Abstract
Objectives
The purpose of this pilot study was to investigate a measure of motor sequencing deficit as a potential endophenotype of speech sound disorder (SSD) in a multigenerational family with evidence of familial SSD.
Methods
In a multigenerational family with evidence of a familial motor-based SSD, affectation status and a measure of motor sequencing during oral motor testing were obtained. To further investigate the role of motor sequencing as an endophenotype for genetic studies, parametric and nonparametric linkage analyses were conducted using a genome-wide panel of 404 microsatellites.
Results
In seven of the ten family members with available data, SSD affectation status and motor sequencing status coincided. Linkage analysis revealed four regions of interest, 6p21, 7q32, 7q36, and 8q24, primarily identified with the measure of motor sequencing ability. The 6p21 region overlaps with a locus implicated in rapid alternating naming in a recent genome-wide dyslexia linkage study. The 7q32 locus contains a locus implicated in dyslexia. The 7q36 locus borders on a gene known to affect component traits of language impairment.
Conclusions
Results are consistent with a motor-based endophenotype of SSD that would be informative for genetic studies. The linkage results in this first genome-wide study in a multigenerational family with SSD warrant follow-up in additional families and with fine mapping or next-generation approaches to gene identification.
Keywords: speech sound disorder, multigenerational family, genome-wide linkage analysis, sequential motor speech task, 6p21, 7q32, 7q36, 8q24
Introduction
Speech sound disorder (SSD) is a childhood disorder affecting the ability to produce speech that is easily understood. Children with SSD can have deficits in articulation, phonological processing, and/or cognitive representation of language (Lewis, et al., 2006). SSD is relatively common, although published prevalence rates vary, in part because of differences in the age of the children in research samples, assessment methodology, and diagnostic criteria. SSD was reported for 15.6% in US preschoolers using a criterion of 75% intelligible speech (Campbell, et al., 2003); 3.8% of US 6-year-olds using a cutoff standard score of – 1.14 in a standardized articulation test (Shriberg, Tomblin, & McSweeny, 1999); and 1.1% of Australian school age children in grades Kindergarten through 6 using a four-stage report procedure that included teacher impressions and a speech-language pathologist's report (McKinnon, McLeod, & Reilly, 2007). Because SSD is typically diagnosed and treated in childhood, it is rarely seen in adolescents and adults and prevalence rates in these populations are unknown.
Various subtype systems have been proposed, for instance classifications based on error types (Dodd, 2005) and suspected etiology (Shriberg, et al., 2005). As summarized in recent reviews (Lewis, et al., 2006; Pennington & Bishop, 2009; Peter, 2010), there is strong evidence of a genetic etiology in SSD but causal genes have not yet been identified. If certain genes were found to be associated with SSD, their identification could lead to a biologically defined SSD subtype classification.
SSD is frequently seen in the presence of language impairment, a disorder affecting comprehension or expression of language in the presence of typical development in other areas, and these co-occurrences are greater than those expected under random conditions (Pennington & Bishop, 2009; Shriberg, et al., 1999). The observation that children with SSD and concomitant language impairment are at higher risk for developing reading disorders in the school years compared to children with isolated SSD (Peterson, Pennington, Shriberg, & Boada, 2009) may imply that the latter group represents a qualitatively different disorder subtype.
Evidence for a genetic etiology in SSD is found in early behavioral genetics studies. In a study of 156 adopted and biological children of parents with and without a history of SSD, 25% of children with a familial risk of speech disorders showed signs of disordered speech, compared to 9% of children without such a risk (Felsenfeld & Plomin, 1997). In a study of 32 monozygotic (MZ) and 25 dizygotic (DZ) same-sex twin pairs where one or both members had speech and/or language deficits, concordance rates were .95 in the MZ pairs, compared to .22 in the DZ pairs (Lewis & Thompson, 1992). Another twin study investigating 79 twin pairs where one or both members had speech and/or language deficits showed evidence of high heritability for SSD (h2= .97) (Bishop, 2002). Whether or not affected members of the same family share similar expressions of the disorder regarding speech error patterns has not yet been investigated. Similarly, the mode of inheritance is unknown. In one study of 45 probands with speech and/or language disorders and their families, likelihood-ratio chi-square testing failed to distinguish between a major gene effect and a multifactorial transmission model (Lewis, Cox, & Byard, 1993). To our knowledge, aggregation or segregation studies in families with SSD under exclusion of language impairment have not been attempted.
To date, four genetics studies of SSD (Miscimarra, et al., 2007; Smith, Pennington, Boada, & Shriberg, 2005; Stein, et al., 2006; Stein, et al., 2004) have focused on the molecular genetics of nonsyndromic SSD. Genomic candidate regions were selected as targets for linkage analysis based on their role in other disorders where disordered speech is also frequently observed, for instance dyslexia, autism, Prader-Willi Syndrome, and Angelman Syndrome. In a linkage analysis in children from 77 families, using markers in a dyslexia candidate region on chromosome (chr) 3, 3p14.2-q13.32, linkage signals were obtained for phonological memory, single-word decoding, and nonsense word reading (Stein, et al., 2004). Another dyslexia candidate region, 1p36-p34, was evaluated for linkage in children from 151 families (Miscimarra, et al., 2007). Linkage signals for measures of articulation and listening comprehension were reported. A candidate region not only for dyslexia but also for autism, Prader-Willi Syndrome, and Angelman Syndrome, 15q14-q21 was evaluated for linkage in children from 151 families (Stein, et al., 2006). Linkage signals were reported for SSD affected status, oral motor function, articulation, and phonological memory. Focusing on three candidate regions for dyslexia, 1p36, 6p22, and 15q21, linkage analyses were conducted in a sample of 111 kindergarten children with SSD and 76 siblings (Smith, et al., 2005). Measures of speech and phonological ability were found to be linked to the dyslexia candidate regions on chrs 6 and 15, and suggestive linkage was reported for the region on chr 1. The 6p22 candidate region for dyslexia has been replicated in multiple studies (Cardon, et al., 1994, 1995; Deffenbacher, et al., 2004; Fisher, et al., 1999; Gayan, et al., 1999; Grigorenko, Wood, Meyer, & Pauls, 2000; Kaplan, et al., 2002; Petryshen, et al., 2001). Recently, a new dyslexia candidate region located closer to the centromere, 6p21, was identified in a genome-wide study in a sample of children with dyslexia and their siblings using a rapid naming task with alternating categories of objects and colors but not with single-category naming tasks (Konig, et al., 2011).
In these molecular studies of SSD genetics, the participants were young children with SSD and their siblings. A genome-wide linkage study using multigenerational families with familial SSD had not been attempted prior to the present study. One reason why older children and adults have not been included in genetic studies of SSD may be the fact that they have typically compensated their disorder and overt manifestations of their disorder can no longer be observed. There is new evidence, however, of residual SSD effects in adults. In a study of adults with and without a history of SSD, those with a positive history had greater difficulty with maximum performance tasks such as imitating nonwords, multisyllabic real words, and tongue twisters, compared to adults without a negative SSD history (Lewis, et al., 2007). This suggests that, although conversational speech normalizes in older children and adults with a history of SSD, residual difficulties remain and can be quantified.
The theoretical framework of the molecular studies published to date was the view that SSD is a common complex disorder inherited as a nonmendelian quantitative trait (Smith, et al., 2005; Stein, et al., 2004). This view is consistent with the common disorder/common variant (CDCV) model, where variants in several genes produce an additive or multiplicative effect to confer disease susceptibility in a given child. Examples of diseases associated with common variants with a major contribution to the disease include several types of autoimmune disease (Fernando, et al., 2008) and late onset Alzheimer's disease (Bird, 2005). Examples of diseases where large numbers of common variants, each of small effect, are thought to produce disease susceptibility include schizophrenia and bipolar disorder (Purcell, et al., 2009). The CDCV model may fit some but not all cases of SSD. For common disorders formerly thought to represent complex, nonmendelian disorders, a paradigm shift toward an alternate framework, the common disease/rare variant (CDRV) model, has begun to take place (McClellan & King, 2010). The CDRV model posits that a single rare variant or very few rare variants cause the disease in a given family and that many cases of disease, such as autism (Bucan, et al., 2009) and hearing loss (Dror & Avraham, 2009; Walsh, et al., 2010), can be explained in aggregate by rare variants in individual families.
There is empirical evidence that the CDRV model may be applicable to SSD. First, in one of the molecular studies reviewed above (Stein, et al., 2004), 34 sib pairs contributed to linkage results for both multisyllabic word repetition (MWR) and nonsense word repetition (NWR), skills that are characteristically impaired in SSD. Nine of these sib pairs were concordantly affected (i.e., both twins in each pair had low MWR and NWR scores) and 2 were concordantly unaffected (i.e., both twins in each pair had typical MWR and NWR scores). The concordantly affected group may represent an SSD subtype with a phonological memory component, and the concordantly unaffected group may represent a different SSD subtype without such a phonological memory component. Second, our group recently showed evidence of the first familial SSD subtype in families with SSD (Peter & Raskind, 2011). Of five participating multigenerational families, two (families 002 and 005) showed evidence of deficits in sequential motor tasks, both in motor speech and keyboard tapping tasks. In the motor speech task, participants produced rapid series of the monosyllable /PA/ and the disyllable /PATA/. Mean syllable durations were converted into z scores and the disyllabic z score was subtracted from the monosyllabic z score. A large positive difference was interpreted as a relative deficit in sequential motor performance. This measure was found to be robust against age effects in adulthood within the sample, even though the norms were based on children age 13 years and younger. Similarly, participants rapidly tapped a computer key repetitively with one finger and in a separate task, they rapidly tapped two computer keys with two fingers in an alternating fashion. As norms across the lifespan are only available for the repetitive tapping task, a durational ratio (alternating/repetitive) of the tap intervals was computed and a ratio < 1 in adults was interpreted as a deficit in sequential hand movements. As shown in the published motor speech norms and our own key tapping data, older children and adults produce shorter intervals in alternating tasks, compared to repetitive tasks, consistent with a speed advantage in alternating tasks. Of note, in the two families with evidence of familial motor sequencing deficits, one or more children had previously received a diagnosis of childhood apraxia of speech (CAS), defined by the American Speech-Language-Hearing Association as a neurological speech disorder that affects “planning and/or programming spatiotemporal parameters of movement sequences results in errors in speech sound production and prosody” (http://www.asha.org/docs/html/PS2007-00277.html).
Considering the CDRV model for at least a subset of families with SSD would explain the discrepancy between the high heritability estimates on one hand and the lack of consistent linkage peaks across studies and an unambiguous mode of inheritance on the other. Under the CDRV model, multiplex families with SSD may cluster into several biologically defined subtypes, each with a characteristic genetic etiology and mode of inheritance.
The purpose of this pilot study was to test the CDRV model in SSD further using an approach never before applied to investigating the molecular genetics of SSD, genome-wide linkage analysis, based on a multigenerational family framework. Given that we identified a familial SSD subtype characterized by a deficit in motor sequencing, we asked whether this trait co-segregates with SSD and, hence, could be modeled as an endophenotype. If so, this may validate inclusion of multigenerational families in genetic studies of SSD, confirm previously posited regions of interest, and identify novel regions of interest. This study was part of a larger ongoing project to investigate SSD genetics in multigenerational families with evidence of familial SSD
Method
This project was conducted with the approval of the University of Washington Human Subjects Division. As previously described (Peter & Raskind, 2011), five families (N = 57) participated, with 39 participants completing all or part of a behavioral test battery that consisted of speech, motor speech, language, verbal and nonverbal processing, and hand motor tasks. Motor speech testing was completed by 34 participants and consisted of rapid repetition of monosyllables (/PA, TA, KA/), disyllables (/PATA, TAKA/) and trisyllables /PATAKA/. Following established procedures (Fletcher, 1972), at least 20 repetitions of each monosyllable, 15 repetitions of each disyllable, and 10 repetitions of the trisyllable were collected. For each syllable type, the average syllable duration was calculated using the freely available software Praat (Boersma, 2001), version 5.1.25. First and last syllables in each breath group were discarded from the analysis to avoid nonlinear rate effects. Average syllable durations were converted into z scores using published norms for ages 2;6 through 6;11 (Robbins & Klee, 1987) and 6 through 13 years (Fletcher, 1972). For the purposes of this study, we quantified deficits in sequential movement relative to repetitive movement in each participant by subtracting the combined average z score from the di- and trisyllables from the combined average z score from the monosyllables. To compare the z score differences in Family 002 with those in the other four families in the project sample (Peter & Raskind, 2011), means were calculated for each family, separately for individuals with, and without, a history of SSD.
Linkage power analyses were conducted for two types of affectation scores, SSD and motor sequencing. Assuming a hypothetical marker with four alleles and an autosomal dominant mode of inheritance with a 90% penetration rate for SSD and a 70% penetration rate for the binary measure of motor sequencing deficit, power analyses were run to compute the maximum log odds (LOD) score for each family, using the GENEHUNTER MODSCORE 3.0 software package (Dietter, et al., 2007; Mattheisen, Dietter, Knapp, Baur, & Strauch, 2008). For this family, an autosomal dominant model was assumed because the proband and all three of his siblings were SSD affected, affectation was found in all four generations and both sexes, and there was an affected father-and-son pair. A penetrance of 90% for SSD and 70% for the motor sequencing deficit were estimated because there was one unaffected obligate risk carrier for both traits, the proband's mother, and because there were fewer family members with positive affectation for the motor sequencing deficit, compared to presence or history of SSD.
Each participant donated a saliva sample using Oragene ® kits. The DNA was extracted using standard laboratory methods, following manufacturer instructions. Samples were incubated at 50 °C for at least one hour. Following admixture of the Oragene-DNA purifier, samples were incubated on ice for 10 minutes and centrifuged at room temperature at 13,000 rpm for 10 minutes. The supernatant was mixed with equal volumes of 95% to 100% ethanol and centrifuged at 13,000 rpm for 2 minutes. The supernatant was discarded and the DNA pellet was washed with 70% ethanol. The pellet was dissolved in 100 μl DNA buffer.
Genotyping was provided by the Mammalian Genotyping Service (MGS), Marshfield, WI, using Weber Screening Set 16 with 404 short tandem repeat polymorphism (STRP) markers, of which 375 were on autosomal chromosomes. The absence of genotyping errors was confirmed using PEDSTATS (Wigginton & Abecasis, 2005) and PedCheck (O'Connell & Weeks, 1998). Only the autosomal markers were entered into the analysis. The average marker distance was 10 centimorgans (cM).
Linkage analyses were conducted using the GENEHUNTER MODSCORE 3.0 software package (Dietter, et al., 2007; Mattheisen, et al., 2008). Both parametric and nonparametric algorithms were used. These two approaches have complementary strengths and weaknesses. Parametric linkage algorithms test for linkage under a mendelian model with a specified mode of inheritance, for instance autosomal dominant or recessive. Parametric linkage analysis is a full likelihood method, consistent with a high level of efficiency, but it requires accurate specification of the inheritance model. Inaccurate assumptions regarding the model of inheritance reduce power to detect linkage. Nonparametric linkage algorithms test for linkage without a mendelian model. They are robust against model misspecifications but less efficient than parametric linkage analysis and also sensitive to pedigree ascertainment biases.
For this exploratory study, the quantitative measure was converted into a binary affectation score. A z score difference > 1 was used to assign positive affectation status with respect to the motor sequencing measure and a negative affectation status was assigned for all other z score differences.
Traditionally, the critical LOD score required to declare linkage at p = .05 is 3.00 for genome-wide linkage analyses, 2.00 for single-chromosome linkage analyses, and 1.00 for single locus linkage analysis or to define a region of interest in a genome-wide analysis (Ott, 1999). Regions of interest were defined as LOD scores > 1.00 and/or nonparametric LOD (NPL) scores > 2.14, as NPL2/4.6 evaluates to a magnitude equivalent to that on the LOD scale (Nyholt, 2000).
Results
Linkage power analyses predicted that, of the five families in the sample, family 002 would provide the highest maximum LOD score, 2.37, for SSD affectation and also the highest maximum LOD score, 1.78, for the motor sequencing measure, followed by family 005 with predicted maximum LOD scores of 1.52 and .79 for SSD affectation and the motor sequencing measure, respectively. Predicted maximum LOD scores of 1.22, .86, and 1.10 were obtained for SSD affection for families 001, 003, and 004, respectively, whereas the motor sequencing measure was uninformative (predicted maximum LOD scores of 0.00). As reported in our previous study (Peter & Raskind, 2011), only families 002 and 005 showed evidence of a motor-based subtype. Based on these considerations, family 002 was selected for a genome-wide linkage analysis.
Family 002 consisted of 21 members, 11 of whom participated in the behavioral testing and donated DNA. Four generations were represented. As previously described (Peter & Raskind, 2011), the proband child was a boy, code 2503, age 8;4 (years;months), with a history of a severe SSD. He had not formally undergone testing for the presence of CAS. His two younger brothers, code 2504, and age 6;5 and code 2505, age 5.5, also had histories of severe SSD and both had received a diagnosis of CAS based on informal measures when they were toddlers. Their younger sister, age 3, also had a suspected SSD but she did not participate in the study. Neither their father, code 2404, nor mother, code 2405, reported speech difficulties in childhood, but the mother's mother, code 2303, reported severe childhood speech difficulties for herself and also for her brother, who did not participate in the study. This brother had a son, code 2401, who reported difficulties with speech in childhood. One of his two children, a girl, code 2502, age 3;7, showed evidence of SSD, whereas the other, a boy, code 2501, age 7;0, did not. The proband's maternal great-grandfather, who was deceased, had a sister, code 2203, who reported speech difficulties in childhood. In total, seven of the 11 active participants had observed or reported SSD histories while the remaining four did not. Figure 1 shows a pedigree graph of the family where SSD affectation is indicated based on clinical evidence, self-report, or report by a family member.
Figure 1.
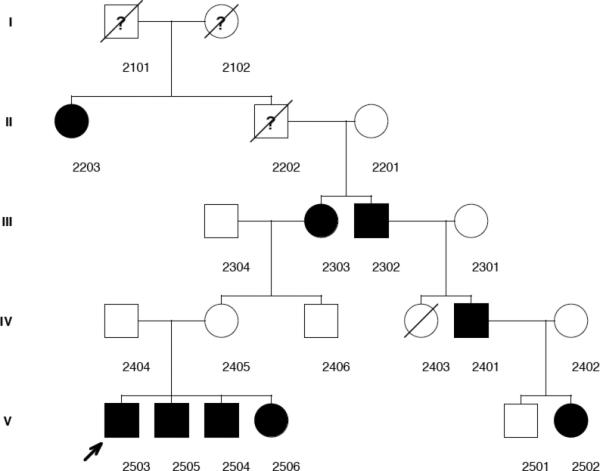
Pedigree graph of family 002
Note: Circles = females, squares = males, filled shapes = SSD affected, open shapes = SSD unaffected, ? = SSD affectation status unknown, / = deceased, arrow = proband child.
Affectation for the motor sequencing measure was applied to the proband child, code 2503, his 6-year-old brother, code 2504, his maternal grandmother, code 2303, his mother's cousin, code 2401, and his great-grandmother, code 2201. For one participant, code 2502, z scores for the multisyllabic repetition task could not be calculated due to her young age. The remaining five participants were labeled as unaffected with respect to the motor sequencing measure. In seven participants, affectation status and motor sequencing status coincided, four with positive affectation and three with negative affectation. One participant without a reported SSD history showed evidence of motor sequencing difficulty and two participants with a history of SSD did not show evidence of motor sequencing difficulty. As previously described in detail (Peter & Raskind, 2011), the project sample contains affected individuals with high z score differences in families 002 and 005, but several high z score differences were also observed among the SSD unaffecteds in these two families. Table 1 summarizes SSD affectation status and the z score difference from the monosyllabic and multisyllabic repetition task for the 11 active participants. Table 2 shows mean scores for the z score difference from the two syllable repetition tasks for all five families in the sample, separately for individuals with, and without, a history of SSD.
Table 1.
Summary of SSD affectation and z score difference between the average z scores for monosyllables and multisyllables
| Code | Father Code | Mother Code | Sex | SSD Affected | Z Score Difference (Monosyllable – Multisyllable) |
|---|---|---|---|---|---|
| 2201 | 0 | 0 | F | No | 1.35 |
| 2203 | 2101 | 2102 | F | Yes | 0.70 |
| 2303 | 2202 | 2201 | F | Yes | 2.09 |
| 2401 | 2302 | 2301 | M | Yes | 2.18 |
| 2404 | 0 | 0 | M | No | -0.18 |
| 2405 | 2304 | 2303 | F | No | 0.52 |
| 2501 | 2401 | 2402 | M | No | 0.75 |
| 2502 | 2401 | 2402 | F | Yes | N/A |
| 2503 | 2404 | 2405 | M | Yes | 1.00 |
| 2504 | 2404 | 2405 | M | Yes | 1.18 |
| 2505 | 2404 | 2405 | M | Yes | 0.73 |
Note: Founder status is coded as 0.
Table 2.
Mean z score difference by family and SSD affectation
| Family | Affected | Unaffected | ||
|---|---|---|---|---|
| Mean | SD | Mean | SD | |
| 001 | -0.29 | 0.44 | 0.64* | N/A |
| 002 | 1.31 | 0.66 | 0.61 | 0.63 |
| 003 | 0.05 | 0.11 | 0.21 | 0.99 |
| 004 | -0.04* | N/A | 0.31 | 0.93 |
| 005 | 0.69 | 0.52 | 0.54 | 0.56 |
Note:
denotes a single observation.
Linkage analyses revealed four regions that met the criteria for regions of interest, 6p21, 7q32, 7q36, and 8q24. All regions of interest were obtained with the binary measure of motor sequencing ability and, in the case of 7q31.q32-34, also with SSD affectation. Table 3 summarizes the regions of interest in terms of the two binary phenotypes, peak LOD and NPL scores, band location, and cM position (all cM positions according to the Marshfield map) of the regions’ boundary markers exceeding the critical LOD or NPL values. Figure 2 shows the LOD and NPL scores for both traits by marker position for chr 6. Figures 3 and 4 show the analogous data for chr 7 and chr 8, respectively.
Table 3.
Peak LOD and NPL scores in regions of interest (cM according to the Marshfield map) by chromosome and trait
| Band | SSD | Motor Sequencing | ||||||
|---|---|---|---|---|---|---|---|---|
| LOD | Position (cM) | NPL | Position (cM) | LOD | Position (cM) | NPL | Position (cM) | |
| 6p21 | < 0.00 | N/A | 0.26 | N/A | 1.10 | 60.60 – 67.00 | 2.90 | 58.20 – 69.00 |
| 7q32 | < 0.00 | N/A | 1.72 | N/A | 0.66 | N/A | 3.51 | 133.40 – 147.32 |
| 7q36 | < 0.00 | N/A | 2.53 | 163.00 – q terminal | 1.35 | 161.4 – q terminal | 4.61 | 158.2 – q terminal |
| 8q24 | < 0.00 | N/A | 1.43 | N/A | 0.57 | N/A | 2.97 | 137.00 – 142.80 |
Figure 2.
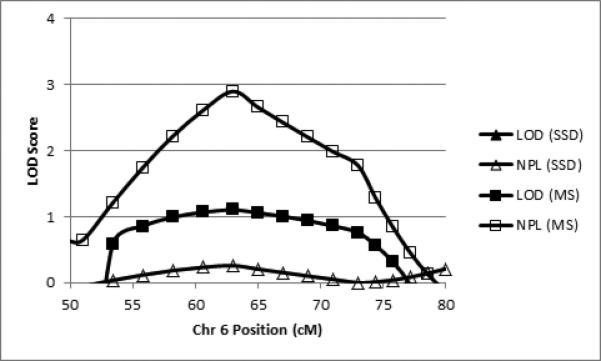
Parametric LOD scores (filled shapes) and NPL scores (open shapes) for SSD (triangles) and motor sequencing (MS; squares) for the linkage peak on chromosome 6 (cM according to the Marshfield map).
Figure 3.
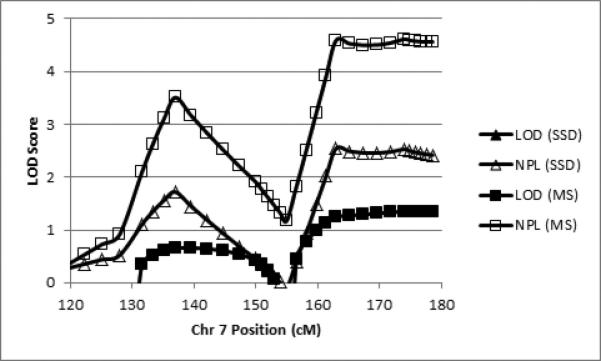
Parametric LOD scores (filled shapes) and NPL scores (open shapes) for SSD (triangles) and motor sequencing (MS; squares) for the two linkage peaks on chromosome 7 (cM according to the Marshfield map).
Figure 4.
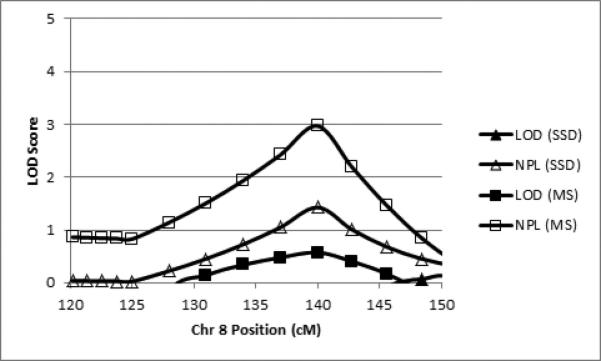
Parametric LOD scores (filled shapes) and NPL scores (open shapes) for SSD (triangles) and motor sequencing (MS; squares) for the linkage peak on chromosome 8 (cM according to the Marshfield map).
Discussion
Based on our previous observation that two of five families with SSD had a demonstrable motor sequencing deficit in a set of motor speech tasks, we investigated the hypothesis that this deficit might define a familial motor-based SSD subtype and would be an informative endophenotype for genetic studies. Such an endophenotype that is observable in adults as well as children would allow inclusion of multigenerational families in linkage analyses, an approach that offers greater power to detect linkage compared to linkage analyses in sib pairs (Wijsman & Amos, 1997). The results from this exploratory study support this hypothesis. Power analyses estimated that, of the five families in our sample, only the two families with a demonstrated motor sequencing deficit would provide LOD scores high enough for suggestive evidence of linkage for a categorical affectation status. Only family 002 was predicted to have the power to detect a region of interest (LOD score > 1.00) with respect to the motor sequencing deficit and was therefore selected for further study. In seven of ten participants with available data in this family, SSD affectation and affectation for motor sequencing ability coincided. The relationship between these two traits is discussed in more detail at the end of this section.
The results from the genome-wide linkage analysis, the first of its kind in SSD, provided further evidence of a motor sequencing endophenotype. All four of the detected regions of interest were identified with the measure of sequential movement ability, whereas only one region, 7q36, was also identified with SSD affectation status. This implies that motor sequencing deficit is a quantifiable residual in individuals with a proposed motor-based form of SSD that may be useful for clinical diagnostics and research purposes. In addition, two of the four regions of interest not only produced sufficiently high NPL scores (> 2.14) to suggest linkage but also LOD scores that met the statistical criteria for a linkage signal of interest (>1.00). This implies that the assumed parameters of autosomal dominant mode of inheritance and selected penetration rates were appropriate for the trait under study, as incorrectly specified models decrease the likelihood of obtaining a linkage signal.
The 6p21 region of interest identified with the parametric and nonparametric linkage analysis using the motor sequencing measure is centromeric to a previously described dyslexia candidate region, DYX2 (Cardon, et al., 1994, 1995; Deffenbacher, et al., 2004; Fisher, et al., 1999; Gayan, et al., 1999; Grigorenko, et al., 2000; Kaplan, et al., 2002; Petryshen, et al., 2001) that was also implicated in children with SSD with respect to measures of speech production and nonword imitation (Smith, et al., 2005). The 6p21 region of interest that resulted from our linkage analysis overlaps with a locus that was recently identified with a genome-wide linkage analysis in a sample of children with dyslexia and their siblings (Konig, et al., 2011). In that study, a rapid naming task with alternating semantic categories was used as the input trait, and its risk interval (44.96 cM – 60.44 cM Marshfield; peak at 60.44 cM) substantially overlaps with our linkage signal (58.2 cM – 69 cM Marshfield). Of note, in the Konig et al. (2011) study, only the alternating naming task produced the signal, whereas the naming tasks without category switches and other dyslexia component traits did not show evidence of linkage. It is possible that the motor sequencing measure developed for the present project captures an endophenotype that affects sequential processing in general. If so, and if influenced by a gene on 6p21, this endophenotype could explain the strong evidence for linkage to a rapid naming task with alternating semantic categories in a sample of children with dyslexia, as the rapid alternating naming task and the measure of motor sequencing in our study both require rapid sequencing of alternating targets.
The 7q32 region of interest contains a locus that has been implicated in two studies investigating measures of written language. In a genome-wide linkage analysis in 11 families with familial dyslexia (Kaminen, et al., 2003), dyslexia affectation was found to be linked to D7S530 at 134.55 cM (Marshfield), which falls into our 7q32 region of interest ranging from 133.40 cM to 147.32 cM with a peak NPL at 137 cM. This marker was also implicated in a genome-wide linkage analysis in 403 families of twins (Bates, et al., 2007) using nonword spelling and irregular spelling as inputs.
The 7q36 region of interest borders on a gene shown to influence common forms of language impairment (Vernes, et al., 2008). As described in recent reviews (Pennington & Bishop, 2009; Weber-Fox, Leonard, Wray, & Tomblin, 2010), children with language impairment show slowed processing speeds in a broad variety of tasks, compared to typical controls, including auditory discrimination, linguistic processing, nonlinguistic processing, and motor tasks (Leonard, et al., 2007; Schul, Stiles, Wulfeck, & Townsend, 2004; Windsor & Hwang, 1999; Windsor, Milbrath, Carney, & Rakowski, 2001). Our findings suggest linkage of a motor sequencing deficit to this general gene region. This trait was defined as substantially slower alternating movement speeds, compared to repetitive movement speeds. It remains to be investigated whether this measure distinguishes between individuals with and without language impairment and also whether CNTNAP2 is a candidate gene for the family described in this study.
To date, chromosome 8 has not been widely implicated in SSD, language impairment, or dyslexia. This region of interest should be further investigated in additional families and with more densely spaced markers.
The fact that the motor sequencing trait was more effective in detecting linkage than SSD affectation leads to questions regarding the relationship between reported SSD histories and performance on the motor speech tasks. Of the three participants with a history of SSD who were not coded as affected with motor sequencing deficits, one was too young for published norms on multisyllable repetitions. Another participant, code 2203, reported childhood speech difficulties but did not meet criteria for affectation with respect to the motor sequencing measure. It is possible that this participant had a different type of speech sound disorder than others in the family, which would constitute a phenocopy. One of the proband's brothers had SSD but missed the cutoff for affectation with motor sequencing deficit by .27 standard deviations. Conversely, one participant, code 2201, the proband's great-grandmother, denied childhood speech difficulties but met criteria as affected with respect to the motor sequencing measure. It is possible that this participant did have speech difficulties but that they went undiagnosed in her childhood. Alternatively, it is possible that she never had SSD but had the motor-based endophenotype. The proband's mother did not show evidence of either the SSD affectation or motor sequencing deficit traits, which would be consistent with nonpenetrance if her carrier status of a risk genotype could be confirmed. The observation that in the two families with evidence of a motor-based SSD subtype, several individuals without reported SSD affectation showed evidence of motor sequencing difficulties whereas this pattern was not observed in the other three families (Table 2) is consistent with patterns of endophenotype expression in other disorders. For instance, unaffected relatives of individuals with schizophrenia show evidence of a neurophysiologic marker of the disease, the failure to suppress an evoked potential during a repeat presentation, at higher rates than unrelated controls (Waldo, Myles-Worsley, Madison, Byerley, & Freedman, 1995). The elevated rates of motor sequencing deficits in the unaffected members of the two families with the motor-based SSD subtype further strengthens our hypothesis of a motor sequencing endophenotype.
Conclusions and Future Studies
Prior to this study, adoption and twin studies and heritability estimates of SSD had produced results that were consistent with a genetic etiology in general. Molecular studies had obtained evidence of linkage to candidate regions selected because of linkage to other disorders that are frequently characterized by impaired speech ability. These studies included children from many families and criteria for SSD affectation were broad. The mode of inheritance was unknown, perhaps in part due to the fact that the samples contained mixed genetic etiologies. Together with the results from our phenotypic study (Peter & Raskind, 2011), the results from this exploratory linkage study in a multigenerational family with SSD support the hypothesis that a deficit in motor sequencing constitutes a motor-based SSD subtype of genetic etiology. A familial subtype with a distinct genetic etiology would be consistent with the CDRV hypothesis for SSD, a hypothesis that should be tested further in additional SSD families. Whereas the mode of inheritance had not been determined in samples containing many different families, the SSD subtype in the family selected for this study appeared to follow an autosomal dominant mode of inheritance. In terms of methodology, our results show that, given a trait that is quantifiable in children as well as adults, it is appropriate to use a genome-wide approach within the framework of a multigenerational family to study the molecular genetics of SSD. This finding thus adds an important methodological insight to the literature on molecular SSD studies to date.
The limitations of our study include the use of binary rather than quantitative measures, a small sample, and a single presumed SSD subtype. The spacing of the markers may not be sufficiently dense to detect chromosomal regions in linkage disequilibrium with the causative variant. Due to the limited size of the family, fine mapping was not possible. Future studies should extend our findings in additional families with a motor-based SSD subtype, using quantitative traits and more densely spaced markers for fine mapping and gene discovery in the regions of interest presented here. They should also identify other familial SSD subtypes and apply a genome-wide approach towards the goal of discovering additional genomic regions of interest and causal genes. Validation of the CDRV model motivates use of other gene identification approaches such as those based on next-generation exome and genome sequencing.
Acknowledgments
The authors thank the families whose participation made this study possible. Many thanks to the following undergraduate and graduate students for their assistance with the data collection and analyses: Leah Anderson, Lynn Bak, Yayin Chen, Erica Gonzales, Mariya Legesse, Amelie Lehmkühler, Jonathan Mahaffie, David Ramm, Elizabeth Sun, and Nancy Yuan. The authors gratefully acknowledge the following funding sources: American Speech-Language-Hearing Foundation New Century Scholars Research Grant (B. Peter), NIDCD T32DC00033 (B. Peter), NIDCD 1R03DC010886 (B. Peter), and R01HD054562 (W. H. Raskind).
Grants: NIDCD T32DC00033 (B. Peter) NIDCD 1R03DC010886-01A1 (B. Peter) American Speech-Language-Hearing Foundation New Century Scholars Research Grant (B. Peter), R01HD054562 (W. H. Raskind).
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bates TC, Luciano M, Castles A, Coltheart M, Wright MJ, Martin NG. Replication of reported linkages for dyslexia and spelling and suggestive evidence for novel regions on chromosomes 4 and 17. Eur J Hum Genet. 2007;15(2):194–203. doi: 10.1038/sj.ejhg.5201739. [DOI] [PubMed] [Google Scholar]
- Bird TD. Genetic factors in Alzheimer's disease. N Engl J Med. 2005;352(9):862–864. doi: 10.1056/NEJMp058027. [DOI] [PubMed] [Google Scholar]
- Bishop DV. Motor immaturity and specific speech and language impairment: evidence for a common genetic basis. Am J Med Genet. 2002;114(1):56–63. doi: 10.1002/ajmg.1630. [DOI] [PubMed] [Google Scholar]
- Boersma P. Praat, a system for doing phonetics by computer. Glot International. 2001;5(9/10):341–345. [Google Scholar]
- Bucan M, Abrahams BS, Wang K, Glessner JT, Herman EI, Sonnenblick LI, et al. Genome-wide analyses of exonic copy number variants in a family-based study point to novel autism susceptibility genes. PLoS Genet. 2009;5(6):e1000536. doi: 10.1371/journal.pgen.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell TF, Dollaghan CA, Rockette HE, Paradise JL, Feldman HM, Shriberg LD, et al. Risk factors for speech delay of unknown origin in 3-year-old children. Child Dev. 2003;74(2):346–357. doi: 10.1111/1467-8624.7402002. [DOI] [PubMed] [Google Scholar]
- Cardon LR, Smith SD, Fulker DW, Kimberling WJ, Pennington BF, DeFries JC. Quantitative trait locus for reading disability on chromosome 6. Science. 1994;266(5183):276–279. doi: 10.1126/science.7939663. [DOI] [PubMed] [Google Scholar]
- Cardon LR, Smith SD, Fulker DW, Kimberling WJ, Pennington BF, DeFries JC. Quantitative trait locus for reading disability: correction. Science. 1995;268(5217):1553. doi: 10.1126/science.7777847. [DOI] [PubMed] [Google Scholar]
- Deffenbacher KE, Kenyon JB, Hoover DM, Olson RK, Pennington BF, DeFries JC, et al. Refinement of the 6p21.3 quantitative trait locus influencing dyslexia: linkage and association analyses. Hum Genet. 2004;115(2):128–138. doi: 10.1007/s00439-004-1126-6. [DOI] [PubMed] [Google Scholar]
- Dietter J, Mattheisen M, Furst R, Ruschendorf F, Wienker TF, Strauch K. Linkage analysis using sex-specific recombination fractions with GENEHUNTER-MODSCORE. Bioinformatics. 2007;23(1):64–70. doi: 10.1093/bioinformatics/btl539. [DOI] [PubMed] [Google Scholar]
- Dodd B. Differential diagnosis and treatment of children with speech disorder. 2nd ed. Whurr; London: Philadelphia: 2005. [Google Scholar]
- Dror AA, Avraham KB. Hearing loss: mechanisms revealed by genetics and cell biology. Annu Rev Genet. 2009;43:411–437. doi: 10.1146/annurev-genet-102108-134135. [DOI] [PubMed] [Google Scholar]
- Felsenfeld S, Plomin R. Epidemiological and offspring analyses of developmental speech disorders using data from the Colorado Adoption Project. J Speech Lang Hear Res. 1997;40(4):778–791. doi: 10.1044/jslhr.4004.778. [DOI] [PubMed] [Google Scholar]
- Fernando MM, Stevens CR, Walsh EC, De Jager PL, Goyette P, Plenge RM, et al. Defining the role of the MHC in autoimmunity: a review and pooled analysis. PLoS Genet. 2008;4(4):e1000024. doi: 10.1371/journal.pgen.1000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher SE, Marlow AJ, Lamb J, Maestrini E, Williams DF, Richardson AJ, et al. A quantitative-trait locus on chromosome 6p influences different aspects of developmental dyslexia. Am J Hum Genet. 1999;64(1):146–156. doi: 10.1086/302190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher SG. Time-by-count measurement of diadochokinetic syllable rate. J Speech Hear Res. 1972;15(4):763–770. doi: 10.1044/jshr.1504.763. [DOI] [PubMed] [Google Scholar]
- Gayan J, Smith SD, Cherny SS, Cardon LR, Fulker DW, Brower AM, et al. Quantitative-trait locus for specific language and reading deficits on chromosome 6p. Am J Hum Genet. 1999;64(1):157–164. doi: 10.1086/302191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigorenko EL, Wood FB, Meyer MS, Pauls DL. Chromosome 6p influences on different dyslexia-related cognitive processes: further confirmation. Am J Hum Genet. 2000;66(2):715–723. doi: 10.1086/302755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminen N, Hannula-Jouppi K, Kestila M, Lahermo P, Muller K, Kaaranen M, et al. A genome scan for developmental dyslexia confirms linkage to chromosome 2p11 and suggests a new locus on 7q32. J Med Genet. 2003;40(5):340–345. doi: 10.1136/jmg.40.5.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan DE, Gayan J, Ahn J, Won TW, Pauls D, Olson RK, et al. Evidence for linkage and association with reading disability on 6p21.3-22. Am J Hum Genet. 2002;70(5):1287–1298. doi: 10.1086/340449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig IR, Schumacher J, Hoffmann P, Kleensang A, Ludwig KU, Grimm T, et al. Mapping for dyslexia and related cognitive trait loci provides strong evidence for further risk genes on chromosome 6p21. Am J Med Genet B Neuropsychiatr Genet. 2011;156B(1):36–43. doi: 10.1002/ajmg.b.31135. [DOI] [PubMed] [Google Scholar]
- Leonard LB, Ellis Weismer S, Miller CA, Francis DJ, Tomblin JB, Kail RV. Speed of processing, working memory, and language impairment in children. J Speech Lang Hear Res. 2007;50(2):408–428. doi: 10.1044/1092-4388(2007/029). [DOI] [PubMed] [Google Scholar]
- Lewis BA, Cox NJ, Byard PJ. Segregation analysis of speech and language disorders. Behav Genet. 1993;23(3):291–297. doi: 10.1007/BF01082469. [DOI] [PubMed] [Google Scholar]
- Lewis BA, Freebairn LA, Hansen AJ, Miscimarra L, Iyengar SK, Taylor HG. Speech and language skills of parents of children with speech sound disorders. Am J Speech Lang Pathol. 2007;16(2):108–118. doi: 10.1044/1058-0360(2007/015). [DOI] [PubMed] [Google Scholar]
- Lewis BA, Shriberg LD, Freebairn LA, Hansen AJ, Stein CM, Taylor HG, et al. The genetic bases of speech sound disorders: evidence from spoken and written language. J Speech Lang Hear Res. 2006;49(6):1294–1312. doi: 10.1044/1092-4388(2006/093). [DOI] [PubMed] [Google Scholar]
- Lewis BA, Thompson LA. A study of developmental speech and language disorders in twins. J Speech Hear Res. 1992;35(5):1086–1094. doi: 10.1044/jshr.3505.1086. [DOI] [PubMed] [Google Scholar]
- Mattheisen M, Dietter J, Knapp M, Baur MP, Strauch K. Inferential testing for linkage with GENEHUNTER-MODSCORE: the impact of the pedigree structure on the null distribution of multipoint MOD scores. Genet Epidemiol. 2008;32(1):73–83. doi: 10.1002/gepi.20264. [DOI] [PubMed] [Google Scholar]
- McClellan J, King MC. Genetic heterogeneity in human disease. Cell. 2010;141(2):210–217. doi: 10.1016/j.cell.2010.03.032. [DOI] [PubMed] [Google Scholar]
- McKinnon DH, McLeod S, Reilly S. The prevalence of stuttering, voice, and speech-sound disorders in primary school students in Australia. Lang Speech Hear Serv Sch. 2007;38(1):5–15. doi: 10.1044/0161-1461(2007/002). [DOI] [PubMed] [Google Scholar]
- Miscimarra L, Stein C, Millard C, Kluge A, Cartier K, Freebairn L, et al. Further evidence of pleiotropy influencing speech and language: analysis of the DYX8 region. Hum Hered. 2007;63(1):47–58. doi: 10.1159/000098727. [DOI] [PubMed] [Google Scholar]
- Nyholt DR. All LODs are not created equal. Am J Hum Genet. 2000;67(2):282–288. doi: 10.1086/303029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63(1):259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott J. Analysis of human genetic linkage. 3rd ed. Johns Hopkins University Press; Baltimore: 1999. [Google Scholar]
- Pennington BF, Bishop DV. Relations among speech, language, and reading disorders. Annu Rev Psychol. 2009;60:283–306. doi: 10.1146/annurev.psych.60.110707.163548. [DOI] [PubMed] [Google Scholar]
- Peter B. New frontiers in understanding speech sound disorder: Unraveling the mysteries of genetic causes. In: Harrison AE, editor. Speech disorders: Causes, treatment and social effect. Nova SciencePublishers; New York: 2010. pp. 119–137. [Google Scholar]
- Peter B, Raskind WH. Evidence for a familial speech sound disorder subtype in a multigenerational study of oral and hand motor sequencing ability. Topics in Language Disorders. 2011;31(2):145–167. doi: 10.1097/TLD.0b013e318217b855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson RL, Pennington BF, Shriberg LD, Boada R. What influences literacy outcome in children with speech sound disorder? J Speech Lang Hear Res. 2009;52(5):1175–1188. doi: 10.1044/1092-4388(2009/08-0024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petryshen TL, Kaplan BJ, Fu Liu M, de French NS, Tobias R, Hughes ML, et al. Evidence for a susceptibility locus on chromosome 6q influencing phonological coding dyslexia. Am J Med Genet. 2001;105(6):507–517. doi: 10.1002/ajmg.1475. [DOI] [PubMed] [Google Scholar]
- Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC, Sullivan PF, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460(7256):748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins J, Klee T. Clinical assessment of oropharyngeal motor development in young children. J Speech Hear Disord. 1987;52(3):271–277. doi: 10.1044/jshd.5203.271. [DOI] [PubMed] [Google Scholar]
- Schul R, Stiles J, Wulfeck B, Townsend J. How ‘generalized’ is the ‘slowed processing’ in SLI? The case of visuospatial attentional orienting. Neuropsychologia. 2004;42(5):661–671. doi: 10.1016/j.neuropsychologia.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Shriberg LD, Lewis BA, Tomblin JB, McSweeny JL, Karlsson HB, Scheer AR. Toward diagnostic and phenotype markers for genetically transmitted speech delay. J Speech Lang Hear Res. 2005;48(4):834–852. doi: 10.1044/1092-4388(2005/058). [DOI] [PubMed] [Google Scholar]
- Shriberg LD, Tomblin JB, McSweeny JL. Prevalence of speech delay in 6-year-old children and comorbidity with language impairment. J Speech Lang Hear Res. 1999;42(6):1461–1481. doi: 10.1044/jslhr.4206.1461. [DOI] [PubMed] [Google Scholar]
- Smith SD, Pennington BF, Boada R, Shriberg LD. Linkage of speech sound disorder to reading disability loci. J Child Psychol Psychiatry. 2005;46(10):1057–1066. doi: 10.1111/j.1469-7610.2005.01534.x. [DOI] [PubMed] [Google Scholar]
- Stein CM, Millard C, Kluge A, Miscimarra LE, Cartier KC, Freebairn LA, et al. Speech sound disorder influenced by a locus in 15q14 region. Behav Genet. 2006;36(6):858–868. doi: 10.1007/s10519-006-9090-7. [DOI] [PubMed] [Google Scholar]
- Stein CM, Schick JH, Gerry Taylor H, Shriberg LD, Millard C, Kundtz-Kluge A, et al. Pleiotropic effects of a chromosome 3 locus on speech-sound disorder and reading. Am J Hum Genet. 2004;74(2):283–297. doi: 10.1086/381562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernes SC, Newbury DF, Abrahams BS, Winchester L, Nicod J, Groszer M, et al. A functional genetic link between distinct developmental language disorders. N Engl J Med. 2008;359(22):2337–2345. doi: 10.1056/NEJMoa0802828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldo M, Myles-Worsley M, Madison A, Byerley W, Freedman R. Sensory gating deficits in parents of schizophrenics. Am J Med Genet. 1995;60(6):506–511. doi: 10.1002/ajmg.1320600605. [DOI] [PubMed] [Google Scholar]
- Walsh T, Shahin H, Elkan-Miller T, Lee MK, Thornton AM, Roeb W, et al. Whole exome sequencing and homozygosity mapping identify mutation in the cell polarity protein GPSM2 as the cause of nonsyndromic hearing loss DFNB82. Am J Hum Genet. 2010;87(1):90–94. doi: 10.1016/j.ajhg.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber-Fox C, Leonard LB, Wray AH, Tomblin JB. Electrophysiological correlates of rapid auditory and linguistic processing in adolescents with specific language impairment. Brain Lang. 2010;115(3):162–181. doi: 10.1016/j.bandl.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigginton JE, Abecasis GR. PEDSTATS: descriptive statistics, graphics and quality assessment for gene mapping data. Bioinformatics. 2005;21(16):3445–3447. doi: 10.1093/bioinformatics/bti529. [DOI] [PubMed] [Google Scholar]
- Wijsman EM, Amos CI. Genetic analysis of simulated oligogenic traits in nuclear and extended pedigrees: summary of GAW10 contributions. Genet Epidemiol. 1997;14(6):719–735. doi: 10.1002/(SICI)1098-2272(1997)14:6<719::AID-GEPI28>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Windsor J, Hwang M. Testing the generalized slowing hypothesis in specific language impairment. J Speech Lang Hear Res. 1999;42(5):1205–1218. doi: 10.1044/jslhr.4205.1205. [DOI] [PubMed] [Google Scholar]
- Windsor J, Milbrath RL, Carney EJ, Rakowski SE. General slowing in language impairment: methodological considerations in testing the hypothesis. J Speech Lang Hear Res. 2001;44(2):446–461. doi: 10.1044/1092-4388(2001/036). [DOI] [PubMed] [Google Scholar]


