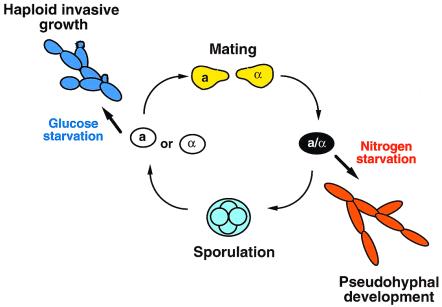
Cells differentiate in response to environmental cues, but different cell types often respond in distinct ways, either in terms of what cues generate a particular response or how the same cue leads to different responses. The molecular mechanisms that underlie the interaction of intrinsic cell properties (cell type) with extrinsic signals (environment) are therefore of great interest. Many examples of such decisions occur in the budding yeast Saccharomyces cerevisiae, which exists in two haploid cell types, a and α, and a diploid cell type, a/α. These three cell types are capable of several differentiation events (Fig. 1) that are controlled by both extrinsic cues and intrinsic determinants (Table 1). During the mating process, a and α cells respond to peptide mating pheromones secreted by the opposite cell type to undergo sexual differentiation and cell–cell fusion to form the a/α cell type. In turn, when simultaneously starved for both carbon and fixed nitrogen, a/α cells undergo meiosis and sporulation to yield four haploid gametes. Like many human pathogenic fungi, solitary S. cerevisiae yeast cells can differentiate into multicellular filaments (1). In diploid a/α cells, nitrogen starvation in the presence of abundant glucose induces the formation of pseudohyphal filaments. Cullen and Sprague (2) report in this issue of PNAS that the opposite signal—glucose starvation in the presence of abundant nitrogen—activates filamentous growth in the haploid a and α cell types. Together with other observations, these studies raise interesting questions about the interplay between cell-intrinsic and cell-extrinsic signals during development.
Figure 1.
Developmental options of S. cerevisiae cell types. Haploid a or α cells differentiate into a filamentous form in response to glucose starvation (see article by Cullen and Sprague (2) in this issue). Haploid cells of the opposite mating type respond to peptide mating pheromones to fuse, forming a/α diploid cells. In contrast to the haploid cell types, a/α cells switch to filamentous growth in response to nitrogen starvation in the presence of abundant glucose. When simultaneously starved for nitrogen and glucose, a/α cells undergo meiosis and sporulation to produce haploid gametes.
Table 1.
Developmental competence of a, α, and a/α cell types of Saccharomyces cerevisiae
| Differentiation event | Environmental cue | Cell
type (presence of the repressor a1-α2)
|
||
|---|---|---|---|---|
| a (absent) | α (absent) | a/α (present) | ||
| Mating | Pheromone | + | + | − |
| Sporulation | Nitrogen and glucose starvation | − | − | + |
| Filamentation (“pseudohyphal development”) | Nitrogen starvation | − | − | + |
| Filamentation (“haploid invasive growth”) | Glucose starvation | + | + | − |
Pluses indicate the ability of the indicated cell type to undergo the indicated differentiation program and environmental cue. Minuses indicate that the cell type is not competent for a particular program.
The pseudohyphal differentiation program of a/α diploid cells is characterized by three visible changes: (i) cells adopt an elongated cell morphology, (ii) cells maintain attachments with each other, and (iii) mother cells tend to bud off daughter cells distally to their previous bud site (3). The triad of elongation, adhesion, and distal budding collaborate to produce multicellular filaments capable of invading the agar substrate. Haploid a and α cells do not form pseudohyphae under conditions of nitrogen starvation. Instead, they bud in a proximal pattern (a.k.a. axial), in which the daughter bud forms adjacent to the site of the previous cell division, a pattern that precludes filamentous growth (3). Surprisingly, haploid cells can form filaments under some conditions (4). Termed haploid invasive growth, this filamentous growth form was first found in haploid yeast strains after several days of incubation on plates with rich media. Invasive growth is assayed by washing plate cultures with water to determine the degree of agar invasiveness. Haploid filaments that penetrate the agar appear similar to the filaments produced during pseudohyphal growth: cells are elongated, they remain attached to each other, and, remarkably, budding pattern is switched such that mother cells give rise to daughter buds at a site distal to the previous bud (4).
Cullen and Sprague (2) now show that glucose starvation is the signal that induces haploid invasive growth. By micromanipulating single cells onto glucose-deficient media, they observed the initiation of filamentous growth in the first cell generation, complete with cell elongation and the critical switch from the axial to a distal budding pattern. Because single cells undergo the switch, it is unlikely that cell–cell signaling is necessary for invasive growth. Taking advantage of the high penetrance of the filamentation phenotype in their assay, Cullen and Sprague found that glucose (and other fermentable carbon sources) completely suppresses haploid filamentous growth whereas manipulating nitrogen levels had no effect. Supporting the view that glucose signaling controls filamentous growth in haploids, the authors found that the key glucose control gene SNF1, which encodes a homologue of the human AMP-activated kinase (5), is necessary for haploid invasive growth. Thus, like the pseudohyphal development of a/α diploids, haploid cells undergo filamentous growth; however, the signals are reversed; glucose starvation induces filamentation in haploid cells, whereas nitrogen starvation does so in diploid cells. Therefore, cell type (a or α versus a/α) determines which signals cause filamentous development. Haploid and diploid cells must be wired up differently—but how? a/α cells differ genetically from a and α cells in one key respect: they express a heterodimeric repressor, a1-α2 (ref. 6; Table 1). This repressor endows diploid cells with the ability to sporulate and form pseudohyphae in response to nitrogen starvation. Therefore, the presence or absence of this transcription factor, both subunits of which are homeodomain proteins, must lie at the heart of why diploid cells filament in response to nitrogen starvation, whereas haploid cells filament in response to glucose starvation.
A formal mechanism can be proposed (Fig. 2A) by which the presence or absence of the a1-α2 repressor controls cellular responsiveness to the two filamentation signals. In the model, a1-α2 represses the expression of one or more components of the glucose sensing pathway. The second key aspect of the model is that there exists a similar repressor (termed “X” in Fig. 2A) that inactivates one or more components of the nitrogen starvation sensing pathway. X itself is repressed by a1-α2. The result of this scheme is that the pathway for sensing glucose starvation is active in a and α cells but not a/α cells, and the nitrogen starvation pathway is active only in a/α cells.
Figure 2.
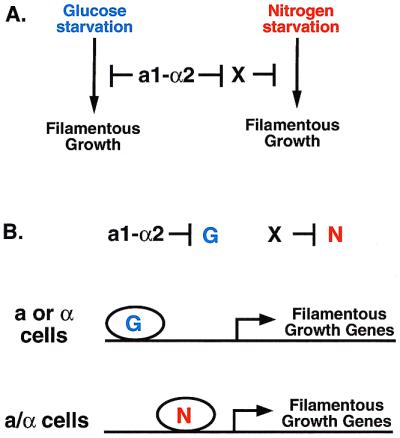
Hypothesis for how cell type determines the response to signals. (A) Formal model. There exist two mechanisms for driving filamentous growth: a glucose starvation pathway and a nitrogen starvation pathway. a1-α2 represses the glucose starvation mechanism whereas a hypothetical factor X represses the nitrogen starvation mechanism. a1-α2 represses the expression of X, restricting its expression to a and α cells. (B) Molecular model. Genes required for filamentous growth are regulated by either a transcription factor that is activated by glucose starvation (G) or a transcription factor activated by nitrogen starvation (N). a1/α2 represses G whereas X represses N.
How might a1-α2 and the hypothetical factor X determine the response of cells to distinct signals? One possibility is that the signaling pathways that activate filamentous growth in haploid and diploid cells are different (6). In this scenario, a1-α2 would repress the haploid-specific pathway and X would repress the diploid-specific pathway. There is precedent for this notion: mating-specific signal transduction components are transcriptionally repressed in a/α diploids by the a1-α2 repressor. Two signaling pathways, the Kss1 mitogen-activated protein kinase (MAPK) pathway and the cAMP-dependent protein kinase pathway (PKA) are required for filamentous growth (reviewed in ref. 1). However, these pathways activate both pseudohyphal development and haploid invasive growth and are therefore not cell type specific. Could different receptors plug into one or both of these pathways in haploid versus diploid cells? Although the receptor for the Kss1 MAPK pathway has not yet been described, a G protein-coupled receptor, Gpr1, acts upstream of the PKA pathway via the Gα homolog Gpa2 (7–9). It has been proposed that Gpr1 is itself a receptor for glucose (10). However, because Gpr1 is required for both pseudohyphal development and haploid invasive growth (10, 11), it cannot account in a simple way for the different responsiveness of yeast cell types to glucose.
Both haploid and diploid cells must be able to sense glucose and fixed nitrogen for functions independent of differentiation. For instance, both cell types presumably alter their general metabolism in similar if not identical ways in response to glucose or nitrogen starvation, yet they clearly respond differently when it comes to filamentous growth. An attractive model would permit cells to integrate cell type and environmental information in several different ways. One way to accomplish such regulatory flexibility would be to integrate information at target promoters that control filamentous growth. For instance, the targets of a1-α2 and X could be a glucose-responsive transcription factor (“G” in Fig. 2B) and a nitrogen-responsive transcription factor (“N” in Fig. 2B), respectively. These factors, in turn, would act specifically on the promoters of genes required for filamentous growth. One good candidate target is the promoter FLO11 gene, which is required for both haploid invasive growth and pseudohyphal growth (12). Analysis of this promoter reveals that it is extraordinarily large and is packed with positive and negative regulatory elements, including those that respond to nitrogen or glucose (13). This hypothesis predicts that X and G would be present only in a or α cells. DNA microarray hybridization studies have revealed all such genes (14), which will permit tests of the model. As with all interesting papers, the findings of Cullen and Sprague raise many new questions and avenues of research for “budding” scientists.
Acknowledgments
I thank Ira Herskowitz for his comments on the manuscript, and Leslie Spector for assistance in manuscript preparation. I am supported by a Burroughs-Wellcome Fund Career Award and a David and Lucile Packard Foundation Fellowship.
Footnotes
See companion article on page 13619.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.011511198.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.011511198
References
- 1.Madhani H D, Fink G R. Trends Cell Biol. 1998;8:348–353. doi: 10.1016/s0962-8924(98)01298-7. [DOI] [PubMed] [Google Scholar]
- 2.Cullen P J, Sprague G F., Jr Proc Natl Acad Sci USA. 2000;97:13619–13624. doi: 10.1073/pnas.240345197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gimeno C J, Ljungdahl P O, Styles C A, Fink G R. Cell. 1992;68:1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- 4.Roberts R L, Fink G R. Genes Dev. 1994;8:2974–2985. doi: 10.1101/gad.8.24.2974. [DOI] [PubMed] [Google Scholar]
- 5.Hardie D G, Carling D, Carlson M. Annu Rev Biochem. 1998;67:821–855. doi: 10.1146/annurev.biochem.67.1.821. [DOI] [PubMed] [Google Scholar]
- 6.Johnson A D. Curr Opin Genet Dev. 1995;5:552–558. doi: 10.1016/0959-437x(95)80022-0. [DOI] [PubMed] [Google Scholar]
- 7.Xue Y, Batlle M, Hirsch J P. EMBO J. 1998;17:1996–2007. doi: 10.1093/emboj/17.7.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lorenz M, Heitman J. EMBO J. 1997;16:7008–7018. doi: 10.1093/emboj/16.23.7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yun C W, Tamaki H, Nakayama R, Yamamoto K, Kumagai H. Biochem Biophys Res Commun. 1997;240:287–292. doi: 10.1006/bbrc.1997.7649. [DOI] [PubMed] [Google Scholar]
- 10.Lorenz M C, Pan X, Harashima T, Cardenas M E, Xue Y, Hirsch J P, Heitman J. Genetics. 2000;154:609–622. doi: 10.1093/genetics/154.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tamaki H, Miwa T, Shinozaki M, Saito M, Yun C W, Yamamoto K, Kumagai H. Biochem Biophys Res Commun. 2000;267:164–168. doi: 10.1006/bbrc.1999.1914. [DOI] [PubMed] [Google Scholar]
- 12.Lo W S, Dranginis A M. Mol Biol Cell. 1998;9:161–171. doi: 10.1091/mbc.9.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rupp S, Summers E, Lo H J, Madhani H, Fink G. EMBO J. 1999;18:1257–1269. doi: 10.1093/emboj/18.5.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galitski T, Saldanha A J, Styles C A, Lander E S, Fink G R. Science. 1999;285:251–254. doi: 10.1126/science.285.5425.251. [DOI] [PubMed] [Google Scholar]