virus isolate American, D380 | 1, 1 | 2(2) | 3 | 100 |
| Human SARS coronavirus (SARS-CoV) | Human SARS coronavirus | 3 | 3(2) | 6 | 67 |
| Human T-cell leukaemia virus (HTCLV) | Human T-cell leukaemia virus type I isolate Caribbean | 2 | 2(1) | 1 | 50 |
| Human T-lymphotropic virus (HTLV) | Human T-lymphotropic virus 1 | 3 | 3(3) | 8 | 33 |
| Human adenovirus (HAdV) | Human adenovirus 2, 7, 9, 31, E, 12, 5, B serotype 3 | 19, 1, 1, 1, 2, 16, 53, 4 | 97(25) | 106 | 70 |
| Human cytomegalovirus (HHV-5) | Human cytomegalovirus, AD169, Towne | 1, 1, 1 | 3(3) | 12 | 100 |
| Human herpesvirus (HHV) | Human herpesvirus 1(17, Patton), 2, 3, 6, 8 | 4(61, 1), 1, 1, 1, 4 | 73(16) | 139 | 63 |
| Human immunodeficiency virus (HIV) | Human immunodeficiency virus type 1 (12, ARV2/SF2, ELI, NDK, HXB3, Lai), 2 isolate BEN | 583(19, 4, 3, 1, 2, 4), 1 | 617(33) | 289 | 48 |
| Human papillomavirus (HPV) | Human papillomavirus type 8, 11, 1a, 31, 45, 58, 6b, 3, 16, 18 | 8, 39, 8, 10, 1, 1, 16, 2, 115, 43 | 243(37) | 153 | 56 |
| Influenza A virus (FLUAV) | Influenza A virus strain A/Turkey/Oregon/71 H7N3, A/Puerto Rico/8/34 H1N1, A/Aichi/2/1968 H3N2 | 1, 10, 1 | 12(7) | 57 | 90 |
| JC polyomavirus (JCPyV) | JC polyomavirus | 2 | 2(1) | 3 | 50 |
| Murine adenovirus (MAV) | Murine adenovirus 1 | 1 | 1(1) | 4 | 100 |
| Murine polyomavirus (MPyV) | Murine polyomavirus strain 3 | 4 | 4(3) | 16 | 100 |
| Poliovirus (PV) | Poliovirus type 1 strain Sabin | 2 | 2(1) | 11 | 100 |
| Rous sarcoma virus (RSV) | Rous sarcoma virus strain Prague C, Schmidt-Ruppin A, Schmidt-Ruppin D | 1, 1, 3 | 5(3) | 3 | 60 |
| Saimiriine herpesvirus (SaHV) | Saimiriine herpesvirus 2 strain 11, 488 | 1, 2 | 3(2) | 49 | 100 |
| Sendai virus (SeV) | Sendai virus strain Fushimi | 1 | 1(1) | 0 | 0 |
| Simian virus (SV) | Simian virus 40, 5 strain W3 | 45, 2 | 47(5) | 121 | 79 |
| Vaccinia virus (VACV/VV) | Vaccinia virus (Copenhagen, Western Reserve/WR, L-IVP) | 3(6, 15, 7) | 31(12) | 62 | 70 |
For hub analysis, two databases were used. First, to analyse human, HIV interactions, we used the NIAID database (Fu et al., 2009) and defined a subset of those interactions relating to direct physical interactions as described (Tastan et al., 2009). Human PPIs were retrieved from the HPRD (Keshava Prasad et al., 2009). Second, to compare all viruses, human PPI data was obtained from the Protein Interaction Network Analysis (PINA) platform (4 March 2010 release) (Wu et al., 2009).
2.3 Curve fitting
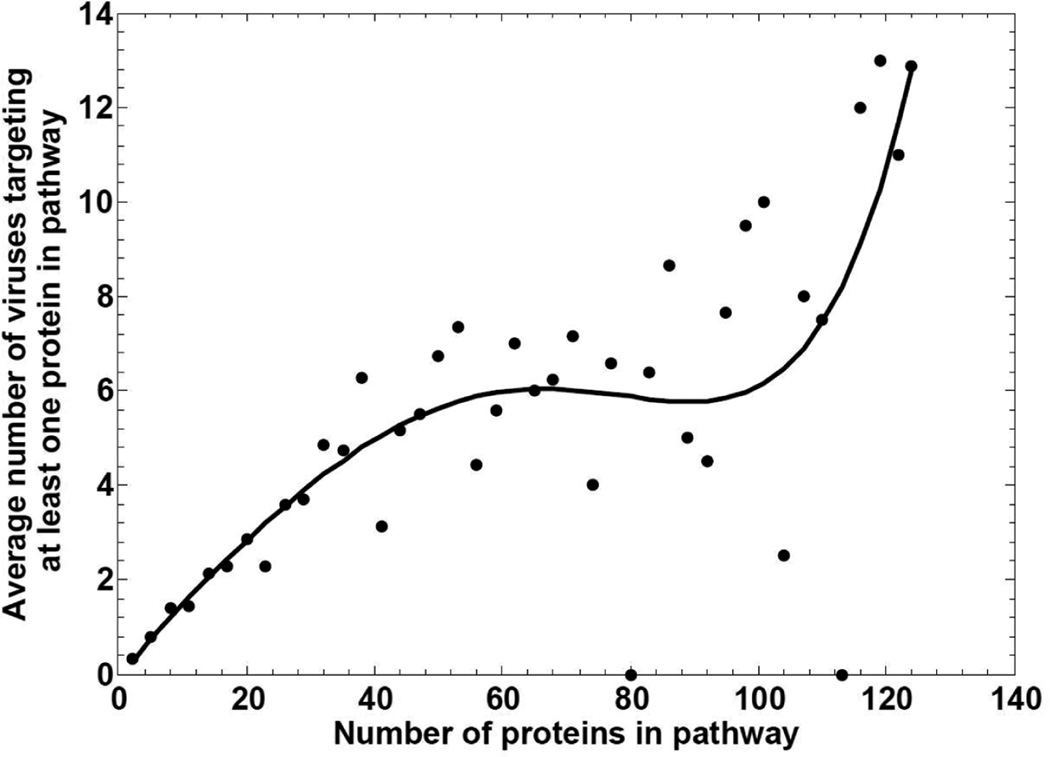
Protein-pathway size dependency curves were fit by Matlab Inc. software using various standard regression techniques, including polynomial, linear, exponential, Weibull, fractional and Gaussian. A comparison of the performance of the fit for different functions and degrees was carried out by plotting the residual error and mean squared error for each fit. The smoothest fit was obtained by a polynomial function of degree 5, which is shown in Figure 2.
Figure 2.
Distribution of the average number of viruses that target at least one protein in a pathway. Bin size was 3, until the number of proteins reached 123, which only includes 7 pathways. The minimum sum of residuals was found for polynomial fit at degree = 5, shown here
2.4 Drug target and protein data
For all virus-targeted human proteins, we checked if they are listed as drug targets in DrugBank (Wishart et al., 2008). The list of drug targets was obtained from DrugBank version 2.5. Additional functional information about the targeted proteins was obtained from the Protein knowledgebase (UniProtKB) (Apweiler et al., 2004).
2.5 Network analysis
We used the Network Modules function in the Ingenuity Pathway Analysis (IPA, Ingenuity® Systems, http://www.ingenuity.com) to identify the enriched networks for the proteins targeted by viruses. These proteins (genes) were clustered into several networks based on their PPI, regulation and other relationships. Enrichment of networks was tested by right-tailed Fisher’s exact test based on the hypergeometric distribution, and its p value was transformed to score by −log10p (Jia et al., 2011; Sun et al., 2010). We used the Merge Networks function in IPA to merge top networks to have a more systematic view into the PPI features for virus targeting. Specifically in this study, we merged the top two networks to gain more insights into the function of genes and their relationships.
3 Results and discussion
An overall summary of the numbers of proteins and pathways investigated in this study with respect to their targeting by viruses with reference to the number of drug targets in the respective groups is provided in Table 1.
Table 1.
Summary of target proteins in the pathways
| Description | Counts |
|---|---|
| No. of proteins in all pathways | 3039 |
| No. of proteins in all pathways targeted by the 28 viruses studied | 413 |
| No. of proteins in all pathways targeted by HIV | 219 |
| No. of proteins in all pathways targeted by two or more viruses | 93 |
| No. of proteins in all pathways that are known drug targets | 710 |
| No. of proteins in all pathways that are known drug targets and are targeted by any virus | 95 |
| No. of proteins in all pathways that are known drug targets and are targeted by HIV | 59 |
3.1 Global virus-pathway interactions
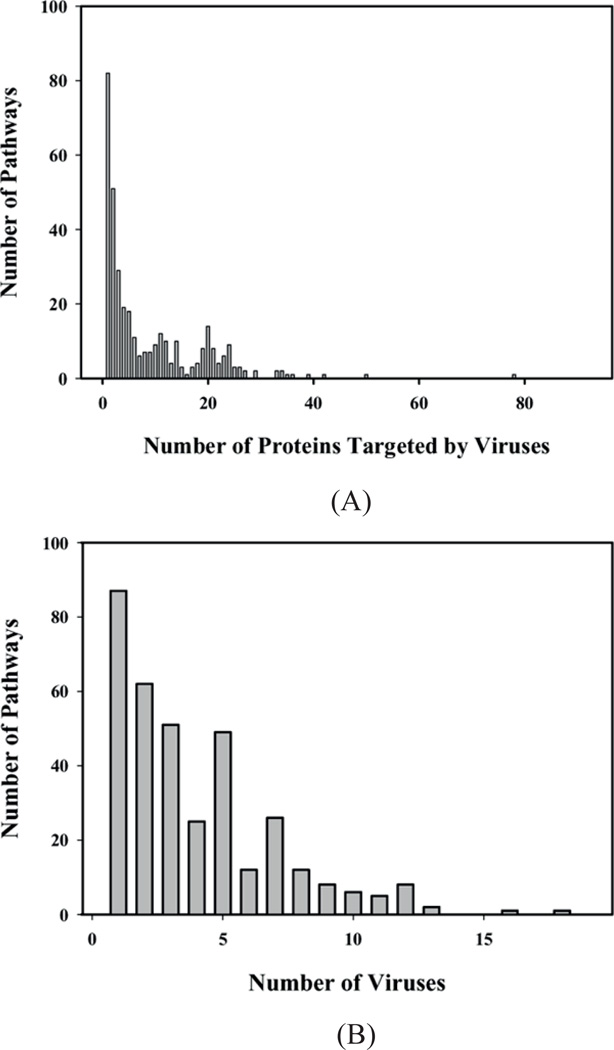
To obtain a general view of how many pathways are potentially targeted by known virus, human interactions, we enumerated the numbers of virus-interacting human proteins present in each pathway. The result is displayed in Figure 1. Figure 1(A) shows the number of pathways vs. the number of proteins targeted in them. A large number of pathways (>80) contain a single known virus binding partner, but the majority of pathways contain more than one protein interacting with a virus. Most pathways include between 2 and 42 virus binding partners, but two pathways display an exceptionally large number of proteins known to interact with viruses. These are the TGF-β and the canonical NF-κB pathways, with 50 and 78 proteins that are known virus binding partners, respectively. These proteins account for 23% and 10% of the proteins present in the corresponding pathway, respectively. Figure 1(B) shows the number of pathways targeted by one or more viruses. The number of pathways targeted by a single virus is of course equal to the number of pathways targeted by a single protein (>80), as in Figure 1(A). Most pathways contain proteins interacting with 2–5 different viruses, but some pathways are potentially targeted by up to 18 of the 31 viruses analysed. The TGF-β signalling pathway and canonical NF-κB pathway are targeted by 18 and 16 viruses, respectively.
Figure 1.
Global analysis of pathways potentially targeted by viruses. (A) Distribution of the number of pathways as a function of virus-interacting human proteins present in each pathway. The number of proteins in a pathway that are targeted by viruses varies for different pathways. This figure quantifies this variation in the number of pathways against the number of proteins that are targeted in those pathways. (B) Number of pathways being targeted by one or more viruses. This graph quantifies the variation in the extent to which different pathways are targeted by viruses
To investigate if pathway targeting is correlated with the size of the pathways, we grouped the pathways according to the number of proteins in the pathways in bins of size 3. Then, we calculated the average number of viruses targeting at least one protein in each pathway group. The results are shown in Figure 2. The results indicate that – as expected – pathways containing more proteins are more likely to be targeted by viruses. This statement is substantiated by fitting of the curve. The best result was obtained with a polynomial fit of degree 5.
3.2 Pathway targeting by different viruses
The 31 virus groups reported to have interactions with human proteins according to VirusMINT are listed in Table 2. For CRPV, HEV and SeV, there are only 5, 3 and 1 interactions reported. These viruses were, therefore, omitted in our analysis. The remaining 28 viruses cover a range of species: 16 have a double-stranded DNA genome, 2 have a single-stranded RNA negative strand genome, 3 have a single-stranded RNA positive strand genome, 6 are retro-transcribing virus and one is a deltavirus. Nine virus groups, HIV, Human papillomavirus (HPV), EBV, Human adenovirus (HadV), Human herpes virus (HHV), Simian virus (SV), Vaccinia virus (VACV), Bovine papillomavirus (BPV) and FLUAV, have the maximum number of known interactions with human proteins. They interact with proteins in 289, 153, 121, 106, 139, 121, 62, 68 and 57 pathways, respectively. In the following, we briefly discuss the PPI of these nine groups, with particular emphasis on the cell surface receptors used by these viruses and the pathways most frequently targeted.
Human immunodeficiency virus-1 (HIV), the causative agent of AIDS, encodes 15 proteins (Frankel and Young, 1998). The maximum number of interactions with human proteins has been reported for this virus. These interactions potentially affect 289 human pathways (Table 2). The largest number of interactions (48) is with the proteins in the canonical NF-κB pathway. HIV initiates infection via the CD4 receptor (Maddon et al., 1986), which participates in several pathways, including the canonical NF-κB pathway, Vpu-mediated degradation of CD4, IL12 signalling mediated by STAT4, IL23-mediated signalling events, downstream TCR signalling, Nef-Mediated CD4 down-regulation, ADP-ribosylation factor 1 pathway, binding and entry of HIV virion, translocation of ZAP-70 to immunological synapse, phosphorylation of CD3 and TCR zeta chains and generation of second messenger molecules pathways. In all of these pathways, HIV targets additional proteins apart from CD4.
Human papillomavirus (HPV) leads to cervical cancer (Schiffman and Castle, 2003). We grouped 10 types of HPV together (Table 2). HPV encodes for 8 or 9 proteins depending on HPV subtype. HPV proteins are reported to be involved in 243 PPIs that potentially perturb 153 signal transduction pathways. TGF-β receptor signalling is the most frequently targeted pathway by HPV. 17 proteins of this pathway have been reported to interact with HPV. Other receptors such as α6 integrin and laminin 5 are also involved in HPV infection. α6 integrin participates in a6b1 and a6b4 Integrin signalling and Integrin cell surface interactions pathways. Laminin 5 is involved in integrin cell surface interactions pathways.
Bovine papillomavirus (BPV) encodes 8 proteins that have been shown to undergo 29 interactions with human proteins. These are predicted to perturb 68 pathways. BPV has its maximum number of interactions with proteins from the Sumoylation by RanBP2 regulates transcriptional repression pathway. 5 proteins of this pathway are targeted by BPV.
Epstein–Barr virus (EBV) belongs to the Herpesviridae family and causes mononucleosis. The EBV strain GD1 genome encodes 30 proteins while EBV strain B95-8 codes for 73 proteins. Taken together, the proteins of the two strains have 145 interactions with human proteins, potentially affecting 121 signal transduction pathways. 23 proteins of GTP hydrolysis and joining of the 60S ribosomal subunit pathway are targeted by EBV. This is the highest number of proteins of any pathway involved in interaction with EBV proteins. CR2 (Complement receptor type 2) and β1 family integrins act as receptors for EBV infection and these receptors participate in a number of different pathways. However, none of the other proteins in them are reported to be binding partners of an EBV protein.
Human adenovirus (HAdV), a virus with 27 proteins, causes respiratory diseases, including croup, bronchitis, pneumonias, keratoconjunctivitis, cystitis and gastroenteritis. 97 interactions of HAdV proteins with human proteins have been reported and are involved in 106 pathways. Among the pathways studied, the TGF-β receptor signalling pathway has the maximum number of interactions with HAdV involving 10 proteins. Coxsackievirus and adenovirus receptor (CXADR) is the cellular receptor for HAdV types A, C, D, E and F, while CD80 and CD86 are receptors for type B and CD46 is the receptor for HAdV B2 and HAdV-3. CXADR appears in the PECAM1 interactions pathway, immunoregulatory interactions between a lymphoid and a non-lymphoid cell pathway, and the formation of platelet plug pathway. These pathways have additional proteins being targeted by other viruses but only CXADR is targeted by HAdV in all these pathways. CD80 and CD86 participate in the following pathways: canonical NF-κB, IL12 signalling mediated by STAT4 and downstream signalling in naive CD8+ T cells. Note that CD80 and CD86 are reported in VirusMINT to interact with HIV but not HAdV.
Human herpesvirus (HHV) encodes 73 proteins and 73 interactions with human proteins have been reported. HHV has maximum interactions with proteins involved in the regulation of telomerase pathway. 15 proteins of this pathway are reported to interact with HHV. Besides heparan sulphate, TNFRSF14 (Tumour necrosis factor receptor superfamily member 14), PVRL1 & PVRL2 (Poliovirus-receptor-related proteins 1 & 2) and Integrin αV-β3 serve as receptors for different HHV subtypes [Note that VirusMINT does not include the interactions with these receptors.]. Integrin αV participates in VEGFR1-specific signals, integrins in angiogenesis, osteopontin-mediated events, S1P3 pathway, PDGFR-alpha signalling pathway, integrin cell surface interactions, S1P1 pathway and PECAM1 interactions pathway. These pathways include other virus-targeted proteins.
Simian virus (SV) encodes 6 proteins, which are known to interact with 47 human proteins across 121 pathways. SV40 is a tumour-causing virus. The TGF-β receptor signalling pathway is the most frequently SV-targeted pathway involving interactions with 7 proteins.
Vaccinia virus (VACV) with 254 proteins participates in 62 interactions with host proteins affecting 70 of analysed pathways. Proteins of canonical NF-κB pathway have the maximum number of interactions (5) with VACV proteins.
Influenza A (FLUAV) causes flu. Its genome encodes 11 proteins that undergo 12 interactions with human proteins affecting 57 pathways. The maximum number of interactions (5) is with proteins of the activation of the pre-replicative complex pathway.
3.3 Comparison of pathways across viruses
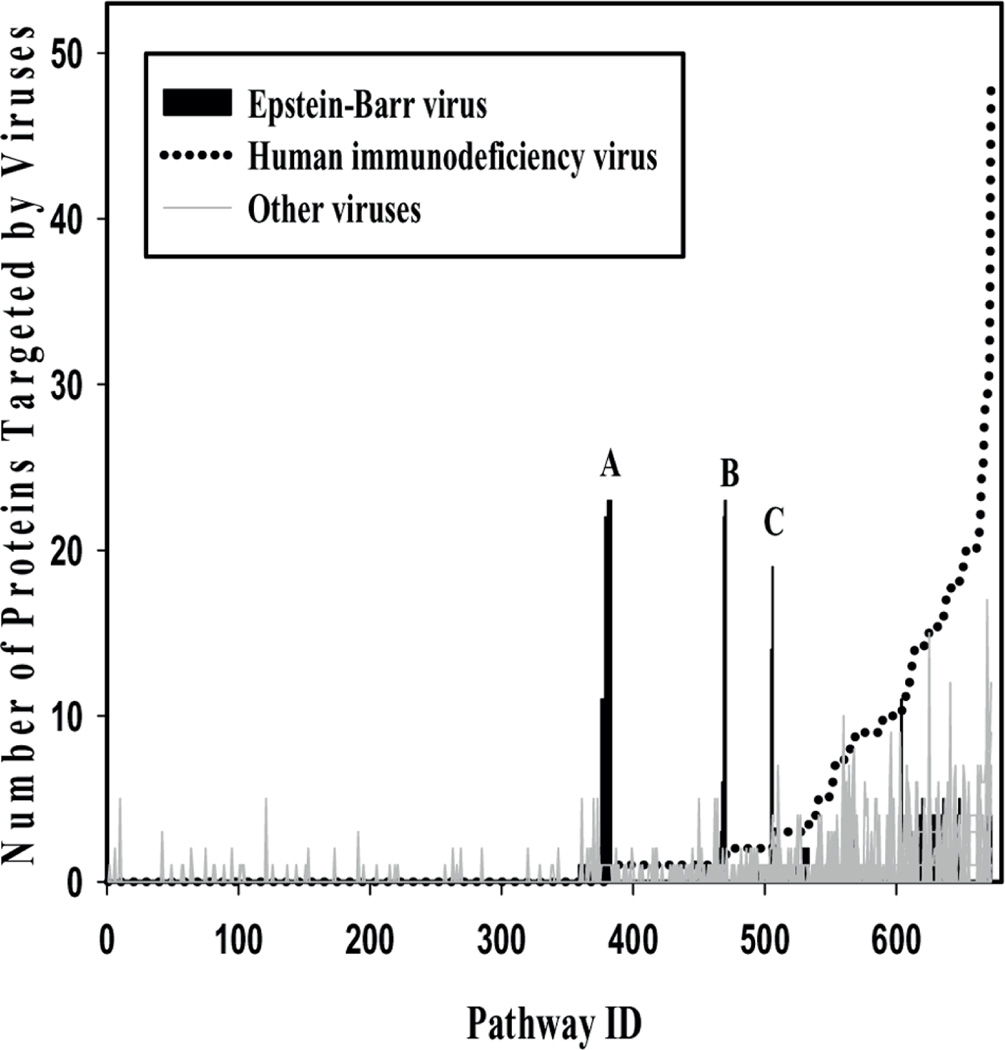
The above-mentioned analysis indicates that despite the differences in cell surface receptors recognised and required for entry, the different viruses subsequently may target overlapping signal transduction pathways. This is also supported by the relatively similar distribution of the numbers of pathways potentially affected by different viruses, shown in Figure 3. There are some significant outliers, however. The graph is sorted according to pathway interactions with HIV. The comparison of the pathways affected by HIV with all other viruses highlights that EBV interacts with proteins in pathways, which other viruses, including HIV, may not affect or for which the corresponding interactions have not yet been studied. Major pathways, which are potentially affected by EBV but not other viruses, are listed in the legend of Figure 3. Further work is required to identify if these outliers reflect a true difference between EBV and other viruses, or if the respective pathways have not yet been investigated for other viruses.
Figure 3.
Variation in proteins targeted by HIV and EBV in pathways. The number of proteins that are targeted by different viruses in the pathways varies. HIV and EBV targeted pathways stand out. In particular, pathways labelled ‘A’ are Activation of the mRNA upon binding of the cap-binding complex and eIFs and subsequent binding to 43S, L13a-mediated translational silencing of Ceruloplasmin expression, Formation of a pool of free 40S subunits, Eukaryotic Translation Termination, Viral mRNA Translation, Translation initiation complex formation, Formation of the ternary complex and subsequently the 43S complex and Ribosomal scanning and start codon recognition. Pathways ‘B’ are GTP hydrolysis and joining of the 60S ribosomal subunit, Peptide chain elongation. Pathways ‘C’ are mRNA Splicing – Major Pathway and Viral Messenger RNA Synthesis
3.4 Analysis at the pathway and protein level
Table 3 lists the proteins that interact with at least 2 viruses and shows the number of pathways in which these proteins participate. If a particular protein is present in many pathways, then its interaction with viral proteins may disrupt multiple pathways simultaneously. Interaction of a viral protein with such human proteins may thus up- or down-regulate many pathways allowing the virus to subvert the host’s cellular machinery for its purpose at a very broad scale. These proteins include proto-oncogenes such as tyrosine-protein kinases LCK, Fyn and Lyn, proteasome subunit β type-6, CREB-binding protein, histone acetyltransferase p300, paxillin and TATA-box-binding protein. Their study can thus be of pharmacological significance. Of the 93 proteins that are targeted by at least 2 proteins, SV interacts with 23 of these and the SV and HPV together have interactions with 17 (from 23) of these proteins. BPV also interacts with a large number (14) of proteins from the set of 93 proteins. BPV and HPV together interact with 13 (from 14) of these proteins. These high overlaps of proteins likely reflect conserved mechanisms of viral communication with their hosts.
Table 3.
List of proteins most frequently targeted by more than two viruses. Column 1: Protein name. Column 2: Uniprot id of the protein. Column 3: Indicates (yes/no) whether the protein is listed as drug target in DrugBank. Column 4: Number of pathways in which the protein participates. Column 5: Number of viruses that interact with the protein. Column 6: List of the viruses targeting the protein via PPI
| Protein name | UniProt ID |
Drug target |
Pathways | Virus target |
Viruses |
|---|---|---|---|---|---|
| Histone acetyltransferase p300 | Q09472 | No | 15 | 6 | HIV,HAdV,SV,MPyV,HPV,BPV |
| Cell division control protein 2 homologue | P06493 | Yes | 18 | 5 | HAdV,HIV,EBV,HPV,SV |
| Serine/threonine-protein phosphatase 2A | P67775 | Yes | 1 | 5 | RSV,SV,MPyV,HIV,HPV |
| CREB-binding protein | Q92793 | No | 16 | 5 | HAdV,SV,HIV,MPyV,HPV |
| TATA-box-binding protein | P20226 | No | 12 | 5 | HAdV,SV,HIV,HPV,BPV |
| Transcription initiation factor IIB | Q00403 | No | 8 | 5 | HHV,HIV,SV,BPV,HPV |
| Transcription initiation factor TFIID | P21675 | No | 8 | 5 | SV,HIV,HAdV,BPV,HPV |
| Cellular tumour antigen p53 | P04637 | No | 7 | 5 | HAdV,HCV,HPV,SV,HIV |
| GTP-binding nuclear protein Ran | P62826 | Yes | 12 | 4 | HAdV,HIV,HPV,SV |
| Histone deacetylase 1 | Q13547 | Yes | 10 | 4 | HAdV,SV,FAdV,HPV |
| Casein kinase II subunit alpha | P68400 | Yes | 7 | 4 | SV,HPV,HIV,EBV |
| Retinoblastoma-associated protein | P06400 | Yes | 4 | 4 | HPV,HAdV,SV,MAV |
| Importin subunit alpha-2 | P52292 | No | 2 | 4 | EBV,HIV,SV,HPV |
| Transcription factor AP-1 | P05412 | Yes | 23 | 3 | VACV/VV,HIV,HPV |
| RAF serine/threonine-protein kinase | P04049 | Yes | 18 | 3 | ASV,HIV,HCV |
| Cell division protein kinase 2 | P24941 | Yes | 17 | 3 | SV,HIV,HPV |
| DNA polymerase catalytic subunit | P09884 | Yes | 8 | 3 | SV,HPV,HHV |
| Histone acetyltransferase PCAF | Q92831 | Yes | 6 | 3 | HAdV,HIV,HPV |
| Tyrosine-protein kinase LCK | P06239 | No | 44 | 3 | SaHV,HCV,HIV |
| Transcription factor Sp1 | P08047 | No | 8 | 3 | HHV,HIV,SV |
| Transcription initiation factor TFIID | P49848 | No | 8 | 3 | HIV,BPV,HPV |
| CCAAT/enhancer-binding protein | P17676 | No | 7 | 3 | HIV,HPV,BPV |
| Importin subunit beta-1 | Q14974 | No | 5 | 3 | HIV,SV,HPV |
| ATP-dependent DNA helicase 2 | P13010 | No | 4 | 3 | HIV,EBV,HHV |
| DNA-dependent protein kinase | P78527 | No | 4 | 3 | HAdV,HIV,HHV |
| Guanine nucleotide-binding protein | P63244 | No | 4 | 3 | EBV,HAdV,HIV |
| Nucleophosmin | P06748 | No | 3 | 3 | HDV,HAdV,HIV |
| Small ubiquitin-related modifier 1 | P63165 | No | 3 | 3 | SARS-CoV,BPV,HPV |
| E3 ubiquitin-protein ligase | Q96J02 | No | 2 | 3 | CeHV,EBV,HIV |
| E3 ubiquitin-protein ligase NEDD4 | P46934 | No | 2 | 3 | RSV,EBOV,HIV |
| Bcl-2 homologous antagonist/killer | Q16611 | No | 1 | 3 | HAdV,VACV/VV,HPV |
| Double-stranded RNA-activated protein kinase | P19525 | No | 1 | 3 | VACV/VV,HCV,HIV |
| Retinoblastoma-like protein 2 | Q08999 | No | 1 | 3 | HIV,HAdV,HPV |
| Tyrosine-protein kinase HCK | P08631 | Yes | 39 | 2 | HIV,HCV |
| Phosphatidylinositol 3-kinase | P27986 | Yes | 38 | 2 | FLUAV,HIV |
| Mitogen-activated protein kinase 1 | P28482 | Yes | 35 | 2 | HHV,HIV |
| Ubiquitin | P62988 | Yes | 27 | 2 | HIV,EBV |
| Signal transducer, activator of transcription 1-alpha/beta | P42224 | Yes | 18 | 2 | HAdV,HCV |
| DNA replication licensing factor MCM7 | P33993 | Yes | 10 | 2 | FLUAV,HPV |
| Histone deacetylase 2 | Q92769 | Yes | 10 | 2 | HHV,HPV |
| Heat shock cognate 71 kDa protein | P11142 | Yes | 4 | 2 | FLUAV,EBV |
| D-isomer specific 2-hydroxyacid dehydrogenase | Q13363 | Yes | 3 | 2 | EBV,HAdV |
| Poly [ADP-ribose] polymerase 1 | P09874 | Yes | 2 | 2 | HHV,SV |
| Tubulin alpha-1B chain | P68363 | Yes | 2 | 2 | HIV,EBV |
| CAD protein | P27708 | Yes | 1 | 2 | HAdV,EBV |
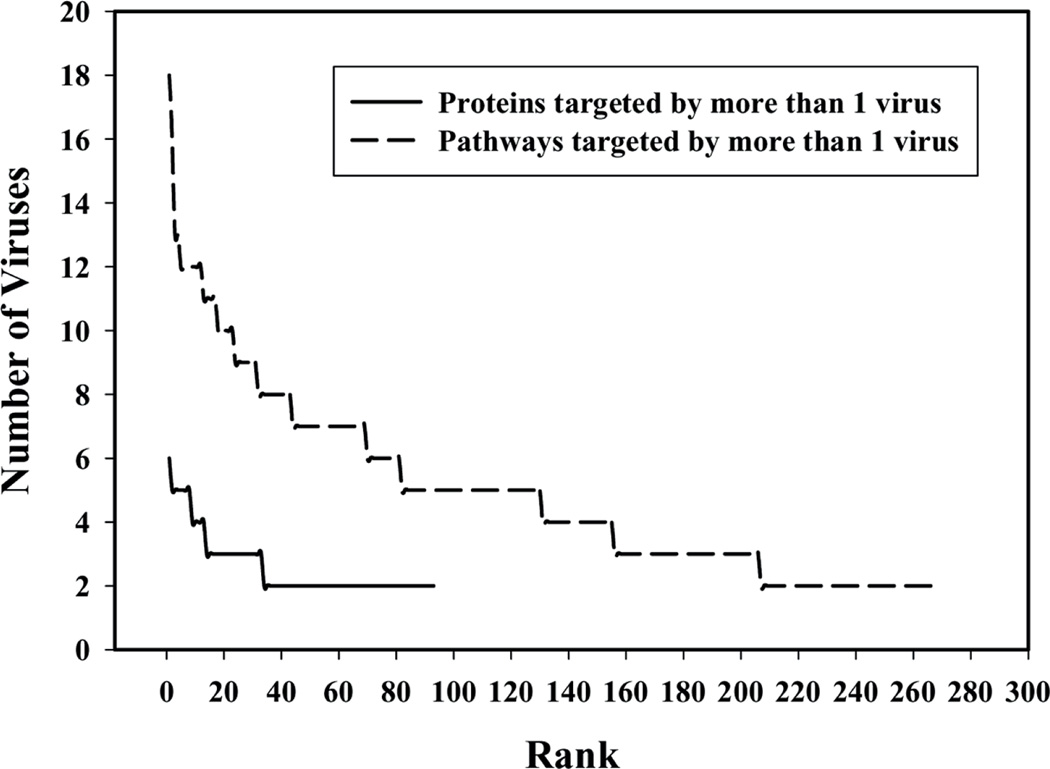
Figure 4 gives the distribution of the number of pathways and number of proteins targeted by more than one virus. One can see that the number of proteins targeted by different viruses is smaller when compared with the number of pathways. This finding suggests that different viruses target a number of the same pathways by binding to different proteins. This implies that interactions may be conserved at the pathway level, rather than the protein level. It may be more critical for a virus to disrupt a particular pathway by any PPI that may do so, rather than restricting itself to a specific PPI. Pathway disruption may be conserved irrespective of the molecular mechanism of disruption.
Figure 4.
Number of viruses targeting proteins and pathways
The above-mentioned analysis indicates that some viral proteins interact more with several host proteins when compared with others. A prime example of this is the TGF-β receptor signalling pathway, which is targeted by the maximum number of viruses (18). One possible explanation is because with 212 proteins this pathway is large (see Figure 2 for the general trend). Even more proteins participate in the canonical NF-κB pathway (739), and this pathway is also frequently targeted (by 16 viruses). However, there are other large pathways that are not frequently targeted, for example the electron transport chain has 78 proteins, yet only one has been shown to interact with a viral protein. Thus, it is likely that the frequent targeting of some pathways is functionally significant. To further investigate this observation, we screened the proteins targeted by viruses (among the 413 proteins with Uniprot ID, 411 could be mapped to gene symbols) using the IPA system (see Section 2). The network analysis resulted in 25 enriched networks whose statistical tests were significant (p < 10−5, score > 5). Table 4 lists 12 most significant networks whose scores were at least 20. We merged networks 1 and 2, both of which had functions related to infectious disease. Figure 5 shows the combined molecular network. As can be seen in the figure, NFKBIA, NFKB1 and NFKB2 are centrally located in the network, providing quantitative support for the finding described earlier, that viruses often target the NF-κB pathway. For many viruses, the largest number of interactions is with the proteins in the canonical NF-κB pathway (see earlier).
Table 4.
List of top 12 networks whose enrichment scores were at least 20 (see Section 2)
| ID | Molecules in network | Score | No. of moleculesa |
Top functions |
|---|---|---|---|---|
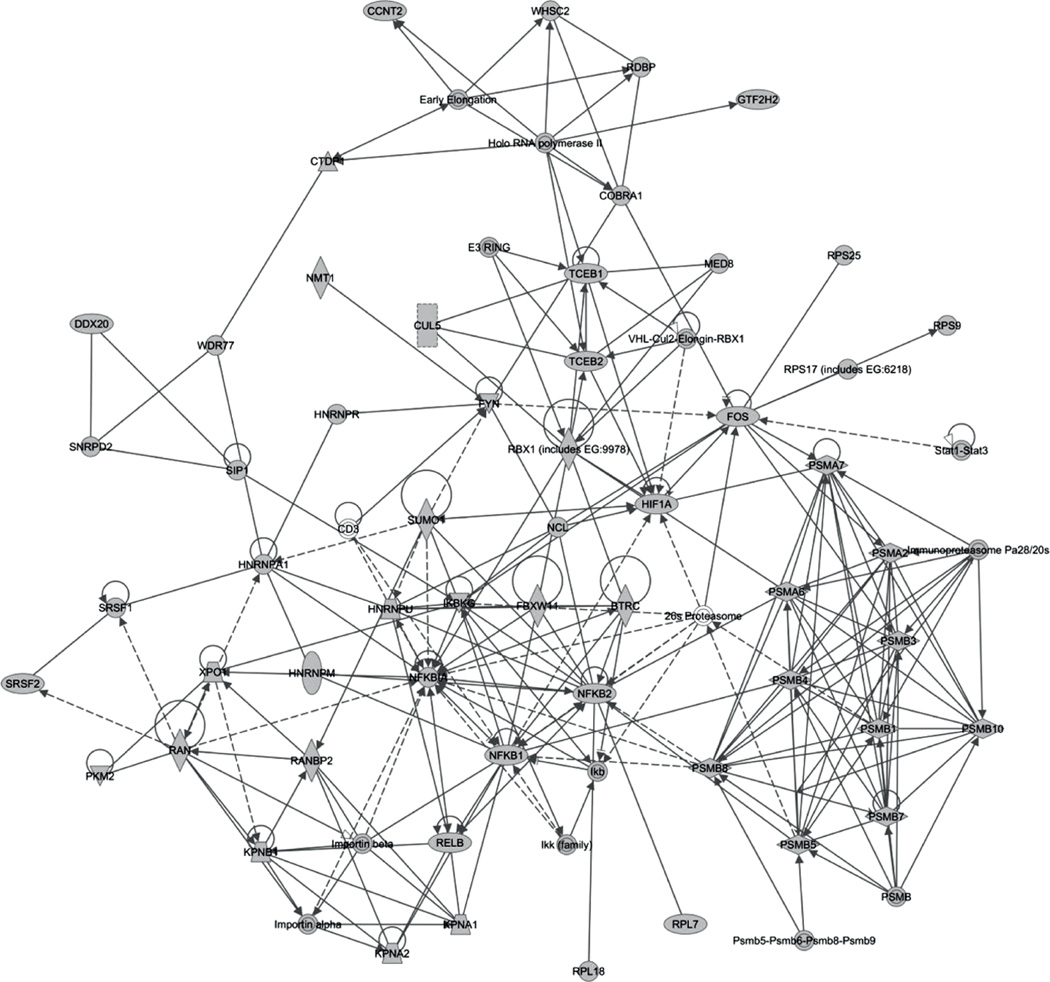
| 1 | BTRC, CD3, DDX20, FBXW11, FYN, HNRNPA1, HNRNPM, HNRNPR, HNRNPU, Ikb, IKBKG, Ikk (family), Importin alpha, Importin beta, KPNA1, KPNA2, KPNB1, NCL, NFKB1, NFKB2, NFKBIA, NMT1, PKM2, RAN, RANBP2, RELB, RPL7, RPL18, SIP1, SNRPD2, SRSF1, SRSF2, SUMO1, WDR77, XPO1 | 50 | 30 | RNA post-transcriptional modification, cancer, infectious disease |
| 2 | 26s Proteasome, CCNT2, COBRA1, CTDP1, CUL5, E3 RING, Early Elongation, FOS, GTF2H2, HIF1A, Holo RNA polymerase II, Immunoproteasome Pa28/20s, MED8, PSMA2, PSMA6, PSMA7, PSMB, PSMB1, PSMB3, PSMB4, PSMB5, PSMB7, PSMB8, PSMB10, Psmb5-Psmb6-Psmb8-Psmb9, RBX1 (includes EG:9978), RDBP, RPS9, RPS25, RPS17 (includes EG:6218), Stat1-Stat3, TCEB1, TCEB2, VHL-Cul2-Elongin-RBX1, WHSC2 | 36 | 26 | Haematological disease, dermatological diseases and conditions, infectious disease |
| 3 | BRCA1-BARD1, CDK2-CyclinE, Cpt, CPT1, CPT2, CPT1A, CPT1B, CPT1C, H2AF, H2AFX, HNRNPA0, Ku, Mre11, NONO, NPM1 (includes EG:4869), PRKDC, Rnr, RPA1, RPS8, RPS13, RPS18, RPS2 (includes EG:6187), RPS3A, Tenascin, TERF2, TNF, TOP1, Trypsin, Ube3, WRN (includes EG:7486), Xrcc, XRCC1, XRCC4, XRCC5, XRCC6 | 34 | 23 | Cell morphology, cellular function and maintenance, DNA replication, recombination, and repair |
| 4 | Angiotensin II receptor type 1, BRCA1, Cbp/p300, CCNT1, CDK7, CDK9, Cyclin T, DHX9, ERCC2, ERK, GTF2B, GTF2H1, GTF2H3, GTF2H4, Hd- neuronal intranuclear inclusions, MNAT1, POLR1A, RNA polymerase I, SUPT16H, Taf, TAF1, TAF4, TAF5, TAF6, TAF9, TAF10, TAF11, TAF13, TAF12 (includes EG:6883), Tak, TBP, TFIIA, TFIIE, TFIIF, TFIIH | 34 | 23 | Gene expression, cell signalling, DNA replication, recombination, and repair |
| 5 | Ahr-aryl hydrocarbon, alcohol group acceptor phosphotransferase, BUB1, BUB3, Casein, CCND3, CCNE1, CDK1, CDK2, CDK1-Cyclin B, CEBPA, collagen, Cyclin B, E2F1, E2F4, EIF2AK2, JUNB, Mmp, PCNA, PKN1, PLAU, PPP1CA, PRKCE, RB1, RBL2, RNA polymerase iii, SF3B1, SF3B2, SF3B3, SL1, Smad2/3, Smad2/3-Smad4, Tgf beta, TOPBP1, UBTF | 28 | 23 | Cell cycle, connective tissue development and function, cellular development |
| 6 | APOA2, Basal transcriptional machinery, BAX, BCL2L11, Caspase, CAV1, CD4, CSNK2A1, Cytochrome c, DFF, DNAJ, Focal adhesion kinase, GATA1, GZMB, HNRNPD, HSP, Hsp70, HSP90AA1, HSPA5, HSPA8, HSPB1, IL12 (complex), KAT2B, MAGED1, MDM2, MHC Class II (complex), Nc2, PP2A, Pro-inflammatory Cytokine, RNA polymerase II, RPL12 (includes EG:6136), SAT1, SP1, TRAF2, YBX1 | 28 | 21 | Cell death, cellular function and maintenance, cellular compromise |
| 7 | 19S proteasome, 20s proteasome, ATM/ATR, ATPase, BANF1, Basc, BLM, DNA-directed DNA polymerase, ERCC3, Gm-Csf Receptor, Jnk, Lamin, Lamin b, LMNA, LMNB1, LMNB2, MAD2L2, MRE11A, NBS1-Rad50-MRE11, POLA1, Proteasome PA700/20s, PSMA4, PSMB2, PSMB6, PSMC, PSMC1, PSMC2, PSMC3, PSMC6, PSMD2, RAD50, Rfc, RFC3, RPA, SMC1A | 27 | 20 | DNA replication, recombination, and repair, cell morphology, cell cycle |
| 8 | Adaptor protein 1, Adaptor protein 2, Alpha tubulin, Ap1 gamma, AP1B1, AP1G1, Ap2 alpha, AP2A2, AP2B1, AP2M1, AP2S1, ARF1, atypical protein kinase C, Beta Arrestin, CD28, Ck2, Clathrin, CLTC, Cofilin, Dynamin, FURIN, Gamma tubulin, Hexokinase, HLA Class I, IL6 RECEPTOR, IL6ST, PACS1, Pld, PPP2CA, PPP2R2A, PTK2, SYK, TUBA1B, TUBA4A, Tubulin | 24 | 18 | Cellular assembly and organisation, nervous system development and function, cell morphology |
| 9 | Alp, APOA1, ATP6V1H, CHEMOKINE, DDB1, Endothelin, FN1, FSH, GLUD1, GNRH, Gsk3, hCG, Histone h3, IKK (complex), IRS1, Lh, MIR124, MYC, NR3C1, P38 MAPK, PAK1, PARP, PPP2CB, PRKACA, PRKCA, RAB11A, Rap1, RELA, RPL22, RPS23, SEC13, Tnf, TP53, TRAF6, Vegf | 24 | 19 | Cell death, haematological system development and function, cell cycle |
| 10 | Actin, ACTN1, Alpha catenin, Cadherin, Collagen type I, Collagen type III, Collagen type IV, Cytochrome c oxidase, Elastase, EZR, Fgf, Fgfr, Gap, GIPC1, GPIIB-IIIA, Hspg, ITGB1, LAMA1, Laminin1, Laminin2, MSN, NUP93, PXN, Rab5, Rho gdi, RPL8, RPL10, RPL13, SDC1, SDC2, SDC3, SDC4, Secretase gamma, SRC, THBS1 | 22 | 17 | Cell-to-cell signalling and interaction, cellular assembly and organisation, cell morphology |
| 11 | Ampa Receptor, CDK1/2, CITED1, Cpla2, Cyclin A/Cdk2, Cyclin E, E2f, EEF1A1, EGR2, EIF3B, EIF5B, Histone H1, Il8r, LRP, MAG, N- cor, NADPH oxidase, NUP54, P4HB, Pdi, PKC (α,β,γ,δ,ε,ι), PKC (α,β,ε,γ), Pkc(s), POU2F1, PRKCB, PRKCG, PRKCH, RPL23, RPS6, RPS16, Rsk, Sod, Thymidine Kinase, TSC2, VDR | 22 | 17 | Protein synthesis, cardiovascular system development and function, cell morphology |
| 12 | Ap1, CDKN1A, CEBPB, Creb, CREBBP, CYP19, E3 HECT, EGR1, ENaC, EP300, Ep300/Pcaf, JUN, LDL, NCOA, NEDD4, NEDD4L, P110, PDGF BB, Rac, RARB, RBPJ, RXRA, Smad, Smad1/5/8, SOCS, STAT, STAT1, STAT3, Stat1 dimer, Stat1/3, TGFA, THRA, UBE2D1, UBE2I, Ubiquitin | 20 | 17 | Gene expression, cellular development, cellular growth and proliferation |
Number of focus molecules in the network.
Figure 5.
The merged molecular network for proteins targeted by viruses. Node shapes represent different molecule types (horizontal diamonds: peptidases; vertical diamonds: enzymes; circles: ‘other’). Solid lines denote protein–protein interactions and dashed lines denote regulation relationships. A few proteins such as NFKBIA, NFKB1 and NFKB2 are centrally located in the network
3.5 Targeting of human hub proteins by viruses
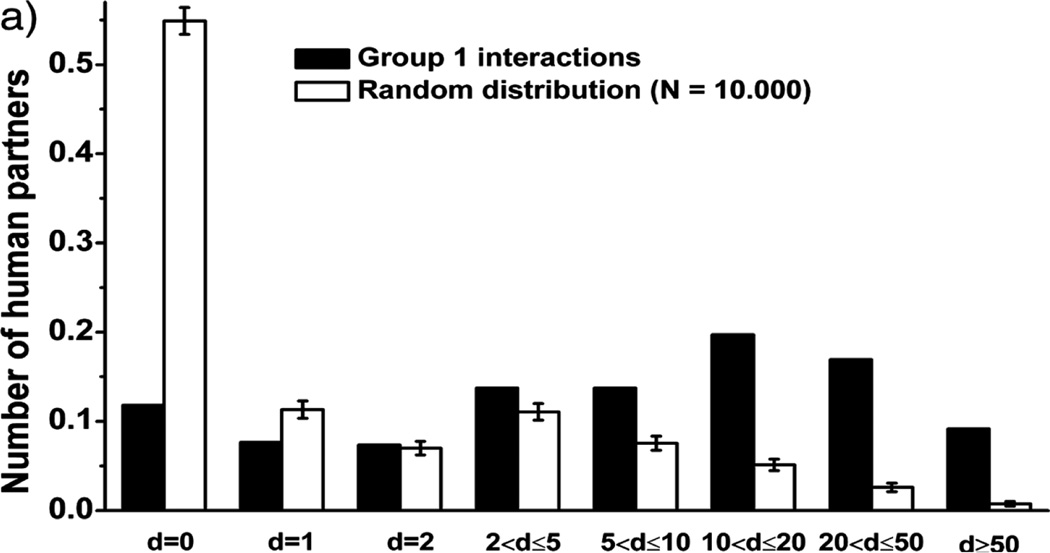
NFKBIA, NFKB1 and NFKB2 are proteins that are generally referred to as hub proteins. Hub proteins are proteins that interact with many other proteins. According to the PINA database (Wu et al., 2009), these three proteins interact with 119, 192 and 195 other human proteins, respectively. To investigate if viruses generally target hub proteins, we first investigated HIV. A comparison between the degree of proteins randomly sampled when compared with those of the proteins interacting with HIV is shown in Figure 6. This comparison shows that especially for proteins with degree above 10 the difference becomes very large. Since there is no standard definition of hub in a network, we defined three groups: hubs, intermediate hubs and non-hubs by degree >20, 10 ≤ degree < 20 and degree < 10, respectively. We then investigated these groups with respect to all virus groups. The results are shown in Table 5. We found a clear inverse relationship between the percentage of proteins (hubs vs. non-hubs) in the pathways targeted by virus or not. This finding confirms that hub proteins are more likely targeted by viruses.
Figure 6.
HIV proteins target human hub proteins. The number of proteins a protein is interacting with is referred to as degree. HIV interactions were retrieved from the NIAID database (Fu et al., 2009) but only those interactions that are more likely related to direct physical interactions are included in this analysis, referred to as ‘group 1’ (for details, see Tastan et al. (2009))
Table 5.
Summary of percentage of hubs, intermediate hubs and non-hubs targeted or not targeted by virus in pathways. Hubs are defined as proteins with interactions >20, non-hubs as those with <10, and intermediate hubs as those in between 10 and 20
| Proteins (counts) | Targeted by a virusa | Not targeted by a virusb |
|---|---|---|
| Hub (746) | 0.2785 | 0.0061 |
| Intermediate hub (645) | 0.0939 | 0.1522 |
| Non-hub (1230) | 0.1091 | 0.3602 |
Fraction of proteins targeted by a virus in pathways.
Fraction of proteins not targeted by a virus in pathways.
3.6 Drug targets in virus-targeted pathways
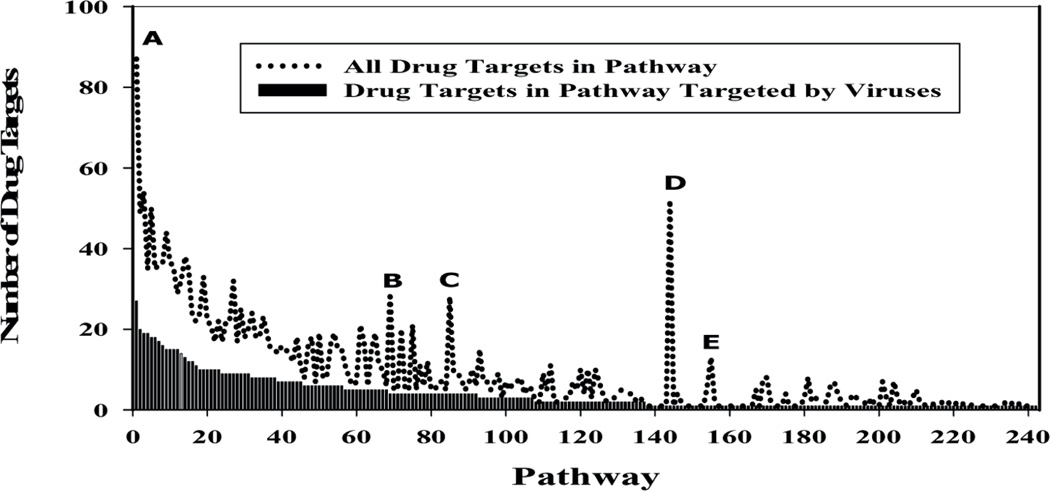
The 671 signal transduction pathways are composed of 3039 unique proteins including 710 drug targets (Table 1). Figure 7 gives a distribution of the known drug targets in the pathways along with the drug targets that have a viral interacting partner. HIV has the maximum interactions with 219 human proteins, followed by HPV. Of the 671 signal transduction pathways investigated, 355 may be targeted by at least one virus. These 355 pathways have 413 proteins that interact with 28 different viruses. Amongst the 413 targeted proteins, 95 are known drug targets with 59 of those interacting with HIV viral proteins. Amongst the 355 targeted pathways, there are 268 that have at least one protein targeted by more than one virus. These 268 pathways have 93 unique proteins with which the various virus proteins interact.
Figure 7.
Distribution of drug targets and drug targets with virus interacting partners in pathways. Approximately half of the drug targets in the pathways appear to be targeted by viruses. However, some pathways have significant higher number of drug targets compared with drug targets with reported virus interactions namely, Pathway ‘A’ Canonical NF-kappaB pathway, Pathway ‘B’ HIF-1-alpha transcription factor network, Pathway ‘C’ Vif-mediated degradation of APOBEC3G, Pathway ‘D’ Electron Transport Chain and Pathway ‘E’ Signalling mediated by p38-alpha and p38-beta
Thus, amongst all 671 pathways, there are 93 human proteins that interact with at least two pathogenic viruses. These 93 proteins are involved in a total of 774 interactions reported across the 268 pathways. We hypothesise that these 93 proteins may be potential drug targets for antiviral drugs against multiple viruses. According to the Drug Bank data, 23 proteins from these 93 proteins are known drug targets, i.e., there are ligands known to interact with these proteins. It would be interesting to test if these compounds have antiviral activity. MAV, HPV, HAdV, FLUAV, EBV, SV, VACV, ASV, RSV, FAdV, MPyV, HCV, HHV and HIV interact with the 23 drug targets from DrugBank.
4 Conclusion
In this paper, we have explored human signal transduction pathways that are potentially targeted by virus-human PPI across different viruses. We find an increase in the number of pathways targeted with the size of the pathway targeted, but some pathways are over-represented in being a virus target. We find that viruses have the potential to target the same pathways, albeit not necessarily via the same PPIs, indicating commonalities in their hijack strategies. One such common strategy is to target hub proteins. It has previously been noted that the EBV targets high degree human proteins (Calderwood et al., 2007) and pathogens in general tend to interact with host proteins with high degrees and betweenness centrality (Dyer et al., 2008). Finally, we need to note that the validity of any global analysis on the protein interaction data is dependent on the quality of the experimental data. The interaction data, in general, are derived through different experiments and experimental conditions and the resulting sets of protein interactions are more likely to include false positive and negative pairs as well as sample bias. Because of this lack of gold standard data sets, any conclusion that is based on protein interaction data sets should be taken with a grain of salt. However, this type of global views has still the potential to lead us to valuable findings regarding the general phenomena of how organisms work and communicate.
Acknowledgements
This work was in part supported by the Bill and Melinda Gates Foundation Grant 51788, the BMBF and ERA-NET ERASysBio+, National Science Foundation CAREER grant CC044917, National Institutes of Health Grants 1R21GM087617-01 and P50GM082251.
Biographies
Zhongming Zhao received his PhD in Human and Molecular Genetics from the University of Texas Health Science Center at Houston and MD Anderson Cancer Center, Houston, Texas, in 2000. Currently, he is an Associate Professor in the Departments of Biomedical Informatics, Psychiatry, and Cancer Biology at the Vanderbilt University Medical Center. His research interests include bioinformatics and systems biology approaches to studying complex diseases, genome-wide or large-scale analysis of genetic variation and methylation patterns, next-generation sequencing data analysis, comparative genomics and biomedical informatics.
Junfeng Xia received his PhD in Bioinformatics from the University of Science and Technology of China in 2010. He is currently a Postdoctoral Researcher in the Department of Biomedical Informatics, Vanderbilt University School of Medicine. His research interests include computational biology, intelligent computing and development and application of bioinformatics methods and algorithms in genomic data sets for complex disease studies.
Oznur Tastan obtained a BSc in Biological Sciences and Bioengineering from Sabanci University, Turkey, in 2004. She received an MSc from the Language Technologies Institute, School of Computer Science at Carnegie Mellon University in 2007. She was a PhD student in the same programme. She is currently a Postdoctoral Researcher with Microsoft Research, New England. Her interests are in machine learning, computational biology and systems biology.
Irtisha Singh received her MSc in Computational Biology from Carnegie Mellon University in 2010. She is currently working as a Bioinformatics Analyst at the Department of Biomedical Informatics at the University of Pittsburgh School of Medicine. Her research includes systems biology, application of bioinformatics methods for drug discovery against HIV and analysis of whole-genome and microRNA array cancer datasets.
Meghana Kshirsagar received her MSc in Computer Science from the Indian Institute of Technology, Bombay. She is currently a Master’s student at the Language Technologies Institute at Carnegie Mellon University and is working on Computational Proteomics. Before joining Carnegie Mellon University, she was working at Yahoo! Labs in the Search Sciences group. Her research interests include machine learning and computational biology.
Jaime Carbonell is the Director of the Language Technologies Institute and Allen Newell Professor of Computer Science at Carnegie Mellon University. He received BS in Physics and Mathematics from Massachusetts Institute of Technology, and MS and PhD in Computer Science from Yale University. His current research includes machine learning, computational proteomics, data mining, text mining and machine translation. He recently invented Proactive Machine Learning, including underlying decision-theoretic framework. He has served on multiple governmental advisory committees and is the Chairman of the Carnegie Speech Company.
Judith Klein-Seetharaman received her PhD in Chemistry from the Massachusetts Institute of Technology in 2000. She is currently Associate Professor at the Department of Structural Biology at the University of Pittsburgh School of Medicine with secondary faculty appointments at the Language Technologies Institute, School of Computer Science, Carnegie Mellon University and Institute for Structural Biology, Research Center Jülich, Germany. Her research is on structure, function and interactions of proteins. Her major research activities include the Biological Language Modelling Project (www.cs.cmu.edu/~blmt), the Data Integration and Bioinformatics Core of the Pittsburgh Center for HIV Protein Interactions (www.hivppi.pitt.edu) and SHIPREC, the Salmonella Host Interactions Project European Consortium (www.erasysbio.net/index.php?index=279).
Contributor Information
Zhongming Zhao, Departments of Biomedical Informatics, Psychiatry, and Cancer Biology, Vanderbilt University Medical Center, Nashville, Tennessee 37232, USA, zhongming.zhao@vanderbilt.edu.
Junfeng Xia, Department of Biomedical Informatics, Vanderbilt University Medical Center, Nashville, Tennessee 37232, USA, junfeng.xia@vanderbilt.edu.
Oznur Tastan, Language Technologies Institute, School of Computer Science, Carnegie Mellon University, Pittsburgh, Pennsylvania 15213, USA, oznur@cs.cmu.edu.
Irtisha Singh, Department of Structural Biology, School of Medicine, University of Pittsburgh, Pittsburgh, Pennsylvania 15260, USA, singhi@upmc.edu.
Meghana Kshirsagar, Language Technologies Institute, School of Computer Science, Carnegie Mellon University, Pittsburgh, Pennsylvania 15213, USA, mkshirsa@cs.cmu.edu.
Jaime Carbonell, Language Technologies Institute, Computer Science Department, Machine Learning Department and Lane Center for Computational Biology, School of Computer Science, Carnegie Mellon University, Pittsburgh, Pennsylvania 15213, USA, jgc@cs.cmu.edu.
Judith Klein-Seetharaman, Department of Structural Biology, School of Medicine, University of Pittsburgh, Pittsburgh, Pennsylvania 15260, USA, judithks@cs.cmu.edu.
References
- Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, Martin MJ, Natale DA, O’Donovan C, Redaschi N, Yeh LS. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2004;Vol. 32:D115–D119. doi: 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakrishnan S, Tastan O, Carbonell J, Klein-Seetharaman J. Alternative paths in HIV-1 targeted human signal transduction pathways. BMC Genomics. 2009;Vol. 10(Suppl. 3):S30. doi: 10.1186/1471-2164-10-S3-S30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderwood MA, Venkatesan K, Xing L, Chase MR, Vazquez A, Holthaus AM, Ewence AE, Li N, Hirozane-Kishikawa T, Hill DE, Vidal M, Kieff E, Johannsen E. Epstein-Barr virus and virus human protein interaction maps. Proc. Natl. Acad Sci. USA. 2007;Vol. 104:7606–7611. doi: 10.1073/pnas.0702332104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatr-Aryamontri A, Ceol A, Peluso D, Nardozza A, Panni S, Sacco F, Tinti M, Smolyar A, Castagnoli L, Vidal M, Cusick ME, Cesareni G. VirusMINT: a viral protein interaction database. Nucleic Acids Res. 2009;Vol. 37:D669–D673. doi: 10.1093/nar/gkn739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll T, Dyer MD, Murali TM, Sobral BW. PIG – the pathogen interaction gateway. Nucleic Acids Res. 2009;Vol. 37:D647–D650. doi: 10.1093/nar/gkn799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer MD, Murali TM, Sobral BW. The landscape of human proteins interacting with viruses and other pathogens. PLoS Pathog. 2008;Vol. 4:e32. doi: 10.1371/journal.ppat.0040032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel AD, Young JA. HIV-1: fifteen proteins and an RNA. Annu. Rev. Biochem. 1998;Vol. 67:1–25. doi: 10.1146/annurev.biochem.67.1.1. [DOI] [PubMed] [Google Scholar]
- Fu W, Sanders-Beer BE, Katz KS, Maglott DR, Pruitt KD, Ptak RG. Human immunodeficiency virus type 1, human protein interaction database at NCBI. Nucleic Acids Res. 2009;Vol. 37:D417–D422. doi: 10.1093/nar/gkn708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia P, S Z, J L, Zheng W, Zhao Z. dmGWAS: dense module searching for genome-wide association studies in protein-protein interaction networks. Bioinformatics. 2011;Vol. 27:95–102. doi: 10.1093/bioinformatics/btq615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerrien S, Alam-Faruque Y, Aranda B, Bancarz I, Bridge A, Derow C, Dimmer E, Feuermann M, Friedrichsen A, Huntley R, Kohler C, Khadake J, Leroy C, Liban A, Lieftink C, Montecchi-Palazzi L, Orchard S, Risse J, Robbe K, Roechert B, Thorneycroft D, Zhang Y, Apweiler R, Hermjakob H. IntAct – open source resource for molecular interaction data. Nucleic Acids Res. 2007;Vol. 35:D561–D565. doi: 10.1093/nar/gkl958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R, Shafreen B, Venugopal A, Balakrishnan L, Marimuthu A, Banerjee S, Somanathan DS, Sebastian A, Rani S, Ray S, Harrys Kishore CJ, Kanth S, Ahmed M, Kashyap MK, Mohmood R, Ramachandra YL, Krishna V, Rahiman BA, Mohan S, Ranganathan P, Ramabadran S, Chaerkady R, Pandey A. Human protein reference database – 2009 update. Nucleic Acids Res. 2009;Vol. 37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederman MM. Host-directed and immune-based therapies for human immunodeficiency virus infection. Ann. Intern. Med. 1995;Vol. 122:218–222. doi: 10.7326/0003-4819-122-3-199502010-00010. [DOI] [PubMed] [Google Scholar]
- Maddon PJ, Dalgleish AG, McDougal JS, Clapham PR, Weiss RA, Axel R. The T4 gene encodes the AIDS virus receptor and is expressed in the immune system and the brain. Cell. 1986;Vol. 47:333–348. doi: 10.1016/0092-8674(86)90590-8. [DOI] [PubMed] [Google Scholar]
- Matthews L, Gopinath G, Gillespie M, Caudy M, Croft D, de Bono B, Garapati P, Hemish J, Hermjakob H, Jassal B, Kanapin A, Lewis S, Mahajan S, May B, Schmidt E, Vastrik I, Wu G, Birney E, Stein L, D’Eustachio P. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 2009;Vol. 37:D619–D622. doi: 10.1093/nar/gkn863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer CF, Anthony K, Krupa S, Buchoff J, Day M, Hannay T, Buetow KH. PID: the Pathway Interaction Database. Nucleic Acids Res. 2009;Vol. 37:D674–D679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffman M, Castle PE. Human papillomavirus: epidemiology and public health. Arch. Pathol. Lab. Med. 2003;Vol. 127:930–934. doi: 10.5858/2003-127-930-HPEAPH. [DOI] [PubMed] [Google Scholar]
- Sun J, Jia P, Fanous AH, Oord Evd, Chen X, Riley BP, Amdur RL, Kendler KS, Zhao Z. Schizophrenia gene networks and pathways and their applications for novel candidate gene selection. PLoS ONE. 2010;Vol. 5:e11351. doi: 10.1371/journal.pone.0011351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tastan O, Qi Y, Carbonell J, Klein-Seetharaman J. Prediction of interactions between HIV-1 and human proteins by information integration. Pacific Symposium on Biocomputing. 2009;Vol. 13:516–527. [PMC free article] [PubMed] [Google Scholar]
- Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, Gautam B, Hassanali M. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;Vol. 36:D901–D906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Vallenius T, Ovaska K, Westermarck J, Makela TP, Hautaniemi S. Integrated network analysis platform for protein-protein interactions. Nat. Methods. 2009;Vol. 6:75–77. doi: 10.1038/nmeth.1282. [DOI] [PubMed] [Google Scholar]
- Zanzoni A, Montecchi-Palazzi L, Quondam M, Ausiello G, Helmer-Citterich M, Cesareni G. MINT: a Molecular INTeraction database. FEBS Lett. 2002;Vol. 513:135–140. doi: 10.1016/s0014-5793(01)03293-8. [DOI] [PubMed] [Google Scholar]