Abstract
Background
Non-small cell lung carcinoma (NSCLC) is a complex malignancy that owing to its heterogeneity and poor prognosis poses many challenges to diagnosis, prognosis and patient treatment. DNA methylation is an important mechanism of epigenetic regulation involved in normal development and cancer. It is a very stable and specific modification and therefore in principle a very suitable marker for epigenetic phenotyping of tumors. Here we present a genome-wide DNA methylation analysis of NSCLC samples and paired lung tissues, where we combine MethylCap and next generation sequencing (MethylCap-seq) to provide comprehensive DNA methylation maps of the tumor and paired lung samples. The MethylCap-seq data were validated by bisulfite sequencing and methyl-specific polymerase chain reaction of selected regions.
Results
Analysis of the MethylCap-seq data revealed a strong positive correlation between replicate experiments and between paired tumor/lung samples. We identified 57 differentially methylated regions (DMRs) present in all NSCLC tumors analyzed by MethylCap-seq. While hypomethylated DMRs did not correlate to any particular functional category of genes, the hypermethylated DMRs were strongly associated with genes encoding transcriptional regulators. Furthermore, subtelomeric regions and satellite repeats were hypomethylated in the NSCLC samples. We also identified DMRs that were specific to two of the major subtypes of NSCLC, adenocarcinomas and squamous cell carcinomas.
Conclusions
Collectively, we provide a resource containing genome-wide DNA methylation maps of NSCLC and their paired lung tissues, and comprehensive lists of known and novel DMRs and associated genes in NSCLC.
Keywords: DNA Methylation, Epigenetics, MethylCap, Next generation sequencing, Non-small cell lung Cancer
Background
Non-small cell lung carcinoma (NSCLC) is a common malignancy characterized by a worldwide high incidence and low survival rate [1]. NSCLC is a heterogenic disease which is broadly classified into three major histopathological subtypes: adenocarcinoma (ADC), squamous cell carcinoma (SCC) and large cell carcinoma (LCC). This heterogeneity poses challenges for diagnosis and treatment, since each subtype presents with a distinctive prognosis [2] and the choice of therapeutic regimen is predominantly based on tumor subtype and staging parameters [3]. The development of personalized diagnostics and therapy is leading the way to a new era that may see us overcome some of the difficulties in treating complex diseases such as NSCLC.
In the past decade, comparative gene expression profiles of tumors have been extensively studied [4-6], yielding useful insights into the molecular hallmarks of carcinogenesis [7,8]. With the advent of next generation sequencing, genome-wide screening has become an attractive tool for profiling tumors versus lung tissues [7,9]. DNA methylation is a very stable epigenetic mark and next generation sequencing studies have recently shown that many genes are aberrantly methylated in cancer [10,11]. Tissue specific DNA methylation patterns are stabilized during embryonic development, and faithfully maintained through cell divisions [12-14]. Nevertheless, established methylation patterns can be reprogrammed, with tumor cells undergoing DNA demethylation and de novo methylation through mechanisms not yet completely understood. CDKN2A and RASSF1 are examples of genes found to be aberrantly methylated in a wide variety of tumors [15-18], and epigenetic silencing of these genes has also been reported in NSCLC [19-22]. The great majority of DNA methylation studies are concentrated on the analysis of CpG islands located in the promoter areas of pre-selected genes. However, differentially methylated areas may be located within genes and at large distances from the nearest neighboring genes [23,24]. Although data on methylated genes in NSCLC are rapidly accumulating, unbiased data concerning specificity of the genome-wide distribution of methylated loci are still scarce.
In this study, we used Methyl-DNA Capture (MethylCap) and high-throughput sequencing (MethylCap-seq, [25]) to perform a genome-wide DNA methylation screening of NSCLC tumors and paired adjacent lung tissues. With this approach, we sought to identify genome-wide aberrant methylation patterns of NSCLC. Specific differentially methylated regions would be promising candidate molecular markers for non-invasive diagnostics using circulating tumor DNA, and increase the number of possible targets for epigenetic therapy.
Results
Methylation profiles in NSCLC-study outline
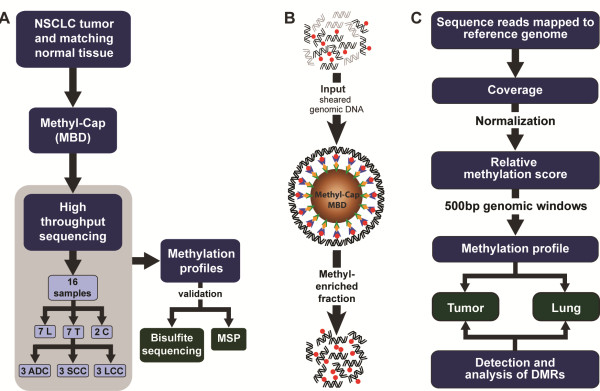
We performed genome-wide DNA methylation analysis of NSCLC using the MethylCap assay followed by high-throughput sequencing: MethylCap-seq (Figure 1A). We used DNA isolated from seven NSCLC tumors and paired lung tissues. Data regarding the samples used in this study can be found in Table 1 and Additional file 1. As controls, we prepared fully methylated and fully unmethylated genomic DNA. The DNA samples were sheared and then enriched for methylated DNA using the MethylCap procedure [25]. This is based on the capture of methylated DNA by biotinylated methyl-binding domain protein (MBD), which is then retrieved by binding to streptavidin-coated beads (Figure 1B). The recovered DNA was directly sequenced using the Illumina Genome Analyzer IIx next generation sequencing platform. For each DNA sample, we performed two independent enrichment procedures and sequence runs. Total numbers of sequence reads, mapped reads and unique reads for each sample are represented in Additional file 2. For th identification of differentially methylated regions (DMRs) we employed a rigorous normalization procedure and a variety of bioinformatics tools (Figure 1C). We used MethylCap-seq data obtained from artificially prepared fully unmethylated and fully methylated DNA samples for normalization of the data and assignment of DMRs. The bioinformatics approach is described in detail in Additional file 3 and Additional file 4. Relative methylation scores were used to build an individual methylation profile for each sample. Subsequently, we compared the profiles to identify highly significant DMRs between tumors and paired lung tissues, and between the subtypes of tumors. Finally, we validated the MethylCap-seq results by bisulfite sequencing and methyl-specific PCR of selected DMRs.
Figure 1.
Experimental design for profiling of DNA methylation patterns in non-small cell lung carcinoma. (A) Overall view of the steps followed to generate the profiles (ADC: adenocarcinoma; LCC: large cell carcinoma; MBD: methyl-binding domain protein; N: Lung; SCC: squamous cell carcinoma; T: tumor), (B) MethylCap using methyl-binding domain proteins: Sheared genomic DNA is used as input fraction (methyl groups in red) and incubated with beads [26], coated with streptavidin (green)-biotin (yellow)) and linked to methyl-binding domain protein (blue) to capture methylated DNA. Captured fragments are subjected to high-throughput sequencing. (C) Summary of the bioinformatics approach used to generate the methylation profiles.
Table 1.
Data from patients used for MethylCap-seq and bisulfite sequencing validation
| Sample ID | Tissue type | Patient ID | Stage | Overall survival (month) | Survival status | Gender | Age at diagnosis | Ethnicity |
|---|---|---|---|---|---|---|---|---|
| 2213 N |
healthy |
SCC1 |
IIB |
2,43 |
Alive |
M |
54,54 |
Caucasian |
| 2214 T |
SCC |
|||||||
| 2235 N |
healthy |
SCC2 |
IIB |
51,23 |
Deceased |
M |
73,73 |
Caucasian |
| 2236 T |
SCC |
|||||||
| 2245 N |
healthy |
ADC1 |
IB |
12,57 |
Alive |
F |
66,26 |
Caucasian |
| 2246 T |
ADC |
|||||||
| 2255 N |
healthy |
ADC2 |
IA |
98,93 |
Deceased |
F |
54,26 |
Caucasian |
| 2256 T |
ADC |
|||||||
| 2257 N |
healthy |
ADC3 |
N/A |
N/A |
N/A |
M |
78,96 |
Caucasian |
| 2258 T |
ADC |
|||||||
| 2261 N |
healthy |
SCC3 |
IIB |
6,77 |
Alive |
M |
70,03 |
Caucasian |
| 2262 T |
SCC |
|||||||
| 22he |
healthy |
LCC1 | IB | 35,73 | Deceased | F | 56,58 | Caucasian |
| 22tu | LCC |
ADC: adenocarcinoma; LCC: large cell carcinoma; N/A: not applicable; SCC: Squamous cell carcinoma.
Global analysis of genome-wide methylation patterns of NSCLC
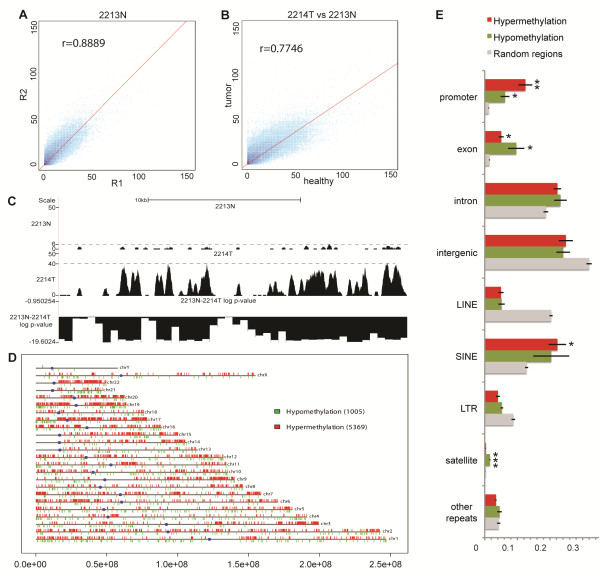
To assess the reproducibility of the MethylCap-seq procedure, we first performed a self/self comparison of the replicate experiments. This yielded an average Pearson’s correlation coefficient of 0.89 (Figure 2A and Additional file 5), indicating excellent reproducibility between independent experiments. Next, we compared methylation signal in tumors with matched healthy lung tissues; the average correlation coefficient between the two was 0.83, indicating a generally high similarity in the methylation patterns of the matched tumor and lung samples (Figure 2B and Additional file 6). Collectively, these analyses establish the high reproducibility and specificity of the MethylCap-seq procedure that we used. To allow visual inspection of the methylation signal and DMRs, raw reads and log P-values of differential methylation were uploaded in the University of California, Santa Cruz (UCSC) Genome browser, where DMRs can be visually inspected (Figure 2C). To focus on the most significant DMRs, we used a very stringent P-value threshold of 10-18, corresponding to a false discovery rate ≤ 10-15 ( Additional file 7). Non-repetitive regions with P-value ≤10-18 were projected onto the genome and visualized as genome-wide maps of differential methylation (Figure 2D and Additional file 8). This revealed that in the non-repetitive fraction of the genome, tumors displayed higher overall hypermethylation than the matching lung tissues. This is consistent with the notion that de novo methylation and hypermethylation of promoter CpG islands are associated with carcinogenesis [27].
Figure 2.
Global analysis of DNA methylation patterns in non-small cell carcinoma. (A) Correlation between experimental replicates. Each point represents the raw methylation signal (mean coverage in 10 bp bins). Density of points (log10 scale) is shown in different shades of blue. Pearson’s correlation coefficient is denoted in the top left corner of each scatter plot. (B) Correlation between tumor (x axis) and paired lung tissue sample (y axis). (C) Raw reads from a lung sample (above panel) and its matching tumor sample (middle panel) in the position chr2:176,716,200 to 176,738,910 viewed in the UCSC genome browser; normalized P-value is depicted in the bottom panel. Y-axis depicts the number of sequence reads in each region per sample. Dashed line represents the maximum number of reads (highest peak) in each sample; (D) Representation of the distribution of hypomethylated (green) and hypermethylated (red) regions across chromosomes in a tumor versus paired lung sample. (E) Composition of hypo- and hypermethylated regions in comparison to randomly sampled regions across the genome. Bars indicate the mean proportions of regions covered by each distinct genomic feature. Error bars denote standard error of the mean. Promoters were defined as regions ± 1 kb from all Ensembl transcription start site. Distinct classes of repeats were retrieved from the UCSC Table Browser (RepeatMasker table for hg18). Repeats were excluded from all subsequent features. Random regions were sampled across the genome independently seven times and each time their number was matched to the number of differentially methylated region in one sample pair. Statistical testing was done against random regions using one-tailed Student’s t-test (*P-≤0.01; **P-≤0.001; ***p≤0.0001).
The DMRs were analyzed according to general hallmarks of their genomic localization (Figure 2E). Compared with the general distribution of randomly sampled regions across the genome (label “Random regions”), we found that promoter areas were particularly enriched among the DMRs, both hypo- (P=5.7 × 10-3) and hypermethylated (p=5.3 × 10-4). Although we removed most signals from repetitive regions by keeping only uniquely mapped reads, the signals from their uniquely mappable reads and flanking regions were still above background and allowed us to test for differential methylation. This revealed that some specific repeat classes such as LINE and satellites were also differentially represented in the DMRs, with satellite repeats being particularly hypomethylated in tumors when compared with lung tissues (P=1.3 × 10-5). By contrast, LINE repeats are relatively underrepresented in the DMRs (hypo and hypermethylation), indicating that the methylation status of the bulk of the LINE elements was similar between the tumors and paired lung counterparts.
Differentially methylated regions in NSCLC
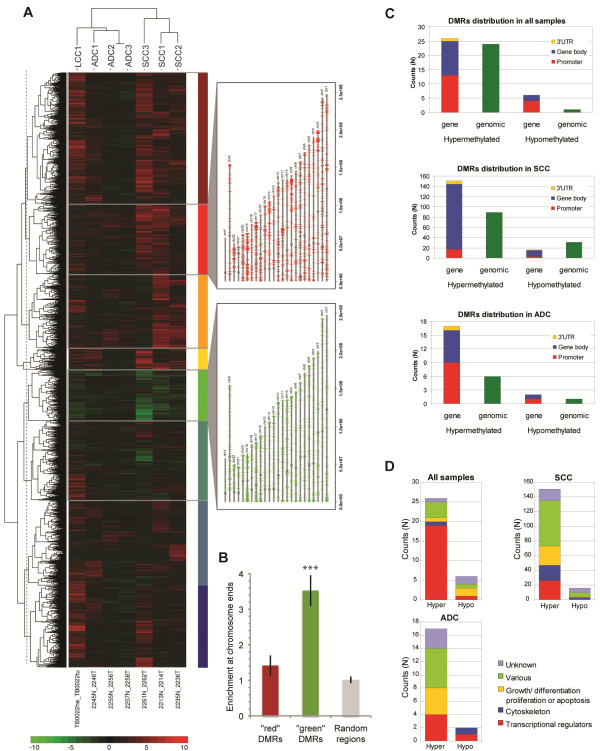
We observed a total of 14,742 DMRs in the seven NSCLC tumor samples ( Additional file 9), that is regions found to be differentially methylated in at least one tumor when compared with its lung counterpart with a two-fold change difference in methylation signal. Performing unsupervised cluster analysis with these DMRs revealed that the samples clustered according to the histological subtype of the tumors (Figure 3). In particular, hypermethylated DMRs were overrepresented in SCC samples. The dendogram of the DMRs indicated the presence of nine major subgroups of DMRs. Two of these were composed of generally hypermethylated (red) or hypomethylated (green) DMRs. These were plotted on the chromosomes, revealing that the ‘red’ DMRs were scattered throughout the chromosomes. By contrast, the ‘green’ DMRs displayed a more localized distribution towards the chromosome ends (Figure 3B), showing on average 3.5-times greater enrichment than random regions (P=6.8 × 10-6). Subtelomeric hypomethylation has been found to be associated with risk of developing cancer [28].
Figure 3.
Differentially methylated region in non-small cell carcinoma versus paired lung tissue samples. (A) Heatmap of 14,742 most significant DMRs, picturing regions with mostly hypermethylated (top), mostly hypomethylated (middle), and mixes (bottom). Color bar at the bottom represents the log ratio of the normalized signal in tumor versus. lung (red= hypermethylation, green= hypomethylation). Dashed line represents cut in the dendogram, generating nine groups of DMRs. The DMRs from the two most distinctive groups are depicted on the chromosomes. Red: cluster containing regions hypermethylated in all samples; green: cluster containing regions hypomethylated in all samples. (B) Bar plot showing mean enrichment of DMRs at chromosome ends calculated as ratio between proportion of 1 Mbp region at chromosome ends and total proportion of each chromosome covered by DMRs. Error bars show standard error of mean of all chromosomes. Statistical testing was done against random regions using one-tailed Student’s t-test (***P≤0.0001). (C) DMRs distribution relative to gene position. Number of hyper- and hypomethylated genes and regions (outside gene area), and their distribution as in promoter, gene body and 3′ UTR (top panel: all seven samples; middle panel: squamous cell carcinomas, bottom panel: adenocarcinoma). (D): Gene function distribution of the differentially methylated genes showing number of hyper- and hypomethylated genes relative to gene classes or function. DMR: differentially methylated regions; UTR: untranslated region.
We found 57 DMRs present in all tumor samples, of which 50 were hypermethylated and 7 were hypomethylated. An example of two DMRs in all samples, including the replicate experiment can be found in Additional file 10. We also analyzed DMRs in relation to NSCLC subtypes. We found 287 DMRs unique for SCC and 26 DMRs unique for ADC (Additioonal File 11). These DMRs were classified by hyper- and hypomethylation and their genomic locations relative to genes (Figure 3B). We found that the ADC-specific DMRs had a genomic distribution similar to that observed for the DMRs present in all seven tumors, while for SCCs, DMRs within genes were concentrated in the gene body. SCC also presented considerably more subgroup-specific DMRs than ADC and ‘All-sample’; this could be explained by the notion that SCC is a relatively homogeneous tumor type.
For the DMRs that fall in gene areas, we analyzed which types of genes were present in each of the three groups (Figure 3C and Additional file 11). We found that for the DMRs present in all tumor samples, more than 75% of the associated genes belonged to the class of transcriptional regulators. For SCC-specific DMRs, we observed a considerable heterogeneity in the functional categories of associated genes. Nevertheless, there was an overrepresentation of genes involved in transcriptional regulation, organization of the cytoskeleton and cell cycle regulation. We note that while some of the DMRs were associated with genes that had been previously reported to be differentially methylated in lung tumors, such as APCCDH13CDKN2ADAPKhMLH1HOX genes, OTX1HOX2 and many others [22,29-32], we also found DMRs which, to the best of our knowledge, have not been previously reported to be associated with methylation in NSCLC or other types of cancer.
Validation of differentially methylated regions in NSCLC by bisulfite sequencing
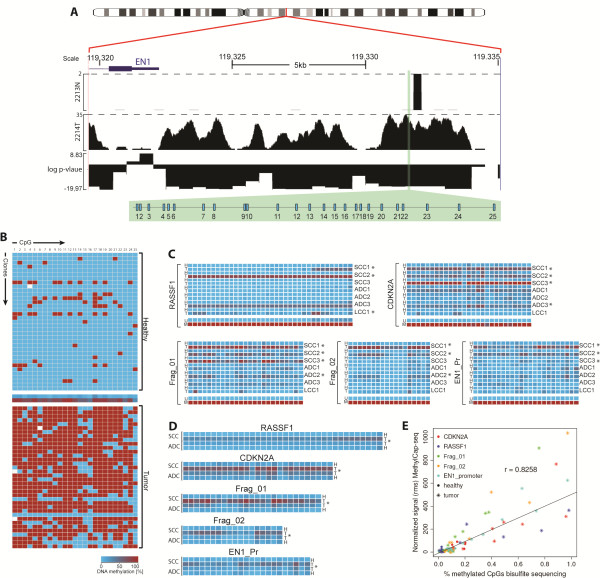
To validate the results obtained with MethylCap-seq, we selected five fragments for analysis by bisulfite sequencing. We chose CDKN2A and RASSF1 since hypermethylation of these genes has been widely reported in a variety of cancers including NSCLC [15,21,22], and the EN1 promoter region since hypermethylation of EN1 had been previously reported in a lung tumor cell line [29] but not yet in primary NSCLC tumors. Since we observed that the region upstream of EN1 was highly hypermethylated in the tumors and involved a region of more than 15 kbp (Figure 4A), we selected two extra fragments within this region, in addition to the EN1 promoter. The other two fragments were located at positions chr2:119,331,343 to 119,331,692 and chr2:119,328,097 to 119,328,400 and we named them Frag_01 and Frag_02, respectively. Selection of the fragments for bisulfite sequencing analysis was based on the MethylCap-seq data (Figure 4).
Figure 4.
Bisulfite sequencing validation for MethylCap-Seq. (A) Sequence reads for the Frag_01 (genomic region located at position Ch2: 119,331,343 to 119,331,692) in a tumor (2214 T) and matching lung tissue (2213 N), plotted in the Genome Browser, showing the distribution of the 25 CpGs contained in the fragment highlighted in green. Dashed lines represent the highest number of reads in 2213 N and 2214 T. (B) Methylation status of each CpG in all 36 individually sequenced clones in the same samples and fragment shown in Figure 4A. The middle row represents the average of methylation in all clones per CpG position. (C) Average of methylation of all clones sequenced per patient in each fragment (M: control totally methylated DNA; N:lung; T:tumor; U:control totally unmethylated DNA). (D) Average of the methylation status of the sum of all clones, grouped per histological subtype; comparison betweenadenocarcinoma and squamous cell carcinoma in all fragments were statistically significant. (E) Correlation between normalized methylation signals from MethylCap-seq and CpG methylation from bisulfite sequence. Different regions are shown in different colors; lung samples are marked by dots and tumors by stars. Pearson’s correlation coefficient is denoted above the linear regression curve. *P≤0.001.
To assure sufficient depth of coverage for quantitative analysis, we sequenced 36 individual clones from each fragment and each bisulfite-converted DNA sample. Figure 4B shows an example of the methylation status for one fragment of a paired sample. The methylation status of the five fragments in all seven paired NSCLC/lung tissue samples is summarized in Figure 4C; full data for all samples is available in Additional files 12 and 13. We also examined potential differences in methylation according to tumor histology. To perform this analysis we grouped the samples in four categories: all clones from the lung tissues of patients (ADC_N and SCC_N) and clones from tumors (ADC_T and SCC_T), (Figure 4D and Additional file 14). We observed a statistically significant difference (P≤0.0001), not only between tumors and lung tissues, but also between ADC and SCC for all fragments analyzed by bisulfite sequencing. This identifies these fragments as candidate markers for differentiating tumors versus paired lung tissues, and also between tumor subtypes. Moreover this finding validates the MethylCap-seq data, since the methylation status determined by bisulfite sequencing correlated quantitatively with the MethylCap-seq data (Figure 4E).
Screening by methylation-specific PCR verifies methylation differences between tumor and paired lung tissues, and between tumor subtypes
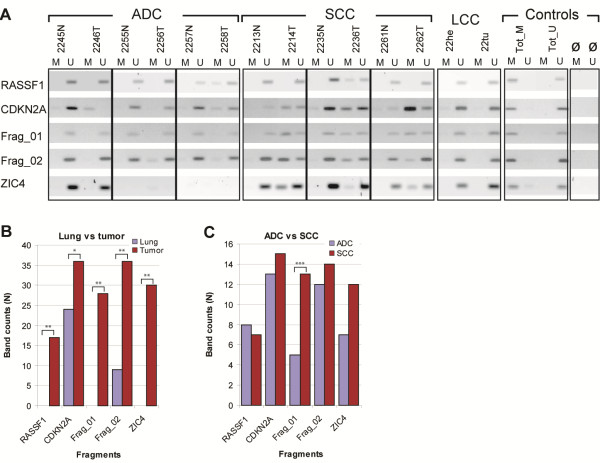
To screen the methylation status of selected DMRs in a larger sample set we used methylation-specific PCR (MSP) [33]. Here, we selected RASSF1, CDKN2A, Frag_01 and Frag_02 because they were the most significant differentially methylated fragments in the bisulphite sequencing experiments. We added ZIC4 because it was one of the DMRs not yet reported as methylated in NSCLC and presented a high difference in relative methylation score between lung and tumor samples. We screened 96 samples (48 NSCLC and paired lung samples), and used totally methylated and totally unmethylated DNA samples as controls for the specificity of the MSP assays. Results for the seven paired samples used in MethylCap-seq are displayed in Figure 5. Data for all 96 samples are summarized in Table 2 and shown in Additional file 15.
Figure 5.
Methylation-specific polymerase chain reaction screening in 48 tumors and matching lung tissues. (A) Cropped gel images for the seven patients used for MethylCap-seq, grouped by histological subtype and controls (M: “methylated primer set”; N: lung; T: tumor; Tot_M: totally methylated control samples; Tot_U: totally unmethylated control sample; U: unmethylated primer set; ø: blank/water). (B) Histogram of the methylation status in lung samples and tumor samples in all 48 patients in the five fragments analyzed. (C) Histogram of the methylation status per histological subtype (statistics were calculated with Fisher’s exact test, two-tailed p-values: *P≤0.02; **P≤0.001 and ***P≤0.01).
Table 2.
Frequency of methylation by methylation-specific polymerase chain reaction in the 96 samples (48 tumor and matching lung tissue) in five fragments
|
CDKN2A |
Frag_01 |
Frag_02 |
RASSF1 |
ZIC4 |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count (n) | Sensitivity (%) | Specificity (%) | Count (n) | Sensitivity (%) | Specificity (%) | Count (n) | Sensitivity (%) | Specificity (%) | Count (n) | Sensitivity (%) | Specificity (%) | Count (n) | Sensitivity (%) | Specificity (%) | |
| Lung |
24/48 |
75 |
27 |
0/47 |
62 |
100 |
9/48 |
75 |
56 |
0/41 |
42 |
100 |
0/39 |
77 |
100 |
| Tumor |
36/48* |
29/47** |
36/48** |
17/41** |
30/39** |
||||||||||
| Adenocarcinoma |
13/17 |
76.5 |
46.4 |
5/16 |
31.3 |
27.7 |
12/17 |
70.6 |
46.1 |
8/15 |
53.3 |
53.3 |
7/13 |
53.9 |
36.8 |
| Squamous cell carcinoma | 15/16 | 93.8 | 53.6 | 13/16 | 81.3** | 72.2 | 14/16 | 87.5 | 53.9 | 7/16 | 43.8 | 46.7 | 12/16 | 75.0 | 63.2 |
Sensitivity is calculated using the ratio between methylated (amplified) sample and the total amount of samples analyzed per fragment; specificity is the ratio of the difference between amplification in tumor and in the matching lung sample (or adenocarcinoma versus squamous cell carcinoma). Statistical significance of the difference between control lung samples and tumors, and between adenocarcinomas and squamous cell carcionmas were calculated using chi-square. *P<0.02; **P0.0001.
MSP is a more qualitative approach than bisulfite sequencing. In MSP, selective amplification depends on the methylation status of only the CpGs present in the designed primers, which were two or three CpGs depending on the fragment. For each methylated (M) primer set, amplification of the fragment is dependent on methylation of the CpGs that are present in the sequence targeted by the “M” primer; while each unmethylated (U) primer set requires unmethylated CpGs in the referred sequence for amplification. In the 48 paired samples analyzed, CDKN2A showed frequent amplification with the M primer set in lung samples (24 out of 48), although the amplified bands were often of low intensity. Tumors had a much larger number displaying amplification with both M and U primer sets. Of the 47 tumors, 36 displayed amplification with the M primer set, and two of the tumors displayed amplification only with the M primer set. For RASSF1 and ZIC4, 41 paired samples were available. Both fragments differentiated between lung and tumor samples, whereas no methylation was detected in lung samples. ZIC4 showed methylation in more than 70% of the tumors, which is much higher than the 41% found for RASSF1. The findings for RASSF1 are in accordance with other studies [18]. Also, Frag_01 presented no amplification with the M primer set in the lung samples, in the Frag_02 M primer set only 5 out of 48 were amplified. In tumors, we observed fragment amplification in 29out of 48 samples for Frag_01 and 36out of 48 samples for Frag_02 with the M primer sets. Differences in methylation status were significant in tumors versus lung samples for all 5 fragments, with a p-value < 0.02 for CDKN2A and P < 0.0001 for the other four fragments (Figure 5B). Frag_01, Frag_02 and ZIC4 present a much lower P-value, indicating they may be better tumor markers than CDKN2A and RASSF1. We also observed that significant differences between SCC and ADC were only found for Frag_01 (Figure 5C). In conclusion, the MSP results validate the MethylCap-seq data, since the results obtained for all five fragments were consistent with the significant DMRs in all tumors, and Frag_01 appeared as part of the SCC-specific DMRs in the MethylCap-seq data.
Discussion
To date, genome-wide methylation studies in NSCLC have concentrated on specific areas of the genome [30], promoter regions [34-36] or sets of pre-selected candidate genes [19,37]. Recently Kwon et al. [38] published a report on genome-wide analysis of DNA methylation in NSCLC. However, they also concentrate the analysis on a selection of candidate genes. In our study, we performed an unbiased genome-wide DNA methylation analysis of seven individual patients with NSCLC and their adjacent lung tissues. We combined MethylCap with high-throughput sequencing (MethylCap-seq) to draw detailed methylome maps of NSCLC tumors and paired lung tissues. The replicate experiments showed that the depth of coverage generated by MethylCap-seq was sufficient to capture the methylation status of the entire genome. Moreover, there was a very high degree of correlation between the replicate experiments, demonstrating the robustness of the MethylCap-seq approach. As controls for normalization and validation purposes we made use of fully methylated and fully unmethylated DNA samples. The MethylCap-seq data of these controls provide a useful resource for benchmarking of the fully methylated versus fully unmethylated status of any particular fragment in the genome. Using the MEDIPS computational tool [39], we generated methylation profiles of the seven paired NSCLC/lung cases which can be viewed in the genome browsers. This broad view of DNA methylation in NSCLC will provide new opportunities for the identification of specific epigenetic markers of NSCLC that could, for instance, be used for early detection of the disease.
The occurrence of methylated CpGs in the fragments selected for bisulfite sequencing showed a high quantitative correlation with the reads found by MethylCap-seq in the same area. In conjunction with the MethylCap-seq data on the fully methylated and fully unmethylated controls, this demonstrates that MethylCap-seq yields reliable quantitative information on the methylation status of the fragments in a particular DNA sample. This is important considering that the tumor cell content of samples to be analyzed will be variable. Provided that sufficient sequencing depth is achieved, it should be possible to use MethylCap-seq for the identification of tumor-specific hypermethylated DNA regions in samples containing only a minority of tumor cells, and possibly in circulating tumor DNA isolated from serum samples. The results obtained with MSP are qualitative in nature, but this PCR-based approach is very sensitive and may therefore be used to detect tumor-specific hypermethylated DNA fragments in serum DNA. As a first step in this direction, we used MSP to investigate the presence of five DMRs in a larger set of paired samples. The results confirmed the MethylCap-seq data and demonstrated that these DMRs are frequently hypermethylated in NSCLC. In particular, the Frag_01 fragment was found to be hypermethylated in 13 out of 16 SCC cases and in none of the ADC, suggesting that this might be a useful marker for this type of NSCLC, and ZIC4 showed 100% specificity for tumors and higher sensitivity (>70%) than RASSF1. In addition, CDKN2A amplification with the M primer was often observed in lung tissue samples, although at much lower amplification levels when compared with tumor samples. This could reflect an early event in the methylation status of CDKN2A in tumorigenesis.
The mapped reads point to differentially methylated areas across the genome, with some marked preferences for hypermethylation or hypomethylation in certain genomic regions. There is a strong correlation between CpG islands and hypermethylation in tumors, with the most significantly hypermethylated regions associated with promoter regions of genes. As previously reported by many groups [40-43], gene promoters are a target of methylation as an epigenetic regulatory mechanism. However, our data also reveal a high degree of hypermethylation outside promoter areas at intergenic regions, and at long distances from genes. This indicates that patterns of DNA methylation may play a role not only by silencing promoters of key tumor suppressor genes, but also by regulating gene expression in a more complex manner through distant regulatory elements such as insulators and enhancers.
We found that satellite regions were particularly hypomethylated in the NSCLC tumors when compared with lung tissues. Recently, Ting et al. [44] have shown that overexpression of satellite repeats is associated with different types of cancer. The observed hypomethylation of these regions in NSCLC would be compatible with their results. We also found that hypomethylated regions are often present in subtelomeric regions, in agreement with the observation that hypomethylation is more frequently located near chromosome ends in lung cancer [45]. While CpG islands and genes are present in the subtelomeric regions of chromosomes, the impact of subtelomeric hypomethylation on the regulation of these genes is currently unclear.
In an alternative approach, we selected the most significant DMRs in the individual NSCLC cases (14,742) and performed unsupervised cluster analysis of the hyper- and hypomethylated regions in the tumor samples. We observed that the samples clustered according to their histological classification. This indicates that there are DMRs that could not only be used as NSCLC markers but also as markers for histological classification of NSCLC tumors. Subsequently we searched for the specific areas that were differentially methylated in all tumors when compared with the paired lung tissues, and specific for the subtypes. We observed that SCCs displayed more DMRs than did ADCs did. This observation could be explained by the heterogeneity of ADCs, which would result in fewer DMRs shared by all three ADC samples. The contrary is seen in SCC, which is a more homogeneous type of NSCLC. This characteristic has been observed previously in gene expression profiling studies [4,46].
Remarkably, the majority (>75%) of the hypermethylated genes present in all seven samples encoded for transcription factors, while the shared hypomethylated genes were spread among many different functional categories. Transcription factors play a central role in maintaining or modifying cell fate, either in normal processes such as development and cell differentiation, or in cancer. Mechanisms of epigenetic regulation via DNA methylation are not yet completely understood; several factors such as DNA methyltransferases, chromatin remodelling proteins and DNA-binding transcription factors are involved [47,48]. Therefore, any perturbation that leads to a decrease in the expression status of those factors may disrupt important pathways for maintenance of the differentiated cellular state. Aberrant methylation can change chromatin structure, rendering DNA binding sites accessible or inaccessible to transcription factors leading to activation or silencing of genes important in cell differentiation and maintenance. It has been recently shown that hypermethylated genes found in lung tumors are associated with cellular morphogenetic differentiation [49]. These cellular mechanisms are orchestrated by transcriptional regulators; therefore changes in the methylation status of their binding sites may disrupt those processes thus contributing to oncogenesis.
Conclusion
Based on the MethylCap-seq data, we generated a list of candidate DMRs and associated genes specific for NSCLC and its subtypes (Table 3). Several of these DMR-associated genes are known to be hypermethylated in NSCLC and other types of cancer, strongly supporting the validity of the data set reported here. For instance, the observed methylation rates of RASSF1 were in agreement with previously reported results [18]. In the MetylCap-seq data, hypermethylation of the genomic region where Frag_01 is located was most significant for the SCC samples. This was supported by the bisulfite sequencing and MSP validation experiments, where this fragment showed significantly more hypermethylation in SCCs when compared with ADCs or LCCs. In conclusion, this list of candidate DMR markers can be used to develop sensitive biological markers for NSCLC, which may enable non-invasive diagnosis and early detection of the disease, and potentially allow histological classification. Collectively, we provide a resource containing genome-wide DNA methylation maps of NSCLC and paired lung tissues, and comprehensive lists of DMRs and associated genes in NSCLC.
Table 3.
List of most differentially methylated genes in non-small cell carcinoma
| Gene | Log2ratio | Gene class/type/ biological function | Gene name | HGNC ID | |
|---|---|---|---|---|---|
| Common in all tumors |
BARX1 |
4.8 |
transcription regulator |
BARX homeobox 1 |
HGNC:955 |
|
PAX9 |
4.3 |
transcription regulator |
paired box 9 |
HGNC:8623 |
|
|
OTX1 |
4.0 |
transcription regulator |
orthodenticle homeobox 1 |
HGNC:8521 |
|
|
NPR3 |
3.8 |
G-protein coupled receptor |
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
HGNC:7945 |
|
|
FGF12 |
3.7 |
growth factor |
fibroblast growth factor 12 |
HGNC:3668 |
|
|
ONECUT2 |
3.6 |
transcription regulator |
one cut homeobox 2 |
HGNC:8139 |
|
|
PRDM14 |
3.5 |
transcription regulator |
PR domain containing 14 |
HGNC:14001 |
|
|
RAX |
3.5 |
transcription regulator |
retina and anterior neural fold homeobox |
HGNC:18662 |
|
|
SHOX2 |
3.3 |
transcription regulator |
short stature homeobox 2 |
HGNC:10854 |
|
|
DMRTA2 |
3.1 |
transcription regulator |
DMRT-like family A2 |
HGNC:13908 |
|
|
FER1L4 |
3.1 |
unknown |
fer-1-like 4 (Caenorhabditis. elegans) pseudogene |
HGNC:15801 |
|
|
SIX6 |
3.1 |
transcription regulator |
SIX homeobox 6 |
HGNC:10892 |
|
|
GATA3 |
3 |
transcription regulator |
GATA binding protein 3 |
HGNC:4172 |
|
|
SKOR1 |
3 |
transcription regulator |
SKI familytranscriptional corepressor 1 |
HGNC:21326 |
|
|
HOXA9 |
3 |
transcription regulator |
homeobox A9 |
HGNC:5109 |
|
|
SALL1 |
2.9 |
transcription regulator |
sal-like 1 (Drosophila) |
HGNC:10524 |
|
|
IRX2 |
2.7 |
transcription regulator |
iroquois homeobox 2 |
HGNC:14359 |
|
|
GRIK2 |
2.7 |
ion channel |
glutamate receptor, ionotropic, kainate 2 |
HGNC:4580 |
|
|
SATB2 |
2.6 |
transcription regulator |
SATB homeobox 2 |
HGNC:21637 |
|
|
MEIS1 |
2.5 |
transcription regulator |
Meis homeobox 1 |
HGNC:7000 |
|
| VAX1 |
2.4 |
transcription regulator |
ventral anterior homeobox 1 |
HGNC:12660 |
|
| TBX15 |
2.3 |
transcription regulator |
T-box 15 |
HGNC:11594 |
|
|
CNTD2 |
−2.4 |
unknown |
cyclin N-terminal domain containing 2 |
HGNC:25805 |
|
|
ZMYND10 |
−2.6 |
unknown |
zinc finger, MYND-type containing 10 |
HGNC:19412 |
|
|
MYC |
−2.9 |
transcription regulator |
v-myc myelocytomatosis viral oncogene homolog (avian) |
HGNC:7553 |
|
|
TSPAN9 |
−2.9 |
Plasma Membrane |
tetraspanin 9 |
HGNC:21640 |
|
|
NAV1 |
−3.1 |
unknown |
neuron navigator 1 |
HGNC:15989 |
|
|
CPEB3 |
−3.4 |
unknown |
cytoplasmic polyadenylation element binding protein 3 |
HGNC:21746 |
|
| Unique for squamous cell carcinomas |
TRAPPC9 |
6.5 |
cell differentiation |
trafficking protein particle complex 9 |
HGNC:30832 |
|
ABHD2 |
6.3 |
hydrolase |
abhydrolase domain containing 2 |
HGNC:18717 |
|
|
CTNND1 |
6.2 |
transcrption regulator |
catenin (cadherin-associated protein), delta 1 |
HGNC:2515 |
|
|
HIST1H2BB |
6.2 |
histone protein |
histone cluster 1, H2bb |
HGNC:4751 |
|
|
EMP1 |
5.8 |
protein binding |
epithelial membrane protein 1 |
HGNC:3333 |
|
|
TBL1XR1 |
5.6 |
transcription regulator |
transducin (beta)-like 1 X-linked receptor 1 |
HGNC:29529 |
|
|
NXPH1 |
5.6 |
protein binding |
neurexophilin 1 |
HGNC:20693 |
|
|
ZIC4 |
5.6 |
transcription regulator |
Zic family member 4 |
HGNC:20393 |
|
|
AOAH |
5.4 |
hydrolase |
acyloxyacyl hydrolase (neutrophil) |
HGNC:548 |
|
|
ACTN4 |
5.4 |
transporter |
actinin, alpha 4 |
HGNC:166 |
|
|
C1orf21 |
5.2 |
unknown |
chromosome 1 open reading frame 21 |
HGNC:15494 |
|
|
PACSIN2 |
5.2 |
transporter |
protein kinase C and casein kinase substrate in neurons 2 |
HGNC:8571 |
|
|
PMM2 |
5.2 |
isomerase |
phosphomannomutase 2 |
HGNC:9115 |
|
|
DOT1L |
5.1 |
histone methyltransferase |
DOT1-like, histone H3 methyltransferase (Sacchormyces. cerevisiae) |
HGNC:24948 |
|
|
WWP2 |
5 |
e3 ubiquitin-protein ligase |
WW domain containing E3 ubiquitin protein ligase 2 |
HGNC:16804 |
|
|
GTF3C1 |
5 |
transcription regulator |
generaltranscription factor IIIC, polypeptide 1, alpha 220 kDa |
HGNC:4664 |
|
|
MDN1 |
5 |
nuclear chaperone |
MDN1, midasin homolog (yeast) |
HGNC:18302 |
|
|
DIDO1 |
5 |
transcrption regulator |
death inducer-obliterator 1 |
HGNC:2680 |
|
|
HIST1H3C |
5 |
histone protein |
histone cluster 1, H3c |
HGNC:4768 |
|
|
ANKRD13B |
4.9 |
unknown |
ankyrin repeat domain 13B |
HGNC:26363 |
|
|
CALCB |
4.9 |
hormone |
calcitonin-related polypeptide beta |
HGNC:1438 |
|
|
PTPRA |
4.9 |
phosphatase |
protein tyrosine phosphatase, receptor type, A |
HGNC:9664 |
|
|
STAT5A |
4.8 |
transcription regulator |
signal transducer and activator of transcription 5A |
HGNC:11366 |
|
|
LIMK1 |
4.8 |
cytoskeleton |
LIM domain kinase 1 |
HGNC:6613 |
|
|
SLC23A2 |
4.7 |
transporter |
solute carrier family 23 (nucleobase transporters), member 2 |
HGNC:10973 |
|
|
BARX1 |
4.5 |
transcription regulator |
BARX homeobox 1 |
HGNC:955 |
|
|
NHS |
4.5 |
unknown |
Nance-Horan syndrome (congenital cataracts and dental anomalies) |
HGNC:7820 |
|
|
MTAP |
4.5 |
glycosyltransferase |
methylthioadenosine phosphorylase |
HGNC:7413 |
|
|
FOXK1 |
4.5 |
transcription regulator |
forkhead box K1 |
HGNC:23480 |
|
|
PCMT1 |
4.4 |
O-methyltransferase activity |
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
HGNC:8728 |
|
|
SETD1A |
4.3 |
transcription regulator |
SET domain containing 1A |
HGNC:29010 |
|
|
CENPP |
4.2 |
centromere protein |
centromere protein P |
HGNC:32933 |
|
|
KIAA1217 |
4.1 |
unknown |
KIAA1217 |
HGNC:25428 |
|
|
SLITRK1 |
4.1 |
enhances neuronal dendrite outgrowth |
SLIT and NTRK-like family, member 1 |
HGNC:20297 |
|
|
RORB |
4 |
transcription regulator |
RAR-related orphan receptor B |
HGNC:10259 |
|
|
DLGAP1 |
4 |
postsynaptic scaffold in neuronal cells |
discs, large (Drosophila) homolog-associated protein 1 |
HGNC:2905 |
|
|
C3orf21 |
4 |
unknown |
chromosome 3 open reading frame 21 |
HGNC:26639 |
|
|
ST6GALNAC1 |
4 |
protein glycosylation |
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 |
HGNC:23614 |
|
|
ZIC3 |
3.9 |
transcription regulator |
Zic family member 3 (odd-paired homolog, Drosophila) |
HGNC:12874 |
|
|
LYPLA1 |
3.8 |
hydrolase |
lysophospholipase I |
HGNC:6737 |
|
|
REST |
3.8 |
transcription regulator |
RE1-silencingtranscription factor |
HGNC:9966 |
|
|
TMEM132D |
-2 |
unknown |
transmembrane protein 132D |
HGNC:29411 |
|
|
MAP1LC3B2 |
-2.1 |
microtubule |
microtubule-associated protein 1 light chain 3 beta 2 |
HGNC:34390 |
|
|
CBY3 |
-2.2 |
unknown |
chibby homolog 3 (Drosophila) |
HGNC:33278 |
|
|
RASIP1 |
-2.3 |
signal transduction |
Ras interacting protein 1 |
HGNC:24716 |
|
|
PRKG1 |
-2.3 |
kinase |
protein kinase, cGMP-dependent, type I |
HGNC:9414 |
|
|
WDR72 |
-2.4 |
unknown |
WD repeat domain 72 |
HGNC:26790 |
|
|
KCNQ2 |
-2.7 |
ion channel |
potassium voltage-gated channel, KQT-like subfamily, member 2 |
HGNC:6296 |
|
|
BAIAP3 |
-2.8 |
G-protein coupled receptor |
BAI1-associated protein 3 |
HGNC:948 |
|
|
MAP3K10 |
-30 |
kinase |
mitogen-activated protein kinase kinase kinase 10 |
HGNC:6849 |
|
| Unique for Adenocarcinomas |
MSC |
4.4 |
transcription regulator |
musculin |
HGNC:7321 |
|
FAM78B |
4.1 |
unknown |
family with sequence similarity 78, member B |
HGNC:13495 |
|
|
HOXA1 |
3.6 |
transcription regulator |
homeobox A1 |
HGNC:5099 |
|
|
SEPT9 |
3.3 |
enzyme |
septin 9 |
HGNC:7323 |
|
|
GAS1 |
3.3 |
Cell cycle/growth |
growth arrest-specific 1 |
HGNC:4165 |
|
|
PTPRN2 |
3.1 |
phosphatase |
protein tyrosine phosphatase, receptor type, N polypeptide 2 |
HGNC:9677 |
|
|
RSPO2 |
2.7 |
Wnt receptor signaling pathway |
R-spondin 2 homolog (Xenopus laevis) |
HGNC:28583 |
|
|
POU3F3 |
2.6 |
transcription regulator |
POU class 3 homeobox 3 |
HGNC:9216 |
|
|
TRPA1 |
2.5 |
transporter |
transient receptor potential cation channel, subfamily A, member 1 |
HGNC:497 |
|
|
SYT6 |
2.3 |
transporter |
synaptotagmin VI |
HGNC:18638 |
|
|
SLC6A2 |
2.3 |
transporter |
solute carrier family 6 (neurotransmitter transporter, noradrenalin), member 2 |
HGNC:11048 |
|
|
LHX1 |
2.1 |
transcription regulator |
LIM homeobox 1 |
HGNC:6593 |
|
|
RAPGEF5 |
2.1 |
small GTPase mediated signal transduction |
Rap guanine nucleotide exchange factor (GEF) 5 |
HGNC:16862 |
|
|
GDF10 |
2.1 |
growth factor |
growth differentiation factor 10 |
HGNC:4215 |
|
|
C3orf45 |
1.6 |
unknown |
chromosome 3 open reading frame 45 |
HGNC:26781 |
|
|
SLIT2 |
1.6 |
differentiation/apoptosis |
slit homolog 2 (Drosophila) |
HGNC:11086 |
|
|
ZNF423 |
−1.6 |
transcription regulator |
zinc finger protein 423 |
HGNC:16762 |
|
| RHOF | −1.8 | actin filament organization | ras homolog gene family, member F (in filopodia) | HGNC:15703 |
Genes were selected based on the average of log2 ratios from the samples in each group. Hypermethylated genes common in all tumors and unique for the ADC groups were selected based on log2 ratio > 2.5; hypomethylation included all genes from those groups. For SCC we selected the first 50 genes with the highest log2 ratios in hypermethylation, and log2 ratio < −2.0 for hypomethylated genes. Gene classes and their correspondent biological functions were retrieved from Gene Ontology (http://www.geneontology.org/) and Protein Knowledgebase UniProtKb (http://www.uniprot.org/).
Methods
Patient samples
Samples were obtained from the patients with NSCLC (n = 48) who had undergone surgical lung resection at Erasmus University Medical Center Rotterdam. Specimens were collected from the tumor and adjacent non-cancerous lung tissue and studied under an anonymous tissue protocol approved by the medical ethical committee of Erasmus University Medical Center Rotterdam. Tissues were snap-frozen within two hours after surgical resection in liquid nitrogen pre-cooled isopentane, and stored at 196 °C or 80 °C until DNA extraction.
Histopathological analysis
Patient samples were independently reviewed by two pathologists. The cohort included 17 with ADC; 16 with SCC, seven with LCC , eight unclassified samples, and paired lung tissues for each tumor sample.
Cell line
The MRC-5 lung fibroblast-like cell line was used as control. Cells were cultured under standard conditions using minimum essential medium supplemented with 10% heat-inactivated fetal bovine serum, 2 mM L-glutamine, 1% non-essential amino acids and penicillin/streptomycin. Cells were harvested when they reached 90% confluence.
DNA isolation
Genomic DNA from patient tissues and cultured cells were extracted by overnight treatment incubation with lysis buffer and proteinase K, followed by phenol-chloroform extraction, ethanol precipitation and RNase digestion.
Artificial demethylation and methylation of genomic DNA
We used DNA extracted from the MRC-5 cell line and commercially available Universal unmethylated DNA (UUD; Millipore(Billerica, MA, USA). Fully unmethylated DNA was obtained by whole-genome amplification using the REPLI-g kit (QiagenQiagen – Germantown, MD, USA) according to manufacturer protocol, followed by phenol-chloroform extraction. Fully methylated DNA was prepared by treating MRC-5 DNA and UUD with M.SssI enzyme (New England Biolabs - Ipswich, MA, USA) according to the manufacturer’s protocol. In short, 10 μg of DNA was incubated for 2 hours at 37°C with 40 U of M.SssI and 640 μM of S-adenosylmethionine. DNA was then treated with phenol-chloroform and recovered by ethanol precipitation.
Methyl-DNA capture
Enrichment of methylated DNA was carried out using the MethylCap technique. DNA samples were sheared by sonication to obtain fragments between 200 and 800 bp. Methylated DNA capture was carried out using an adapted protocol from the MethylMiner Methylated DNA Enrichment kit (Invitrogen – Carlsbad, CA, USA). Two adaptations were made. Firstly, 20 μL of beads and 14 μL (7 μg) of MBD-biotin protein were used for 5 μg of sheared genomic DNA. Secondly, a single elution fraction was obtained by resuspending the beads in 200 μL of 1X binding/washing buffer containing 2 μL of proteinase K (20 mg/ml). The samples were then incubated for 90 minutes at 57 °C with 800 rpm agitation. Remaining steps were performed following the manufacturer’s protocol scaled up to 5 μg of DNA. Before the samples were used for high-throughput sequencing, methyl-enrichment was tested by quantitative (q) PCR (data not shown). qPCR was carried out using SYBR r green, Phire Hot Start DNA polymerase (Finnzymes - Vantaa, Finland)), and 1 μL of the enriched samples. Primer sequences are available in Additional file 16. qPCR was performed in triplicate and enrichment levels were calculated as previously described [50].
High-throughput sequencing
High-throughput sequencing (HTS) was carried out on 18 samples: seven tumors (three ADC, three SCC and one LCC), seven paired lung tissues and four controls (two artificially fully methylated DNA and two fully unmethylated DNA). For each sample, DNA recovered from two independent Methyl Capture experiments was sequenced. The Illumina Genome Analyzer IIx platform (San Diego, CA, USA was used for both replicates, one sample per lane, according to the manufacturer’s protocol. In short: after MethylCap, fragments of the methyl-enriched fraction were end-repaired and ligated to single-read adaptors. Samples were size-selected to an average of 320 bp, PCR-amplified and 36 bp sequenced.
The Illumina Casava pipeline was used for base calling, alignment and quality control. Results were mapped against the reference Human_UCSChg18_AllChromosomes using eland_extended by Illumina pipeline 1.6.0.
Bioinformatic analysis
Detailed information about the evaluation and selection of normalization parameters, and identification, characterization and selection of DMRs can be found in Additional file 3.
Bisulfite sequencing
Bisulfite sequencing was carried out on the same 16 samples used for MethylCap HTS. Detailed information is provided in Additional file 3. In short: bisulfite conversion was carried out using the Epitect kit from Qiagen following the manufacturer’s protocol. Samples were amplified using primers specific for the fragments of interest ( Additional file 16). Amplified fragments were loaded on 2% agarose gels and extracted from the gel using a NucleoSpin Extract II kit (Machery-Nagel – Düren, Germany). Fragments were then ligated to pGEM-t easy vector (Promega - Madison, WI, USA) and cloned into DH12-β competent cells. Colony PCR was performed on 36 colonies and PCR fragments sequenced. Analysis of methylated and unmethylated CpGs was executed using two online programs: BISMA (http://biochem.jacobs-university.de/BDPC/BISMA) and QUMA (http://quma.cdb.riken.jp).
Methylation-specific PCR
To determining the methylation status of the selected regions, we used MSP [33] in 96 samples (48 tumors and matching lung tissues). Samples were bisulfite converted as described for bisulfite sequencing, and amplified using two different set of primers designed for the methylated and unmethylated sequences ( Additional file 16). The fully methylated and fully unmethylated DNA samples were used as controls, and a water blank reaction used as control for contamination. After amplification, products were resolved on 2% agarose gels containing ethidium bromide and visualized under UV transillumination.
Statistical analysis
Statistical analysis for bisulfite sequencing was provided by the BISMA program. In short, Fisher’s exact test was used for the statistical significance between two bisulfite sequence groups at each CpG, and a Mann-Whitney U-test for the statistical significance between two groups of the entire set of CpG sites. Two-tailed P-value of Fisher’s exact test was calculated from the 2 × 2 tables at each CpG site. A Mann-Whitney U-test was used for the statistical significance of the entire set of CpG sites between the tumor and lung groups. For determination of significance in the MSP, we used chi-square distribution. Two-tailed P-values were determined by the counts of the amplified bands for each primer in the group of samples lung/tumor or SCC/ADC. Groups that showed a P-value < 0.05 were considered significantly different.
Abbreviations
ADC, adenocarcinoma; Bp, base pair; DMR, differentially methylated region; HTS, high-throughput sequencing; LCC, large cell carcinoma; LINE, long interspersed element; MBD, methyl-binding domain; MSP, methylation-specific PCR; NSCLC, non-small cell lung carcinoma; PCR, polymerase chain reaction; SCC, squamous cell carcinoma; UUD, Universal unmethylated DNA; UTR, untranslated region.
Competing interests
The authors declare no that they have no competing interests.
Authors’ contributions
RHC, VH, BL and SP participated in the concept and writing of the manuscript. RHC, WvI and CK executed specific experiments. RHC, VH, JH, ST, TvG, MvV, RB, ER, BL and SP were involved in the analysis of the data. RHC, VH, JA, BL and SP substantially contributed for interpretation of data. RHC, AS, JF and SP participated in the design of the experiments. RHC, JA, FG, and SP participated in editing or revising the manuscript. All authors have read and approved the final manuscript.
Supplementary Material
TableS1. Data from all samples used in this study.
TableS2. High-throughput sequencing reads from all samples and replicate experiments.
FigureS1. Selection of normalization method. Distribution of Pearson’s correlation coefficients between replicates for different normalization methods.
FigureS2. Correlation between technical replicates. Scatter plots comparing raw MethylCap-seq signal from two technical replicates (R1 and R2) for all 14 samples. Each point represents one 10 bp genomic bin. Density of points (log10 scale) is shown in different shades of blue. Pearson’s correlation coefficient is denoted in the top left corner of each scatter plot.
FigureS3. Correlation between lung and tumor samples. Scatter plots comparing raw MethylCap-seq signal from healthy and tumor tissue for six pairs of samples. Each point represents one 10 bp genomic bin. Density of points (log10 scale) is shown in different shades of blue. Pearson’s correlation coefficient is denoted in the top left corner of each scatter plot.
FigureS4. Selection of P-value threshold. Number of significant hypo- and hypermethylated regions for different P-value thresholds. Y-axis shows number of significant regions on log10 scale for decreasing P-value threshold, shown on the x-axis.
FigureS5. along each chromosome for each paired sample. Representation of the log-ratio of relative methylation scores for tumors vs. paired lung samples for each sample pair showing DMRs along each chromosome. Green colors marks hypomethylation in tumor, negative log-ratio ≤2, and red colors marks hypermethylation in tumor, positive log-ration >2.
TableS3. Table with all DMRs found in this study. 14472 DMRs with chromosomal coordinates, log2 ratio, position where DMR is located regarding to genes and gene identities for DMRs that are in gene areas.
FigureS6. MethlCap-seq reads of two DMRs in all seven samples and replicate experiments. All 14 samples and two replicate experiments for two DMRs that were present in all tumors. (A) Sequence reads plotted in the UCSC genome browser spanning 100kbp in chromosome 2:63,090,001 to 63,190,000; (B) Sequence reads spanning 100 kbp in chromosome 14: 60,000,001 to 60,100,000. Reproducibility can be observed by the similarity between R1 and R2 of each sample. Red bar highlight hypermethylated DMRs in each chromosome found to be present in all seven tumor samples.
FileS1. Tables with hyper- and hypomethylated regions separated in “All tumors”, SCC and ADC subtypes. Tables show hyper- and hypomethylated regions found present in all tumors or that were either unique for SCC or ADC, separated per tumor subtype and DMR type.
FigureS7. Methylation status of CpGs per paired sample in all five fragments. A) Model of analysis. Blue = unmethylated; red = methylated. CpGs are horizontally ordered and clones vertically ordered. Per paired samples in each fragment, the average of methylation of each CpG is calculated and represented from blue to red depending on the percentage of clones that were methylated in the specific CpG. Top panel represents lung samples, bottom panel: tumor samples. B) All paired analyses for CDKN2A, RASSF1, Frag_01, Frag_02 and EN1.
FileS2. Tables with bisulfite sequence analysis of tumor versus lung. Counts and statistics per CpG for all clones in CDKN2A, RASSF1, Frag_01, Frag_02 and EN1 fragments.
FileS3. Tables with bisulfite sequencing statistic analysis of SCC versus ADC. Statistics per CpG for all pooled SCC samples versus pooled ADC samples.
FigureS8. MSP screening. Cropped gel images for all 48 samples grouped by histological subtype in the five fragments analyzed: 17 ADC, 16 SCC, 7 LCC, 8 others (other types and unclassified NSCLC) M: primer for methylated CpG; N: normal/healthy; T: tumor; *: samples used in MethylCap-seq; TM: Totally methylated control sample; TU: Totally unmethylated control sample; ø: blank/water; U: primer for umethylated CpG.
TableS4. Primers. Sequence and conditions of primers used for Bisulfite sequencing and MSP.
Contributor Information
Rejane Hughes Carvalho, Email: r.hughescarvalho@erasmusmc.nl.
Vanja Haberle, Email: vanja.haberle@gmail.com.
Jun Hou, Email: j.hou@erasmusmc.nl.
Teus van Gent, Email: t.vangent@erasmusmc.nl.
Supat Thongjuea, Email: supat.thongjuea@bccs.uib.no.
Wilfred van IJcken, Email: w.vanijcken@erasmusmc.nl.
Christel Kockx, Email: c.kockx@erasmusmc.nl.
Rutger Brouwer, Email: r.w.w.brouwer@erasmusmc.nl.
Erikjan Rijkers, Email: h.rijkers@erasmusmc.nl.
Anieta Sieuwerts, Email: a.sieuwerts@erasmusmc.nl.
John Foekens, Email: j.foekens@erasmusmc.nl.
Mirjam van Vroonhoven, Email: m.vandenhout@erasmusmc.nl.
Joachim Aerts, Email: j.aerts@erasmusmc.nl.
Frank Grosveld, Email: f.grosveld@erasmusmc.nl.
Boris Lenhard, Email: boris.lenhard@bccs.uib.no.
Sjaak Philipsen, Email: j.philipsen@erasmusmc.nl.
Acknowledgements
We thank Peter Riegman and Monique Oomen for providing patient samples stored in the Erasmus MC tissue bank, and Ilya Grigorev for the MRC-5 cell line. We thank Nynke Gillemans for technical assistance and Thamar van Dijk, Cíntia Bombardieri, Ricardo Leite, Vojka Gouka and Marco Gouka for helpful discussions during the preparation of this manuscript. We also thank Alex Pindyurin for this help with the R script. This work was supported by the Netherlands Genomics Initiative.
References
- Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- Little AG. No nodes is good nodes. Ann Thorac Surg. 2006;82:4–5. doi: 10.1016/j.athoracsur.2006.03.051. [DOI] [PubMed] [Google Scholar]
- Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. 2008;83:584–594. doi: 10.4065/83.5.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J, Aerts J, den Hamer B, van Ijcken W, den Bakker M, Riegman P, van der Leest C, van der Spek P, Foekens JA, Hoogsteden HC, Grosveld F, Philipsen S. et al. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS One. 2010;5:e10312. doi: 10.1371/journal.pone.0010312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen JE, Pavey SJ, Passmore LH, Bowman R, Clarke BE, Hayward NK, Fong KM. Expression profiling defines a recurrence signature in lung squamous cell carcinoma. Carcinogenesis. 2007;28:760–766. doi: 10.1093/carcin/bgl207. [DOI] [PubMed] [Google Scholar]
- Liang P, Pardee AB. Analysing differential gene expression in cancer. Nat Rev Cancer. 2003;3:869–876. doi: 10.1038/nrc1214. [DOI] [PubMed] [Google Scholar]
- Kameyama K, Huang CL, Liu D, Masuya D, Nakashima T, Sumitomo S, Takami Y, Kinoshita M, Yokomise H. Reduced ING1b gene expression plays an important role in carcinogenesis of non-small cell lung cancer patients. Clin Cancer Res. 2003;9:4926–4934. [PubMed] [Google Scholar]
- Petty RD, Nicolson MC, Kerr KM, Collie-Duguid E, Murray GI. Gene expression profiling in non-small cell lung cancer: from molecular mechanisms to clinical application. Clin Cancer Res. 2004;10:3237–3248. doi: 10.1158/1078-0432.CCR-03-0503. [DOI] [PubMed] [Google Scholar]
- Ding L, Mardis ER, Li S, Larson DE, Chen K, Wallis JW, Harris CC, McLellan MD, Fulton RS, Fulton LL, Abbott RM, Hoog J, Dooling DJ, Koboldt DC, Schmidt H, Kalicki J, Zhang Q, Chen L, Lin L, Wendl MC, McMichael JF, Magrini VJ, Cook L, McGrath SD, Vickery TL, Appelbaum E, Deschryver K, Davies S, Guintoli T, Lin L. et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464:999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Bird AP, Wolffe AP. Methylation-induced repression–belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- Leonhardt H, Page AW, Weier HU, Bestor TH. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell. 1992;71:865–873. doi: 10.1016/0092-8674(92)90561-p. [DOI] [PubMed] [Google Scholar]
- Agathanggelou A, Honorio S, Macartney DP, Martinez A, Dallol A, Rader J, Fullwood P, Chauhan A, Walker R, Shaw JA, Hosoe S, Lerman MI, Minna JD, Maher ER, Latif F. Methylation associated inactivation of RASSF1A from region 3p21.3 in lung, breast and ovarian tumours. Oncogene. 2001;20:1509–1518. doi: 10.1038/sj.onc.1204175. [DOI] [PubMed] [Google Scholar]
- Kohonen-Corish MR, Sigglekow ND, Susanto J, Chapuis PH, Bokey EL, Dent OF, Chan C, Lin BP, Seng TJ, Laird PW, Young J, Leggett BA, Jass JR, Sutherland RL. Promoter methylation of the mutated in colorectal cancer gene is a frequent early event in colorectal cancer. Oncogene. 2007;26:4435–4441. doi: 10.1038/sj.onc.1210210. [DOI] [PubMed] [Google Scholar]
- Liu L, Broaddus RR, Yao JC, Xie S, White JA, Wu TT, Hamilton SR, Rashid A. Epigenetic alterations in neuroendocrine tumors: methylation of RAS-association domain family 1, isoform A and p16 genes are associated with metastasis. Mod Pathol. 2005;18:1632–1640. doi: 10.1038/modpathol.3800490. [DOI] [PubMed] [Google Scholar]
- Pfeifer GP, Yoon JH, Liu L, Tommasi S, Wilczynski SP, Dammann R. Methylation of the RASSF1A gene in human cancers. Biol Chem. 2002;383:907–914. doi: 10.1515/BC.2002.097. [DOI] [PubMed] [Google Scholar]
- Feng Q, Hawes SE, Stern JE, Wiens L, Lu H, Dong ZM, Jordan CD, Kiviat NB, Vesselle H. DNA methylation in tumor and matched normal tissues from non-small cell lung cancer patients. Cancer Epidemiol Biomarkers Prev. 2008;17:645–654. doi: 10.1158/1055-9965.EPI-07-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu HS, Chen TP, Hung CH, Wen CK, Lin RK, Lee HC, Wang YC. Characterization of a multiple epigenetic marker panel for lung cancer detection and risk assessment in plasma. Cancer. 2007;110:2019–2026. doi: 10.1002/cncr.23001. [DOI] [PubMed] [Google Scholar]
- Vaissiere T, Hung RJ, Zaridze D, Moukeria A, Cuenin C, Fasolo V, Ferro G, Paliwal A, Hainaut P, Brennan P, Tost J, Boffetta P, Herceg Z. Quantitative analysis of DNA methylation profiles in lung cancer identifies aberrant DNA methylation of specific genes and its association with gender and cancer risk factors. Cancer Res. 2009;69:243–252. doi: 10.1158/0008-5472.CAN-08-2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammann R, Li C, Yoon JH, Chin PL, Bates S, Pfeifer GP. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat Genet. 2000;25:315–319. doi: 10.1038/77083. [DOI] [PubMed] [Google Scholar]
- Clark SJ. Action at a distance: epigenetic silencing of large chromosomal regions in carcinogenesis. Hum Mol Genet. 2007;16 Spec No 1:R88–R95. doi: 10.1093/hmg/ddm051. [DOI] [PubMed] [Google Scholar]
- Mayor R, Casadome L, Azuara D, Moreno V, Clark SJ, Capella G, Peinado MA. Long-range epigenetic silencing at 2q14.2 affects most human colorectal cancers and may have application as a non-invasive biomarker of disease. Br J Cancer. 2009;100:1534–1539. doi: 10.1038/sj.bjc.6605045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman AB, Simmer F, Ma K, Kaan A, Zhu J, Stunnenberg HG. Whole-genome DNA methylation profiling using MethylCap-seq. Methods. 2010;52:232–236. doi: 10.1016/j.ymeth.2010.06.012. [DOI] [PubMed] [Google Scholar]
- Teodoridis JM, Hardie C, Brown R. CpG island methylator phenotype (CIMP) in cancer: causes and implications. Cancer Lett. 2008;268:177–186. doi: 10.1016/j.canlet.2008.03.022. [DOI] [PubMed] [Google Scholar]
- Keshet I, Schlesinger Y, Farkash S, Rand E, Hecht M, Segal E, Pikarski E, Young RA, Niveleau A, Cedar H, Simon I. Evidence for an instructive mechanism of de novo methylation in cancer cells. Nat Genet. 2006;38:149–153. doi: 10.1038/ng1719. [DOI] [PubMed] [Google Scholar]
- Rauch TA, Pfeifer GP. DNA methylation profiling using the methylated-CpG island recovery assay (MIRA) Methods. 2010;52:213–217. doi: 10.1016/j.ymeth.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch T, Wang Z, Zhang X, Zhong X, Wu X, Lau SK, Kernstine KH, Riggs AD, Pfeifer GP. Homeobox gene methylation in lung cancer studied by genome-wide analysis with a microarray-based methylated CpG island recovery assay. Proc Natl Acad Sci U S A. 2007;104:5527–5532. doi: 10.1073/pnas.0701059104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch TA, Zhong X, Wu X, Wang M, Kernstine KH, Wang Z, Riggs AD, Pfeifer GP. High-resolution mapping of DNA hypermethylation and hypomethylation in lung cancer. Proc Natl Acad Sci U S A. 2008;105:252–257. doi: 10.1073/pnas.0710735105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt B, Liebenberg V, Dietrich D, Schlegel T, Kneip C, Seegebarth A, Flemming N, Seemann S, Distler J, Lewin J, Tetzner R, Weickmann S, Wille U, Liloglou T, Raji O, Walshaw M, Fleischhacker M, Witt C, Field JK. SHOX2 DNA methylation is a biomarker for the diagnosis of lung cancer based on bronchial aspirates. BMC Cancer. 2010;10:600. doi: 10.1186/1471-2407-10-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zochbauer-Muller S, Fong KM, Virmani AK, Geradts J, Gazdar AF, Minna JD. Aberrant promoter methylation of multiple genes in non-small cell lung cancers. Cancer Res. 2001;61:249–255. [PubMed] [Google Scholar]
- Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M, Lin Z, Zhou L, Chudin E, Garcia EW, Wu B, Doucet D, Thomas NJ, Wang Y, Vollmer E, Goldmann T, Seifart C, Jiang W, Barker DL, Chee MS, Floros J, Fan JB. High-throughput DNA methylation profiling using universal bead arrays. Genome Res. 2006;16:383–393. doi: 10.1101/gr.4410706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano A, Kage H, Sugimoto K, Kitagawa H, Aki N, Goto A, Fukayama M, Nakajima J, Takamoto S, Nagase T, Yatomi Y, Ohishi N, Takai D. A second-generation profiling system for quantitative methylation analysis of multiple gene promoters: application to lung cancer. Oncogene. 2007;26:6518–6525. doi: 10.1038/sj.onc.1210483. [DOI] [PubMed] [Google Scholar]
- Shames DS, Girard L, Gao B, Sato M, Lewis CM, Shivapurkar N, Jiang A, Perou CM, Kim YH, Pollack JR, Fong KM, Lam CL, Wong M, Shyr Y, Nanda R, Olopade OI, Gerald W, Euhus DM, Shay JW, Gazdar AF, Minna JD. A genome-wide screen for promoter methylation in lung cancer identifies novel methylation markers for multiple malignancies. PLoS Med. 2006;3:486. doi: 10.1371/journal.pmed.0030486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang R, Song H, Huang G, Yi J, Zheng Y, Wang J, Chen L. Methylation of multiple genes as a candidate biomarker in non-small cell lung cancer. Cancer Letters. 2011;303:21–28. doi: 10.1016/j.canlet.2010.12.011. [DOI] [PubMed] [Google Scholar]
- Kwon YJ, Lee SJ, Koh JS, Kim SH, Lee HW, Kang MC, Bae JB, Kim YJ, Park JH. Genome-wide analysis of DNA methylation and the gene expression change in lung cancer. J Thorac Oncol. 2012;7:20–33. doi: 10.1097/JTO.0b013e3182307f62. [DOI] [PubMed] [Google Scholar]
- Chavez L, Jozefczuk J, Grimm C, Dietrich J, Timmermann B, Lehrach H, Herwig R, Adjaye J. Computational analysis of genome-wide DNA methylation during the differentiation of human embryonic stem cells along the endodermal lineage. Genome Res. 2010;20:1441–1450. doi: 10.1101/gr.110114.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung JH, Lee HJ, Kim BH, Cho NY, Kang GH. DNA methylation profile during multistage progression of pulmonary adenocarcinomas. Virchows Arch. 2011;459:201–211. doi: 10.1007/s00428-011-1079-9. [DOI] [PubMed] [Google Scholar]
- Esteller M. Epigenetic gene silencing in cancer: the DNA hypermethylome. Hum Mol Genet. 2007;16 Spec No 1:R50–R59. doi: 10.1093/hmg/ddm018. [DOI] [PubMed] [Google Scholar]
- Liu Y, Pang JC, Dong S, Mao B, Poon WS, Ng HK. Aberrant CpG island hypermethylation profile is associated with atypical and anaplastic meningiomas. Hum Pathol. 2005;36:416–425. doi: 10.1016/j.humpath.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Marsit CJ, Houseman EA, Christensen BC, Eddy K, Bueno R, Sugarbaker DJ, Nelson HH, Karagas MR, Kelsey KT. Examination of a CpG island methylator phenotype and implications of methylation profiles in solid tumors. Cancer Res. 2006;66:10621–10629. doi: 10.1158/0008-5472.CAN-06-1687. [DOI] [PubMed] [Google Scholar]
- Ting DT, Lipson D, Paul S, Brannigan BW, Akhavanfard S, Coffman EJ, Contino G, Deshpande V, Iafrate AJ, Letovsky S, Rivera MN, Bardeesy N, Maheswaran S, Haber DA. Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science. 2011;331:593–596. doi: 10.1126/science.1200801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer GP, Rauch TA. DNA methylation patterns in lung carcinomas. Semin Cancer Biol. 2009;19:181–187. doi: 10.1016/j.semcancer.2009.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J, Lambers M, den Hamer B, den Bakker M, Hoogsteden MC, Grosveld F, Hegmans J, Aerts J, Philipsen S. Expression profiling-based subtyping identifies novel non-small cell lung cancer subgroups and implicates putative resistance to pemetrexed therapy. J Thorac Oncol. 2012;7:105–114. doi: 10.1097/JTO.0b013e3182352a45. [DOI] [PubMed] [Google Scholar]
- Chatagnon A, Ballestar E, Esteller M, Dante R. A role for methyl-CpG binding domain protein 2 in the modulation of the estrogen response of pS2/TFF1 gene. PLoS One. 2010;5:e9665. doi: 10.1371/journal.pone.0009665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouaire T, de Las Heras JI, Merusi C, Stancheva I. Recruitment of MBD1 to target genes requires sequence-specific interaction of the MBD domain with methylated DNA. Nucleic Acids Res. 2010;38:4620–4634. doi: 10.1093/nar/gkq228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helman E, Naxerova K, Kohane IS. DNA hypermethylation in lung cancer is targeted at differentiation-associated genes. Oncogene. 2011;31:1181–1188. doi: 10.1038/onc.2011.307. [DOI] [PubMed] [Google Scholar]
- Karlen Y, McNair A, Perseguers S, Mazza C, Mermod N. Statistical significance of quantitative PCR. BMC Bioinformatics. 2007;8:131. doi: 10.1186/1471-2105-8-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: A language and environment for statistical computing. Reference index version: 2.11.1. R Foundation for Statistical Computing, Vienna, Austria; 2010. http://www.R-project.org. [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J. et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Down TA, Rakyan VK, Turner DJ, Flicek P, Li H, Kulesha E, Graf S, Johnson N, Herrero J, Tomazou EM, Thorne NP, Bäckdahl L, Herberth M, Howe KL, Jackson DK, Miretti MM, Marioni JC, Birney E, Hubbard TJ, Durbin R, Tavaré S, Beck S. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat Biotechnol. 2008;26:779–785. doi: 10.1038/nbt1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelizzola M, Koga Y, Urban AE, Krauthammer M, Weissman S, Halaban R, Molinaro AM. MEDME: an experimental and analytical methodology for the estimation of DNA methylation levels based on microarray derived MeDIP-enrichment. Genome Res. 2008;18:1652–1659. doi: 10.1101/gr.080721.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
TableS1. Data from all samples used in this study.
TableS2. High-throughput sequencing reads from all samples and replicate experiments.
FigureS1. Selection of normalization method. Distribution of Pearson’s correlation coefficients between replicates for different normalization methods.
FigureS2. Correlation between technical replicates. Scatter plots comparing raw MethylCap-seq signal from two technical replicates (R1 and R2) for all 14 samples. Each point represents one 10 bp genomic bin. Density of points (log10 scale) is shown in different shades of blue. Pearson’s correlation coefficient is denoted in the top left corner of each scatter plot.
FigureS3. Correlation between lung and tumor samples. Scatter plots comparing raw MethylCap-seq signal from healthy and tumor tissue for six pairs of samples. Each point represents one 10 bp genomic bin. Density of points (log10 scale) is shown in different shades of blue. Pearson’s correlation coefficient is denoted in the top left corner of each scatter plot.
FigureS4. Selection of P-value threshold. Number of significant hypo- and hypermethylated regions for different P-value thresholds. Y-axis shows number of significant regions on log10 scale for decreasing P-value threshold, shown on the x-axis.
FigureS5. along each chromosome for each paired sample. Representation of the log-ratio of relative methylation scores for tumors vs. paired lung samples for each sample pair showing DMRs along each chromosome. Green colors marks hypomethylation in tumor, negative log-ratio ≤2, and red colors marks hypermethylation in tumor, positive log-ration >2.
TableS3. Table with all DMRs found in this study. 14472 DMRs with chromosomal coordinates, log2 ratio, position where DMR is located regarding to genes and gene identities for DMRs that are in gene areas.
FigureS6. MethlCap-seq reads of two DMRs in all seven samples and replicate experiments. All 14 samples and two replicate experiments for two DMRs that were present in all tumors. (A) Sequence reads plotted in the UCSC genome browser spanning 100kbp in chromosome 2:63,090,001 to 63,190,000; (B) Sequence reads spanning 100 kbp in chromosome 14: 60,000,001 to 60,100,000. Reproducibility can be observed by the similarity between R1 and R2 of each sample. Red bar highlight hypermethylated DMRs in each chromosome found to be present in all seven tumor samples.
FileS1. Tables with hyper- and hypomethylated regions separated in “All tumors”, SCC and ADC subtypes. Tables show hyper- and hypomethylated regions found present in all tumors or that were either unique for SCC or ADC, separated per tumor subtype and DMR type.
FigureS7. Methylation status of CpGs per paired sample in all five fragments. A) Model of analysis. Blue = unmethylated; red = methylated. CpGs are horizontally ordered and clones vertically ordered. Per paired samples in each fragment, the average of methylation of each CpG is calculated and represented from blue to red depending on the percentage of clones that were methylated in the specific CpG. Top panel represents lung samples, bottom panel: tumor samples. B) All paired analyses for CDKN2A, RASSF1, Frag_01, Frag_02 and EN1.
FileS2. Tables with bisulfite sequence analysis of tumor versus lung. Counts and statistics per CpG for all clones in CDKN2A, RASSF1, Frag_01, Frag_02 and EN1 fragments.
FileS3. Tables with bisulfite sequencing statistic analysis of SCC versus ADC. Statistics per CpG for all pooled SCC samples versus pooled ADC samples.
FigureS8. MSP screening. Cropped gel images for all 48 samples grouped by histological subtype in the five fragments analyzed: 17 ADC, 16 SCC, 7 LCC, 8 others (other types and unclassified NSCLC) M: primer for methylated CpG; N: normal/healthy; T: tumor; *: samples used in MethylCap-seq; TM: Totally methylated control sample; TU: Totally unmethylated control sample; ø: blank/water; U: primer for umethylated CpG.
TableS4. Primers. Sequence and conditions of primers used for Bisulfite sequencing and MSP.