Abstract
We recently reported a novel curcuminoid 4-[3,5-bis(2-chlorobenzylidene-4-oxo-piperidine-1-yl)-4-oxo-2-butenoic acid] or CLEFMA as a potent anti-proliferative agent, and showed that it induces autophagic cell death in lung cancer cells. We are now reporting a drug-in-CD-in-liposome approach to formulate CLEFMA liposomes that could be labeled with Tc-99m radionuclide for non-invasive imaging of their biodistribution. CLEFMA encapsulation was enabled by hydroxypropyl-β-cyclodextrin. In vitro studies showed that CLEFMA possessed more potent anti-proliferative activity in lung adenocarcinoma H441 cells than naturally occurring curcumin. At the same time, it had no effect on the proliferative capacity of normal lung fibroblasts. CLEFMA liposomes retained the antiproliferative potency of free CLEFMA, while maintaining its non-toxic nature in normal lung fibroblasts. In nude rats bearing xenograft H441 tumors, the tumor volume significantly reduced after i.v. treatment with CLEFMA liposomes (p<0.05); the tumor inhibition was determined to be 94%. The anti-tumor activity of CLEFMA liposomes was confirmed by the observation that F-18-fluorodeoxyglucose uptake in tumors of treated rats was reduced as compared to those of control rats. Tc-99m-labeled CLEFMA liposomes accumulated in liver (33.7%); spleen showed the largest accumulation on per gram tissue basis (6.2 %/g). Upon histopathological examination of liver, lung and kidney, we found no apparent toxicity from multiple CLEFMA liposome administrations. The results demonstrate the utility of liposomes to serve as a carrier for CLEFMA. This study is the first to demonstrate the efficacy of novel curcuminoid CLEFMA in a preclinical model.
Keywords: CLEFMA, Curcumin, Cancer, Imaging, Liposomes, HPβCD
1. INTRODUCTION
Lung cancer is one of the leading causes of cancer-related deaths in the world. According to the report from the U.S. Cancer Statistics Group, 158,683 people in the United States died from lung cancer in the year 2007 [1]. About 60% people with lung cancer die within one year of being diagnosed with the disease. In non-small cell lung carcinoma (NSCLC), surgery is the only curative treatment modality [2]. Meta-analysis of clinical data suggests that up to 85% of the NSCLC patients depend on systemic chemotherapy as part of the overall management [3]. However, despite advances in understanding of cancer biology and potential molecular targets, lung cancer chemotherapy remains palliative in nature. The poor clinical response to anticancer drugs in lung cancer patients has led to a continuous search for newer and more potent compounds. We synthesized a novel synthetic second generation curcumin analog, CLEFMA, and reported its efficacy as a potent anti-proliferative agent in various cancer cell lines [4]. CLEFMA is a derivative of a previously reported diphenyldifluoroketone called EF24 [5], and is chemically named as 4-[3,5-bis(2-chlorobenzylidene-4-oxo-piperidine-1-yl)-4-oxo-2-butenoic acid].
The pre-formulation studies on CLEFMA depicted it as a highly hydrophobic drug with octanol-water partition coefficient of 5.66. We hypothesized that liposomes might serve as a vehicle for CLEFMA preparation meant for parenteral administration. Parenteral liposome formulations of lipophilic drugs are challenging. Incorporation of a lipid-partitioning drug in liposomes may produce structurally instable liposomes, resulting in burst-release of drug, rapid clearance from the body, and/or overall poor efficacy. The entrapment of cyclodextrin (CD)-based inclusion complexes (IC) inside the liposomes can be a reliable and effective method of encapsulation of hydrophobic drugs. This strategy was first reported by Gregoriades [12], and has recently been used for the encapsulation of isotretinoin [13].
Arguably, the CD-based IC themselves could be used as a water soluble product for intravenous administration. However, encapsulating drug-CD IC inside liposomes has several potential advantages. Encapsulation not only modifies the biodisposition of IC, but also prolongs its clearance. If desired, the outer lipid layer of liposomes could be used to attach ligands to target drug delivery. Liposomes are relatively benign drug delivery vehicles, and many liposomal products are in clinical use [6, 7]. An enhanced permeability and retention (EPR) effect enables liposomes to be selectively trapped in the tumor tissue for an extended period because of high vascular density, larger intracellular space and high vascular permeability [8, 9]. Besides EPR-mediated accumulation in tumor, liposomal carriers modify the pharmacokinetics and tissue distribution of incorporated drugs. Evidently, the size, surface charge and composition of lipid bilayer have strong influences on liposome kinetics [10]. The altered behavior of liposome-encapsulated drug in biological system is often manifested as enhanced efficacy as well as reduced side-effects of drugs [11].
Recently, we reported optimization of the drug-in-CD-in-liposome approach for EF24, a close analog of CLEFMA [15]. The liposomes were designed to facilitate labeling with single photon-emitting radionuclide Tc-99m for imaging of their biodistribution after administration. In this presented work, we investigated the in vivo efficacy of CD-enabled liposomal CLEFMA in a lung cancer xenograft model in nude rats. We employed positron emission tomography (PET) as a non-invasive imaging tool for confirming anti-proliferative efficacy of CLEFMA liposomes. At the same time, we labeled CLEFMA liposomes with Tc-99m for single photon emission tomography (SPECT). The results of this investigation suggest that liposomal formulation provides an acceptable platform for parenteral delivery of curcuminoids such as CLEFMA.
2. MATERIALS AND METHODS
2.1 Formation of CLEFMA Inclusion Complex (IC)
CLEFMA was synthesized and characterized according to the methods described elsewhere [4]. We performed a phase solubility analysis of CLEFMA according to the published method [14], and prepared ICs of CLEFMA with HPβCD in the solution phase as reported previously [15]. Briefly, an excess of CLEFMA was added to 5 ml of HPβCD solution (1 g/ml). The mixture was continuously agitated on a shaker incubator at 25°C for 72 h. After allowing settling of coarse particulate matter for 6 h, the supernatant was centrifuged at 14,000 rpm for 15 min to obtain a clear supernatant free of any insoluble material. The solution was passed through a sterile 0.22 μm cellulose acetate filter, and the amount of HPβCD-solubilized CLEFMA was estimated spectrophotometrically at 320 nm. The phase solubility diagram was constructed by plotting the estimated CLEFMA concentration against HPβCD concentrations. The stability constant was calculated from the phase diagram using the following equation-
The complexation efficiency (CE) was calculated as a product of S0 and K1:1
2.2 Preparation of Liposomes
CLEFMA liposomes were prepared by high pressure homogenization [16]. A lipid composition of 1, 2-disteroyl-sn-glycero-3-phosphatidylcholine: cholesterol: dipalmitoylphosphatidyl glycerol (DSPC: CHO: DMPG as 50:50:5 mol%) was dissolved in a mixture of chloroform: methanol (2:1) and transferred to a round bottom flask. The solvent mixture was evaporated at 58 °C on an R-210 rotavapor (Buchi Corporation, New Castle, DE) to obtain a thin film of lipids. Any residual traces of organic solvent were removed by keeping the film under high vacuum for 12 h. The phospholipid film was rehydrated with HypureTM endotoxin-free, cell culture grade water (Hyclone, Logan, UT), maintaining the total phospholipid concentration to 12 mM (total lipid 2 g/dL). The resulting suspension of multilamellar vesicles was subjected to eight freeze-thaw (FT) cycles. An FT cycle consisted of snap-freezing the suspension in liquid nitrogen followed by immediate thawing in a 58 °C water bath. The liposome suspension was lyophilized for 48 h in a Triad lyophilizer (Labconco, Kansas city, MO). The dried mass was rehydrated with sterile aqueous solution of CLEFMA-HPβCD IC.
To enable Tc-99m labeling, the rehydration mixture also contained glutathione (100 mM, pH 6.8); the lipid concentration during rehydration was maintained at approximately 12 mM. The liposome suspension thus formed was homogenized in Emulsiflex C3 homogenizer (Avestin Inc, Canada). A three-step homogenization process was adopted, where the mixture was subjected to pressures of 15K PSI (1 cycle), 20K PSI (2 cycles), and 23K PSI (1 cycle). Each step was separated by a cooling period of 15 min during which the preparation was kept at 4° C. The liposomes were separated from any un-entrapped material by ultracentrifugation at 50,000 rpm and 4 °C for 40 min (Beckman Optima L-100 XP, Fullerton, CA). The liposome pellet was washed three times with PBS (pH 7.4), before re-suspending in PBS. Strict aseptic conditions were maintained during the entire processing.
A control preparation of liposomes was also prepared identically, except that the rehydration solution was devoid of CLEFMA, but contained HPβCD and glutathione.
2.3 Characterization of CLEFMA Liposomes
CLEFMA inside the liposomes was determined by digesting a measured aliquot of liposome suspension in methanol, and spectrophotometrically estimating CLEFMA at 310 nm after appropriate dilution with methanol. Control liposomes (liposomes identical to the CLEFMA-liposomes in all respects but devoid of CLEFMA) were also digested in methanol and used as estimation blank.
Phospholipid concentration in the liposomes was determined by Stewart assay [17]. A 10 μl aliquot of liposomes was vigorously mixed with a binary system consisting of 2 ml each of chloroform and ferrothiocyanate reagent. The aqueous ferrothiocyanate reagent contained 27.03 g/L ferric chloride hexahydrate and 30.4 g/L ammonium thiocyanate. The color in the chloroform phase was measured at 485 nm and compared to a set of DSPC standards treated in an identical fashion.
The particle size of the liposomes was determined by photon correlation spectroscopy using a Brookhaven particle size analyzer equipped with Mas Option software. Zeta potential of preparations was measured in a Zeta PLUS Zeta potential analyzer (Brookhaven Instruments Corp, Holtsville, NY). For zeta potential, the liposomes (~ 40 μg of phospholipid) in 1.5 ml of 0.22 μm filtered de-ionized water were scanned at 25 °C for 10 runs, each run consisting of 20 cycles. Zeta potential values were obtained as millivolt ± standard error of mean.
The Transmission Electron Microscopy (TEM) was performed at the University of Oklahoma (Norman, OK). An ultradilute liposome suspension was stained with a solution consisting of 2.5% phosphotungstic acid and 2.5% trehalose (pH 7). A drop of liposome suspension was first applied to the copper grid, allowed to adsorb on the grid for 2 min, and then blotted with a filter paper. A drop of the stain was added to the wet grid and immediately blotted. TEM images were recorded on a Zeiss 10 electron microscope.
2.4 In vitro Anti-proliferative Activity of CLEFMA and Liposomes
Human lung adenocarcinoma cell line NCI-H441 (ATCC # HTB-174) and normal lung fibroblasts LL-24 (ATCC #CCL-151) were obtained from American Type Culture Collection (Manassas, VA). The cells were maintained at 37°C with 5% CO2 in RPMI 1640 medium (Invitrogen, Carlsbad, CA). The medium was supplemented with 10% heat-inactivated fetal bovine serum and 50 μg/ml of gentamicin (GIBCO Laboratories, Grand Island, NY). For cell proliferation assay, the cells were seeded in 96-well flat-bottom tissue culture plates at a density of 0.5-1 × 104 cells per well. The cells were allowed to adhere and grow overnight, followed by treatment with CLEFMA or CLEFMA-liposomes. The concentration of CLEFMA was maintained in the range of 0.1-25 μM in 100 μl of RPMI 1640 medium. For comparison, the cells were also treated with 1 and 10 μM natural curcumin and EF24. EF24 is a potent synthetic curcuminoid and a close analog of CLEFMA [5]. All solutions of plain drugs were made in dimethylsulfoxide (DMSO). The concentration of DMSO in the treatment medium was maintained at 0.1%. The control wells received equivalent amounts of DMSO or phospholipid-matched control liposomes. Inhibition of cell proliferation was determined by measuring a decrease in hexosaminidase activity as described by Landegren [18]. Para-nitrophenol-N-acetyl-beta-D-glucosaminide (60 μl per well) was used as the substrate for the hexosaminidase enzyme. The results of hexosaminidase assay were also confirmed by visualization of cultured cells under a microscope.
2.5 Rat Model of Xenograft Lung Cancer
The animal experiments were performed according to the NIH Animal Use and Care Guidelines and were approved by the Institutional Animal Care Committee of the University of Oklahoma Health Sciences Center. Athymic nude rats (n=12, 200-225 g) were obtained from Harlan Laboratories (Indianapolis, IN), and housed in controlled environment with 12 h day/night cycle. The animals were allowed to acclimatize at least one week before inoculation of H441 cells. On the day of tumor implantation, the rats were anesthetized with 2-3% isoflurane in oxygen stream. H441 cell suspension in PBS (0.2 ml, 50 million cells/ml) was subcutaneously injected in the left dorsal thigh region. The rats were returned to their cages, and the tumor was allowed to grow till a visible and palpable tumor was observed in about 15 days post-implantation (Table 1).
Table 1.
Treatment schedule of xenograft tumor in nude rats. The tumor was implanted on day 1 and allowed to grow for 15 days. Intravenous treatment (T) with CLEFMA liposomes was started on day 15 and continued up to day 33. PET imaging of FDG accumulation was carried out on day 36, followed next day by the biodistribution study.
| Procedure | Day | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 to 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | 30 | 31 | 32 | 33 | 34 | 35 | 36 | 37 | |||
| Tumor implant | Implant | ||||||||||||||||||||||||
| Treatment | T | T | T | T | T | T | |||||||||||||||||||
| PET imaging | PET | ||||||||||||||||||||||||
| Biodistribution | BIO | ||||||||||||||||||||||||
2.6 Drug Treatment
The tumor bearing rats were randomized in two groups of six animals each. Intravenous treatment with CLEFMA liposomes was started 15 days post-implantation. The animals were treated with CLEFMA liposomes (treated) or control liposomes (control) every fourth day for a total of six injections (Table 1). The CLEFMA dose was maintained at 40 μg per administration (approximately 0.2 mg/Kg body weight). The control animals received equivalent amounts of phospholipid as control liposomes. On each injection day, the tumor growth was determined by measuring two dimensions using the Vernier calipers to obtain tumor volume = (Length × Width2)/2. Finally, the percent tumor growth inhibition as of 20th day of treatment was determined by using the following formula- % Tumor Growth Inhibition = [(1−T/C)*100], where T and C are mean tumor volume in the treated and control groups, respectively.
2.7 Positron Emission Tomography
After the treatment schedule, the animals were recruited for PET using F-18-fluorodeoxyglucose (FDG) as a biomarker of tumor growth (Table 1). The PET imaging was performed on day 22 of the treatment. FDG (100 μCi, 0.3 ml) was intravenously injected in the tail vein of the rats anesthetized with 2-3% isoflurane in oxygen. The animals were allowed to wake up and returned to their cages for distribution of FDG to take place and the background radioactivity to clear. After 2 h, the animals were anesthetized again and placed inside the PET detector of X-O-PET machine (Gamma Medica-Ideas, Northridge, CA, USA). Static images of FDG accumulation were acquired for 20 min, followed by a fly-mode computed tomography (CT). The PET image was reconstructed using filtered back projection algorithm and fused with the CT image. The fused image was employed for visualization using Amira 3.1 software (Visage Image Inc., San Diego, CA, USA).
2.8 Biodistribution of CLEFMA Liposomes
After the PET imaging, the rats were allowed to go back into their cages in order to let the F-18 radioactivity (T1/2 110 min) decay completely. The next day, the rats were recruited for biodistribution study (Table 1). For biodistribution, CLEFMA liposomes were labeled with Tc-99m radionuclide. The radiolabeling was performed using a lipophilic chelate Tc-99m-hexamethyl propylene amine oxime or HMPAO (OUHSC-Nuclear Pharmacy, Oklahoma City, OK) and according to the method described previously [19-21]. Approximately 0.2 ml liposomes were mixed with equal volume of Tc-99m-HMPAO and allowed to stand at room temperature for 30-45 min. Lipophilic Tc-99m-HMAPAO partitions into the liposomes and gets entrapped secondary to its glutathione-mediated conversion into a hydrophilic species [19-21]. The Tc-99m-labeled CLEFMA liposomes were separated from any free Tc-99m-HMPAO by gel exclusion chromatography using a PD-10 column.
The Tc-99m-labeled CLEFMA liposomes were evaluated for distribution in tumor bearing nude rats (n=10). On the day of the experiment, the rats were anesthetized with isoflurane gas (2% in oxygen at 2 L/min). A 25G butterfly was secured in the tail vein for administration of radiolabeled preparation. Tc-99m-CLEFMA liposomes (250 μCi, 0.15 ml, 0.26 mg phospholipid) were infused through the tail vein. The rats were randomized into two groups (5 rats each) to study biodistribution at 6 and 24 h post-injection. After specific times, the rats were euthanized by an intraperitoneal overdose of Euthasol (Virbac Corp., Fort Worth, TX). Various organs were excised, washed with saline, weighed and appropriate tissue samples were counted in an automated gamma counter (Perkin-Elmer, Boston, MA). Total blood volume, bone and muscle mass were estimated as 5.7%, 10% and 40% of body weight, respectively [22, 23]. A diluted sample of injected Tc-99m-CLEFMA liposomes served as a standard for comparison. The accumulation of injected preparation in various organs was also calculated as percent of injected radioactivity. All data were corrected for decay of Tc-99m radioactivity (T1/2 = 6 h) and background-subtracted.
For imaging of Tc-99m-CLEFMA liposomes distribution in vivo, we used single photon emission tomography (SPECT). The tumor-bearing rats were intravenously injected with 1 mCi of Tc-99m-CLEFMA liposomes. Longitudinal images were acquired in a NanoSPECT machine (Bioscan, Washington DC) over a 12 h-duration.
2.9 Histopathology
After the animals were euthanized, a part of the liver, lung and kidney were fixed in 10% formalin. The formalin-preserved tissues were used for histology by staining 5 μm sections of paraffin-embedded tissues with H&E stain. The sections were read by a veterinary pathologist (OUHSC, Oklahoma City) in a blinded fashion for the presence of necrosis, hemorrhage, inflammation or any other pathological changes.
2.10 Data Analysis
The biological data was analyzed for significance of difference at p < 0.05 using Prism 5.0 (GraphPad Software, Inc., La Jolla, CA). The biodistribution data was presented as percent injected dose per gram tissue. Both control and treated groups lost one rat each during the course of investigation. Therefore, the data were censored to allow the dead rats to contribute to the overall results for the entire length of time they were followed, but to statistically remove them after the death was recorded.
3. RESULTS
3.1 CLEFMA Inclusion Complex and Liposomes
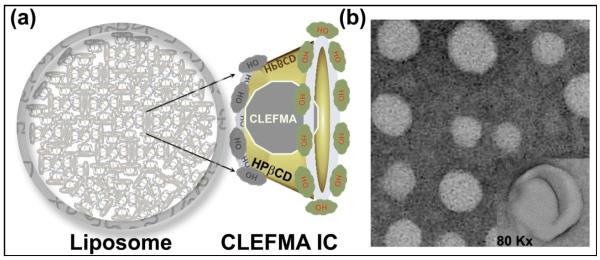
The phase solubility analysis of CLEFMA in the presence of HPβCD revealed that CLEFMA forms a 1:1 complex with HPβCD on a molar basis. The stability constant of the complex was found to be 0.0126 M−1and the complexation efficiency was approximately 0.57. A stable inclusion complex was obtained with a HPβCD:CLEFMA molar ratio of 53:74. We formulated a liposome preparation using CLEFMA-in CD-in liposome approach (Figure 1a). The liposomes were prepared using an aqueous solution of the inclusion complex to hydrate the dry lipid powder. The final liposome preparation had approximately 8.05 mM CLEFMA; the CLEFMA: phospholipid molar ratio was 0.133. The phospholipid concentration in the final preparation was 60.3 mg/ml. The % of CLEFMA encapsulated was approximately 19.4%. The size of the liposomes was measured to be 310.3 ± 4.7 and the zeta potential was −43 ± 1.91 mV. The transmission electron micrographs of the liposomes carrying ICs showed uniformly-dispersed spherical structures of liposomes (Figure 1b).
Figure 1.
(a) Schematic illustration of CLEFMA liposomes using a ‘Drug-in CD-in liposome’ approach. The inclusion complex of CLEFMA with HPβCD was encapsulated inside the liposomes consisting of distearoylphosphatidylcholine: cholesterol: dimyristoylphosphatidyl glycerol (50:50:5 mol%). (b) Transmission electron micrograph of CLEFMA liposomes (inset, 80,000 X).
3.2 In vitro Activity of CLEFMA and Liposomes in Cultured Cells
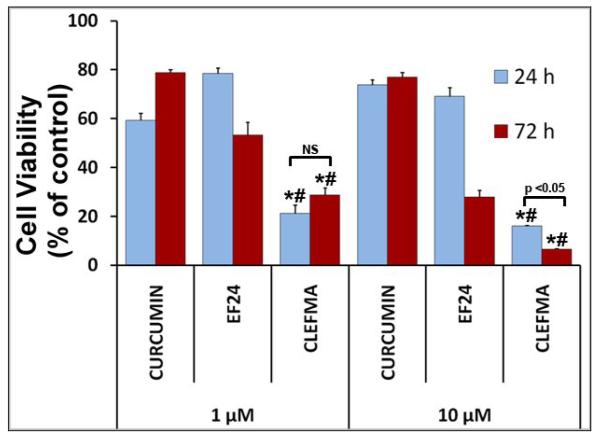
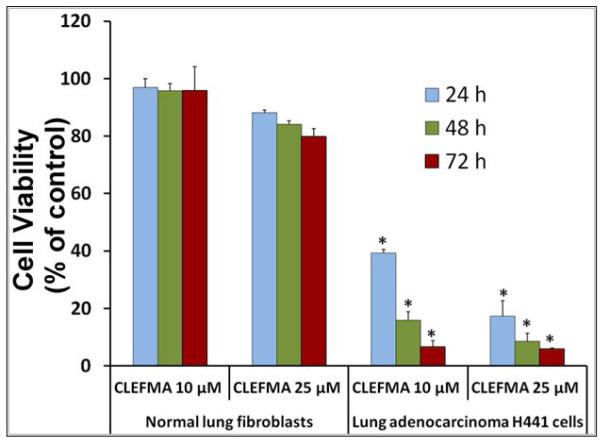
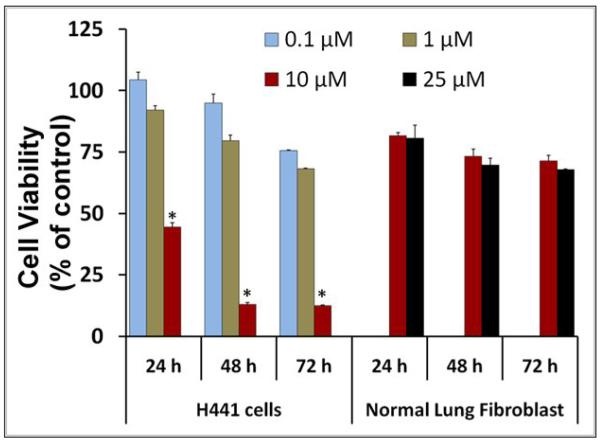
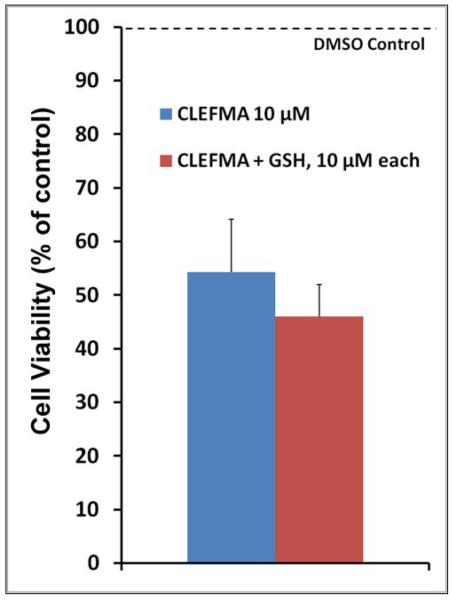
As shown in Figure 2a, CLEFMA is more potent than curcumin and another synthetic curcuminoid EF24 in suppressing the proliferation of H441 cells. We further investigated whether CLEFMA is selective in its action on cancer cells. It is clear from the data in Figure 2b that CLEFMA is antiproliferative to the H441 cancer cells, but is substantially less active against normal lung fibroblasts. Less than 40% of H441 cells survived the 24 h treatment with 10 μM CLEFMA. On the other hand, normal lung fibroblasts essentially remained unscathed under the identical treatment conditions. Liposomal CLEFMA retained the potency of CLEFMA against cancer cells, and maintained the relative innocuousness toward normal lung fibroblasts (Figure 2c). Since CLEFMA liposomes contained glutathione as a constituent, we also investigated the effect of glutathione on the activity of CLEFMA in H441 cells. Glutathione was not anti-proliferative by itself, but it potentiated CLEFMA activity in a statistically insignificant manner (Figure 2d).
Figure 2.
Cell viability of H441 cells treated with various preparations. (a) CLEFMA, EF24 and curcumin were compared for anti-proliferative efficacy (* and # are p< 0.05 against curcumin and EF24, respectively). (b) CLEFMA potently suppresses growth of H441 cells, but not that of normal lung fibroblasts LL-24 (* p < 0.05 against respective normal lung fibroblast cells). (c) Liposomal CLEFMA retains the antiproliferative activity of free CLEFMA while remaining non-toxic to normal lung fibroblasts. (d) Presence of glutathione (GSH) inside the liposomes does not have significant impact on anti-proliferative activity of CLEFMA. The data is presented as the mean ± sem of results from at least 3 individual experiments performed in triplicates.
3.3 In vivo Efficacy of CLEFMA Liposomes in Xenograft Tumor Model
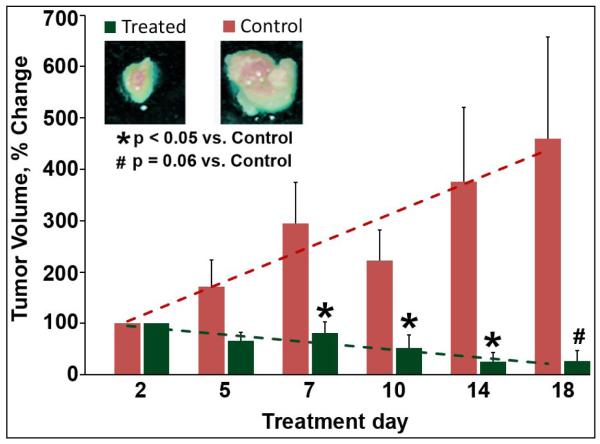
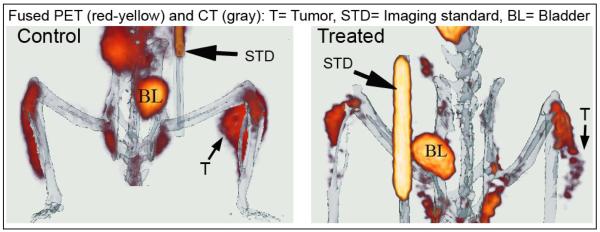
The anti-proliferative efficacy of CLEFMA liposomes was investigated in a nude rat model of xenografted tumor. Figure 3a shows the effect of CLEFMA liposome treatment on tumor volume as measured by Vernier calipers. Because of the variability in the initial size of the tumor, the data were calculated as a percent reduction in size compared to the initial tumor size in the same animal. Over a period of 20 days, the tumor volume almost quadrupled in control rats, but reduced to approximately half the initial size in rats receiving CLEFMA liposomes. The percent tumor inhibition was calculated to be approximately 94%. After the last scheduled treatment, the rats were subjected to PET imaging with F-18-FDG. A higher accumulation of FDG signifies a growing tumor. As shown in a representative picture (Figure 3b), the rat treated with control liposomes accumulated significant amounts of FDG in tumor, but the rat treated with CLEFMA liposomes showed minimal FDG uptake in the tumor.
Figure 3.
(a) Change in tumor volume in response to liposomal CLEFMA therapy. The insets show the representative pictures of excised tumors upon necropsy. The dashed lines are the trend line fits to the plotted data. The data is presented as the mean ± sem of results from experiments on n=4 (treatment) and n=3 (control) rats. (b) A representative set of fused PET/CT images of F-18-FDG accumulation in control and treated rats. The images clearly show that the treated rat has significantly less accumulation of FDG than the control animal.
3.4 Biodistribution
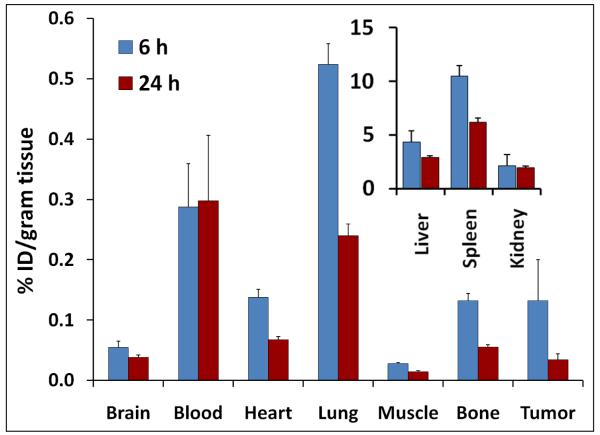
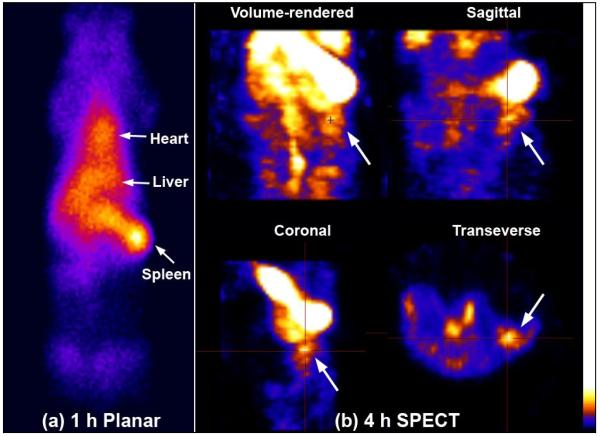
After PET imaging, the rats were subjected to a biodistribution study of CLEFMA liposomes labeled with Tc-99m radionuclide. The labeling efficiency of CLEFMA liposomes exceeded 70% before column separation. After gel exclusion chromatography, more than 95% of radioactivity was found associated with the liposomes. Tc-99m-CLEFMA liposomes were intravenously injected and the rats were euthanized after 6 and 24 h (n=5 each) to collect various organs for counting tissue-associated radioactivity. Figure 4 shows the accumulation of CLEFMA liposomes in various organs of the rats, suggesting liver as the organ of major uptake. Substantial amounts of injected dose were also found in spleen. There was measurable accumulation of CLEFMA liposomes in the tumors, but the tumor-to-blood ratio was found to be less than unity. Because of the non-stealth nature of the CLEFMA liposomes, it was not surprising to observe their rapid elimination from blood circulation; approximately 5% of injected dose remained in circulation by 6 h post-injection. The differences in 6 and 24 h uptake values of Tc-99m-CLEFMA liposomes in blood, liver and kidney were not significant. Figure 4b demonstrates the application of imaging in drug delivery. Tc-99m-CLEFMA liposomes were seen accumulating in tumor tissue in the left flank of the rat. The images also confirmed the biodistribution data that the majority of administered liposomes accumulate in liver and spleen.
Figure 4.
Biodistribution of Tc-99m-labeled CLEFMA liposomes in rats. (a) Injected dose per gram of tissue. The data is presented as the mean ± sem of results from experiments involving n=5 each at 6 h and 24 h. (b) SPECT images of a tumor-bearing rat injected with Tc-99m-labeled CLEFMA liposomes. H441 cells were injected in this rat in the left flank (arrow).
The weight of the excised tumor also decreased after the treatment with CLEFMA liposomes; the average weight of tumor in treated rats was 0.28 ± 0.14 g compared to 0.49 ± 0.25 g in control rats. Upon necropsy, it was observed that much of the weight and volume in treated tumors was contributed by the accumulation of necrotic fluid because of potent cell death induced by CLEFMA.
3.5 Histopathology
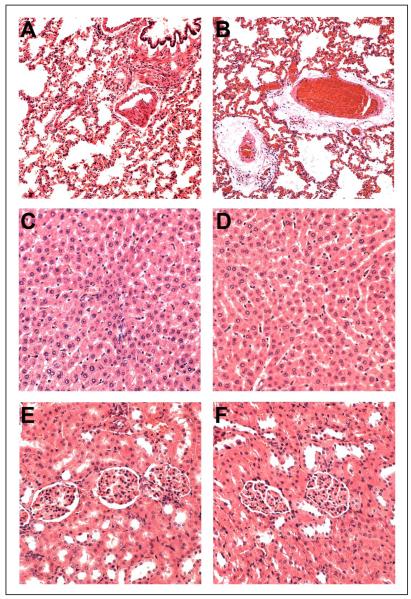
Histopatholgical examination of lung, liver and kidney showed insignificant differences between the control and treated groups (Figure 5). Both control and treated rats had moderate to marked pulmonary congestion with no evidence of capillary leakage or acute inflammation, with occasional intra-alveolar macrophage present. Mild intra-alveolar edema and perivascular edema was evident in one rat belonging to the treatment group. These mild changes in lung might have been caused by prolonged gaseous anesthesia before the rats were euthanized. The liver sections of both control and treated rats showed no mitotic figures or apoptotic bodies within the 50 high-power fields. Mild to moderate diffuse hepatocellular vacuolation with mild congestion was occasionally seen in both control and treated liver sections. Similarly, in kidney sections mild corticomedullary congestion with no evidence of tubular or glomerular damage or acute inflammation was observed in either of the groups.
Figure 5.
Histology of lung (A and B), liver (C and D) and kidney (E and F) from control (A, C and E) and treated (B, D and F) rats. As described in the text, no significant observations differentiating treated from control rats were made by the veterinary pathologist.
4. DISCUSSION
Chemotherapy has definite palliative benefits in symptomatic relief and improvement in quality of life in lung cancer. A small window of survival and short life-expectancy often necessitates aggressive therapy with a cocktail of highly toxic drugs, radiotherapy and surgery. Availability of medicines with absolute selectivity to cancer is ideal, but an elusive goal. The molecularly-targeted drugs (Bevacizumab, Erlotinib, etc.) show promise, but are riddled with their own genuine problems [38]. For instance, heterogeneity in the expression of selective target resulted in the failure of clinical trials of epidermal growth factor receptor inhibitors [39]. Even if tumor heterogeneity and emergence of resistance are disregarded, targeted therapies succeed only when patient population is segregated into target-selective subpopulations in a timely fashion [40]. Not surprisingly, a consensus is building that targeted therapies do not dramatically change clinical outcome for most patients [41, 42]. In this article we describe a liposome formulation of CLEFMA for potential therapy of lung cancer.
CLEFMA is a curcumin analog, and it has its origin in a systematic structure activity relationship performed on a potent anticancer curcuminoid 3,5-bis-(2-fluorobenzylidene)-4-piperidone or EF24 [4, 24]. EF24 was first created in Emory University (Atlanta, GA) [25-29], and it has been shown to inhibit IkappaB kinase [26]. It induces apoptosis in A549 lung cancer cells; the apoptosis was synergistically increased by inhibition of p38 MAPK [42]. Compared to EF24, CLEFMA appears to possess more potent anti-proliferative activity in H441 cells (Figure 2a). The experimental data collected in our laboratory also suggests that CLEFMA is comparable to other clinical anticancer drugs in potency, such as doxorubicin, paclitexal and gemcitabine (data not shown). In a parallel investigation, we investigated the molecular basis of CELFMA’s anti-proliferative action and found evidence that CLEFMA induces autophagy in H441 cells [4]. For the cells with deficient apoptotic machinery, alternative modes of cell death, including macroautophagy assume importance [43]. Like the majority of lung cancer cells, H441 cells carry k-Ras mutation that confers anti-apoptotic advantage to the cells [44].
In order to enable intravenous administration of poorly water soluble CLEFMA, we employed an approach where CLEFMA was first solubilized in aqueous HPβCD solution as 1:1 IC, and the IC was encapsulated within the liposomes. Using EF24 as a model drug, we recently reported standardization of encapsulation parameters and stability of resultant preparation in an accompanying report [15]. The strategy was successful for CLEFMA also, because even after several days of storage at 4 °C, there was no sign of liposome destabilization. The instability of liposomes would have resulted in the leakage of co-encapsulated glutathione, culminating in a significant drop in Tc-99m labeling efficiency. We did not find any decrease in efficiency of Tc-99m-labeling over the storage period of >20 days.
An intravenous parenteral preparation of CLEFMA could also be accomplished simply by the use of aqueous HPβCD-CLEFMA IC. We chose to make liposomes of IC because of several reasons, most importantly the propensity of HPβCD IC towards rapid renal clearance. In a separate work in a rat model, we observed renal clearance of IC accounting for >50% elimination within the first 10 min after administration (data not shown). Liposomes on the other hand are eliminated through a relatively slower reticuloendothelial route- a metabolic pathway that can be delayed by altering the liposome composition [30]. Potentially, CLEFMA liposomes could be modified to circulate in blood and make drugs bioavailable over a prolonged time without affecting the efficacy of the encapsulated drug. We have previously shown the utility of poly(ethylene glycol) or PEG for prolonging circulation as well as reducing its toxicity [30, 31]. Once encapsulated, the innate pharmacokinetic and metabolic profiles of the liposomes take precedence over that of the drug. Other beneficial features of liposomes are the generic liposome characteristics, such as biodegradability, preferential EPR-dependent accumulation in tumor and potential for targeting.
Although not targeted for delivery, CLEFMA appears to possess selective anti-proliferative activity in cancer cells. Histopatholgical examination of liver lung and kidney showed no toxicities in these tissues. This observation was in agreement with the in vitro observation that CLEFMA was not anti-proliferative to the normal lung fibroblasts. While the fundamental basis of this selectivity towards cancer cells is not clear, we have found CLEFMA to be equally potent in controlling proliferation of other cancer cells, such as pancreatic (MiaPaCa-2 and Panc-1), colon (HCT-116) and prostate (PC-3) cancer cells [4, 24]. In this work, we found that liposome-encapsulated CLEFMA not only retained the potency of free CLEFMA, but was also effective in curtailing the growth of H441 tumor when administered in rats carrying xenografts of human lung adenocarcinoma H441 cells (Figure 3). In 2/6 treated animals, there was complete remission of tumor. These observations were confirmed by PET imaging of FDG accumulation in tumor. Relative to untreated tumors in control rats, the reduced FDG uptake in tumors of CLEFMA liposome-treated rats is a molecular indicator of suppressed tumor growth.
An attractive aspect of CLEFMA liposomes is the ability to radiolabel them with an imageable probe without affecting the availability, kinetics and efficacy of the encapsulated drug [15, 20, 30, 32-34]. In a Critical Path Initiative, the FDA and NCI have emphasized the role of imaging in hastening drug development [35, 36]. Imaging has the ability to provide drug accumulation profile in a non-invasive and longitudinal fashion under physiologic conditions. Indeed, we show this capability of CLEFMA liposomes by imaging their accumulation in various organs and tumor (Figure 4). From the biodistribution studies, it was clear that most of the CLEFMA-liposomes accumulated in liver. In SPECT images, the spleen also appears to carry large amount of administered dose. As a member of reticuloendothelial system, splenic and hepatic macrophages are specialized for particle uptake. These observations are in line with our previous observations in experiments involving liposomes without a PEG-linked lipid in the bilayer [30]. The circulation persistence of these liposomes was also relatively short because of the absence of PEG-lipid.
In summary, we have shown that CLEFMA can be encapsulated inside the liposomes using aqueous HPβCD as a solubilizing agent. The resultant liposomes appear to be stable upon storage under refrigerated conditions. In vitro and in vivo efficacy results of this study, coupled with the absence of any apparent toxicity, imply that CLEFMA has a potential to be developed as a drug against lung cancer. The presented liposome formulation not only carries an anticancer drug, but could also be monitored by imaging using non-invasive SPECT. We believe that the potency of CLEFMA liposomes may be enhanced further by incorporating poly(ethylene glycol)-linked phospholipid in the liposome formulation. The potent efficacy, histopathologic non-toxicity, and selective anti-proliferative action in cancer cells are beneficial outcomes of CLEFMA therapy and warrant further investigations.
RESEARCH HIGHLIGHTS.
CLEFMA is a potent antiproliferative curcuminoid derivative
Liposome containing CLFEMA could be produced using hydroxypropyl-β-cyclodextrin
Intravenously injected CLEFMA liposomes accumulated in reticuloendothelial organs
CLEFMA liposomes suppressed growth of lung cancer xenograft tumor in a rat model
ACKNOWLEDGEMENTS
The authors acknowledge the help from Prachi Vilekar for in vitro cell culture work. Imaging studies were performed in the Small Animal Imaging Facility of the College of Pharmacy, and technical contribution from Sandra Bryant is acknowledged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1]. Available at: http://www.cdc.gov/uscs.
- [2].Burdett SS, Stewart LA, Rydzewska L. Cochrane Database Syst. Rev. 2007 doi: 10.1002/14651858.CD006157.pub2. CD006157. [DOI] [PubMed] [Google Scholar]
- [3].Akerley W, Choy H. Semin. Radiat. Oncol. 1999;9:85. [PubMed] [Google Scholar]
- [4].Lagisetty P, Vilekar P, Sahoo K, Anant S, Awasthi V. Bioorg. Med. Chem. 2010;18:6109. doi: 10.1016/j.bmc.2010.06.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Adams BK, Ferstl EM, Davis MC, Herold M, Kurtkaya S, Camalier RF, Hollingshead MG, Kaur G, Sausville EA, Rickles FR, Snyder JP, Liotta DC, Shoji M. Bioorg. Med. Chem. 2004;12:3871. doi: 10.1016/j.bmc.2004.05.006. [DOI] [PubMed] [Google Scholar]
- [6].Samad A, Sultana Y, Aqil M. Curr. Drug Delivery. 2007;4:297. doi: 10.2174/156720107782151269. [DOI] [PubMed] [Google Scholar]
- [7].Schwendener RA. Advances in Experimental Medicine and Biology. 2007;620:117. doi: 10.1007/978-0-387-76713-0_9. [DOI] [PubMed] [Google Scholar]
- [8].Kaasgaard T, Andresen TL. Expert Opin. Drug Deliv. :7225. doi: 10.1517/17425240903427940. [DOI] [PubMed] [Google Scholar]
- [9].Maeda H, Greish K. J. Fang, Adv. Polym. Sci. 2006;193:103. [Google Scholar]
- [10].Gregoriadis G. In: Stealth Liposomes. Lasic DD, Martin F, editors. CRC Press; Boca Raton, FL: 1995. Chapter 2. [Google Scholar]
- [11].Huwyler J, Drewe J, Krahenbuhl S. Int. J. Nanomedicine. 2008;3:21. [PMC free article] [PubMed] [Google Scholar]
- [12].McCormack B, Gregoriadis G. J. Drug Targeting. 1994;2:449. doi: 10.3109/10611869408996821. [DOI] [PubMed] [Google Scholar]
- [13].Kaur N, Puri R, Jain SK. AAPS PharmSciTech. 2010;11:528. doi: 10.1208/s12249-010-9411-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Higuchi T, Connors K. Adv. Anal. Chem. Instrum. 1965;4:127. [Google Scholar]
- [15].Agashe H, Lagisetty P, Sahoo K, Bourne D, Grady B, Awasthi V. J. Nanopart. Res. 2010 doi: 10.1007/s11051-010-0154-5. In press. DOI 10.1007/s11051-010-0154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Agashe H, Lagisetty P, Awasthi S, Awasthi V. Colloids Surf., B. 2009;75:573. doi: 10.1016/j.colsurfb.2009.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Stewart JC. Anal. Biochem. 1980;104:10. doi: 10.1016/0003-2697(80)90269-9. [DOI] [PubMed] [Google Scholar]
- [18].Landegren U. J. Immunol. Methods. 1984;67:379. doi: 10.1016/0022-1759(84)90477-0. [DOI] [PubMed] [Google Scholar]
- [19].Phillips WT, Rudolph AS, Goins B, Timmons JH, Klipper R, Blumhardt R. Nucl. Med. Biol. 1992;19:539. doi: 10.1016/0883-2897(92)90149-s. [DOI] [PubMed] [Google Scholar]
- [20].Awasthi V, Goins B, McManus L, Klipper R, Phillips WT. Nucl. Med. and Biol. 2003;30:159. doi: 10.1016/s0969-8051(02)00419-5. [DOI] [PubMed] [Google Scholar]
- [21].Awasthi VD, Garcia D, Goins BA, Phillips WT. Int. J. Pharm. 2003;253:121. doi: 10.1016/s0378-5173(02)00703-2. [DOI] [PubMed] [Google Scholar]
- [22].Frank DW. In: Handbook of Laboratory Animal Science. Melby ECJ, editor. CRC Press; Boca Raton, FL: 1976. p. 23. [Google Scholar]
- [23].Petty C. Research Techniques in the Rats. Charles C. Thomas; Springfield, IL: 1982. [Google Scholar]
- [24].Lagisetty P, Powell DR, Awasthi V. J. Mol. Struct. 2009;936:23. [Google Scholar]
- [25].Adams BK, Cai J, Armstrong J, Herold M, Lu YJ, Sun A, Snyder JP, Liotta DC, Jones DP, Shoji M. Anticancer Drugs. 2005;16:263. doi: 10.1097/00001813-200503000-00005. [DOI] [PubMed] [Google Scholar]
- [26].Kasinski AL, Du Y, Thomas SL, Zhao J, Sun SY, Khuri FR, Wang CY, Shoji M, Sun A, Snyder JP, Liotta D, Fu H. Mol. Pharmacol. 2008;74:654. doi: 10.1124/mol.108.046201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Selvendiran K, Tong L, Vishwanath S, Bratasz A, Trigg NJ, Kutala VK, Hideg K, Kuppusamy P. J. Biol. Chem. 2007;282:28609. doi: 10.1074/jbc.M703796200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Subramaniam D, May R, Sureban SM, Lee KB, George R, Kuppusamy P, Ramanujam RP, Hideg K, Dieckgraefe BK, Houchen CW, Anant S. Cancer Res. 2008;68:1962. doi: 10.1158/0008-5472.CAN-07-6011. [DOI] [PubMed] [Google Scholar]
- [29].Sun A, Shoji M, Lu YJ, Liotta DC, Snyder JP. J. Med. Chem. 2006;49:3153. doi: 10.1021/jm051141k. [DOI] [PubMed] [Google Scholar]
- [30].Awasthi VD, Garcia D, Klipper R, Goins BA, Phillips WT. J. Pharmacol. Exp. Ther. 2004;309:241. doi: 10.1124/jpet.103.060228. [DOI] [PubMed] [Google Scholar]
- [31].Awasthi VD, Goins B, Phillips WT. Am. J. Pharmacol. Toxicol. 2007;2:98. [Google Scholar]
- [32].Awasthi V, Goins B, Klipper R, Loredo R, Korvick D, Phillips WT. J. Nucl. Med. 1998;39:1089. [PubMed] [Google Scholar]
- [33].Awasthi VD, Goins B, Klipper R, Phillips WT. J. Drug Targeting. 2002;10:419. doi: 10.1080/1061186021000001878. [DOI] [PubMed] [Google Scholar]
- [34].Awasthi V, Yee SH, Jerabek P, Goins B, Phillips WT. J. Appl. Physiol. 2007;103:28. doi: 10.1152/japplphysiol.00136.2006. [DOI] [PubMed] [Google Scholar]
- [35].Altar CA. Clin. Pharmacol. Ther. 2008;83:361. doi: 10.1038/sj.clpt.6100471. [DOI] [PubMed] [Google Scholar]
- [36].Woodcock J, Woosley R. Ann. Rev. Med. 2008;59:1. doi: 10.1146/annurev.med.59.090506.155819. [DOI] [PubMed] [Google Scholar]
- [37].Schettino C, Bareschino MA, Maione P, Rossi A, Ciardiello F, Gridelli C. Curr. Genomics. 2008;9:252. doi: 10.2174/138920208784533665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Danesi R, Pasqualetti G, Giovannetti E, Crea F, Altavilla G, Del Tacca M, Rosell R. Adv. Drug Delivery Rev. 2009;61:408. doi: 10.1016/j.addr.2009.03.001. [DOI] [PubMed] [Google Scholar]
- [39].Ricciardi S, Tomao S, de Marinis F. Clin. Lung Cancer. 2009;10:28. doi: 10.3816/CLC.2009.n.004. [DOI] [PubMed] [Google Scholar]
- [40].Dempke WC, Suto T, Reck M. Lung Cancer. 2010;67:257. doi: 10.1016/j.lungcan.2009.10.012. [DOI] [PubMed] [Google Scholar]
- [41].Gridelli C, Ardizzoni A, Douillard JY, Hanna N, Manegold C, Perrone F, Pirker R, Rosell R, Shepherd FA, De Petris L, Di Maio M, de Marinis F. Lung Cancer. 2010;68:319. doi: 10.1016/j.lungcan.2009.11.018. [DOI] [PubMed] [Google Scholar]
- [42].Thomas SL, Zhao J, Li Z, Lou B, Du Y, Purcell J, Snyder JP, Khuri FR, Liotta D, Fu H. Biochem. Pharmacol. 2010;80:1309. doi: 10.1016/j.bcp.2010.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Bergmann A. Cell. 2007;131:1032. doi: 10.1016/j.cell.2007.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Meylan E, Dooley AL, Feldser DM, Shen L, Turk E, Ouyang C, Jacks T. Nature. 2009;462:104. doi: 10.1038/nature08462. [DOI] [PMC free article] [PubMed] [Google Scholar]