Abstract
Several associations have been described between the frequency of human leukocyte antigen (HLA) class I genes in certain populations and the risk of developing nasopharyngeal carcinoma (NPC). Associations between ethnic background and geographic distribution, and relative disease incidence have been reported. . Populations in geographical areas at higher risk of developing NPC display HLA distribution patterns different and sometimes opposite from areas of low incidence, whereas populations in areas with intermediate incidence display a totally independent pattern. Two main reasons may explain this association between HLA phenotype distribution and the risk of developing NPC in various populations. First, given the fact that expression of Epstein-Barr Virus (EBV) proteins by cancer cells is tightly linked with NPC development, HLA may influence the development of NPC by modulating the expression of EBV proteins. This explanation is, however, based primarily on theoretical assumptions given that no clear definition of HLA binding pattern of EBV epitopes has been directly shown to significantly alter the recognition of EBV proteins and the risk of developing the disease. Alternatively, HLA may represent a genetic marker flagging the presence of a NPC predisposition locus in close linkage disequilibrium with the HLA class I region. A critical review of known HLA associations in various geographical areas and their interpretation will be presented in this review.
Keywords: Nasopharyngeal carcinoma, human leukocyte antigen, epstein - barr virus, disease associations
INTRODUCTION
Nasopharyngeal carcinoma (NPC) with its characteristic geographic distribution, familial clustering, and multi-factorial etiology has attracted much debate about whether a genetic predisposition exists which may place particular populations at risk. According to Globocan 2002 in International Agency for Research on cancer (IARC), 78,802 new NPC cases were diagnosed worldwide in that year, of which 83% were from Asian Countries (Fig. 1). Three areas with different incidence were classified: a high incidence area (10–35/100,000 person-years) in Eastern Asia and South-eastern Asia where the highest incidence of NPC was found in the Southern Chinese population, an intermediate incidence area (1–10/100,000 person-years) in North Africa and the Arctic region and, a low incidence area (under 1/100,000 person-years) which included most Western Countries. Several epidemiological studies indicate that more than 10% of the NPC patients have a familiar predisposition. A study from Guangdong, China reported that the risk of developing NPC was 9.31 times higher in first degree relatives of patients with NPC than in the first degree relatives of their spouses. The separation ratio and hereditability were 0.0588 (0.0182, 0.0994) and 68.08% respectively, indicating a significant and powerful genetic trend toward familial clustering of NPC among Cantonese [1]. In addition, a significantly higher incidence of NPC is maintained in emigrant populations that have migrated from high incidence areas such as Malaysia, Singapore, Vietnam and the Philippines, to areas of low incidence such as Western Countries [2]. Therefore, it is likely that genetic predispositions may add to the oncogenic properties of different strains of Epstein Barr Virus (EBV), environmental conditions, and socioeconomic status toward the development of NPC.
Fig. (1). Estimated new cases/per year of NPC worldwide.
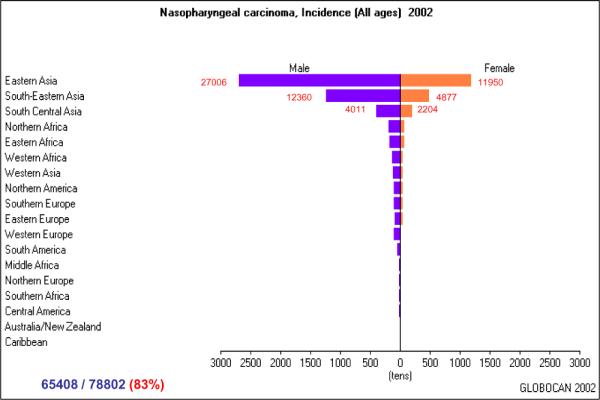
Calculated by the software “Globocan 2002” downloaded from http://www-dep.iarc.fr/GLOBOCAN/downloads.htm International Agency for Research on cancer (IARC).
As with other complex diseases, many genes may contribute to define the genetic predisposition to acquire NPC following an accumulative and multi-factorial model of inheritance. A candidate susceptibility locus was identified and mapped to Chromosome 4(4p15.1–q12) in a subset of NPC families of Guangdong [3], and significant single nucleotide polymorphisms (SNPs) were further identified within or near this region [4]. In addition, 14 of 319 alleles included in 34 microsatellites spanning an 18-megabase region of chromosome 4 were found to be associated with NPC prevalence or chronic immunoglobulin A production (p=0.001–0.03) [5]. These observations suggest the presence of a susceptibility locus in this region. A locus on Chromosome 3p21 was linked to NPC with a maximum logarithm of odds for linkage score of 4.18. Fine mapping has located this locus to a 13.6-cM region on 3p21.31–21.2 where a tumor suppressor gene cluster may exist [6–8] Polymorphisms of genes coding for the T-cell receptor, [9, 10], glutathione Stransferase MI [11, 12], Cytochrome P450 2E1 [13–15], polymeric immunoglobulin receptor [16, 17], stress protein HSP70-2 [18], DNA repair enzymes XRCC1 and hOGG1 [19] are among several other associations with risk of developing NPC.
The Human leukocyte antigen (HLA) complex is located on Chromosome 6p21.3 (Fig. 2) and it has been repeatedly documented to be associated with the prevalence of NPC in several populations. HLA was originally shown to be associated with high prevalence of NPC among Singapore Chinese in 1974 [20]. This observation has been subsequently confirmed through many association studies. Moreover, a strong association with some HLA class I alleles has been demonstrated in Chinese patients with NPC [21–24]. Similar association studies have been carried out in other ethnic groups. In this review, we will summarize the current information regarding positive and negative HLA associations with NPC prevalence within geographical areas characterized by different incidence of NPC. We will then critically discuss the possible significance of such associations in terms of HLA related immune functions or potential genetic linkage disequilibrium between HLA genes and a putative NPC predisposition gene located in close proximity. Some summary data will be presented that have been recently published by our group in a recent Meta analysis.
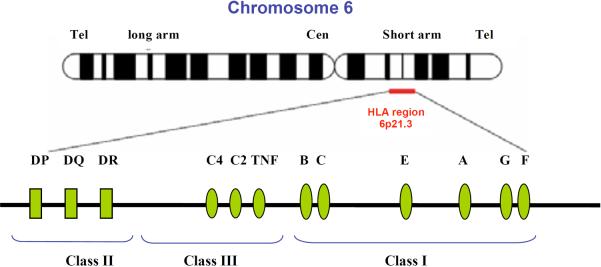
Fig. (2). The HLA region in Chromosome 6.
The HLA region located on 6p21.3 is composed of Class II, Class III, and Class I genes located in this order from the centromeric to the telomeric end. The Class I molecules are involved in binding and presenting endogenously derived peptides to CD8+T cell, whereas the class II molecules are involving in binding and presenting exogenously derived peptides to CD4+T cell. The Class III molecules are involved in the innate immune / inflammatory response.
HLA ALLELES ASSOCIATED WITH NPC
The HLA system is characterized by high polymorphism at the gene and haplotype level which may be associated with several diseases [25–28]. It has been observed that the frequency of some HLA alleles and/or haplotypes is increased (susceptible) or decreased (protective) in patients with NPC compared with normal individuals, and that the pattern of associated alleles or haplotyes varied in different geographical areas characterized by differences in prevalence of NPC. This suggests that some HLA alleles/haplotypes may be linked to the development of this disease either in terms of immune functions or genetic predisposition according to geographical and ethnic distribution.
HLA Alleles Associated with Prevalence of NPC in Geographical Areas with High Incidence
Alleles Associated with Increased Susceptibility
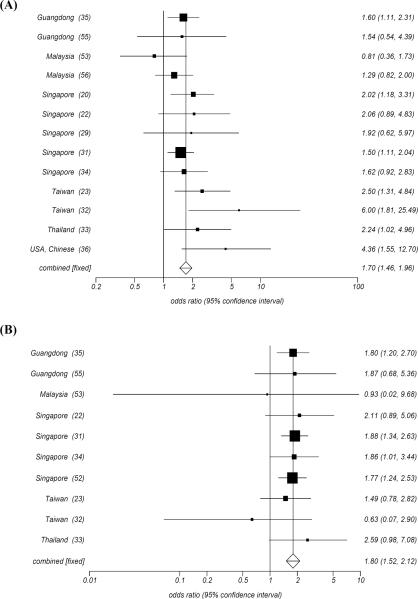
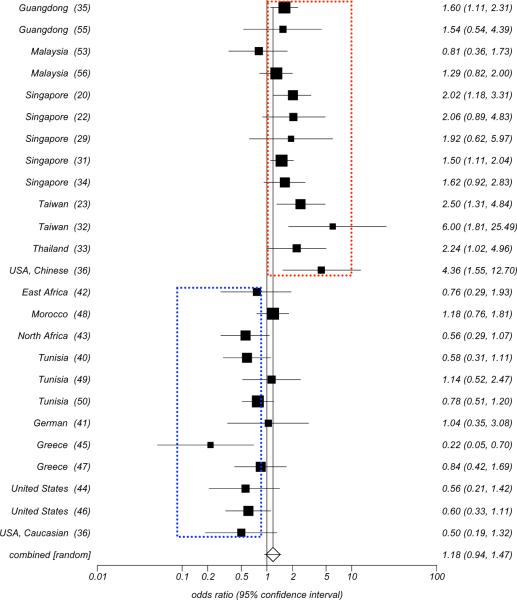
HLA-A2 and HLA-B46 have been described most consistently as the HLA class I alleles whose frequency are increased in patients with NPC from high incidence areas, especially in the Chinese population. In 1974, an HLA-A2 and an HLA-B haplotype “blank” allele was reported based on low resolution serological testing in patients with NPC from the Singapore Chinese population [19]. The profile consisted of an increased frequency of HLA-A2 and a lack of a detectable HLA antigen in the second locus. This high-risk haplotype had a much higher frequency than other haplotypes including HLA-A2 but lacking the associated HLA-B blank allele, reaching a statistical p-value (p=0.00006) and a relative risk (RR= 4.5). The blank locus was identified as singpapore-2 in 1975 [29] and officially named HLA-Bw46 in 1977 [30]. Subsequently, Chan SH [31] HLA typed 336 patients with NPC and 368 matched normal controls in the Chinese population from Singapore and confirmed that HLA-A2 (RR = 1.5, p=0.00008) and HLA-Bw46 (RR = 1.88, p=0.004) were associated with increased risk of developing NPC. In addition, the relative risk was higher when both alleles from the two loci occurred simultaneously (RR=2.4, P<0.01). His finding was gradually supported by studies in Taiwan [23, 32], Thailand [33], other Singapore Chinese populations [20, 22, 29, 34], Chinese from the mainland [35], and US Chinese [36], (Fig. 3A, B). Recently, using high resolution molecular technologies for HLA typing, Hildesheim A et al. [37] reported in 2002 an increased risk to develop NPC in individuals bearing the HLA-A*0207 (OR=2.3, 95%CI=1.5–3.5) and the HLA-B*4601 (OR=1.8, 95%CI=1.2–2.5) alleles and an increased risk (OR=2.8, 95%CI=1.7 to4.4) in individuals bearing the HLAA*0207/-B*4601 haplotype. In addition, they could extend the definition of the haplotype to the centromeric region of chromosome 6 where the HLA Class II loci are located (HLA-A*0207-B*4601-DRB1*0901-DQB1*0303-DPB1*0501)[38]. These observations strongly support the significance of the association between HLA-A2/-B46 and the risk of developing NPC in high risk Asian populations.. Pimtanothai et al. [39] in 2002, also observed a significantly higher frequency of HLA-B*4601 among patients with NPC (OR=3.8, 95%CI=1.34–11.82) in Thailand. To explore the genetic associations in more detail and test the independency of the two alleles in determining the risk of developing NPC, Lu CC [23] applied two-locus analysis and found that HLA-A2 could increase the risk for NPC significantly in populations bearing the HLA-B46 allele (OR=20.2, Pc=0.011) but insignificantly in those lacking this allele. Moreover, the HLA-B46 bearing status did not affect the risk of developing NPC in the population bearing the HLA-A2 allele. Finally, a significantly higher odds ration (OR) was again observed in cases bearing both HLA-A2 and HLA-B46 (OR=2.6, Pc=0.039) compared to individuals not bearing HLA-A2/-B46. It was, therefore, concluded that HLA-B46 alone might be insufficient to confer a high risk of NPC but it may play a “cofactor-like” role, in conjunction with HLA-A2, in determining an individual's predisposition to develop NPC among Chinese (Taiwanese and Singaporean Chinese). Interestingly, HLA-A2 and HLA-B46 did not appear to be associated with increased risk of developing NPC in geographical areas with intermediate and low prevalence of the disease. Conversely, HLA-A2 appeared to exert a protective effect in these areas. In 1977 Jing J et al. [36] demonstrated that HLA-A2 was significantly associated with an increased risk of developing NPC in US Chinese but not in US Caucasians, and several other studies in intermediate/low incidence areas subsequently confirmed this observation [36, 40–50] (Fig. 4). Two possible explanations have been suggested for this observation. First, the HLA-A2 allele HLA-A*0207 is common among Chinese (>40%) and absent in Caucasians, while HLA-A*0201 is highly prevalent (90%) among Caucasians bearing the HLAA2 serological type. The consistent association between HLA-A*0207 (OR=2.3, 95%CI=1.5 to 3.5) but not HLA-A*0201 (OR=0.79, 95%CI=0.55 to 1.2) and NPC observed among Chinese [37] might possibly result in the positive association between HLA-A2 and NPC specifically in high incidence areas since this allele is not present in low incidence areas. Second, HLA-A2,which is not associated with the risk of developing NPC in intermediate and low incidence areas [44, 46, 51], may not be significantly associated with NPC, but rather may behave as a protective allele (OR=0.46, 95%CI=0.19–0.99) in the absence of HLAB46 in high incidence area [31]..
Fig. (3). Odd ratio Meta analysis plot in high incidence area.
A: HLA-A2, B: HLA-B46, C: HLA-A11, D: HLA-B13.
Odds ratio meta-ananlysis is calculated by fixed effect model (Mantel-Haenszel, Robins-Breslow -Greenland) and Random effects (DerSimonian-Laird) model according to the Cochran's Q test.
Fig. (4).
Odd ratio Meta analysis plot (random effects) for HLA-A2 in high, intermediate, and low incidence areas.
Other less consistently reported associations between HLA class I allele, such as HLA-B17 and HLA-B18, and the risk of developing NPC have been reported. .A significant association between HLA-B17 and the prevalence of NPC was observed [52] in 1981 and was confirmed in a subsequent study in 1983 [31] by Chan SH et al. among Singapore Chinese. and with a stronger association subsequently reported (OR=3.36, 95%CI=1.36–8.08) among Malaysians[53]. Moreover, a highly significant association was found between HLA-B18 and prevalence of NPC (OR=4.39, 95%CI=1.93–9.84) in the same Malaysian population. Although, these two associations could not be confirmed by other studies, most investigators observed a non-statistically significant increase in frequency of these two alleles in patients with NPC [20, 29, 31–33, 35, 36, 54], suggesting the possibility that these studies were underpowered.
Alleles Associated with Decreased Risk of NPC
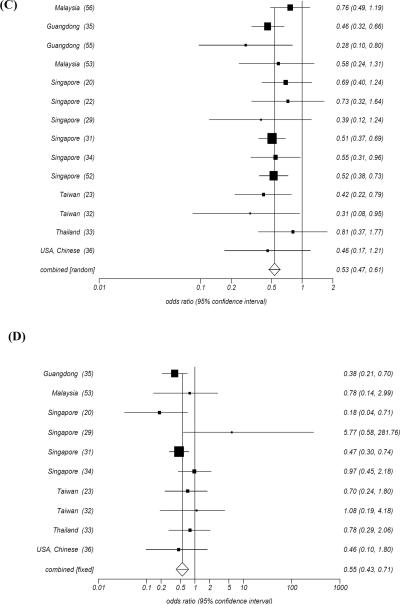
HLA-A11, HLA-B13 and HLA-B27 have been reported to be associated with decreased risk of developing NPC in high incidence areas. In 1981 Chan SH et al. [52] observed a low frequency of HLA-A11 among Chinese patients with NPC (133/313) compared with normal individuals (193/330) (OR=0.52, p<0.003). This was confirmed (OR=0.5, P=0.00008) by an eight-year long follow up study [31]. Moreover, an association was found between HLA-A13 and the survival of patients with NPC that was also associated with a lower risk of developing the disease (OR=0.47, P<0.02). Among a total of twelve studies done subsequently in Guangdong [55], Singapore [20, 22, 29, 31, 34, 52], Taiwan [23, 32], Thailand [33], and Malaysia [53, 56], eight confirmed a significantly negative association between HLA-A11 and prevalence of NPC with an OR ranging from 0.28 to 0.55. The remaining studies consistently indicated a trend toward a low frequency of HLA-A11 in patient with NPC (Fig. 3C), suggesting a protective role for this allele. Similarly, the low frequency of HLA-B13 was recognized by most studies so far reported from geographic areas characterized by high prevalence of NPC (Fig. 3D). In particular, a large study in which 247 NPC cases vs. 274 controls among Cantonese were HLA typed confirmed the protective association between HLA-B13 and the prevalence of NPC (OR=0.38, P<0.005) [35]. HLA-B27 seemed to be insignificantly associated with NPC in this study; however, most studies consistently showed a low frequency of this allele in patients with NPC. Conversely, the protective effects of these three alleles, especially HLA-A11, were not observed in geographical areas with intermediate or low prevalence of NPC (Fig. 5).
Fig. (5).
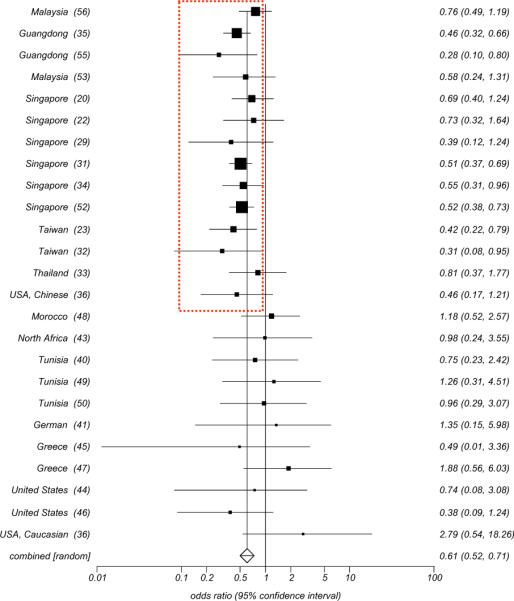
Odd ratio Meta analysis plot (random effects) for HLA-A11 In high, intermediate, and low incidence areas.
It is noteworthy that the frequencies of HLA-B22 and HLA-B55 were consistently low in NPC patients regardless of high, intermediate and low incidence areas although the associations never reached significance in individual studies. However, a meta-analysis proposed a significant association in high incidence but not in intermediate and low incidence areas possibly due to the relatively limited number of studies and the small sample size analyzed in individual studies from intermediate and low incidence areas. Therefore, further studies will be required on this subject.
HLA-A31, a rarer allele in high incidence area, was recently discovered to bear a strong protective effect toward the development of NPC. Lu CC et al. [23] reported a low frequency of HLA-A31 in NPC patients compared with normal controls among Taiwanese. A subsequent study done by Hu SP et al. [35] demonstrated a protective effect (OR=0.00, P<0.005) for HLA-A31 in the development of NPC, and a much higher significance (OR=0.00, P<0.0000) after combining data from the other two studies.
HLA Alleles Associated with Prevalence of NPC in Geographical Areas of Intermediate Incidence
Associations between incidence of NPC and HLA phenotype have been less intensively studied in geographical areas characterized by intermediate or low prevalence of the disease. Among the areas characterized by intermediate prevalence, North Africa and East Africa have been the most studied [40, 42, 43, 48–50]. Some associations reported have been completely different from those reported in high incidence areas [40]. These observations suggest that the associations between HLA and NPC prevalence may represent markers of a genetic link to a predisposition gene in close proximity to the HLA class I loci rather than due to intrinsic functional characteristics of individual HLA molecules coded by difference alleles.
Alleles Associated with Increased Risk for NPC
It can be concluded from the results of six studies that HLA-A10, -B13, -B51, and -B18 are associated with an increased risk of developing NPC. HLA-A10 was observed with higher frequency in patients with NPC among Tunisians (18% vs. 9%) [40], Algerians (22.4% vs. 12.6%) [43], and Moroccans (OR=1.80, P<0.05) [48]. Three other studies, however, did not confirm this association. The relationship between HLA-B13 and risk of developing NPC has been more consistently observed. A high frequency was found in NPC patients in five studies [40, 42, 43, 48, 50] and a significant association was reported in Tunisians (15% vs. 4%, OR=4.17, p<0.05)[49]. HLA-B51 was reported as associated with risk of developing NPC in three Tunisian populations [40, 49, 50]. HLA-B18 was also reported at high frequency in Tunisians [40, 50] and Moroccans with NPC (OR=4.14, P<0.001) [48]. However, other studies have provided inconsistent results [42, 43, 49].
Alleles Associated with Decreased Risk of Developing NPC
HLA-B14 was observed to be associated with a decreased risk of developing NPC in geographical areas with intermediate prevalence of NPC (Fig. 6) such as in Tunisians (OR=0.09, P=0.0001) [50] and North Africans (OR=0.07, P=0.002) [43]. Moreover, the frequency of this allele was consistently lower in patients with NPC compared with normal controls in all other studies performed in areas with intermediate incidence of NPC [40, 42, 48, 49]. In addition, a low frequency of this allele in patients with NPC was also observed in some geographical areas characterized by low incidence of the disease with the exception of Greek [45] and German [41] populations. A trend toward a low frequency of HLA-A33 was also reported in several studies [43, 49, 50].
Fig. (6).
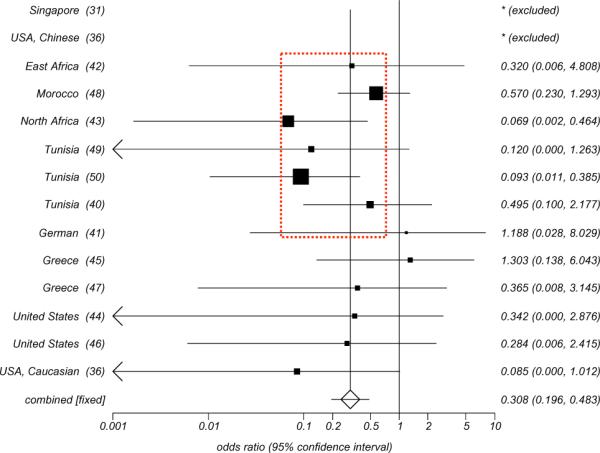
Odd ratio Meta analysis plot (Fixed effects) for HLA-B14.
We have recently reported the results of a sequence-based high resolution analysis [50] comparing HLA class I frequencies in a cohort of 136 Tunisian patients with NPC matched for gender, age and geographical residence to 148 normal Tunisians. In addition, we compared the HLA allele frequencies in normal Tunisians with those of Northern Moroccan Berbers to evaluate whether the Tunisian population in our study could be considered representative of other Maghrebian populations. This study confirmed previous reports indicating significantly positive associations between HLA-B18 and HLA-B51 and the risk of developing NPC. In addition, we observed that HLA-B14 and HLA-Cw08 were negatively associated with NPC (OR=0.09 and 0.18 respectively, Fisher P2-value=0.0001 and =0.003). The HLA-B14-Cw8 haplotype frequency was 0.007 in NPC patients compared to 0.057 in both Tunisian (OR=0.12, P2-value=0.001) and Moroccan controls. Our result strongly supported that HLA-B14 may indeed represent an important marker of a predisposition to develop NPC in intermediate incidence area.
HLA Alleles Associated with Prevalence of NPC in Geographical Areas of Low Incidence
Since several associations between HLA alleles and NPC were implicated by studies in high incidence areas, especially in Chinese populations, several studies have been carried out in low incidence area simultaneously, including North American Caucasians, Germans and Greeks to determine whether these associations could also be observed in these populations [36].
Alleles Associated with Increased Susceptibility to Develop NPC
Compared to areas with high or intermediate incidence of NPC, HLA-A1 and HLA-B5 have been more consistently associated with increased risk for NPC in low incidence areas. Jing J et al. [36] pointed out that HLA-A1, one of the most common antigens of the HLA-A locus, was more frequent (51.3% vs. 28.6%) in Caucasian patients with NPC although association did not reach significance. A higher frequency of HLA-B5 (15.4% vs. 12.2) in NPC patients was also reported in this study. Subsequently, other studies have reported a consistent and similar trend for the two alleles in two Greek [45, 47] and two USA Caucasian populations [44, 46]. Among these studies, the HLA-A1 positive association with NPC was reported as significant (OR=2.4, P<0.05) in one of the Greek population studies [45]. A significant association between HLA-B5 and NPC (OR=2.67, p<0.005) has been reported in another Greek population [47]. These two alleles were also reported in high and low incidence area but the data have been less consistent.
Alleles Associated with Decreased Incidence of NPC
HLA-A2, which is associated with high risk of developing NPC in high incidence areas, was observed to be conversely associated with the decreased risk in low incidence areas (Fig. 4) and this observation has been consistently reported by several studies [36, 41, 44–47].
Meta Analyses
With the exception of the high incidence areas, NPC is a relatively uncommon disease worldwide with an incidence ranging from 0.5–35/100,000 person-years. Although there have been a number of investigations on HLA association with the prevalence of NPC, most individual studies have been limited in number of cases/controls analyzed and, therefore, may have lacked the statistical power to identify potentially real associations. Meta-analysis, as a way to combine data from several studies has, therefore, been applied in various epidemiological and evidence-based studies to overcome this problem.
Four Meta analyses have evaluated HLA/NPC associations. In 1994, Burt et al. [51] confirmed the association between HLA-A2 and the increased risk of developing NPC in non-Chinese populations, contrasting with a protective effect in Chinese. In a larger follow-up meta analysis published in 1996 [46] where the previous study was combined with others that had been published through 1995 it was concluded that significant associations existed between HLA-A2, -A11, -A28, -B5 (serological family including the -B51 and -B52 splits), -B8 and -DR2. Opposite HLA-A2 associations with NPC were confirmed again comparing the Chinese (OR=1.65, 95%CI=1.28–2.13) with non-Chinese population (OR=0.63, 95%CI=0.48–0.82). HLA-A11 was again associated with a protective effect when four studies in Chinese populations were combined (OR=0.48, 95%CI=0.37–0.62) and a weak protective association in intermediate and low incidence areas. HLA-B5 was significantly associated with increased risk for NPC only in Caucasians (OR=2.81, 95%CI=1.76–5.12). In 2002, a meta-analysis [57] was undertaken on 13 previous studies from Southern Chinese using both fixed-effects and random-effects models as appropriate. Evidence for positive associations between NPC and HLA-A2, -B14, -B46 was found, and negative associations were identified for HLA-A11, -B13 and -B22. Recently, Hu et al. [35] performed an embedded mini-meta-analysis confirming the known HLA-A2, -A11, -B46 and -B13 associations and identifying new ones including HLA-A31, -A33, -B27, -B38, -B39, -B55, and -B58 associated with increased risk of developing NPC among Cantonese. These results strongly support the existence of a southern Chinese-restricted, recessive gene closely linked to the HLA class I region that may play a major role in determining the genetic predisposition to develop NPC in Chinese.
With the purpose of globally describing the profile of HLA associations with NPC in geographical areas with different prevalence of NPC incidence, we reported a meta-analysis based on fixed effects or random effects models including all known previously published studies from 1974 to 2007. Among them, 13 were from high incidence areas, 6 from intermediate incidence areas, and 5 from low incidence areas. Table 1 summarizes alleles significantly associated with NPC including HLA-A11 (OR=0.53), -A2 (OR=1.69), -B13 (OR=0.55), -B27 (OR=0.38), -B46 (OR=1.96), -B18 (OR=2.42), and -B55 (OR=0.23) in high incidence area. In contrast, HLA-A10 (OR=1.98), -A33 (OR=0.34), -B14 (OR=0.27) and -B51 (OR=2.08) were found to be significantly associated in intermediate incidence areas. Lastly, HLA-A1 (OR=1.62), -A2 (OR=0.60), and -B5 (OR=2.54) were associated with NPC in low incidence areas. In addition to the distinct effects of HLA-A2 and -A11 in different incidence areas, diversity in HLA allele associations was observed globally. Noticeably, HLA-B14 exerted a strong protective effect in intermediate incidence. HLA-B13 appeared associated with a protective effect in high incidence areas but was associated with enhanced risk in intermediate incidence areas.
Table 1.
Association Between HLA Alleles with NPC (Odds Ratio Meta-Analysis, 1974–2007)
| HLA allele | No. of study | Summary | Different incidence areas | ||
|---|---|---|---|---|---|
| High | Intermediate | Low | |||
| A1 | 21 | 1.25* | 1.18 | 1.09 | 1.62*** |
| A10 | 15 | 1.28 | 0.89 | 1.98**** | 0.94 |
| A11 | 24 | 0.58****** | 0.53****** | 1.03 | 0.96 |
| A2 | 26 | 1.22**** | 1.69****** | 0.84 | 0.60*** |
| A23 | 7 | 0.55** | - | 0.48 | 0.57 |
| A26 | 7 | 1.21 | 1.44 | 2.35* | 0.39* |
| A31 | 7 | 0.59 | 0.07*** | 0.19 | 2.22 |
| A33 | 10 | 0.92 | 1.20 | 0.34**** | 1.49 |
| A9 | 13 | 0.73 | 0.78 | 0.79 | 0.43* |
| B13 | 22 | 1.02 | 0.55****** | 2.07*** | 1.41 |
| B14 | 12 | 0.31****** | - | 0.27****** | 0.36* |
| B18 | 17 | 1.66** | 2.42**** | 1.86 | 1.02 |
| B22 | 13 | 0.62*** | 0.61*** | 0.59 | 0.79 |
| B27 | 21 | 0.65*** | 0.38****** | 1.33 | 0.86 |
| B38 | 6 | 1.36 | 1.66* | - | 0.76 |
| B39 | 8 | 0.65 | 0.22** | 0.49 | 1.63 |
| B40 | 14 | 0.80* | 0.77* | 0.73 | 1.53 |
| B46 | 12 | 1.99****** | 1.96****** | - | - |
| B5 | 18 | 1.21 | 0.98 | 1.01 | 2.54** |
| B51 | 4 | 1.45* | 0.98 | 2.08*** | - |
| B55 | 5 | 0.30*** | 0.23**** | - | 0.33 |
| B8 | 16 | 1.17 | 0.79 | 0.98 | 1.57* |
| DR4 | 6 | 0.50**** | 0.39** | - | 0.75 |
| A2;B46 | 3 | 2.08**** | 2.08**** | - | - |
1. P< 0.05,
P< 0.01,
P< 0.005,
P< 0.001,
P< 0.0005,
P< 0.0001.
2. Odds ratio meta-analysis is calculated by fixed effects (Mantel-Haenszel, Robins-Breslow. -Greenland) and Random effects (DerSimonian-Laird) model according to the Cochran's Q test.
3. If some catalog is less than 12, Conditional maximum likelihood is calculated.
4. Mark "-" if the number of study is less than 2.
5. The Statsdirect program was used.
BIOLOGICAL RELEVANCE OF HLA ASSOCIATIONS WITH RISK OF DEVELOPING NPC
The HLA-Dependent Immunoescape in NPC
Several lines of evidences suggest that the oncogenesis of NPC is associated with the expression of specific Epstein - Barr virus (EBV) proteins by cancer cells, which could be recognized by EBV-specific T cells when processed and presented in association with HLA class I alleles. Like other Herpes viruses, EBV may down-regulate the expression of HLA alleles to decrease the recognition and killing of EBV-expressing cancer cells. Recently two studies have suggested down-regulation of HLA class I molecules on the cancer cell surface as a mechanism of immune escape, providing indirect evidence that the expression of EBV epitopes in association with some HLA alleles may play a relevant role in the immune surveillance against the onset of NPC. In 2006, Sengupta et al. [58] published a genome-wide expression profile analysis of 31 NPC tumors and 10 healthy nasopharyngeal epithelium tissues collected from Taiwan, observing an EBV-associated inhibition of HLA class I expression in NPC. Among the human genes whose inhibition was most strongly correlated with increased EBV gene expression were MHC class I HLA genes involved in regulation of immune response via antigen presentation. Subsequently Ogino et al. [59] observed that CD8+T cell infiltration in tumor lesions significantly correlated with HLA class I molecule expression (r=0.34) suggesting that some HLA alleles may be necessary for the control of NPC growth and that down-regulation of HLA may be an important component of immune escape.
Polymorphism of the HLA genes is concentrated in the antigen-binding cleft of the molecule on the surface of the cell membrane. It is believed that polymorphism results in the expression of different alleles in different populations which has been determined through selective pressure during evolution to protect individuals residing in particular geographical areas from pathogens endemic to that area. The extent of HLA polymorphism probably provides a heterozygous advantage because it allows presentation of diverse antigenic epitopes to T cells, therefore, broadening the immune response to pathogens. Some polymorphisms appear to affect generic aspects of antigen presentation such as class I assembly and rate of transport to the cell surface [60, 61]. Zernich D et al. [61] have reported that HLA polymorphism can control the antigen presentation pathways. For example, a single amino acid variant distinguishing HLA-B*4402 (Asp 116) from -B*4405 (Tyr 116) permitted HLA-B*4405 to acquired peptides independent of transporter associated with Ag presentation (TAP) function. TAP-independent antigen presentation is, in turn, less susceptible to viral interference aimed at diminishing TAP function, theoretically making HLA-B*4405 a more efficient allele in the presentation of viral epitopes than HLA-B*4402. Therefore, it was proposed that antigen processing and presentation may be influenced by HLA molecules which may determine higher or lower risk of developing NPC. This hypothesis has been tested in previous studies which have mapped EBV epitopes recognized by T cells in association with specific HLA alleles [62–66]. HLA-A2-restricted epitopes from the latent membrane protein (LMP)-1 of EBV were first investigated in healthy, EBV-seropositive individuals [66]. Lin HJ et al. [64] reported an epitope variant with a mutation involving one of the anchor residues of the HLA molecule in position 2 (125 L to F) (YFL) and an additional mutation in position 5 (129 M to I) (YLL). Subsequent functional assays [63] have indicated the this variant was associated with a lower efficiency of epitope biding to HLA-A2 molecules which resulted in the abrogation of CTL recognition, and that this finding preferentially pertained to patients with NPC. This observation suggests that HLA-A2-restricted “epitope loss variants” of LMP1 dominated in the EBV isolates from NPC of southern China and Taiwan and that they may allow EBV-infected cells to escape from immune recognition in high incidence areas.
LMP-2 (another EBV protein commonly expressed by NPC cells), has subsequently been observed to include the majority of EBV epitopes recognized by T cells in association with HLA-A2. Lee et al. [67] identified an HLA-A*0201-restricted CTL response to a defined epitope in LMP-2 (residues 426–434) in 6 of 12 HLA-A*0201-positive donors tested and later identified two other CTL epitopes whose recognition was associated, though with different efficiency, with other HLA-A2 subtypes (A0201, A0206, A0203, and A0207)[68]. However, the relevance of epitope recognition in experiments utilizing allogeneic T-cells still remains to be elucidated when a partial mismatch between effector and target cells is present [69]. Moreover, the functional relevance of HLA class I dependent antigen presentation in determining the development of NPC remains to be a challenge due to the lack of a reproducible identification of candidate epitopes presented in association with HLA class I alleles which predispose to the risk of developing NPC in different geographical areas. The EBV-specific epitopes restricted to HLA-A11 and some other important alleles significantly associated with the prevalence of NPC have been so far less extensively investigated. In a recent comprehensive epitope mapping of the LMP-2 protein, we failed to identify significant differences in epitope immunogenicity among different HLA alleles belonging to the HLA-A2 family and expressed with different frequency in different ethnic groups and geographical areas [70].
We have found in our study a predominant recognition of LMP-2 viral products in association with HLA-A*0201 [70] by healthy EBV-experienced individuals. Of 64 CD8+ T cell responses directed against LMP-2, 39 occurred in PBMC from the HLA-A*0201 expressing donors. This finding may explain why the strongest associations between NPC incidence and HLA phenotype have been associated with HLA-A2, and why HLA-A2 may be associated with higher risk of developing NPC in Southern Asia where the prevalence of HLA-A*0201 is relatively low compared with other HLA-A2 family members. In particular, the Chinese population may lack the powerful immune response elicited by LMP-2 in the context of HLA-A*0201 because the most frequent HLA-A alleles in Southern Asia include HLA-A*0203 and HLA-A*0207. The predominant role that HLA-A*0201 may play in the response to LMP-2 (if confirmed) may partly explain in functional terms the epidemiologic observation that NPC is predominant in Chinese patients expressing HLA-A*0207 [37]. Therefore, a comprehensive comparison of responses to LMP-2 in the context of the HLA-A2 super family in different incidence areas is still warranted in the future.
Innate Immunity: NK-KIR-HLA
HLA class I alleles play an additional role in modulating natural killer (NK) cell biology which are part of a potentially powerful anti-cancer innate effector mechanism. Interaction of NK cells with normal levels of expression of HLA class I molecules by cancer or healthy cells triggers the activation of Killer-immunoglobulin like receptors (KIRs) whose activation exerts inhibitory effects of the function of NK cells. This recognition, however, is HLA class I allele dependent and different HLA class I alleles can inhibit the function of select KIR molecules on the surface of individual NK cells. The KIR family is highly polymorphic and consists of activating receptors (KIR2DS1, 2, 3, 4, 5 and KIR3DS1) and inhibitory receptors (KIR2DL1, 2/3, 4, 5A/B and KIR3DL1, 2, 3, 4) [71]. Recently, our knowledge about their relationship with various HLA alleles has increased and data suggests that it may modulate the immune surveillance against Hepatitis C Virus infection [72–74], Human Immunodeficiency Virus infection [75–80], malignant melanoma [81, 82], cervical cancer [83] and leukemia [84–86]. Several interactions between KIRs and HLA alleles, such as KIR3DS1 and KIR3DL1 with HLA-Bw4, KIR3DL1 with HLA-B57, KIR2DS1, KIR2DL3 and KIR2DS2 with HLA-Cw1 have been clearly defined [71]. A hypothesis has recently been proposed by Butsch Kovacic M. et al. [87] which attempts to link KIRs and HLA-Cw to the incidence of NPC. An increased number of activating KIRs was found in NPC patients, especially in individuals seropositive for anti-EBV antibodies known to be linked to NPC susceptibility (Ptrend= 0.07). A 3.4-fold increased risk of NPC (95% CI = 0.74–15.7) was found in EBV exposed individuals carrying ≥5 activating KIRs compared with individuals without functional activating KIRs. Meanwhile, individuals expressing HLA-Cw*0401 were observed to be at a significantly reduced risk to develop NPC (OR=0.46, 95%CI=0.23–0.92). These results are consistent with other findings examining KIR expression in HIV related cervical neoplasia[83].
Prevalence of NPC Determined by a Susceptibility Locus Linked to HLA
An alternative and, perhaps, more likely explanation for the associations between HLA and the prevalence of NPC could be the presence of a locus in the HLA region in strong linkage disequilibrium with HLA genes characterized by a genetic propensity to promote the development of NPC [21, 35].
The first linkage study was published by Lu SJ et al. [21] in 1990, based on affected sibling pairs from Singapore, Hong Kong, and South China. This study first suggested the existence of a recessive NPC susceptibility gene or locus in strong linkage disequilibrium with the HLA class I region. The maximum likelihood estimated a relative risk of approximately 21. Subsequently, Ooi et al. [22] have analyzed several microsatellite markers (D6S273, D6S276, and D6S1624) around the tumor necrosis factor- gene (also defined as the HLA class III region) as well as the loci of HLA-A, -B, -DR in 55 NPC patients and 62 normal Chinese controls from Singapore. This study located this putative NPC susceptibility gene within the centromeric end of the HLA class I region and the telomeric end of the HLA class-III region near the D6S1624 locus (Fig. 7). The presence of allele 4 of the microsatellite conferred a 3.5-fold increase in the risk of developing NPC; a risk higher than that associated with specific HLA loci. Subsequently, this hypothesis has been strengthened by two other studies in Taiwanese patients. In 2003, Lu CC et al. [23] reported a microsatellite marker D6S211(Fig. 7) located 97 kb telomeric to HLA-A (OR = 3.97, p= 0.0042),. Moreover, Lu CC et al. [24] carried out a further study in which eight polymorphic microsatellite markers distributed over a one megabase region surrounding the HLA-A locus were subjected to genetic analysis. This study narrowed down the likelihood of an NPC susceptibility locus to a 132 Kb segment situated between the D6S211 and D6S510 close to the HLA-A locus (Fig. 7). Recently Hu et al. have provided further evidence for an HLA-related recessive mutation associated with NPC among Chinese [35]. These results facilitated further positional cloning of the susceptibility locus within this region. However, positional cloning has proven to be a challenge in this region due to the presence of 23 pseudogenes among a total of 24 genes. In order to evaluate whether this region is associated with NPC, we have applied an association study on a Chinese population by means of detection of tag single nucleotide polymorphisms (SNPs) and found that several tag SNPs in this region were simultaneously associated with an increased risk of developing NPC (unpublished observation). Among these tag SNPs, one was located at the 5′ terminus of a pseudogene adjacent to HLA-A locus. Computational prediction and electrophoresis mobility shift assay (EMSA) verified the presence of a specific transcription binding site when an A to G variant was present. This association between non HLA loci and the prevalence of NPC expands the possible explanation for such associations. Although HLA-A may represent an important gene that predisposes to the occurrence of NPC because of specific immunological functions, it is more likely that another susceptibility locus or gene in the same region may predispose to the development of NPC through different functional pathways. Moreover, it is possible that pseudogenes in linkage with the HLA-A locus may bear direct or indirect functional effects. Recently Zheng et al. have extensively examined the transcriptional activity of pseudogenes in the ENCyclopedia of DNA Elements (ENCODE) project and have demonstrated that at least one fifth of 201 pseudogenes in ENCODE regions are transcribed in one or more cell lines and tissues [88, 89]. The discovery that the Oct4 pseudogenes (Oct4-pg5 and Oct4-pg1) were transcribed in different cancer cells and tissues suggests a possible regulatory function that may be pertinent to carcinogenesis [90]. Finally, Yano et al. [91] have reported that pseudogenes transcribed as non-coding RNA could play a new role in the regulation of homologous messenger RNA stability. Therefore, the exact role of pseudogenes in this region should be investigated although such investigation is currently challenged by the lack of efficient technologies to assess the downstream functional effects.
Fig. (7).
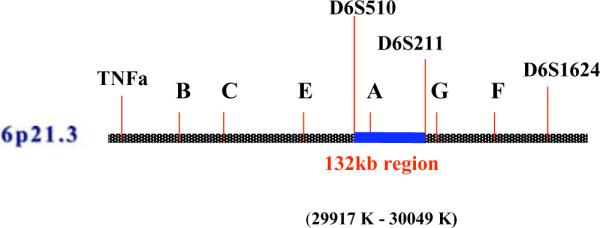
Susceptibility region in HLA.
CONCLUSION
Genetic susceptibility and EBV infection have been consistently reported as etiological co-factors responsible for the risk of developing NPC in various geographical areas at various level of risk. Other dietary, social and environmental factors may be more specific to different areas and ethnic groups. Investigations aimed at the identification of genes conferring susceptibility to NPC have focused on the HLA region in the short arm of chromosome 6 due to the consistency in which associations between specific HLA alleles and the prevalence of NPC have been reported especially in the Chinese population. Although the reasons for these associations still remain elusive, HLA is undoubtedly playing an important role in NPC predisposition, either by playing a functional role in modulating an innate and adaptive immune response against EBV, or as a marker of an unrelated predisposition locus in close linkage.
Therefore, further investigations should be encouraged to: 1) Extensively explore the relationship between specific HLA restricted epitopes and EBV associated peptides; 2) Further investigate the relationship between KIR and HLA phenotypes associated with the development of NPC; 3) Expand the search for NPC susceptibility loci within or close to the HLA region; and 4) Improve computational prediction approaches and downstream functional technologies to explore the functional effects of genetic variances detected in this region.
REFERENCES
- [1].Huang T, Liu Q, Huang H, Cao S. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2002;19:134–7. [PubMed] [Google Scholar]
- [2].Grulich AE, McCredie M, Coates M. Br. J. Cancer. 1995;71:400–8. doi: 10.1038/bjc.1995.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Feng BJ, Huang W, Shugart YY, Lee MK, Zhang F, Xia JC, Wang HY, Huang TB, Jian SW, Huang P, Feng QS, Huang LX, Yu J, Li D, Chen LZ, Jia WH, Fang Y, Huang HM, Zhu JL, Liu XM, Zhao Y, Liu WQ, Deng MQ, Hu WH, Wu SX, Mo HY, Hong MF, King MC, Chen Z, Zeng YX. Nat. Genet. 2002;31:395–9. doi: 10.1038/ng932. [DOI] [PubMed] [Google Scholar]
- [4].Jiang RC, Qin HD, Zeng MS, Huang W, Feng BJ, Zhang F, Chen HK, Jia WH, Chen LZ, Feng QS, Zhang RH, Yu XJ, Zheng MZ, Zeng X. Cancer Res. 2006;66:693–700. doi: 10.1158/0008-5472.CAN-05-2166. [DOI] [PubMed] [Google Scholar]
- [5].Guo XC, Scott K, Liu Y, Dean M, David V, Nelson GW, Johnson RC, Dilks HH, Lautenberger J, Kessing B, Martenson J, Guan L, Sun S, Deng H, Zheng Y, de The G, Liao J, Zeng Y, O'Brien SJ, Winkler CA. Hum. Genomics. 2006;2:365–75. doi: 10.1186/1479-7364-2-6-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Xiong W, Zeng ZY, Xia JH, Xia K, Shen SR, Li XL, Hu DX, Tan C, Xiang JJ, Zhou J, Deng H, Fan SQ, Li WF, Wang R, Zhou M, Zhu SG, Lu HB, Qian J, Zhang BC, Wang JR, Ma J, Xiao BY, Huang H, Zhang QH, Zhou YH, Luo XM, Zhou HD, Yang YX, Dai HP, Feng GY, Pan Q, Wu LQ, He L, Li GY. Cancer Res. 2004;64:1972–4. doi: 10.1158/0008-5472.can-03-3253. [DOI] [PubMed] [Google Scholar]
- [7].Yau WL, Lung HL, Zabarovsky ER, Lerman MI, Sham JS, Chua DT, Tsao SW, Stanbridge EJ, Lung ML. Int. J. Cancer. 2006;119:2821–6. doi: 10.1002/ijc.22232. [DOI] [PubMed] [Google Scholar]
- [8].Zeng Z, Zhou Y, Zhang W, Li X, Xiong W, Liu H, Fan S, Qian J, Wang L, Li Z, Shen S, Li G. Genet. Med. 2006;8:156–60. doi: 10.1097/01.gim.0000196821.87655.d0. [DOI] [PubMed] [Google Scholar]
- [9].Chen Y, Chan SH. Int. J. Cancer. 1994;56:830–3. doi: 10.1002/ijc.2910560613. [DOI] [PubMed] [Google Scholar]
- [10].Chen Y, Chew CT, Chan SH. Br. J. Cancer. 1995;72:117–22. doi: 10.1038/bjc.1995.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Nazar-Stewart V, Vaughan TL, Burt RD, Chen C, Berwick M, Swanson GM. Cancer Epidemiol. Biomarkers Prev. 1999;8:547–51. [PubMed] [Google Scholar]
- [12].Tiwawech D, Srivatanakul P, Karalak A, Ishida T. Asian Pac. J. Cancer Prev. 2005;6:270–5. [PubMed] [Google Scholar]
- [13].He ZM, Wang SL, Yuan JH, Chen ZC. Ai Zheng. 2002;21:597–600. [PubMed] [Google Scholar]
- [14].Hildesheim A, Chen CJ, Caporaso NE, Cheng YJ, Hoover RN, Hsu MM, Levine PH, Chen IH, Chen JY, Yang CS. Cancer Epidemiol. Biomarkers Prev. 1995;4:607–10. [PubMed] [Google Scholar]
- [15].Kongruttanachok N, Sukdikul S, Setavarin S, Kerekhjanarong V, Supiyaphun P, Voravud N, Poovorawan Y, Mutirangura A. BMC Cancer. 2001;1:4. doi: 10.1186/1471-2407-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hirunsatit R, Kongruttanachok N, Shotelersuk K, Supiyaphun P, Voravud N, Sakuntabhai A, Mutirangura A. BMC Genet. 2003;4:3. doi: 10.1186/1471-2156-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Fan Q, Jia WH, Zhang RH, Yu XJ, Chen LZ, Feng QS, Zeng YX. Ai Zheng. 2005;24:915–8. [PubMed] [Google Scholar]
- [18].Jalbout M, Bouaouina N, Gargouri J, Corbex M, Ben Ahmed S, Chouchane L. Cancer Lett. 2003;193:75–81. doi: 10.1016/s0304-3835(02)00697-3. [DOI] [PubMed] [Google Scholar]
- [19].Cho EY, Hildesheim A, Chen CJ, Hsu MM, Chen IH, Mittl BF, Levine PH, Liu MY, Chen JY, Brinton LA, Cheng YJ, Yang CS. Cancer Epidemiol. Biomarkers Prev. 2003;12:1100–4. [PubMed] [Google Scholar]
- [20].Simons MJ, Day NE, Wee GB, Shanmugaratnam K, Ho HC, Wong SH, Ti TK, Yong NK, Darmalingam S, De-The G. Cancer Res. 1974;34:1192–5. [PubMed] [Google Scholar]
- [21].Lu SJ, Day NE, Degos L, Lepage V, Wang PC, Chan SH, Simons M, McKnight B, Easton D, Zeng Y, et al. Nature. 1990;346:470–1. doi: 10.1038/346470a0. [DOI] [PubMed] [Google Scholar]
- [22].Ooi EE, Ren EC, Chan SH. Int. J. Cancer. 1997;74:229–32. doi: 10.1002/(sici)1097-0215(19970422)74:2<229::aid-ijc16>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- [23].Lu CC, Chen JC, Jin YT, Yang HB, Chan SH, Tsai ST. Int. J. Cancer. 2003;103:745–51. doi: 10.1002/ijc.10861. [DOI] [PubMed] [Google Scholar]
- [24].Lu CC, Chen JC, Tsai ST, Jin YT, Tsai JC, Chan SH, Su IJ. Int. J. Cancer. 2005;115:742–6. doi: 10.1002/ijc.20946. [DOI] [PubMed] [Google Scholar]
- [25].Brewerton DA. Curr. Opin. Rheumatol. 2003;15:369–73. doi: 10.1097/00002281-200307000-00001. [DOI] [PubMed] [Google Scholar]
- [26].Larsen CE, Alper CA. Curr. Opin. Immunol. 2004;16:660–7. doi: 10.1016/j.coi.2004.07.014. [DOI] [PubMed] [Google Scholar]
- [27].Ahmad T, Marshall SE, Jewell D. World J. Gastroenterol. 2006;12:3628–35. doi: 10.3748/wjg.v12.i23.3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ghodke Y, Joshi K, Chopra A, Patwardhan B. Eur. J. Epidemiol. 2005;20:475–88. doi: 10.1007/s10654-005-5081-x. [DOI] [PubMed] [Google Scholar]
- [29].Simons MJ, Wee GB, Chan SH, Shanmugaratnam K, Day NE, de-The G. IARC. Sci. Publ. 1975:249–58. [PubMed] [Google Scholar]
- [30].Bull. WHO. 1978;56:461–5. 1977. [Google Scholar]
- [31].Chan SH, Day NE, Kunaratnam N, Chia KB, Simons MJ. Int. J. Cancer. 1983;32:171–6. doi: 10.1002/ijc.2910320206. [DOI] [PubMed] [Google Scholar]
- [32].Wu SB, Hwang SJ, Chang AS, Hsieh T, Hsu MM, Hsieh RP, Chen J. Anticancer Res. 1989;9:1649–53. [PubMed] [Google Scholar]
- [33].Pimtanothai N, Kangwanshiratada O, Charoenwongse P. J. Med. Assoc. Thai. 2003;86(Suppl 2):S237–41. [PubMed] [Google Scholar]
- [34].Simons MJ, Wee GB, Singh D, Dharmalingham S, Yong NK, Chau JC, Ho JH, Day NE, De-The G. Natl. Cancer Inst. Monogr. 1977;47:147–51. [PubMed] [Google Scholar]
- [35].Hu SP, Day NE, Li DR, Luben RN, Cai KL, Ou-Yang T, Li B, Lu XZ, Ponder BA. Br. J. Cancer. 2005;92:967–70. doi: 10.1038/sj.bjc.6602347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Jing J, Louie E, Henderson BE, Terasaki P. Natl. Cancer Inst. Monogr. 1977;47:153–6. [PubMed] [Google Scholar]
- [37].Hildesheim A, Apple RJ, Chen CJ, Wang SS, Cheng YJ, Klitz W, Mack SJ, Chen IH, Hsu MM, Yang CS, Brinton LA, Levine PH, Erlich A. J. Natl. Cancer Inst. 2002;94:1780–9. doi: 10.1093/jnci/94.23.1780. [DOI] [PubMed] [Google Scholar]
- [38].Simons MJ. ASHI quarterly, Second quarter. 2003:52–55. [Google Scholar]
- [39].Pimtanothai N, Charoenwongse P, Mutirangura A, Hurley CK. Tissue Antigens. 2002;59:223–5. doi: 10.1034/j.1399-0039.2002.590308.x. [DOI] [PubMed] [Google Scholar]
- [40].Betuel H, Camoun M, Colombani J, Day NE, Ellouz R, de-The G. Int. J. Cancer. 1975;16:249–54. doi: 10.1002/ijc.2910160207. [DOI] [PubMed] [Google Scholar]
- [41].Kruger J, I.V., Dahr W. Cancer Campaign, Naspharyngeal carcinoma. 1981;5:201–205. [Google Scholar]
- [42].Hall PJ, Levin AG, Entwistle CC, Knight SC, Wasunna A, Kung'u A, Brubaker G. Hum. Immunol. 1982;5:91–105. doi: 10.1016/0198-8859(82)90055-6. [DOI] [PubMed] [Google Scholar]
- [43].Herait P, Tursz T, Guillard MY, Hanna K, Lipinski M, Micheau C, Sancho-Garnier H, Schwaab G, Cachin Y, Degos L. Tissue Antigens. 1983;22:335–41. doi: 10.1111/j.1399-0039.1983.tb02262.x. [DOI] [PubMed] [Google Scholar]
- [44].Moore SB, Pearson GR, Neel HB, 3rd, Weiland LH. Tissue Antigens. 1983;22:72–5. doi: 10.1111/j.1399-0039.1983.tb01168.x. [DOI] [PubMed] [Google Scholar]
- [45].Zervas J. GS, Tourlas D, Varlegidis E, Pantazopoulos P. Panminerva Medica. 1983;25:255–7. [PubMed] [Google Scholar]
- [46].Burt RD, Vaughan TL, McKnight B, Davis S, Beckmann AM, Smith AG, Nisperos B, Swanson GM, Berwick M. Cancer Epidemiol. Biomarkers Prev. 1996;5:879–87. [PubMed] [Google Scholar]
- [47].Daniilidis M, Fountzilas G, Fleva A, Daniilidis J, Tourkantonis A. Oncology. 1997;54:185–92. doi: 10.1159/000227686. [DOI] [PubMed] [Google Scholar]
- [48].Dardari R, Khyatti M, Jouhadi H, Benider A, Ettayebi H, Kahlain A, Mansouri A, El Gueddari B, Benslimane A. Int. J. Cancer. 2001;92:294–7. doi: 10.1002/1097-0215(200102)9999:9999<::aid-ijc1177>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- [49].Mokni-Baizig N, Ayed K, Ayed B, Ayed S, Sassi F, Ladgham A, Bel Hadj O, El May A. Oncology. 2001;61:55–8. doi: 10.1159/000055353. [DOI] [PubMed] [Google Scholar]
- [50].Li X, Ghandri N, Piancatelli D, Adams S, Chen D, Robbins FM, Wang E, Monaco A, Selleri S, Bouaouina N, Stroncek D, Adorno D, Chouchane L, Marincola FM. J. Transl. Med. 2007;5:22. doi: 10.1186/1479-5876-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Burt RD, Vaughan TL, Nisperos B, Swanson M, Berwick M. Int. J. Cancer. 1994;56:465–7. doi: 10.1002/ijc.2910560402. [DOI] [PubMed] [Google Scholar]
- [52].Chan SH, D. N. E., Khor TH, Kunaratnam N, Chia KB. Cancer Campaign, Vol 5, Naspharyngeal carcinoma. Gustav Fischer Verlag. Stuttgart. New York: 1981. pp. 205–11. [Google Scholar]
- [53].Chan SH, Chew CT, Prasad U, Wee GB, Srinivasan N, Kunaratnam N. Br. J. Cancer. 1985;51:389–92. doi: 10.1038/bjc.1985.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Simons MJ, Day NE. Natl. Cancer Inst. Monogr. 1977;47:143–6. [PubMed] [Google Scholar]
- [55].Ou BJ, Y. H. Y., Fang Y, Liu LM, Li ZQ, Zhang F. Ai Zheng. 1985;4:5–7. [Google Scholar]
- [56].Lee LK, Tan EL, Gopala K, Sam CK. Singapore Med. J. 2007;48:632–4. [PubMed] [Google Scholar]
- [57].Goldsmith DB, West TM, Morton R. Clin. Otolaryngol. Allied Sci. 2002;27:61–7. doi: 10.1046/j.0307-7772.2001.00529.x. [DOI] [PubMed] [Google Scholar]
- [58].Sengupta S, den Boon JA, Chen IH, Newton MA, Dahl DB, Chen M, Cheng YJ, Westra WH, Chen CJ, Hildesheim A, Sugden B, Ahlquist P. Cancer Res. 2006;66:7999–8006. doi: 10.1158/0008-5472.CAN-05-4399. [DOI] [PubMed] [Google Scholar]
- [59].Ogino T, Moriai S, Ishida Y, Ishii H, Katayama A, Miyokawa N, Harabuchi Y, Ferrone S. Int. J. Cancer. 2007;120:2401–10. doi: 10.1002/ijc.22334. [DOI] [PubMed] [Google Scholar]
- [60].Parham P, Ohta T. Science. 1996;272:67–74. doi: 10.1126/science.272.5258.67. [DOI] [PubMed] [Google Scholar]
- [61].Zernich D, Purcell AW, Macdonald WA, Kjer-Nielsen L, Ely LK, Laham N, Crockford T, Mifsud NA, Bharadwaj M, Chang L, Tait BD, Holdsworth R, Brooks AG, Bottomley SP, Beddoe T, Peh CA, Rossjohn J, McCluskey J. J. Exp. Med. 2004;200:13–24. doi: 10.1084/jem.20031680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Jurgens LA, Khanna R, Weber J, Orentas RJ. J. Clin. Immunol. 2006;26:22–32. doi: 10.1007/s10875-006-6532-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Lin HJ, Cherng JM, Hung MS, Sayion Y, Lin JC. J. Biomed. Sci. 2005;12:925–36. doi: 10.1007/s11373-005-9017-y. [DOI] [PubMed] [Google Scholar]
- [64].Lin JC, Cherng JM, Lin HJ, Tsang CW, Liu YX, Lee SP. J. Gen. Virol. 2004;85:2023–34. doi: 10.1099/vir.0.19696-0. [DOI] [PubMed] [Google Scholar]
- [65].Lin CL, Lo WF, Lee TH, Ren Y, Hwang SL, Cheng YF, Chen CL, Chang YS, Lee SP, Rickinson AB, Tam PK. Cancer Res. 2002;62:6952–8. [PubMed] [Google Scholar]
- [66].Khanna R, Burrows SR, Nicholls J, Poulsen LM. Eur. J. Immunol. 1998;28:451–8. doi: 10.1002/(SICI)1521-4141(199802)28:02<451::AID-IMMU451>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- [67].Lee SP, Thomas WA, Murray RJ, Khanim F, Kaur S, Young LS, Rowe M, Kurilla M, Rickinson AB. J. Virol. 1993;67:7428–35. doi: 10.1128/jvi.67.12.7428-7435.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Lee SP, Tierney RJ, Thomas WA, Brooks JM, Rickinson AB. J. Immunol. 1997;158:3325–34. [PubMed] [Google Scholar]
- [69].Weng X, Liang Z, Lu X, Zhong M, Lu S, Zhang C, Deng J, Wu X, Gong F. Sci. China C. Life Sci. 2007;50:203–11. doi: 10.1007/s11427-007-0036-y. [DOI] [PubMed] [Google Scholar]
- [70].Provenzano M, Selleri S, Jin P, Wang E, Werden R, Slezak S, Adams SD, Panelli MC, Leitman SF, Stroncek DF, Marincola FM. Cancer Immunol. Immunother. 2007;56:1047–63. doi: 10.1007/s00262-006-0246-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Williams AP, Bateman AR, Khakoo SI. Mol. Interv. 2005;5:226–40. doi: 10.1124/mi.5.4.6. [DOI] [PubMed] [Google Scholar]
- [72].Singh R, Kaul R, Kaul A, Khan K. World J. Gastroenterol. 2007;13:1770–87. doi: 10.3748/wjg.v13.i12.1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Rauch A, Laird R, McKinnon E, Telenti A, Furrer H, Weber R, Smillie D, Gaudieri S. Tissue Antigens. 2007;69(Suppl 1):237–40. doi: 10.1111/j.1399-0039.2006.773_4.x. [DOI] [PubMed] [Google Scholar]
- [74].Khakoo SI, Thio CL, Martin MP, Brooks CR, Gao X, Astemborski J, Cheng J, Goedert JJ, Vlahov D, Hilgartner M, Cox S, Little AM, Alexander GJ, Cramp ME, O'Brien SJ, Rosenberg WM, Thomas DL, Carrington M. Science. 2004;305:872–4. doi: 10.1126/science.1097670. [DOI] [PubMed] [Google Scholar]
- [75].Pascal V, Yamada E, Martin MP, Alter G, Altfeld M, Metcalf JA, Baseler MW, Adelsberger JW, Carrington M, Anderson SK, McVicar DW. J. Immunol. 2007;179:1625–33. doi: 10.4049/jimmunol.179.3.1625. [DOI] [PubMed] [Google Scholar]
- [76].Martin MP, Qi X, Yamada E, Martin JN, Pereyra F, Colombo S, Brown EE, Shupert WL, Phair J, Goedert JJ, Buchbinder S, Kirk GD, Telenti A, Connors M, O'Brien SJ, Walker BD, Parham P, Deeks SG, McVicar DW, Carrington M. Nat. Genet. 2007;39:733–40. doi: 10.1038/ng2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Ravet S, Scott-Algara D, Bonnet E, Tran HK, Tran T, Nguyen N, Truong X, Theodorou I, Barre-Sinoussi F, Pancino G, Paul P. Blood. 2007;109:4296–305. doi: 10.1182/blood-2006-08-040238. [DOI] [PubMed] [Google Scholar]
- [78].Gaudieri S, Nolan D, McKinnon E, Witt CS, Mallal S, Christiansen FT. Mol. Immunol. 2005;42:557–60. doi: 10.1016/j.molimm.2004.07.041. [DOI] [PubMed] [Google Scholar]
- [79].De Maria A, Mavilio D, Costa P, Dignetti P, Fogli M, Mingari MC. Immunol. Lett. 2000;72:179–82. doi: 10.1016/s0165-2478(00)00186-3. [DOI] [PubMed] [Google Scholar]
- [80].De Maria A, Moretta L. Hum. Immunol. 2000;61:74–81. doi: 10.1016/s0198-8859(99)00169-x. [DOI] [PubMed] [Google Scholar]
- [81].Naumova E, Mihaylova A, Ivanova M, Mihailova S. Cancer Immunol. Immunother. 2007;56:95–100. doi: 10.1007/s00262-006-0151-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Naumova E, Mihaylova A, Stoitchkov K, Ivanova M, Quin L, Toneva M. Cancer Immunol. Immunother. 2005;54:172–8. doi: 10.1007/s00262-004-0575-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Carrington M, Martin MP. In: Current topics in microbiology and immunology: activating and inhibitory NK cell receptors. Vivier E, Colonna M, editors. Springer; Heidelberg (German): 2005. [Google Scholar]
- [84].Savani BN, Mielke S, Adams S, Uribe M, Rezvani K, Yong AS, Zeilah J, Kurlander R, Srinivasan R, Childs R, Hensel N, Barrett AJ. Leukemia. 2007;21:2145–52. doi: 10.1038/sj.leu.2404892. [DOI] [PubMed] [Google Scholar]
- [85].Sconocchia G, Lau M, Provenzano M, Rezvani K, Wongsena W, Fujiwara H, Hensel N, Melenhorst J, Li J, Ferrone S, Barrett AJ. Blood. 2005;106:3666–72. doi: 10.1182/blood-2005-02-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Verheyden S, Bernier M, Demanet C. Leukemia. 2004;18:2002–7. doi: 10.1038/sj.leu.2403525. [DOI] [PubMed] [Google Scholar]
- [87].Butsch Kovacic M, Martin M, Gao X, Fuksenko T, Chen CJ, Cheng YJ, Chen JY, Apple R, Hildesheim A, Carrington M. Cancer Epidemiol. Biomarkers Prev. 2005;14:2673–7. doi: 10.1158/1055-9965.EPI-05-0229. [DOI] [PubMed] [Google Scholar]
- [88].Zheng D, Frankish A, Baertsch R, Kapranov P, Reymond A, Choo W, Lu Y, Denoeud F, Antonarakis SE, Snyder M, Ruan Y, Wei CL, Gingeras R, Guigo R, Harrow J, Gerstein MB. Genome Res. 2007;17:839–51. doi: 10.1101/gr.5586307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Zheng D, Gerstein MB. Genome Biol. 2006;7(Suppl 1):S13, 1–10. doi: 10.1186/gb-2006-7-s1-s13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Suo G, Han J, Wang X, Zhang J, Zhao Y, Zhao Y, Dai J. Biochem. Biophys. Res. Commun. 2005;337:1047–51. doi: 10.1016/j.bbrc.2005.09.157. [DOI] [PubMed] [Google Scholar]
- [91].Yano Y, Saito R, Yoshida N, Yoshiki A, Wynshaw-Boris A, Tomita M, Hirotsune S. J. Mol. Med. 2004;82:414–22. doi: 10.1007/s00109-004-0550-3. [DOI] [PubMed] [Google Scholar]