Abstract
The correct number and shape of teeth are critical factors for an aesthetic and functional dentition. Understanding the molecular mechanisms regulating tooth number and shape are therefore important in orthodontics. Mice have only one incisor and three molars in each jaw quadrant that are divided by a tooth-less region, the diastema. Although mice lost teeth in the diastema during evolution, the remnants of the evolutionary lost teeth are observed as transient epithelial buds in the wild-type diastema during early stages of development. Shh and Fgf signaling pathways that are essential for tooth development have been shown to be repressed in the diastema. It remains unclear however how Wnt signaling, that is also required for tooth development, is regulated in the diastema. In this study we found that in the embryonic diastema, Wnt5a expression was observed in mesenchyme, whereas Wnt4 and Wnt10b were expressed in epithelium. The expression of Wnt6 and Wnt11 was found in both tissues. The Wnt co-receptor, Lrp6, was weakly expressed in the diastema overlapping with weak Lrp4 expression, a co-receptor that inhibits Wnt signaling. Secreted Wnt inihibitors Dkk1, Dkk2, and Dkk3 were also expressed in the diastema. Lrp4 mutant mice develop supernumerary teeth in the diastema that is accompanied by upregulation of Wnt signaling and Lrp6 expression. Wnt signaling is thus usually attenuated in the diastema by these secreted and membrane bound Wnt inhibitors.
Introduction
An abnormal dentition could be caused by many factors such as micrognathia, macrognathia, abnormal tooth shape including microdontia and macrodontia, and abnormal tooth number including fused teeth, missing teeth, and extra teeth. The correct position, shape, and number of teeth are thus vital factors for establishment of aesthetic and functional dentitions. Since the number and shape of teeth are genetically determined, understanding the molecular mechanisms regulating tooth number and shape is of fundamental importance to orthodontics.
Teeth develop through sequential and reciprocal interactions between oral epithelium and neural crest-derived mesenchyme. The first morphological sign of tooth development is a narrow band of thickened epithelium on the developing jaw primodia. The thickened epithelium progressively takes the form of the bud, cap, and bell configurations as differentiation proceeds. Subsequently, epithelial cells and mesenchymal cells (dental papilla) differentiate into enamel-producing ameloblasts and dentin-producing odontoblasts, respectively. It is known that many signaling pathways such as Bmp, Fgf, Wnt, and Shh play critical roles in regulating tooth position, number, and shape (Thesleff and Sharpe, 1997; Thesleff and Åberg, 1999)
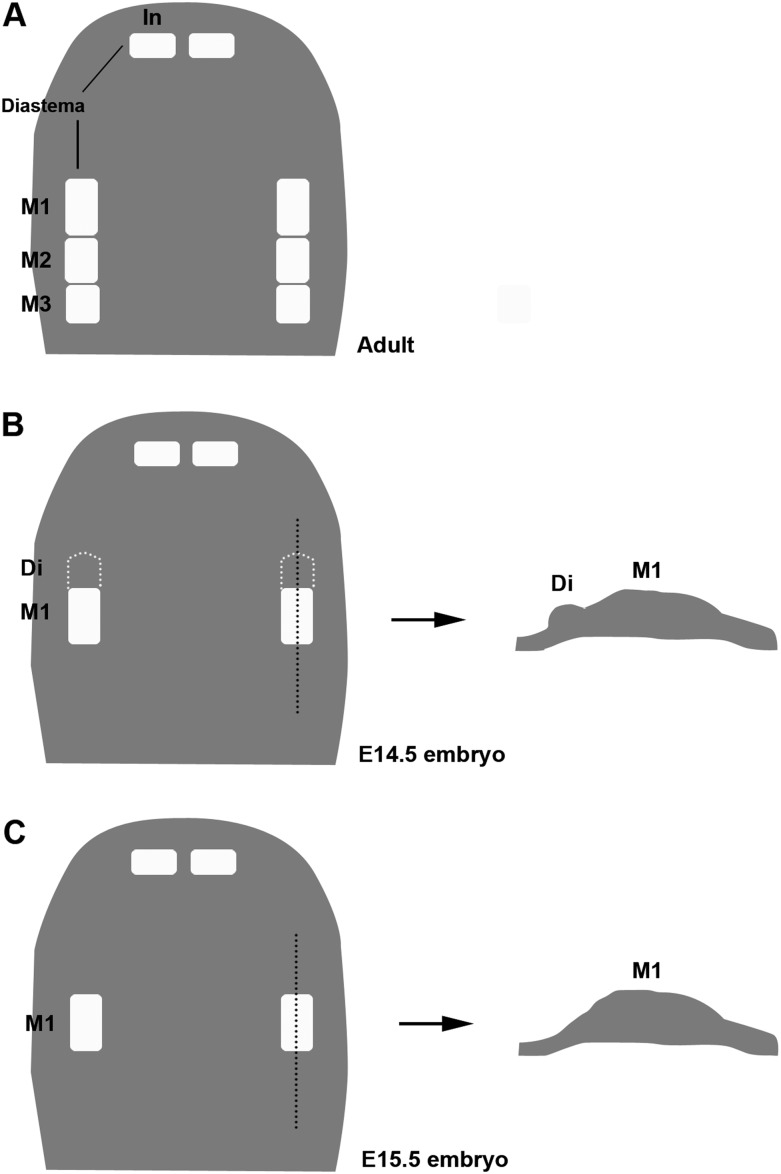
Mice are the commonly studied mammals for investigating the mechanisms of tooth development; however, they have a highly derived dentition; one incisor and three molars in each jaw quadrant that are divided by a tooth-less region, the diastema. It is known that mice lost teeth in the diastema during evolution approximately 100 Mya since modern placental mammals have evolved from a common ancestor with three incisors, one canine, four premolars, and three molars (Butler, 1939; Peyer, 1968; Meng et al., 1994; Ji et al., 2002). In the diastema of mice, rudimentary tooth primordia develops through the initial stages of tooth development as remnants of the evolutionary lost tooth but cease before the cap stage and regress by apoptosis (Peterkova et al., 2000; Figure 1).
Figure 1.
Schematic diagrams showing tooth buds in rodent maxilla. Schematic representations of maxilla from wild-type mouse at adult (A), E14.5 (B), and E15.5 (C). Right diagrams of B and C showing sagittal view of tooth germs. In, incisor; M1, first molar; M2, second molar; M3, third molar; Di, rudimentary tooth bud.
Negative feedback regulators of Fgf signaling, Sprouty2, and Sprouty4 are expressed in the embryonic diastema and mutant mice show supernumerary teeth in the diastema (Hacohen et al., 1998; Kim and Bar-Sagi, 2004; Klein et al., 2006). The Shh inhibitor, Gas1, is expressed in the diastema and Gas1 mutant mice also have extra teeth in the diastema (Ohazama et al., 2009). Fgf and Shh signalings are thus inhibited in the diastema to prevent embryonic tooth buds from developing into the teeth. The data from these mutants also suggest that mice have retained the genetic potential for the development of the evolutionary lost teeth.
Wnt signaling plays critical roles in many biological processes such as cell fate determination, cell polarity, cell proliferation, cell differentiation, and apoptosis in many organs including liver, kidney, brain, and bone during development. It has been shown that canonical Wnt signaling is one of major signaling pathways regulating tooth development. Transgenic mice ectopically expressing the Wnt inhibitor, Dkk1, in epithelial cells show arrest of tooth development (Andl et al., 2002). Mutation of Lef1 (a central transcription mediator of Wnt signaling) also causes arrested tooth development (van Genderen et al., 1994). Conversely, the constitutive activation of β-catenin in embryonic oral epithelium results in continuous supernumerary tooth formation from multiple regions of the jaw (Järvinen et al., 2006; Wang et al., 2009). Upregulation of Lef1 in the oral epithelium shows numerous tooth-like epithelial invaginations in non-tooth regions of the mouth (Zhou et et al., 1995). Canonical Wnt signaling thus regulates initiation of tooth development, but it is unclear how Wnt signaling is inhibited in the diastema.
We show here expression in the diastema of Wnt ligands and Wnt co-receptors known to be expressed during tooth development. Lrp4 mutant mice showed supernumerary tooth formation in the diastema that was accompanied by upregulation of canonical Wnt signaling. Wnt signaling is thus attenuated in the diastema by secreted and membrane bound inhibitors.
Materials and methods
Production and analysis of transgenic mice
Lrp4 hypomorphic mice (Lrp4hypo/hypo) were generated from the mouse line produced by Johnson et al. (2005). Lrp4 null mice were generated by deletion of the transcription start site at exon 1, which encodes the signal peptide and the initiating ATG. To trace canonical Wnt activity, BAT-gal mice were crossed into the Lrp4hypo/hypo line. BAT-gal mice were produced as described by Maretto et al. (2003).
The day on which vaginal plugs were found was considered as embryonic day (E) 0.5. To accurately assess the age of embryos, somite pairs were counted and the stage confirmed using morphological criteria, such as relative size of maxillary and mandibular primordia, extent of nasal placode invagination, and the size of limb buds. Harvested embryos were genotyped using polymerase chain reaction and Southern blot analysis using standard techniques. Lrp4 mutant and wild-type mouse heads were fixed in 4 percent paraformaldehyde (PFA), embedded, and serially sectioned at 8 μm. Sections were split over 4–10 slides and prepared for histology and radioactive in situ hybridization. Decalcification using 0.5 M EDTA was performed after fixation of newborn mice.
In situ hybridization
Radioactive in situ hybridization with 35S-UTP-radiolabelled riboprobes was performed as described previously by Ohazama et al. (2008).
Wnt activity detection
Tissues were fixed in 1 percent PFA and 0.2 percent glutaraldehyde for 1 hour at 4°C. Explants were then assayed for β-gal activity by staining with X-Gal staining solution overnight at 37°C.
Results
Tooth-related gene expression in the diastema
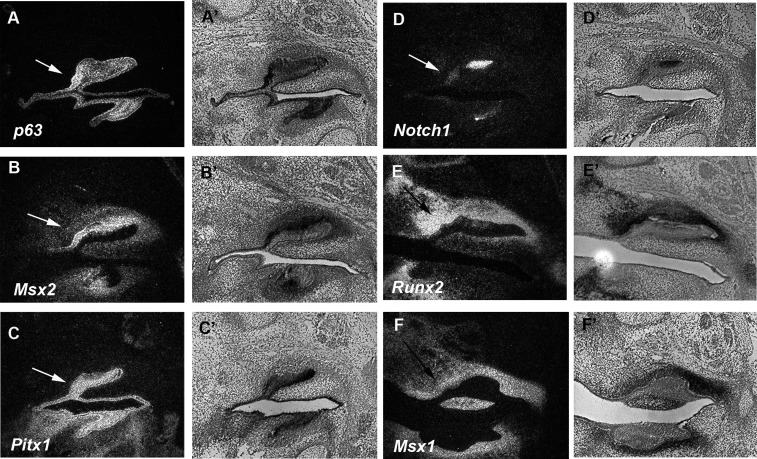
Vestigial tooth primordia are observed in the diastema from E12 to E14.5 (Peterkova et al., 2006). Sprouty2 mutant mice show extra teeth in the diastema and the tooth germs of the extra teeth are distinct from E14.5 (Peterkova et al., 2009). Tooth-related gene expression has been seen in the diastema at E12 and E13, but it is unclear whether these molecules are expressed in the diastema at E14.5 (Tureckova et al., 1995; Keränen et al., 1999). We examined p63, Msx2, Pitx1, and Notch1 that are known to be expressed in tooth epithelium and Runx2 and Msx1 that have been shown to be expressed in tooth mesenchyme (MacKenzie et al., 1992; Jowett et al., 1993; Mitsiadis et al., 1995; D’Souza et al., 1999; St Amand et al., 2000; Laurikkala et al., 2006). The expression of p63, Msx2, Pitx1, and Notch1 was observed in diastema epithelium, whereas Runx2 and Msx1 were expressed in diastema mesenchyme, suggesting that tooth-related genes retained their expression in the diastema at E14.5 Figure 2). Their expression patterns were similar to those in the first molars.
Figure 2.
Tooth -related genes in the diastema. Dark field (A, B, C, D, E, and F) and light field (A’, B’, C’, D’, E’, and F’) images of radioactive in situ hybridization on sagittal sections in wild-type mice at E14.5. The expression of p63 (A and A’), Msx2 (B and B’), Pitx1 (C and C’), and Notch1 (D and D’) were found in upper diastema epithelium. Runx2 (E and E’) and Msx1 (F and F’) expression were found in upper diastema mesenchyme. Both black and white arrows indicate rudimentary tooth bud.
Expression of Wnt ligands in the diastema
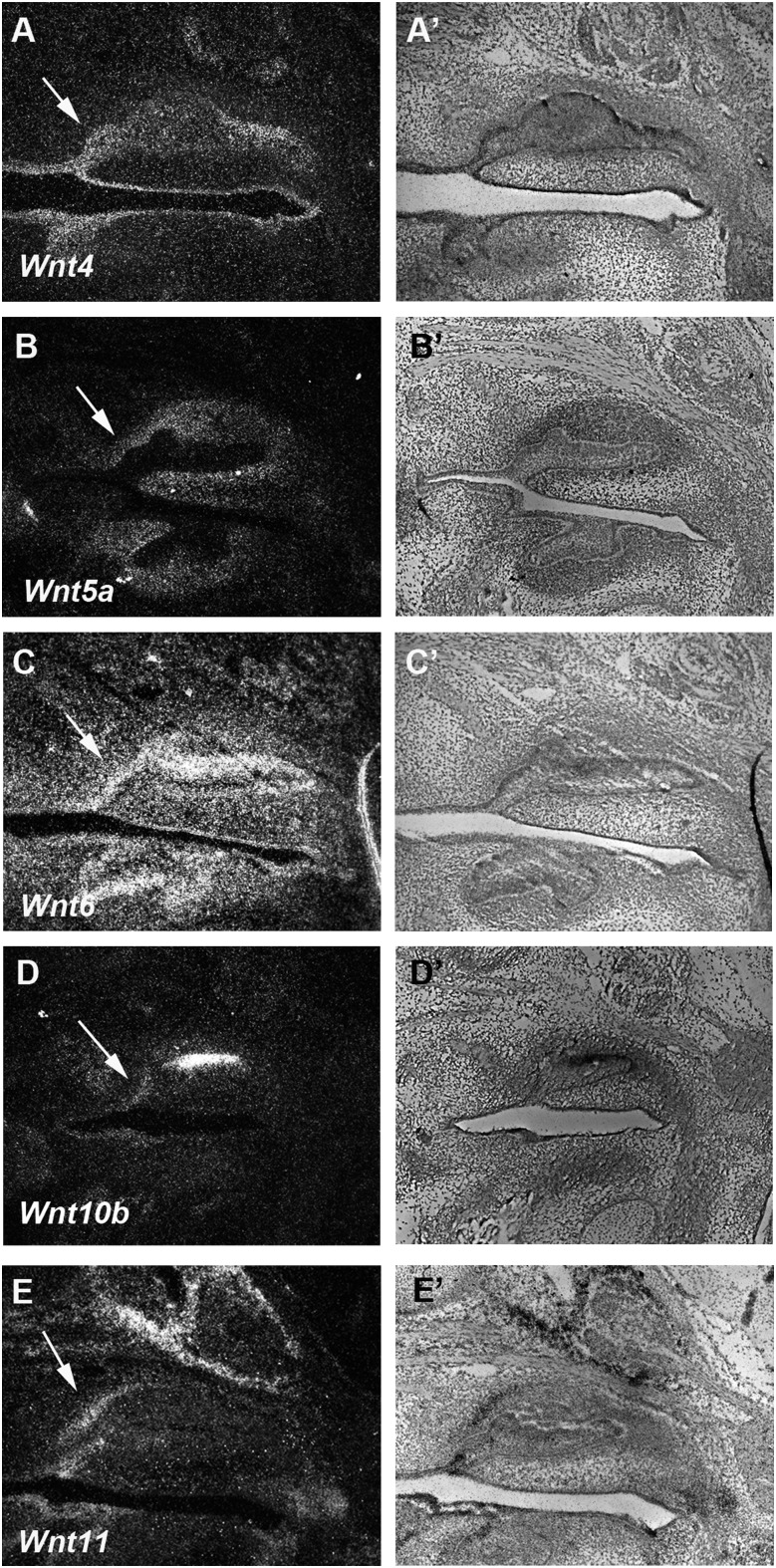
A comparison of the gene expression of Wnt signaling-related molecules including ligands and their receptors is necessary as a starting point to understand how Wnt signaling is regulated in the diastema. We examined the expression of Wnt ligands that have been shown to be expressed in tooth development as well as in the development of other craniofacial ectodermal organs, such as palate and tongue papillae (Sarkar and Sharpe 1999; Lee et al., 2008; Kim et al., 2009). Wnt6 and Wnt11 were expressed in both diastema epithelium and mesenchyme, whereas weak Wnt4 and Wnt10b expression could be detected in diastema epithelium at E14.5 (Figure 3A, 3A’, and 3C–3E’). The expression of Wnt5a was observed in diastema mesenchyme at E14.5 (Figure 3B and 3B’).
Figure 3.
Wnt ligands in the diastema. Dark field (A, B, C, D, and E) and light field (A’, B’, C’, D’, and E’) images of radioactive in situ hybridization on sagittal sections in wild-type mice at E14.5. Weak expression of Wnt4 and Wnt10b (A, A’, D, and D’) was detected in diastema epithelium. Wnt5a expression (B and B’) was observed in diastema mesenchyme. Wnt6 and Wnt11 (C, C’, E, and E’) were expressed in both diastema epithelium and mesenchyme. Arrows indicate rudimentary tooth bud.
Lrp4 and Lrp6 expression in the diastema
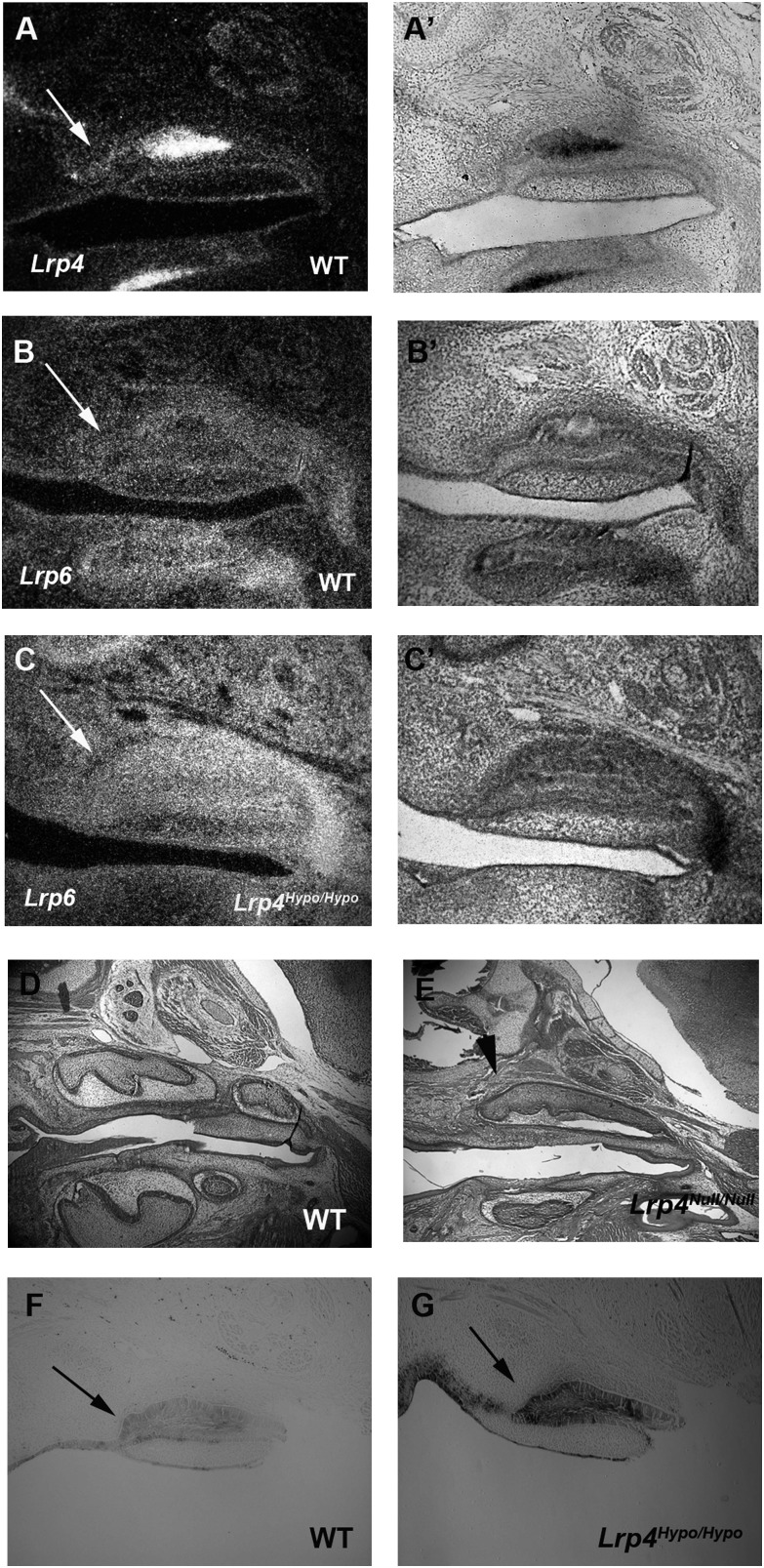
The Wnt co-receptor, Lrp6, is an essential molecule in canonical Wnt signaling and was found to be weakly expressed in both mesenchyme and epithelium of the diastema at E14.5 (Figure 4B and 4B’). Lrp4, a co-receptor that inhibits Wnt signaling, was also weakly expressed in both mesenchyme and epithelium in the diastema at E14.5 (Figure 4A–4A’; Ohazama et al., 2008). The expression of Lrp4 overlapped with Lrp6 expression in the diastema at E14.5. In common with Lrp4Hypo/Hypo mice, Lrp4 null mutant mice also showed extra teeth in the diastema. The mutant extra teeth showed upregulation of Lrp6 expression and Wnt activity in the diastema at E14.5 (Figure 4C, 4C’, and 4G: Ohazama et al., 2008).
Figure 4.
Wnt signaling in the diastema. Dark field (A, B, and C) and light field (A’, B’, and C’) images of radioactive in situ hybridization on sagittal sections in wild-type mice at E14.5. Weak expression of Lrp4 (A and A’) and Lrp6 (B and B’) was observed in both upper diastema epithelium and mesenchyme. Upregulation of Lrp6 expression was detected in Lrp4Hypo/Hypo mutants (C and C’). Supernumerary tooth in the diastema was found in Lrp4Null/Null mutants at E18.5 (arrowhead in E). The first and the second molars were present in wild-type at E18.5 (D). Upregulation of ß-gal activity in the diastema was shown in Lrp4Hypo/Hypo; BAT-gal mice at E14.5 (arrow in G) compared with β-gal activity in the diastema of wild-type at E14.5 (arrow in F). Both black and white arrows indicate rudimentary tooth bud.
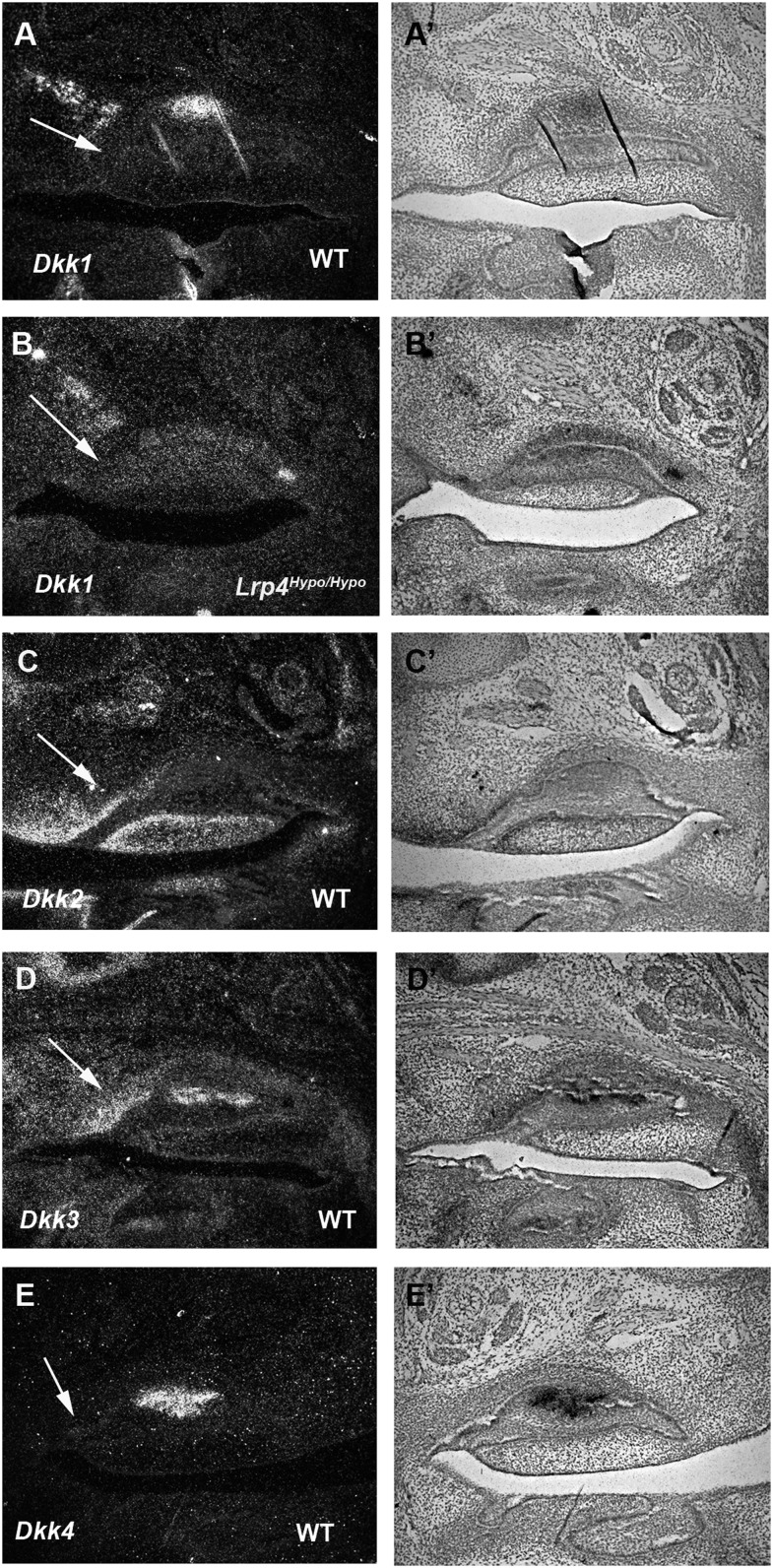
Expression of Dkks in the diastema
Dickkopfs (Dkks) are secreted proteins that typically antagonize Wnt/β-catenin signaling (Bafico et al., 2001; Nusse, 2001; Niehrs, 2006). The Dkk family comprises four evolutionarily conserved gene members (Dkk1–Dkk4). In the diastema, Dkk2 and Dkk3 were expressed in mesenchyme, whereas Dkk4 showed no expression (Figure 5C–5E’). Dkk1 was expressed in presumptive maxillary bone adjacent to the diastema (Figure 5A and 5A’). No obvious differences in Dkk1 expression could be detected in Lrp4 mutant diastema (Figure 5B and 5B’).
Figure 5.
Dkk expression in the diastema. Dark field (A, B, C, D, and E) and light field (A’, B’, C’, D’, and E’) images of radioactive in situ hybridization on sagittal sections in wild-type mice at E14.5. The expression of Dkk1 (A and A’) was found in presumptive maxillary bone adjacent to the diastema in wild-type. Normal expression of Dkk1 (B and B’) was detected in Lrp4Hypo/Hypo mutant. Dkk2 (C and C’) and Dkk3 (D and D’) were expressed in upper diastema mesenchyme. Dkk4 expression (E and E’) could not be detected in the diastema. Arrows indicate rudimentary tooth bud.
Wnt signaling-related molecules including ligands, co-receptors, and inhibitors are thus expressed in the diastema and upregulation of Wnt activity is observed in Lrp4 mutant diastema.
Discussion
Canonical Wnt signaling comprises a complex molecular network. In the absence of Wnt ligands, β-catenin is phosphorylated by a destruction complex that contains the scaffolding protein Axin and glycogen synthase kinase 3 (Gsk3). In the presence of the ligands, the binding of Wnt ligands to Frizzled/Lrp5/6 recruits a complex formed by Dishevelled-Axin-Gsk3β to Frizzled. Gsk3β then phosphorylates Lrp6 that allows Axin binding to Lrp6. β-catenin becomes stabilized, accumulates in the cytoplasm, and translocates to the nucleus where it mediates the transcription of Wnt target genes (Logan and Nusse, 2004; Nusse, 2005; Clevers, 2006). Lrp4 and Lrp6 belong to the low-density lipoprotein receptor family that is a group of evolutionarily conserved transmembrane receptors found in mammals and other organisms. Recent findings have shown that these receptors function in a broad spectrum of diverse biological processes, including lipid and vitamin homeostasis, brain development, synaptic plasticity, and signal transduction (Herz and Bock, 2002; Nykjaer and Willnow, 2002). Although there is structural similarity between Lrp4 and Lrp5/6, the cytoplasmic domain of Lrp4 lacks the Axin-binding motifs that are present in Lrp5 and Lrp6. Therefore, Lrp4 does not transmit canonical Wnt signaling, whereas Lrp5 and Lrp6 are essential molecules in the canonical Wnt signaling cascade (Pinson et al., 2000; Tamai et al., 2000; Wehrli et al., 2000). Overlapping expression of Lrp4 and Lrp6 in the diastema suggests that the presence of Lrp4 results in reduction of ligand binding to Frizzled/Lrp5/6, which leads to attenuation of Wnt signaling in the diastema.
Wise (Ectodin, Sostdc1, and USAG-1) is a secreted Wnt modulator that is expressed in the developing diastema (Itasaki et al., 2003; Kassai et al., 2005; Ohazama et al., 2008). Although Wise and Wnt ligands share the same binding domain of Lrp6, Wise does not activate Wnt signalling (Itasaki et al., 2003). The presence of Wise therefore results in the reduction of ligands binding to Frizzled/Lrp5/6, which leads to attenuation of Wnt activity in the diastema. Wise mutant mice show extra teeth in the diastema, although upregulation of Wnt signaling has not been reported in the Wise mutant diastema (Kassai et al., 2005; Murashima-Suginami et al., 2008). Dkks are secreted molecules that inhibit Wnt signaling. Dkk1 inhibits Wnt signaling by a different mechanism to Wise since Dkk1 interacts with a different domain from Wise-binding region on Lrp6 (Itasaki et al., 2003). Dkk1 was expressed in the diastema, suggesting that Wnt signaling can be reduced by both secreted molecules, Dkk1 and Wise in the diastema. Dkk2 and Dkk3 expression was also observed in the diastema, although Dkk2 and Dkk3 can either activate or inhibit Wnt pathway depending on the tissue context, the level of Lrp5/6, and the presence of Dkk receptor Kremlin2 (Brott and Sokol, 2002; Li et al., 2002; Mao and Niehrs, 2003; Hoang et al., 2004; Hackam, 2005; Yue et al., 2008). It is conceivable that these molecules are involved in switching on/off Wnt signaling to induce and regress the vestigial epithelial buds in the diastema.
Supernumerary tooth formation in the diastema has been found in mice with mutations of Lrp4, Sprouty2/4, Wise, Polaris, and Gas1 (Zhang et al., 2003; Kassai et al., 2005; Klein et al., 2006; Murashima-Suginami et al., 2008; Ohazama et al., 2008, 2009). It has been shown that upregulation of Shh and Fgf signaling causes extra tooth formation in Polaris/Gas1 and Sprouty2/4 mutants, respectively (Zhang et al., 2003; Klein et al., 2006; Ohazama et al., 2009). Upregulation of Wnt signaling was observed in Lrp4 mutant mice and ectopic teeth in the diastema were also observed in mice misexpressing ectodysplasin A1 (EdaA1) and EdaA1 receptor (Edar; Grüneberg, 1966; Mustonen et al., 2003; Pispa et al., 2003; Tucker et al., 2004; Peterková et al., 2005). It has however not been shown which signaling pathways are involved in EdaA1/Edar for tooth formation in the diastema. EdaA1 and Edar belong to the TNF ligand family and TNF receptor family, respectively, which are responsible for X-linked hypohidrotic ectodermal dysplasia in humans (Kere et al., 1996). Spontaneous mutation of the EdaA1 in mice shows the presence of supernumerary teeth in the diastema and over-expression of EdaA1 or Edar also induces extra tooth formation in the diastema (Grüneberg, 1966; Mustonen et al., 2003; Pispa et al., 2003; Tucker et al., 2004; Peterková et al., 2005). These results suggest that fine tuning of EdaA1/Edar is necessary to inhibit tooth formation in the wild-type diastema. It is known that there is a cross talk between EdaA1/Edar and Wnt/β-Catenin in hair follicles, suggesting the possibility that alteration of EdaA1/Edar induces Wnt signaling activity in the diastema (Zhang et al., 2009).
Signaling pathways such as Wnt, Fgf, Bmp, and Shh play critical roles in development. Secreted or membrane bound antagonists play a role in regulating the activity of these pathways by controlling the extracellular concentration of these ligands. We found that Wnt ligands, Dkks, Lrp4, and Lrp6, are expressed in the diastema, suggesting that Wnt signaling is involved in the repression of diastema tooth formation, modulated by these molecules.
Funding
W J B Houston Memorial Research Scholarship (European Orthodontic Society); Medical Research Council. AO is an RCUK Fellow. J.H. is supported by grants from the National Institutes of Health and is the recipient of a Wolfgang Paul Award from the Alexander-von-Humboldt Foundation.
References
- Andl T, Reddy ST, Gaddapara T, Millar SE. Wnt signals are required for the initiation of hair follicle development. Developmental Cell. 2002;2:643–653. doi: 10.1016/s1534-5807(02)00167-3. [DOI] [PubMed] [Google Scholar]
- Bafico A, Liu G, Yaniv A, Gazit A, Aaronson SA. Novel mechanism of wnt signalling inhibition mediated by dickkopf-1 interaction with lrp6/arrow. Nature Cell Biology. 2001;3:683–686. doi: 10.1038/35083081. [DOI] [PubMed] [Google Scholar]
- Brott BK, Sokol SY. Regulation of wnt/lrp signaling by distinct domains of dickkopf proteins. Molecular and Cellular Biology. 2002;22:6100–6110. doi: 10.1128/MCB.22.17.6100-6110.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler PM. Studies of the mammalian dentition-differentiation of the post-canine dentition. Proceedings of the Zoological Society of London. 1939;B109:1–36. [Google Scholar]
- Clevers H. Wnt/β-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- D’Souza RN, et al. Cbfa1 is required for epithelial-mesenchymal interactions regulating tooth development in mice. Development. 1999;126:2911–2920. doi: 10.1242/dev.126.13.2911. [DOI] [PubMed] [Google Scholar]
- Grüneberg H. The molars of the tabby mouse, and a test of the ‘single-active x-chromosome’ hypothesis. Journal of Embryology and Experimental Morphology. 1966;15:223–244. [PubMed] [Google Scholar]
- Hackam AS. The wnt signaling pathway in retinal degenerations. IUBMB Life. 2005;57:381–388. doi: 10.1080/15216540500137586. [DOI] [PubMed] [Google Scholar]
- Hacohen N, Kramer S, Sutherland D, Hiromi Y, Krasnow MA. Sprouty encodes a novel antagonist of fgf signaling that patterns apical branching of the drosophila airways. Cell. 1998;92:253–263. doi: 10.1016/s0092-8674(00)80919-8. [DOI] [PubMed] [Google Scholar]
- Herz J, Bock HH. Lipoprotein receptors in the nervous system. Annual Review of Biochemistry. 2002;71:405–434. doi: 10.1146/annurev.biochem.71.110601.135342. [DOI] [PubMed] [Google Scholar]
- Hoang BH, et al. Dickkopf 3 inhibits invasion and motility of saos-2 osteosarcoma cells by modulating the wnt-β-catenin pathway. Cancer Research. 2004;64:2734–2739. doi: 10.1158/0008-5472.can-03-1952. [DOI] [PubMed] [Google Scholar]
- Itasaki N, et al. Wise, a context-dependent activator and inhibitor of wnt signalling. Development. 2003;130:4295–4305. doi: 10.1242/dev.00674. [DOI] [PubMed] [Google Scholar]
- Järvinen E, Salazar-Ciudad I, Birchmeier W, Taketo MM, Jernvall J, Thesleff I. Continuous tooth generation in mouse is induced by activated epithelial wnt/β-catenin signaling. Proceedings of the National Academy of Sciences. 2006;103:18627–18632. doi: 10.1073/pnas.0607289103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Q, Luo Z-X, Yuan C-X, Wible JR, Zhang J-P, Georgi JA. The earliest known eutherian mammal. Nature. 2002;416:816–822. doi: 10.1038/416816a. [DOI] [PubMed] [Google Scholar]
- Johnson EB, Hammer RE, Herz J. Abnormal development of the apical ectodermal ridge and polysyndactyly in megf7-deficient mice. Human Molecular Genetics. 2005;14:3523–3538. doi: 10.1093/hmg/ddi381. [DOI] [PubMed] [Google Scholar]
- Jowett AK, Vainio S, Ferguson MW, Sharpe PT, Thesleff I. Epithelial-mesenchymal interactions are required for msx 1 and msx 2 gene expression in the developing murine molar tooth. Development. 1993;117:461–470. doi: 10.1242/dev.117.2.461. [DOI] [PubMed] [Google Scholar]
- Kassai Y, et al. Regulation of mammalian tooth cusp patterning by ectodin. Science. 2005;309:2067–2070. doi: 10.1126/science.1116848. [DOI] [PubMed] [Google Scholar]
- Keränen SVE, Kettunen P, Åberg T, Thesleff I, Jernvall J. Gene expression patterns associated with suppression of odontogenesis in mouse and vole diastema regions. Development Genes and Evolution. 1999;209:495–506. doi: 10.1007/s004270050282. [DOI] [PubMed] [Google Scholar]
- Kere J, et al. X-linked anhidrotic (hypohidrotic) ectodermal dysplasia is caused by mutation in a novel transmembrane protein. Nature Genetics. 1996;13:409–416. doi: 10.1038/ng0895-409. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Bar-Sagi D. Modulation of signalling by sprouty: a developing story. Nature Reviews Molecular Cell Biology. 2004;5:441–450. doi: 10.1038/nrm1400. [DOI] [PubMed] [Google Scholar]
- Kim JY, et al. Shh and ROCK1 modulate the dynamic epithelial morphogenesis in circumvallate papilla development. Developmental Biology. 2009;325:273–280. doi: 10.1016/j.ydbio.2008.10.034. [DOI] [PubMed] [Google Scholar]
- Klein OD, Minowada G, Peterkova R, Kangas A, Yu BD, Lesot H. Sprouty genes control diastema tooth development via bidirectional antagonism of epithelial-mesenchymal fgf signaling. Developmental Cell. 2006;11:181–190. doi: 10.1016/j.devcel.2006.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurikkala J, Mikkola ML, James M, Tummers M, Mills AA, Thesleff I. p63 regulates multiple signalling pathways required for ectodermal organogenesis and differentiation. Development. 2006;133:1553–1563. doi: 10.1242/dev.02325. [DOI] [PubMed] [Google Scholar]
- Lee JM, et al. Wnt11/Fgfr1b cross-talk modulates the fate of cells in palate development. Developmental Biology. 2008;314:341–350. doi: 10.1016/j.ydbio.2007.11.033. [DOI] [PubMed] [Google Scholar]
- Li L, Mao J, Sun L, Liu W, Wu D. Second cysteine-rich domain of dickkopf-2 activates canonical wnt signaling pathway via lrp-6 independently of dishevelled. Journal of Biological Chemistry. 2002;277:5977–5981. doi: 10.1074/jbc.M111131200. [DOI] [PubMed] [Google Scholar]
- Logan CY, Nusse R. The wnt signaling pathway in development and disease. Annual Review of Cell and Developmental Biology. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- MacKenzie A, Ferguson MW, Sharpe PT. Expression patterns of the homeobox gene, Hox-8, in the mouse embryo suggest a role in specifying tooth initiation and shape. Development. 1992;115:403–420. doi: 10.1242/dev.115.2.403. [DOI] [PubMed] [Google Scholar]
- Mao B, Niehrs C. Kremen2 modulates dickkopf2 activity during wnt/lrp6 signaling. Gene. 2003;302:179–183. doi: 10.1016/s0378-1119(02)01106-x. [DOI] [PubMed] [Google Scholar]
- Maretto S, Cordenonsi M, Dupont S, Braghetta P, Broccoli V, Hassan AB. Mapping wnt/β-catenin signaling during mouse development and in colorectal tumors. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:3299–3304. doi: 10.1073/pnas.0434590100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng J, Wyss AR, Dawson MR, Zhai R. Primitive fossil rodent from inner mongolia and its implications for mammalian phylogeny. Nature. 1994;370:134–136. doi: 10.1038/370134a0. [DOI] [PubMed] [Google Scholar]
- Mitsiadis TA, Lardelli M, Lendahl U, Thesleff I. Expression of Notch 1, 2 and 3 is regulated by epithelial-mesenchymal interactions and retinoic acid in the developing mouse tooth and associated with determination of ameloblast cell fate. Journal of Cell Biology. 1995;130:407–418. doi: 10.1083/jcb.130.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashima-Suginami A, Takahashi K, Sakata T, Tsukamoto H, Sugai M, Yanagita M. Enhanced bmp signaling results in supernumerary tooth formation in usag-1 deficient mouse. Biochemical and Biophysical Research Communications. 2008;369:1012–1016. doi: 10.1016/j.bbrc.2008.02.135. [DOI] [PubMed] [Google Scholar]
- Mustonen T, Pispa J, Mikkola ML, Pummila M, Kangas AT, Pakkasjärvi L. Stimulation of ectodermal organ development by ectodysplasin-a1. Developmental Biology. 2003;259:123–136. doi: 10.1016/s0012-1606(03)00157-x. [DOI] [PubMed] [Google Scholar]
- Niehrs C. Function and biological roles of the dickkopf family of wnt modulators. Oncogene. 2006;25:7469–7481. doi: 10.1038/sj.onc.1210054. [DOI] [PubMed] [Google Scholar]
- Nusse R. Developmental biology: making head or tail of dickkopf. Nature. 2001;411:255–256. doi: 10.1038/35077199. [DOI] [PubMed] [Google Scholar]
- Nusse R. Wnt signaling in disease and in development. Cell Research. 2005;15:28–32. doi: 10.1038/sj.cr.7290260. [DOI] [PubMed] [Google Scholar]
- Nykjaer A, Willnow TE. The low-density lipoprotein receptor gene family: a cellular swiss army knife? Trends in Cell Biology. 2002;12:273–280. doi: 10.1016/s0962-8924(02)02282-1. [DOI] [PubMed] [Google Scholar]
- Ohazama A, et al. Primary cilia regulate shh activity in the control of molar tooth number. Development. 2009;136:897–903. doi: 10.1242/dev.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohazama A, Johnson EB, Ota MS, Choi HJ, Porntaveetus T, Oommen S. Lrp4 modulates extracellular integration of cell signaling pathways in development. PLoS ONE. 2008;3:e4092. doi: 10.1371/journal.pone.0004092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterkova R, Churava S, Lesot H, Rothova M, Prochazka J, Peterka M. Revitalization of a diastemal tooth primordium in spry2 null mice results from increased proliferation and decreased apoptosis. Journal of Experimental Zoology Part B: Molecular and Developmental Evolution. 2009;312B:292–308. doi: 10.1002/jez.b.21266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterkova R, Lesot H, Peterka M. Phylogenetic memory of developing mammalian dentition. Journal of Experimental Zoology Part B: Molecular and Developmental Evolution. 2006;306B:234–250. doi: 10.1002/jez.b.21093. [DOI] [PubMed] [Google Scholar]
- Peterková R, Lesot H, Viriot L, Peterka M. The supernumerary cheek tooth in tabby/eda mice—a reminiscence of the premolar in mouse ancestors. Archives of Oral Biology. 2005;50:219–225. doi: 10.1016/j.archoralbio.2004.10.020. [DOI] [PubMed] [Google Scholar]
- Peterkova R, Peterka M, Viriot L, Lesot H. Dentition development and budding morphogenesis. Journal of Craniofacial Genetics and Developmental Biology. 2000;20:158–172. [PubMed] [Google Scholar]
- Peyer B. Comparative odontology. Illinois: University of Chicago Press.Chicago; 1968. [Google Scholar]
- Pinson KI, Brennan J, Monkley S, Avery BJ, Skarnes WC. An ldl-receptor-related protein mediates wnt signalling in mice. Nature. 2000;407:535–538. doi: 10.1038/35035124. [DOI] [PubMed] [Google Scholar]
- Pispa J, Mikkola ML, Mustonen T, Thesleff I. Ectodysplasin, edar and tnfrsf19 are expressed in complementary and overlapping patterns during mouse embryogenesis. Gene Expression Patterns. 2003;3:675–679. doi: 10.1016/s1567-133x(03)00092-9. [DOI] [PubMed] [Google Scholar]
- Sarkar L, Sharpe PT. Expression of Wnt signalling pathway genes during tooth development. Mechanism of Ageing and Development. 1999;85:197–200. doi: 10.1016/s0925-4773(99)00095-7. [DOI] [PubMed] [Google Scholar]
- St Amand TR, et al. Antagonistic signals between BMP4 and FGF8 define the expression of Pitx1 and Pitx2 in mouse tooth-forming anlage. Developmental Biology. 2000;217:323–332. doi: 10.1006/dbio.1999.9547. [DOI] [PubMed] [Google Scholar]
- Tamai K, et al. Ldl-receptor-related proteins in wnt signal transduction. Nature. 2000;407:530–535. doi: 10.1038/35035117. [DOI] [PubMed] [Google Scholar]
- Thesleff I, Åberg T. Molecular regulation of tooth development. Bone. 1999;25:123–125. doi: 10.1016/s8756-3282(99)00119-2. [DOI] [PubMed] [Google Scholar]
- Thesleff I, Sharpe P. Signalling networks regulating dental development. Mechanisms of Development. 1997;67:111–123. doi: 10.1016/s0925-4773(97)00115-9. [DOI] [PubMed] [Google Scholar]
- Tucker AS, Headon DJ, Courtney JM, Overbeek P, Sharpe PT. The activation level of the tnf family receptor, edar, determines cusp number and tooth number during tooth development. Developmental Biology. 2004;268:185–194. doi: 10.1016/j.ydbio.2003.12.019. [DOI] [PubMed] [Google Scholar]
- Tureckova J, Sahlberg C, Aberg T, Ruch JV, Thesleff I, Peterkova R. Comparison of expression of the msx-1, msx-2, bmp-2 and bmp-4 genes in the mouse upper diastemal and molar tooth primordia. International Journal of Developmental Biology. 1995;39:459–468. [PubMed] [Google Scholar]
- van Genderen C, et al. Development of several organs that require inductive epithelial-mesenchymal interactions is impaired in lef-1-deficient mice. Genes & Development. 1994;8:2691–2703. doi: 10.1101/gad.8.22.2691. [DOI] [PubMed] [Google Scholar]
- Wang XP, et al. Apc inhibition of wnt signaling regulates supernumerary tooth formation during embryogenesis and throughout adulthood. Development. 2009;136:1939–1949. doi: 10.1242/dev.033803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrli M, et al. Arrow encodes an ldl-receptor-related protein essential for wingless signalling. Nature. 2000;407:527–530. doi: 10.1038/35035110. [DOI] [PubMed] [Google Scholar]
- Yue W, et al. Downregulation of dkk3 activates beta-catenin/tcf-4 signaling in lung cancer. Carcinogenesis. 2008;29:84–92. doi: 10.1093/carcin/bgm267. [DOI] [PubMed] [Google Scholar]
- Zhang Q, et al. Loss of the tg737 protein results in skeletal patterning defects. Developmental Dynamics. 2003;227:78–90. doi: 10.1002/dvdy.10289. [DOI] [PubMed] [Google Scholar]
- Zhang Y, et al. Reciprocal requirements for eda/edar/nf-kappab and wnt/beta-catenin signaling pathways in hair follicle induction. Developmental Cell. 2009;17:49–61. doi: 10.1016/j.devcel.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Byrne C, Jacobs J, Fuchs E. Lymphoid enhancer factor 1 directs hair follicle patterning and epithelial cell fate. Genes & Development. 1995;9:700–713. doi: 10.1101/gad.9.6.700. [DOI] [PubMed] [Google Scholar]