Abstract
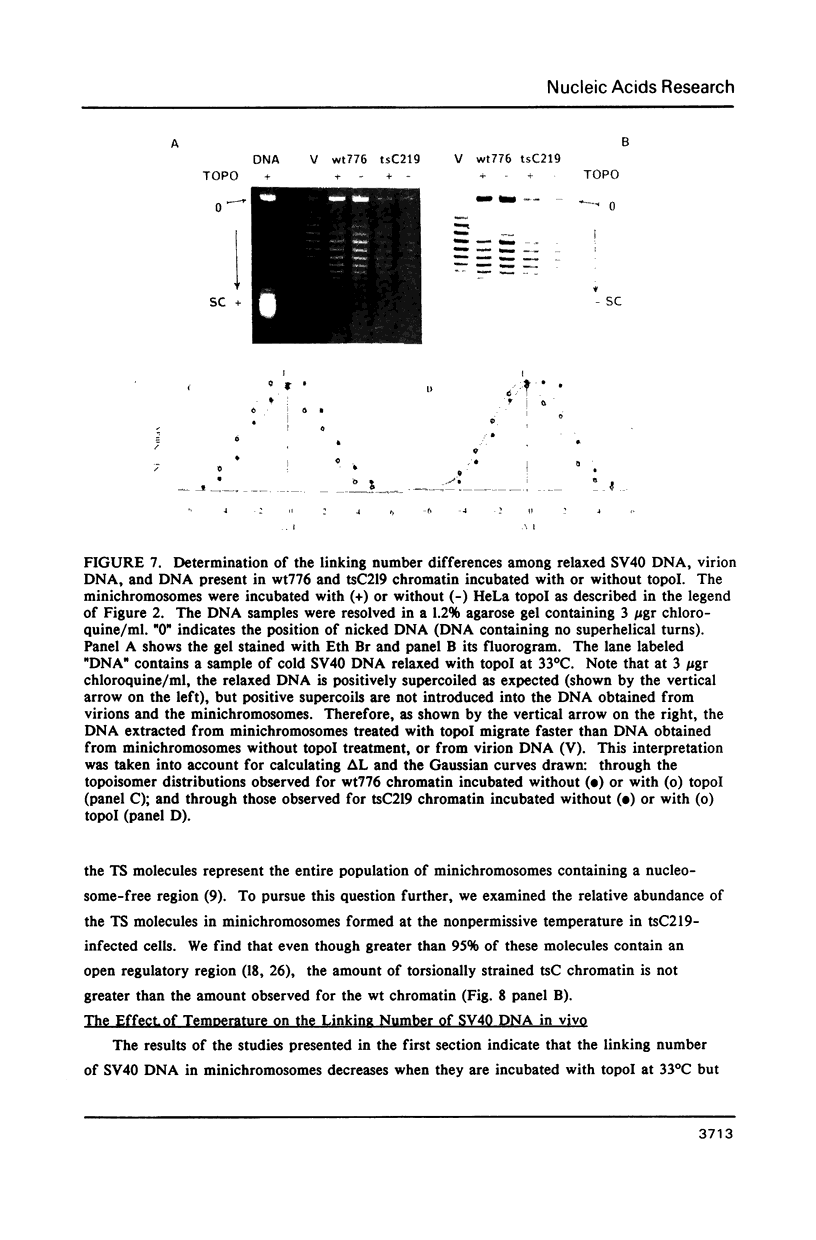
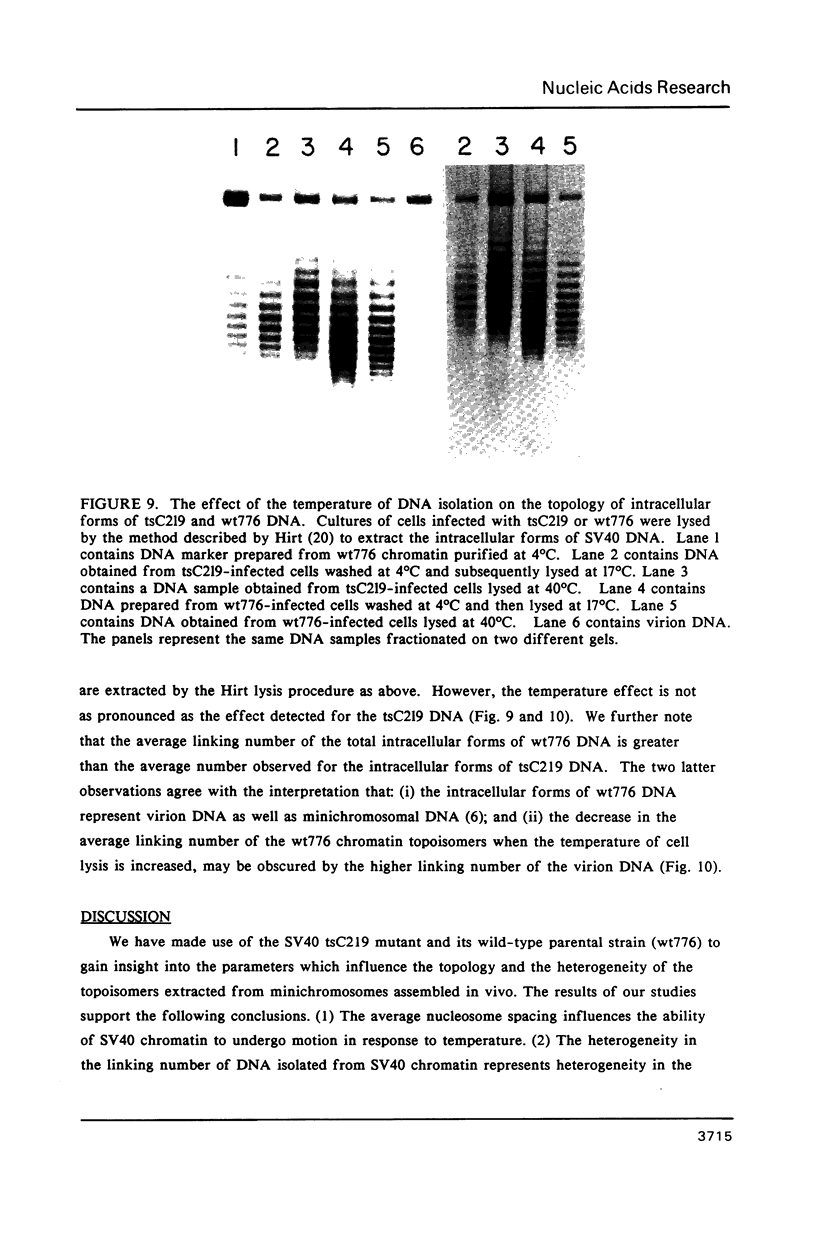
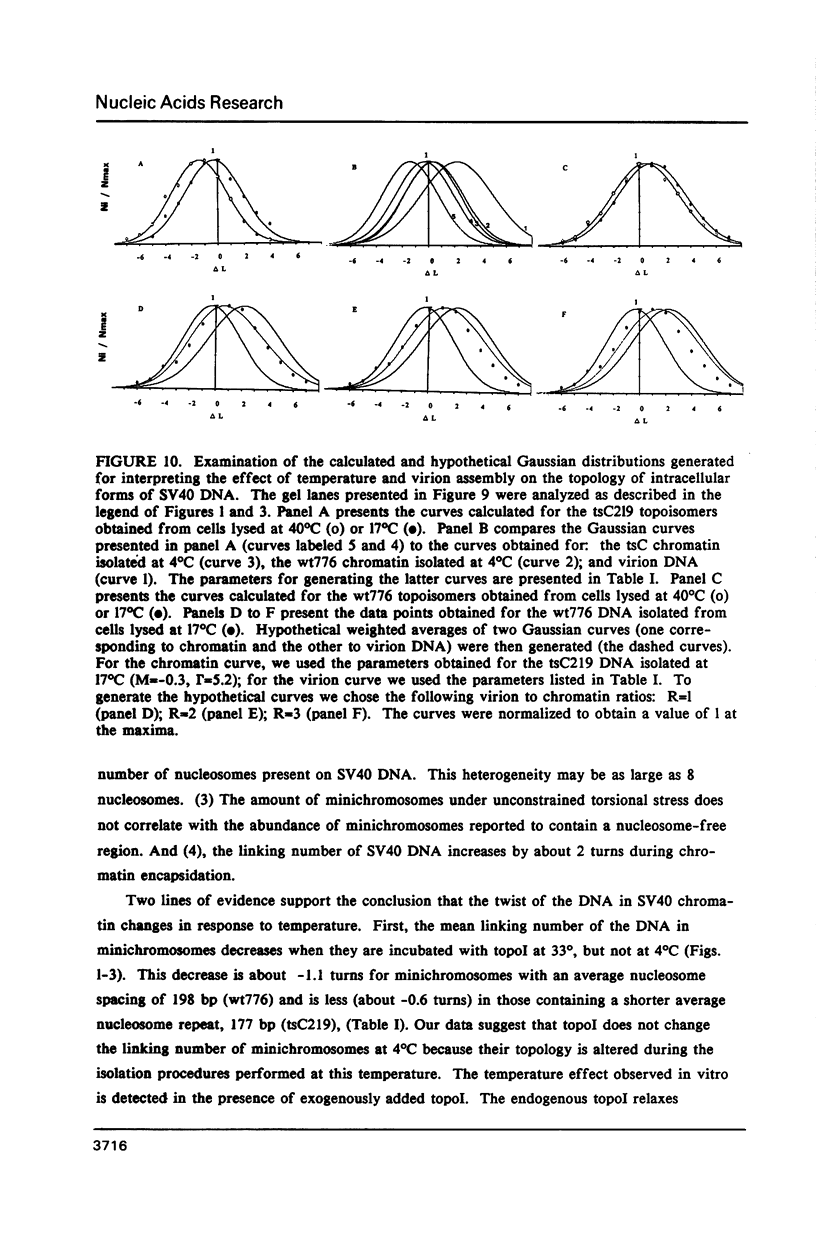
The linking number of DNA in minichromosomes increases by 2 turns during SV40 assembly. Changes in temperature also influence the average linking number of the total intracellular forms of SV40 DNA. When the isolated minichromosomes assembled in vivo are incubated with topoisomerase I at 33 degrees C in vitro, the linking number of SV40 DNA decreases. This decrease is about: -1.1 turns for minichromosomes with an average nucleosome spacing of 198 base pairs (bp), wt776; and -0.6 turns for minichromosomes containing a shorter average nucleosome repeat (177 bp), tsC219. The difference between the average linking number of naked SV40 DNA relaxed with topoI at 33 degrees C and minichromosomes relaxed with the enzyme at the same temperature indicates that SV40 chromatin contains on the average 26 nucleosomes. However, the results of studies obtained both on DNA flexibility in chromatin and in naked DNA, and on the shape of the topoisomer distribution curves, indicate that all of the minichromosomes, regardless of their overall structure, do not contain the same number of nucleosomes; this heterogeneity may be as large as 8 nucleosomes. We find no apparent correlation between the amount of minichromosomes containing unconstrained torsional stress and the abundance of the molecules with a structure characteristic of transcriptionally active chromatin.
Full text
PDF















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ambrose C., Blasquez V., Bina M. A block in initiation of simian virus 40 assembly results in the accumulation of minichromosomes containing an exposed regulatory region. Proc Natl Acad Sci U S A. 1986 May;83(10):3287–3291. doi: 10.1073/pnas.83.10.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsoum J., Berg P. Simian virus 40 minichromosomes contain torsionally strained DNA molecules. Mol Cell Biol. 1985 Nov;5(11):3048–3057. doi: 10.1128/mcb.5.11.3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bina-Stein M., Simpson R. T. Specific folding and contraction of DNA by histones H3 and H4. Cell. 1977 Jul;11(3):609–618. doi: 10.1016/0092-8674(77)90078-2. [DOI] [PubMed] [Google Scholar]
- Bina M., Beecher S., Blasquez V. Stability and components of mature simian virus 40. Biochemistry. 1982 Jun 22;21(13):3057–3063. doi: 10.1021/bi00256a004. [DOI] [PubMed] [Google Scholar]
- Bina M., Blasquez V., Ng S. C., Beecher S. SV40 morphogenesis. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):565–569. doi: 10.1101/sqb.1983.047.01.066. [DOI] [PubMed] [Google Scholar]
- Blasquez V., Beecher S., Bina M. Simian virus 40 morphogenetic pathway. An analysis of assembly-defective tsB201 DNA protein complexes. J Biol Chem. 1983 Jul 10;258(13):8477–8484. [PubMed] [Google Scholar]
- Blasquez V., Stein A., Ambrose C., Bina M. Simian virus 40 protein VP1 is involved in spacing nucleosomes in minichromosomes. J Mol Biol. 1986 Sep 5;191(1):97–106. doi: 10.1016/0022-2836(86)90425-0. [DOI] [PubMed] [Google Scholar]
- Chen S. S., Hsu M. T. Evidence for variation of supercoil densities among simian virus 40 nucleoprotein complexes and for higher supercoil density in replicating complexes. J Virol. 1984 Jul;51(1):14–19. doi: 10.1128/jvi.51.1.14-19.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depew D. E., Wang J. C. Conformational fluctuations of DNA helix. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4275–4279. doi: 10.1073/pnas.72.11.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg J. C., Cartwright I. L., Thomas G. H., Elgin S. C. Selected topics in chromatin structure. Annu Rev Genet. 1985;19:485–536. doi: 10.1146/annurev.ge.19.120185.002413. [DOI] [PubMed] [Google Scholar]
- Germond J. E., Hirt B., Oudet P., Gross-Bellark M., Chambon P. Folding of the DNA double helix in chromatin-like structures from simian virus 40. Proc Natl Acad Sci U S A. 1975 May;72(5):1843–1847. doi: 10.1073/pnas.72.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Jakobovits E. B., Bratosin S., Aloni Y. A nucleosome-free region in SV40 minichromosomes. Nature. 1980 May 22;285(5762):263–265. doi: 10.1038/285263a0. [DOI] [PubMed] [Google Scholar]
- Keller W. Determination of the number of superhelical turns in simian virus 40 DNA by gel electrophoresis. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4876–4880. doi: 10.1073/pnas.72.12.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller W., Müller U., Eicken I., Wendel I., Zentgraf H. Biochemical and ultrastructural analysis of SV40 chromatin. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):227–244. doi: 10.1101/sqb.1978.042.01.025. [DOI] [PubMed] [Google Scholar]
- Luchnik A. N., Bakayev V. V., Yugai A. A., Zbarsky I. B., Georgiev G. P. DNAaseI-hypersensitive minichromosomes of SV40 possess an elastic torsional strain in DNA. Nucleic Acids Res. 1985 Feb 25;13(4):1135–1149. doi: 10.1093/nar/13.4.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchnik A. N., Bakayev V. V., Zbarsky I. B., Georgiev G. P. Elastic torsional strain in DNA within a fraction of SV40 minichromosomes: relation to transcriptionally active chromatin. EMBO J. 1982;1(11):1353–1358. doi: 10.1002/j.1460-2075.1982.tb01322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse R. H., Cantor C. R. Nucleosome core particles suppress the thermal untwisting of core DNA and adjacent linker DNA. Proc Natl Acad Sci U S A. 1985 Jul;82(14):4653–4657. doi: 10.1073/pnas.82.14.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petryniak B., Lutter L. C. Topological characterization of the simian virus 40 transcription complex. Cell. 1987 Jan 30;48(2):289–295. doi: 10.1016/0092-8674(87)90432-6. [DOI] [PubMed] [Google Scholar]
- Pulleyblank D. E., Shure M., Tang D., Vinograd J., Vosberg H. P. Action of nicking-closing enzyme on supercoiled and nonsupercoiled closed circular DNA: formation of a Boltzmann distribution of topological isomers. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4280–4284. doi: 10.1073/pnas.72.11.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saavedra R. A., Huberman J. A. Both DNA topoisomerases I and II relax 2 micron plasmid DNA in living yeast cells. Cell. 1986 Apr 11;45(1):65–70. doi: 10.1016/0092-8674(86)90538-6. [DOI] [PubMed] [Google Scholar]
- Saragosti S., Moyne G., Yaniv M. Absence of nucleosomes in a fraction of SV40 chromatin between the origin of replication and the region coding for the late leader RNA. Cell. 1980 May;20(1):65–73. doi: 10.1016/0092-8674(80)90235-4. [DOI] [PubMed] [Google Scholar]
- Shure M., Pulleyblank D. E., Vinograd J. The problems of eukaryotic and prokaryotic DNA packaging and in vivo conformation posed by superhelix density heterogeneity. Nucleic Acids Res. 1977;4(5):1183–1205. doi: 10.1093/nar/4.5.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. T., Thoma F., Brubaker J. M. Chromatin reconstituted from tandemly repeated cloned DNA fragments and core histones: a model system for study of higher order structure. Cell. 1985 Oct;42(3):799–808. doi: 10.1016/0092-8674(85)90276-4. [DOI] [PubMed] [Google Scholar]
- Sogo J. M., Stahl H., Koller T., Knippers R. Structure of replicating simian virus 40 minichromosomes. The replication fork, core histone segregation and terminal structures. J Mol Biol. 1986 May 5;189(1):189–204. doi: 10.1016/0022-2836(86)90390-6. [DOI] [PubMed] [Google Scholar]
- Wang J. C., Peck L. J., Becherer K. DNA supercoiling and its effects on DNA structure and function. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):85–91. doi: 10.1101/sqb.1983.047.01.011. [DOI] [PubMed] [Google Scholar]