Abstract
Conjugation of ubiquitin to proteins (ubiquitylation) has emerged to be one of the most crucial post-translational modifications controlling virtually all cellular processes. What was once regarded as a mere signal for protein degradation has turned out to be a major regulator of molecular signalling networks. Deregulation of ubiquitin signalling is closely associated with various human pathologies. Here, we summarize the current knowledge of ubiquitin signalling in immune deficiencies and cancer as well as the available therapeutic strategies targeting the ubiquitin system in combating these pathogenic conditions.
Keywords: apoptosis, cell death, inflammation, NF-κB, ubiquitin
Introduction
The modification of proteins by post-translational changes represents a key regulatory mechanism of signal transduction that allows a rapid and dynamic cellular response to distinct environmental stimuli as well as to intracellular signals (Deribe et al, 2010). The addition of chemical moieties or proteins alters the structure, localization, activity and thereby the function of the modified protein. Once a protein is modified by a post-translational modification (PTM), the signal is decoded by unique recognition motifs within the same protein or in others leading to changes in the constituents of molecular complexes in the cells (Seet et al, 2006). The dynamic processes of assembly and disassembly of complexes mediated by PTMs form the central core of the signal transduction machinery driving various cellular functions. Recent studies have revealed that more than 200 forms of protein modifications are present in the cells and we have only started to understand the biological significance of the complex cross talk between them in regulating various processes (Mann & Jensen, 2003). In the early 1980s, covalent conjugation of ubiquitin, a 76 amino acid polypeptide of the family of ubiquitin-like (UBL) proteins, to the target proteins was linked to their degradation via the proteasome (Hershko & Ciechanover, 1998). Today, ubiquitin is recognized as a most versatile form of PTM as it can be conjugated either as a monomer or as polyubiquitin chains of various kinds (Ikeda & Dikic, 2008). Similar to other PTMs such as phosphorylation or acetylation, ubiquitylation is a reversible and usually inducible event. Conjugation of ubiquitin to a substrate protein leads to various consequences in the cell such as proteolytical degradation by proteasomes or lysosomes, altered subcellular localization or activity and interaction with other proteins (Bergink & Jentsch, 2009; Hirsch et al, 2009; Mukhopadhyay & Riezman, 2007; Varshavsky, 2005; Vucic et al, 2011). As ubiquitylation represents a general regulatory mechanism of signal transduction, it is not surprising that misregulation of ubiquitin signalling is associated with initiation and progression of various human diseases including inflammation and cancer (Hoeller & Dikic, 2009). Thus, targeting components of the ubiquitin machinery has evolved as a valid strategy for therapeutic interventions. The clinical successes of the proteasome inhibitor bortezomib (PS-341) in treating haematological malignancies such as multiple myeloma and mantle-cell lymphoma has encouraged scientists to further pursue development of proteasome inhibitors to treat other kinds of cancer as well (Chen et al, 2011). In this review, we will provide the basic description of the ubiquitin machinery, summarize the role of ubiquitin in molecular medicine approaches in the field of immune disorders and cancer as well as the current therapeutic strategies targeting these pathologies.
Principles of ubiquitin signalling
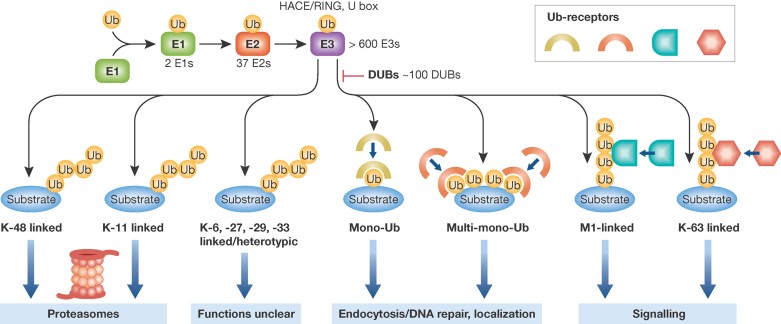
The attachment of ubiquitin to target-proteins, i.e. the ubiquitylation process, usually occurs on lysine residues and proceeds via a three-step procedure involving three different types of enzymes. In the first step, an E1-activating enzyme forms a thioester bond with ubiquitin in an ATP-dependent manner. In the second step, ubiquitin is transferred to an E2 enzyme by trans-thiolation (Schulman & Harper, 2009). Finally, the E3 ubiquitin ligases catalyze the transfer of ubiquitin from the E2 to the ε-amino group of a lysine residue in a target-specific manner (Fig 1). The human genome encodes two E1 enzymes, 37 E2 enzymes and more than 600 E3 ligases. The three classes of E3 ligases (RING, HECT, U-box) are responsible for the recognition of substrates (Grabbe et al, 2011). RING-type ubiquitin E3 ligases contain the RING domain, a zinc-binding protein–protein interaction motif, while HECT E3 ligases harbour a motif with a catalytic Cys residue, which becomes part of a thioester intermediate when ubiquitin is transferred to its substrate. E2 ubiquitin-conjugating enzymes have recently emerged as key mediators of chain assembly by controlling the switch from ubiquitin chain initiation to elongation and by regulating the processivity of chain formation as well as the topology of assembled chains (Grabbe et al, 2011). Further, deubiquitinases (DUBs, approximately 100) add another layer of complexity by editing or removing ubiquitin from substrates (Clague et al, 2012; Haglund & Dikic, 2005). Proteins can be modified with an addition of one ubiquitin molecule on a single lysine (monoubiquitylation) or on several lysines (multi-monoubiquitylation). This type of ubiquitin modification has been associated with processes like deoxyribonucleic acid (DNA) repair, histone regulation and endocytosis (Haglund & Dikic, 2005). Further, ubiquitin itself possesses seven lysines (6, 11, 27, 29, 33, 48, 63), which could serve as an acceptor for ubiquitin chains (Fig 1). Recent studies revealed that head-to-tail linear ubiquitin chains (M1-linked) could also be synthesized in vivo by dedicated E3 ubiquitin ligases (Iwai & Tokunaga, 2009; Walczak et al, 2012). The lysine 48-linked chain is the prototypic ubiquitin signal for degradation via the proteasome. By comparison, linkage through lysine 63 or M1 (linear) chains represents a typical non-degradative modification primarily contributing to assemblage of protein complexes and signal transduction (Ikeda & Dikic, 2008). The physiological roles of atypical ubiquitin chains are just emerging and this remarkable diversity influences almost all aspects of cellular physiology. Ubiquitin is recognized by more than 20 kinds of ubiquitin binding domains, which exhibit specificity for the mode of ubiquitylation and bind non-covalently to ubiquitin (Fig 1) (Dikic et al, 2009). These UBL receptors finally determine the functional outcomes of the entire process.
Figure 1. Principles of ubiquitin signalling.
Ubiquitylation is mediated by the sequential activity of a set of enzymes including activating (E1), conjugating (E2) and ligating (E3) enzymes. This leads to the conjugation of monoubiquitin or polyubiquitin chains of different lengths and link ages to target proteins. Depending on the differents types of ubiquitin chains, proteins are subsequently degraded via the proteasome or participate in various cell ularfunctions including signalling, DNA repair or endocytosis. See text for more details.
Glossary
DUB
Deubiquitinases are proteases responsible for cleaving ubiquitin from substrate proteins. They also process ubiquitin precursors to maintain ubiquitin homeostasis.
E3 ligases
Enzymes responsible for catalysing the transfer of ubiquitin to a lysine residue in the substrate protein.
F-box
A protein domain of ∼50 amino acids involved in mediating protein–protein interactions. F-box proteins function as substrate recognition subunits in cullin-ring ubiquitin ligases.
HECT
A protein domain present in many ubiquitin ligases. These domains possess a catalytic Cys residue that forms a thioester intermediate during the conjugation of ubiquitin to the substrate protein.
Proteasome
A multisubunit protein complex responsible for ATP-dependent degradation of ubiquitin tagged proteins. Inhibitors of proteasome are pursued as cancer chemotherapeutic drugs to kill tumour cells.
RING
A zinc-binding protein–protein interaction motif that binds to the E2-ubiquitin thioester and thereby promotes the conjugation of ubiquitin to substrate proteins.
UBD
A short (∼40 amino acids) sequence motif that mediates ubiquitin binding.
Ubiquitylation
A posttranslational modification where ubiquitin is covalently conjugated in a three step enzymatic cascade to a lysine residue in the modified protein. Ubiquitin conjugated proteins are recognized by ubiquitin receptors, which determine the functional outcomes.
Ubiquitin signalling in immune disorders and inflammation
Protein ubiquitylation has also emerged as one of the key mechanisms that control innate or adaptive immune responses. Ubiquitin signalling has a broad role in these processes by controlling the development of the immune system, as well as several phases of the immune response, ranging from initiation, propagation and termination of the immune response (Bhoj & Chen, 2009; Wertz & Dixit, 2010). Innate immunity represents the first line of defence against invading microorganisms that is highly effective and conserved throughout evolution (Takeuchi & Akira, 2010). Pathogen-associated molecular patterns (PAMPs) serve as a signal for recognition by pattern recognition receptors (PRRs) in the host (Janeway & Medzhitov, 2002). Three classes of PRRs have been identified and extensively characterized: TOLL-like receptors (TLRs), non-obese diabetic (NOD)-like receptors (NLRs) and RIG-1-like receptors (RLRs).
Common downstream pathways that are activated by all these receptors comprise the NF-κB and the mitogen-activated protein (MAP) kinase signalling cascades. Ubiquitylation plays a fundamental role in the activation of the transcriptional factor NF-κB, which controls various cellular processes including immunity, inflammation and cell death. The activation of NF-κB is mediated by canonical and non-canonical pathways. The classical (canonical) pathway is activated by a wide range of stimuli including ligation of TNFRI by its cognate ligand tumour-necrosis factor-α (TNFα), which leads to the phosphorylation and degradation of nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha (IκBα) resulting in the nuclear translocation of p65 and p50 subunits and transcriptional activation of target genes (Perkins, 2007). The phosphorylation of IκBα is regulated by the activation of IKK complex, which comprises of inhibitor of NF-κB kinase subunit alpha (IKKα), inhibitor of NF-κB kinase subunit beta (IKKβ) and an essential regulatory subunit NF-κB essential modulator (NEMO)/IKKγ. Phosphorylated IκBα is recognized by the multi-subunit E3 ubiquitin ligase complex βTrCP, which catalyzes the conjugation of Lys48-linked ubiquitin chains, thereby targeting it for proteosomal degradation. The activation of the IKK complex requires upstream kinases TGFbeta activated kinase 1 (TAK1) and receptor-interacting protein 1 (RIP1) and involves the contribution of both lysine-linked and linear ubiquitin chains (Grabbe et al, 2011). Recent studies have revealed an essential role for the linear ubiquitin assembly complex (LUBAC) in the regulation of the NF-κB pathway (Gerlach et al, 2011; Ikeda et al, 2011; Tokunaga et al, 2011). Mice deficient in the LUBAC component SHANK-associated RH domain interactor (SHARPIN) suffer from multi-organ inflammation including chronic dermatitis (Seymour et al, 2007). Apart from being conjugated with linear ubiquitin chains, NEMO can also bind to linear chains in vivo with its ubiquitin-binding domain UBAN (ubiquitin binding in A20-binding inhibitor of NF-κB activation (ABIN) and NEMO) (Rahighi et al, 2009). The amino acids in the UBAN domain of NEMO responsible for this interaction are mutated in patients suffering from X-linked ectodermal dysplasia revealing the patho-physiological relevance of this interaction (Rahighi et al, 2009). In the absence of inhibitor of apoptosis (IAPs), stimulation with TNFα often leads to the formation of complex II where caspase-8 is recruited for its activation leading to apoptosis (Wang et al, 2008). TNFα-induction can result in the formation of a ‘necrosome complex’ involving RIP1 and RIP3 when caspase-8 activation is inhibited and cIAPs are depleted, leading to a form of programmed cell death (PCD) with features of necrosis, termed necroptosis (Vandenabeele et al, 2010). Recent studies with various mouse models revealed a crucial tissue-specific role of RIP1-RIP3 in regulating necroptosis-mediated inflammation (Bonnet et al, 2011; Duprez et al, 2011; Gunther et al, 2011; Kaiser et al, 2011; Welz et al, 2011). Further, loss of cIAPs has also been shown to spontaneously trigger the formation of RIP1, FAS-associated via death domain (FADD) and caspase-8-containing Ripoptosome complex in the cytosol (approximately 2MDa complex) to mediate apoptosis (Feoktistova et al, 2011; Tenev et al, 2011). The signalling machinery driving necroptosis and its role in regulating various inflammation-associated disorders (like systemic inflammatory response syndrome (SIRS)) is intensively studied in several labs around the world and the readers are referred to recent excellent reviews (Han et al, 2011; Vandenabeele et al, 2010).
The non-canonical pathway is primarily characterized by the proteolytic processing of p100 subunit leading to the formation of p52 fragment in a NIK-dependent manner (Dejardin, 2006). cIAP1/2 E3 ligases in combination with TRAF2/3 contribute to the constitutive polyubiquitylation and proteosomal degradation of NIK (Vallabhapurapu et al, 2008; Zarnegar et al, 2008). B-cell-activating factor (BAFF) is one of the well-studied physiological stimuli known to activate this pathway in B-cells. Upon binding of BAFF to its receptor, the NIK-degrading complex is disrupted, resulting in NIK accumulation, which in turn phosphorylates IKKα (Dejardin, 2006). In both pathways, the constant cross talk between ubiquitylation (of various kinds) and phosphorylation fine-tunes the signal cascade contributing to NF-κB activation.
Also, RLRs and some TLRs can activate interferon regulatory factors (IRF), which act in concert with NF-κB to initiate the production of interferon-1 as effector molecules. Of note, ubiquitylation has been reported to play a critical role in the signalling cascades triggered by all three classes of innate immune response receptors including NLRs such as NOD2 and RLRs such as RIG-1. IAP proteins, TRAFs and Pellino proteins act as ubiquitin ligases that mediate ubiquitylation of critical substrate proteins (such as IRAKs, RIPs, NEMO) that in turn leads to activation of TAK1 and/or IKK complexes. Further, IAP proteins have also been shown to function as direct E3 ubiquitin ligases of RIP2 contributing to NOD1/2-mediated production of inflammatory cytokines and chemokines (Bertrand et al, 2009). In addition, ubiquitylation also serves as the signal for the clearance of pathogenic bacteria in the host cells by xenophagy, a form of selective autophagy (Kirkin et al, 2009).
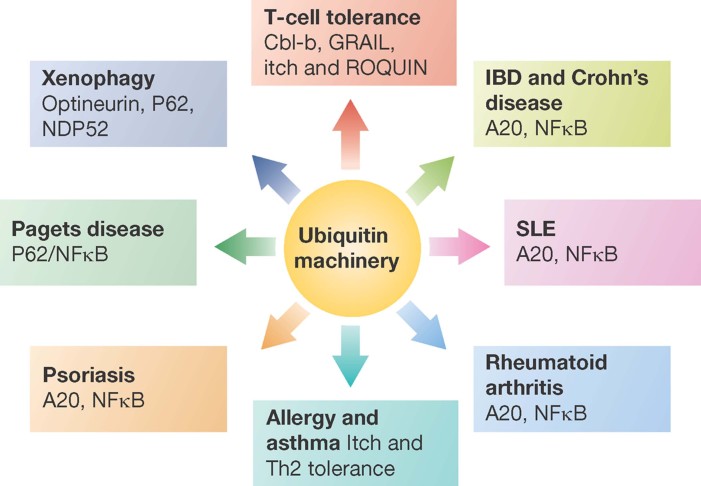
Ubiquitin chains attached to substrates can be removed by DUBs. This removal of ubiquitin molecules by the deubiquitylating enzymes provides a mechanism to dampen the immune response. Several DUBs play a crucial role in the immune system and have been shown to be associated with various immune disorders summarized below (Fig 2). Cylindromatosis (CYLD) was one of the first DUBs implicated in NF-κB activation and has been shown to cleave ubiquitin chains required for NF-κB activation including Lys63-linked and linear chains (Kovalenko et al, 2003; Reiley et al, 2005). Using an ribonucleic acid (RNA) interference screen directed against DUBs, CYLD was identified as a key regulator of the transcription factor NF-κB (Brummelkamp et al, 2003). CYLD contains an ubiquitin hydrolase domain at its C-terminus that is able to remove ubiquitin chains from NF-κB signalling molecules including TNF receptor-associated factor 2 (TRAF2), TNF receptor-associated factor 6 (TRAF6) and RIP1 (Kovalenko et al, 2003; Reiley et al, 2005). In addition, CYLD controls the Bcl-3 pathway by de-ubiquitylating Bcl-3, thereby inhibiting its translocation to the nucleus and the Bcl-3-mediated transcriptional activation of proliferative genes (Massoumi et al, 2006). CYLD is mutated in familial cylindromatosis, a hereditary disorder with tumours of the skin appendages (Bignell et al, 2000). Furthermore, CYLD plays an important role in the regulation of immune response and inflammation (Sun, 2010). For example, CYLD knockout mice show dysregulation of thymocyte development and activation of T-cells associated with bowel inflammation and autoimmune responses (Reiley et al, 2006, 2007; Zhang et al, 2006). CYLD-deficient mice were reported to display a reduced number of single positive thymocytes in peripheral T-cells, while early-stage thymocytes were produced normally (Reiley et al, 2006). Since CYLD positively regulated lymphocyte-specific protein-tyrosine kinase (LCK), a proto-oncogene tyrosine-protein kinase (SRC)-family protein tyrosine kinase involved in T-cell receptor-proximal signalling events and thymocyte development (Molina et al, 1992), CYLD-deficient thymocytes were described to harbour a defect in the interaction of LCK and zeta-chain-associated protein kinase 70 (ZAP70) and associated downstream signalling events (Reiley et al, 2006). Furthermore, CYLD has been reported to act as a crucial regulator of B-cell function (Sun, 2008). Lack of CYLD expression was reported to lead to constitutive activation of the canonical NF-κB signalling cascase in B-cells as shown by higher expression levels of typical NF-κB target genes (Jin et al, 2007). Furthermore, CYLD-deficient B-cells have been described to exhibit defects in B-cell maturation and homeostasis including hyperproduction of marginal zone B-cells and expansion of B-cells in peripheral lymphoid organs. Similarly, abnormalities of B-cell function were reported in mice with a shorter isoform of CYLD (sCYLD), which is a product of a natural splicing variant of the CYLD gene lacking the two exons 7 and 8 (Hovelmeyer et al, 2007).
Figure 2. Deregulation of ubiquitin signalling in immune disorders.
Pathological alterations in ubiquitin signalling pathways can cause various disorders of the immune system. See text for more details.
Moreover, CYLD has been implicated in the innate immune response following bacterial or viral infection. For example, CYLD has been shown to downregulate the inflammatory response following bacterial infection with Escherichia coli by negatively regulating the innate immune response via inhibition of NF-κB signalling (Lim et al, 2008). Accordingly, CYLD-deficient mice were hypersusceptible to E. coli-mediated pneumonia with higher rates of mortality (Lim et al, 2008). In contrast, CYLD-deficient mice were shown to be protected from Streptococcus pneumoniae-mediated acute lung injury and lethality (Lim et al, 2007). In this model, CYLD enhanced acute lung injury and mortality following S. pneumoniae infection by inhibiting MKK3-p38 kinase-mediated expression of plasminogen activator inhibitor-1 (Lim et al, 2007). These findings indicate that CYLD can differentially regulate signalling pathways during the innate immune response to bacterial infections, depending at least in part on the pathogen. In addition, CYLD has been shown to act as a negative regulator of the anti-viral response by removing polyubiquitin chains from the RNA helicase retinoic acid inducible gene I (RIG-1) (Friedman et al, 2008; Zhang et al, 2008). RIG-1 activates the IRF3 signalling pathway to induce interferon type I gene expression in response to viral infections.
A20 (TNFAIP3) acts as a negative feedback regulator of NF-κB signalling in response to numerous inflammatory stimuli, including TNFα, IL-1β and pathogens. A20 possesses both E3 ligase and DUB activities and attenuates TNFα signalling by removing Lys63-linked chains from RIP1 and promotes Lys48-linked chains for proteosomal degradation (Wertz et al, 2004). A20-deficient mice show profound activation of the immune system leading to multiorgan inflammation autoimmunity, characterized by spontaneous production of inflammatory cytokines such as TNFα, IL-1β and IL-6 (Matmati et al, 2011). Further, loss of A20 in keratinocytes leads to hyperkeratosis, which is a common feature in psoriasis. A20 is associated with various immune-pathologies in humans such as rheumatoid arthritis, systemic lupus erythematousus (SLE), type I diabetes, Crohn's disease and psoriasis (Vereecke et al, 2009). Interestingly, efficiency of A20 in myeloid cells has recently been shown to promote erosive polyarthritis that resembles rheumatoid arthritis in a mouse model, pointing to a crucial and cell-specific function of A20 in the pathogenesis of rheumatoid arthritis (Matmati et al, 2011). Furthermore, A20 has been characterized as a susceptibility gene for several autoimmune disorders (Fig 2) (Vereecke et al, 2009). The roles of CYLD and A20 in cancer are discussed below.
Though increased NF-κB activity is associated with inflammation-associated disorders, recent studies revealed that inhibition of NF-κB in non-immune cells such as epithelia leads to spontaneous inflammation in a TNFα-dependent manner (Pasparakis, 2009). However, in chronic inflammatory diseases it has been shown to be quiet useful to reduce the effects of the pro-inflammatory cytokines, especially TNFα. Anti-TNFα therapeutics (e.g. infliximab) have successfully been employed in treating several autoimmune disorders including rheumatoid arthritis, Crohn's diseases, ulcerative colitis and SLE. A better understanding of the mechanisms and the tissue-specific roles is clearly warranted to pursue therapeutics that target NF-κB for the treatment of inflammatory disorders.
Ubiquitin system in cancer
Numerous evidence indicate that the deregulation of ubiquitin pathways can directly and indirectly contribute to the development and progression of human cancers (Hoeller & Dikic, 2009) (Table 1). One of the most studied aspects is linked to defective quality control that is essential for the removal of damaged organelle, misfolded or aggregated proteins. For example, the deregulation of proteasomal degradation can lead to the accumulation of mutated or oncogene-encoded proteins. Perturbing the fine balance between the tumour suppressors and proto-oncogenes would already predispose normal cells towards oncogenic transformation. Similarly, accumulation of damaged mitochondria due to deficient autophagy, for example in cells deficient for the autophagy regulator Beclin1, may contribute to oncogenic load by an increase in the oxidative radical levels and enhanced DNA damage (Mathew & White, 2007). Therefore, alterations in ubiquitylation of numerous cellular proteins can contribute to tumour formation and progression. Here, we discuss several examples that are instructive for molecular medicine.
Table 1.
E3 ligases and DUBs associated with cancers
| Enzymes | Targets | Cancer association |
|---|---|---|
| E3 ligases | ||
| MDM2 (HDM2) | p53 | Over-expressed in multiple cancers including soft tissue sarcoma and lung cancer (Anderson et al, 1999; Lind et al, 2006; Menin et al, 2006) |
| CBL | RTKs, e.g. FLT3, c-Kit | c-Cbl point mutation (Cbl-R420Q) was detected in AML and myoproliferative disorders (Grand et al, 2009) |
| FBW7 | Myc, Jun, cyclin E, KLF5, Notch1 and TGIF1, Mcl-1 | Deleted or mutated in various cancers including T-ALL (Inuzuka et al, 2011; Wertz et al, 2011) |
| FBX011 | Bcl-6 | Deleted or inactivated in diffuse large B-cell lymphoma (Duan et al, 2012) |
| IAPs | Various substrates | Over-expressed in various cancers. C-IAP2 is associated with MALT-lymphoma (Dierlamm et al, 1999; Fulda & Vucic, 2012) |
| Deubiquitinases | ||
| CYLD | Various substrates including RIP1 and Bcl3 | Mutated in familial cylindromatosis, inactivated in skin cancers, hepatocellular and cervical carcinoma (Bignell et al, 2000; Massoumi et al, 2006; Strobel et al, 2002) |
| USP7 | MDM2, PTEN, FOXO4 and others | Downregulation reported in non-small cell lung cancer (Masuya et al, 2006) |
| A20 | RIP1, RIP2, TRAF2, TRAF6, UBCH5, NEMO and others | Frequent inactivation in B-cell lymphomas (Kato et al, 2009) |
| Usp9x | Mcl-1, β-catenin and others | Over-expressed in follicular lymphomas and diffuse large B-cell lymphomas, multiple myeloma (Schwickart et al, 2010) |
| Usp10 | P53 | Downregulated in renal cell carcinomas (Yuan et al, 2010) |
| DUB3 | Cdc25A | Overexpression in breast cancers (Pereg et al, 2010) |
| Others | ||
| PTEN | Promoted ubiquitylation of EGFR through formation of EGFR–CBL complex | Inactivated in various cancers (Trotman et al, 2007) |
Several well-known E3 ligases are either aberrantly activated or display reduced functions in human cancers. For example, mouse double minute 2 (MDM2) is the E3 ligase that ubiquitylates the tumour suppressor protein p53 and thus delivers p53 for proteasomal degradation (Miliani de Marval & Zhang, 2011). Increased MDM2 activity antagonizes the tumour suppressor function of p53 resulting in loss of function of p53. Overexpression of MDM2, for example due to genomic amplification, has been identified in a variety of human cancers, for example in soft tissue sarcoma or lung cancer (Anderson et al, 1999; Lind et al, 2006; Menin et al, 2006). Recently, TRIP12/ULF was identified as a novel E3 ligase of Alternate Reading Frame (ARF of the INK4a locus), a tumour suppressor that regulates ubiquitin-dependent degradation (Chen et al, 2010). An additional mechanism of how the p53 pathway may be regulated involves USP10, a DUB that promotes the stabilization of p53 in response to DNA damage in an ATM-dependent signalling pathway (Yuan et al, 2010). After DNA damage, USP10 is stabilized by ATM-mediated phosphorylation and a pool of USP10 translocates to the nucleus to deubiquitylate and activate p53 (Yuan et al, 2010). Notably, downregulation of USP10 expression was found in a high percentage of renal cell carcinomas, which are known to have few p53 mutations (Yuan et al, 2010), indicating that USP10 downregulation may represent an alternative mechanism of inactivating the p53 pathway in cancers.
Casitas B-lineage lymphoma (CBL) is another well-studied ubiquitin E3 ligase that has been implicated in cancer pathogenesis (Lipkowitz & Weissman, 2011). CBL is involved in the downregulation of receptor tyrosine kinases, e.g. of FLT3 or c-KIT, via multiple ubiquitylation events (Schmidt & Dikic, 2005). Deregulation of CBL has been identified in various cancers including acute myeloid leukaemia (AML), lymphoma and gastric carcinoma and has been linked to insufficient termination of receptor tyrosine kinase signalling (Casas et al, 2003). For example, CBL was found to be involved in the regulation of FLT3 signalling in AML. c-Cbl has been shown to physically interact with FLT3 and to undergo tyrosine phosphorylation upon binding of FLT3-ligand (Sargin et al, 2007). The involvement of c-Cbl in Flt3 signalling was further confirmed by overexpression of a dominant-negative form of c-Cbl (Cbl-70Z) that blocked FLT3-ligand-induced FLT3 ubiquitylation and internalization (Sargin et al, 2007). Interestingly, a c-Cbl point mutation (Cbl-R420Q) was detected in primary AML cells that similarly inhibited FLT3 ubiquitylation (Sargin et al, 2007). Also, CBL mutations were identified in myeloproliferative disorders (Grand et al, 2009). Of note, mutant variants of CBL that are defective in their E3 ligase activity were shown to promote c-KIT- or FLT3-mediated transformation (Bandi et al, 2009).
The stem cell factor (SCF) complex is a multi-subunit ubiquitin ligase complex composed of S-phase kinase-associated protein 1 (SKP1), CUL1 and a variable, substrate-specific F-box protein (Cardozo & Pagano, 2004). F-box and WD repeat domain-containing 7 (FBW7), a F-box protein, is the substrate-specific component of this composite E3 ligase (Cardozo & Pagano, 2004). FBW7 binds to phospho-degrons, i.e. phosphorylated regions of the substrate proteins, leading to their polyubiquitylation and subsequent proteasomal degradation (Cardozo & Pagano, 2004). Target proteins of FBW7 comprise various oncogenes as well as key signalling mediators of cell growth and proliferation, i.e. Myc, Jun, cyclin E, krueppel-like factor 5 (KLF5), Notch homolog 1, translocation-associated (Drosophila) (Notch1) and TGFβ-induced factor 1 (TGIF1) (Cardozo & Pagano, 2004). Since overexpression of several FBW7 target proteins such as Jun, Myc or Notch 1 results in increased cell death in addition to elevated proliferation, it has been unclear until recently how FBW7-deficient cells can evade programmed cell death under these conditions. Therefore, the recent identification of MCL1 as a FBW7 target protein has provided a plausible explanation for this open question (Inuzuka et al, 2011; Wertz et al, 2011). MCL1 belongs to the pro-survival proteins of the BCL2 family that block mitochondrial apoptosis (Adams & Cory, 2007). Furthermore, sensitivity to microtubule-targeted agents such as Taxol and vincristine has been demonstrated to be regulated by MCL1 and FBW7 (Wertz et al, 2011). During mitotic arrest, MCL1 protein levels decline post-translationally through FBW7-mediated destruction via the proteasome (Wertz et al, 2011). The interaction of MCL1 with FBW7 is promoted upon phosphorylation of MCL1 in mitotically arrested cells, enhancing polyubiquitylation of MCL1 and its subsequent proteasomal degradation (Wertz et al, 2011). Findings in primary tumour samples showing elevated MCL1 levels and concomitant FBW7 inactivation underscored the roles of both proteins in oncogenesis (Inuzuka et al, 2011; Wertz et al, 2011). Furthermore, probable ubiquitin carboxyl-terminal hydrolase FAF-X (USP9X) was recently identified as a MCL1-specific DUB that removes ubiquitin from the anti-apoptotic protein MCL1, thereby promoting the stability of MCL1 and opposing apoptosis (Schwickart et al, 2010). USP9X binds directly to MCL1 and de-ubiquitylates Lys 48-linked ubiquitin chains that label MCL1 for proteasomal degradation (Schwickart et al, 2010). Of note, overexpression of USP9X correlates with increased MCL1 protein expression in human cancers, i.e. in follicular lymphomas and diffuse large B-cell lymphomas (Schwickart et al, 2010). In patients with multiple myeloma, increased expression of USP9X correlated with a poor prognosis (Schwickart et al, 2010).
Interestingly, aurora kinase A has recently been reported to stabilize N-Myc by competing for FBW7-binding (Otto et al, 2009). This implies that aurora kinase A inhibitors could suppress proliferation by restoring FBW7-mediated degradation of c-Myc. Loss of FBW7 is frequently detected in various malignancies, including breast or colon cancer and T-cell acute lymphoblastic leukaemia (Crusio et al, 2010). Genetic inactivation of FBW7 in mouse T-cells has been shown to promote lymphomagenesis (Onoyama et al, 2007), validating FBW7 as a tumour suppressor gene. Along these lines, it is interesting to point out that the gene-encoding another F-box containing protein FBXO11 is deleted or inactivated in diffuse B-cell lymphomas (DLBCL) and Bcl-6 has recently been identified as a target protein. Loss of FBX011 leads to increased stabilization of Bcl-6, thereby contributing to pathogenesis of human B-cell lymphomas (Duan et al, 2012).
Phosphatase and tensin homologue deleted in chromosome 10 (PTEN) is a tumour suppressor protein that is inactivated in many human cancers (Georgescu, 2010). PTEN is well known as a phosphatidylinositol 3,4,5-triphosphatase that negatively regulates the PIK/Akt/mTOR cascade (Georgescu, 2010). More recently, PTEN has been shown to be also involved in ubiquitin signalling. It was reported that PTEN promotes ubiquitylation and downregulation of receptor tyrosine kinases such as epidermal growth factor receptor (EGFR) by enhancing the formation of the EGFR–CBL complex (Vivanco et al, 2010). Thus, by targeting EGFR for degradation, PTEN promotes the termination of EGFR signalling. Furthermore, PTEN itself is regulated by ubiquitin-dependent mechanisms in cancer. Monoubiquitylation of PTEN has been demonstrated to be required for its nuclear localization and tumour suppression (Trotman et al, 2007). Lysine residue K289 was identified as one of the major monoubiquitylation sites that are essential for PTEN import (Trotman et al, 2007). Interestingly, the PTEN lysine mutant K289E, which retains catalytic activity but cannot be monoubiquitylated, is defective in nuclear import and accumulation (Trotman et al, 2007). This lysine mutant of PTEN has been associated with Cowden syndrome, a cancer-susceptibility syndrome with inherited PTEN mutation (Trotman et al, 2007).
Furthermore, ubiquitylation regulates PTEN stability. The HECT-domain protein neural precursor cell expressed, developmentally down-regulated 4-1 (NEDD4-1) has been identified as the E3 ligase that catalyzes PTEN polyubiquitylation and triggers proteasomal degradation of PTEN (Wang et al, 2007). Importantly, high levels of NEDD4-1 were detected in tumour samples of various malignancies with low PTEN protein expression on normal genetic background of PTEN (Wang et al, 2007). This implies that upregulation of NEDD4-1 in cancers can suppress PTEN on a post-translational level. In turn, the activity of NEDD4-1 is opposed by the tyrosine kinase RAK. It has been shown that RAK physically interacts with PTEN and phosphorylates PTEN on Tyr336, thereby protecting it from ubiquitin-mediated degradation (Yim et al, 2009). Silencing of RAK led to increased binding of PTEN to NEDD4-1 and enhanced polyubiquitylation and degradation of PTEN (Yim et al, 2009). The de-ubiquitylation enzyme herpes-virus-associated ubiqutin-specific protease (HAUSP, also known as USP7) catalyzes the de-ubiquitylation of both MDM2 and p53 and can thereby modify the link between MDM2 and p53. Downregulation of HAUSP in human cancers therefore leads to inactivation of the p53 signalling pathway, an event that has been reported, e.g. in non-small cell lung carcinoma (Masuya et al, 2006). Further, HAUSP also regulates the de-ubiquitylation of PTEN thereby favouring its exclusion from the nucleus (Song et al, 2008).
Another interesting class of E3 ligases found to be directly associated with cancers include IAP proteins that have been shown to be highly expressed in several human cancers (Fulda & Vucic, 2012). For example, c-IAP1 can function as an oncogene in hepatocellular carcinoma and is part of 11q21-q23 amplicons in human cancers (Zender et al, 2006). In multiple myeloma, deletions of c-IAP1 and cellular inhibitor of apoptosis 2 (c-IAP2) were identified that lead to stimulation of the non-canonical NF-κB pathway (Annunziata et al, 2007; Keats et al, 2007). This finding is consistent with the known function of c-IAP1 and c-IAP2 as E3 ligases that constitutively trigger degradation of NIK via the proteasome (Vallabhapurapu et al, 2008; Zarnegar et al, 2008). Further, gene rearrangements t(11;18) (q21;21) leading to the formation of fusion protein c-IAP2/MALT are associated with 50% of mucosa associated lymphatic tissue (MALT) lymphomas (Dierlamm et al, 1999). The chimeric protein constitutively activated NF-κB contributing to B-cell transformation. Recently, IAP proteins have also been shown to regulate tumour cell migration by functioning as the direct E3 ubiquitin ligases of the RhoGTPase Rac1 (Oberoi et al, 2012).
Besides E3 ligases, deregulation of de-ubiquitylation enzymes (DUBs) can contribute to tumorigenesis by stabilizing oncoproteins or proteins that promote proliferation or block cell death. For example, the de-ubiquitylation enzyme CYLD represents a well-known tumour suppressor. Mutation in CYLD is found in familial cylindromatosis, a hereditary disorder with skin tumours (Bignell et al, 2000). Furthermore, decreased or absent expression of CYLD occurs in other skin cancers, for example basal cell carcinoma, squamous cell carcinoma of the skin as well as hepatocellular and cervical carcinoma (Massoumi et al, 2006; Strobel et al, 2002). Recent studies revealed that A20 can also function as a tumour suppressor in lymphomas. Frequent inactivation of A20 is detected primarily in B-cell lymphomas leading to uncontrolled NF-κB signalling and tumorigenesis (Kato et al, 2009). Further, SCF ligase, which is responsible for degradation of IκBα, has recently been reported to be mutated in a high number of human cancers (Lee et al, 2009), thereby contributing to aberrant NF-κB signalling in human malignancies.
Finally, oncogenes have also been shown to be activated by non-degradative ubiquitylation. An example for such non-degradative ubiquitin signalling in cancer is lysine 63-linked polyubiquitylation of Akt via TRAF6, which facilitates membrane localization of Akt and its subsequent activation (Yang et al, 2009). This mechanism of Akt ubiquitylation represents a step that is involved in oncogenic Akt activation, since human cancer-associated mutant variants of Akt showed elevated Akt ubiquitylation, thereby promoting its membrane localization, phosphorylation and activation (Yang et al, 2009).
Therapeutic perspective of targeting the ubiquitin system
The growing understanding of the molecular mechanisms and biological consequences of the ubiquitin system provides the basis for the development of drug-like inhibitors of enzyme targets within this system. The surprising efficiency of bortezomib in treating multiple myeloma facilitated faster FDA approval in 2003 and there are several proteosomal inhibitors in various stages of clinical development (Table 2). In 2006, bortezomib was approved for treating mantel cell lymphoma. Bortezomib is also pursued in clinical trials in combination with other chemotherapeutic drugs for treating solid tumours. Furthermore, bortezomib showed synergistic activities together with the death receptor ligand TNF-related apoptosis-inducing ligand (TRAIL) to trigger apoptosis in various human cancer (Naumann et al, 2011; Sayers & Murphy, 2006; Unterkircher et al, 2011). Following bortezomib, new classes of inhibitors including carfilzomib, Salinospororamidine A (NPI-002), CEP-18770, PR-957 and Ritonavir are in various stages of preclinical/clinical development (Ruschak et al, 2011). The novel proteasome inhibitors like Carfilzomib differ from bortezomib primarily in targeting the specific protease activity of the 20S proteasome (Ruschak et al, 2011).
Table 2.
Anti-cancer drugs in clinical trials targeting the ubiquitin machinery
| Drugs | Properties | Source | Stage of clinical development |
|---|---|---|---|
| Bortezomib | 20S proteasome inhibitor | Millennium Pharmaceuticals | Approved for multiple myeloma, mantle cell lymphoma |
| MLN9708 | Oral proteasome inhibitor | Millennium Pharmaceuticals | Phase I |
| Carfilzomib/PR-171 | Proteasome inhibitor derived from epoxomycin | Onyx Pharmaceuticals | Phase I/II |
| NPI-0052 | Irreversible 20S proteasome inhibitor | Nereus Pharmaceuticals | Phase I |
| CEP-18770 | Orally active proteasome inhibitor | Ethical Oncology Science | Phase I |
| ONYX 0912 | Oral proteasome inhibitor | Onyx Pharmaceuticals | Phase I |
| RO5503781, RO5045337 | Small molecule MDM2 antagonist | Hoffmann-La Roche | Phase I |
| MLN4924 | NEDD8 inhibitor | Millenium Pharmaceuticals | Phase I |
| JNJ-26854165 | MDM2 inhibitor | Johnson & Johnson Pharmaceutical Research & Development, LLC | Phase I |
| GDC-0152 | IAP inhibitor | Genentech | Phase I |
| AT-406 | IAP inhibitor | Ascenta Therapeutics | Phase I |
| LCL-161 | IAP antagonist | Novartis Pharmaceuticals | Phase I |
| AEG-35156 | XIAP antagonist | Aegera Therapeutics | Phase I/II |
| TL32711 | IAP antagonist | TetraLogic Pharmaceuticals | Phase I/II |
| HGS1029 | IAP antagonist | Human Genome Sciences | Phase I |
Source: http://www.clinicaltrials.gov.
Since the substrate selectivity of the ubiquitin system primarily resides in the specificity of the several hundreds of E3 ligases, these enzymes constitute promising targets for therapeutic intervention. One prominent target is MDM2, as its inhibition results in the activation of the p53 pathway, thereby leading to cell cycle arrest and cell death. Accordingly, small-molecule MDM2 inhibitors have been developed that specifically target the E3 ligase activity of MDM2 (Yang et al, 2005). HLI98 represents a small-molecule MDM2 inhibitor of the first generation that blocks the E3 ligase activity of MDM2 leading to stabilization of p53, p53-dependent transcription and induction of cell death (Di et al, 2011; Yang et al, 2005). A structural study confirmed the mode of action of small-molecule MDM2 antagonists nutlins showing that these compounds bind MDM2 via the p53-binding pocket, thereby preventing its interaction with p53 (Vassilev et al, 2004). RITA (2,5 bis(5-hydroxy-methyl-2thienyl)furan) targets the interaction between p53 and MDM2. However, both RITA and nutlins are shown to exhibit off-target effects. Currently, Johnson and Johnson reported benzodiazepinedione inhibitors that selectively kill p53 wild-type cells (Koblish et al, 2006) and a phase I clinical trial with the lead compound JNJ-26854165 has just been completed.
S-phase kinase-associated protein 2 (SKP2) is another E3 ligase that is considered as a promising cancer drug target. SKP2 is a substrate-specific subunit of the SCF ligase complex that targets cell cycle regulators such as p27 for proteasomal degradation (Bedford et al, 2011). Inhibition of SKP2 is expected to result in increased levels of the cell cycle inhibitor p27, restraining cell proliferation. Indeed, targeting SKP2 has been reported to lead to p27- and SKP2-mediated cell cycle arrest (Chen et al, 2008). The discovery of NEDD8 modification of cullins for E3 ligase activity prompted millenium pharmaceutics to develop a NEDD-8-specific E1 inhibitor (ML4924). ML4924 is currently in phase I clinical trials for treatment of hematologic malignancies and melanoma.
Inhibitor of apoptosis proteins represent another well-known family of E3 ligases that are involved in the regulation of cell death and NF-κB signalling. Since IAP proteins are expressed at aberrantly high levels in various cancers and have been associated with poor treatment response, they are currently undergoing evaluation as cancer drug targets (Straub, 2011). Accordingly, several approaches to neutralize IAP expression and function have been developed in recent years which are currently tested in early clinical trials (Fulda & Vucic, 2012). For example, Smac mimetics that bind to IAP proteins such as X-linked inhibitor of apoptosis protein (XIAP) and cIAPs in a manner similar to the endogenous mitochondrial protein Smac have been shown to neutralize IAP-imposed inhibition of caspases and to trigger their autoubiquitylation and degradation (Fulda & Vucic, 2012). Treatment of tumour cells with Smac mimetic compounds has been shown to sensitize tumour cells both in vitro and in vivo and many of these compounds are already in clinical trials (Fulda & Vucic, 2012; Table 2).
Apart from targeting the E3 ligases, attempts have also been made to target the E1 activation and ubiquitin-UBD interactions. PYR-41 was found in a small-molecule screen as an inhibitor of E1 activity though no effects on anti-tumour activity were reported (Yang et al, 2007). Ubistatins are the first class of compounds identified to disrupt the interaction between ubiquitin and the ubiquitin-binding domains (Verma et al, 2004). However, they are not pursued further, as they are unable to penetrate cell membranes. Despite the existing problems, these pilot studies have already opened up an avenue for targeting protein–protein interactions to regulate ubiquitin signalling. Improved structural analysis accompanied by a better understanding of the molecular mechanisms driving the function of ubiquitylating enzymes will help to develop more selective inhibitors.
Conclusions and future perspectives
The last decade has seen a phenomenal surge in the interest on ubiquitin signalling which has launched PTMs via ubiquitin moieties as a central regulator of virtually all cellular processes (Grabbe et al, 2011). The clinical success of bortezomib has proved the importance of targeting the UPS machinery and several proteasome inhibitors are being currently pursued in clinical trials. Efforts to target conjugation enzymes (E1-3) have been pursued with some success. There are a growing number of E3 ligases being implied in various tumours and drugs targeting some of these enzymes like IAP antagonists or MDM2 inhibitors are already in clinical trials. The role of NF-κB in the pathology of autoimmune disorders has been well established and efforts have been made to circumvent uncontrolled inflammation by attenuating cytokine responses. Infliximab is a successful drug of this kind and more effective inhibitors are being developed to inhibit downstream signalling.
Like in many cases, these drugs exhibit side effects and only further insights into the regulation of signal transduction pathways that are deregulated in cancer and autoimmune disorders will allow us to develop more specific therapeutic agents. Studies involving in vivo mouse disease models continue to prove their worth albeit limitations. The recent advancements in mass spectrometric analyses and structural studies have significantly improved our understanding of the ubiquitin machinery. Only in 2006, we have uncovered the ligases responsible for synthesizing linear ubiquitin chains and quickly in vivo studies have revealed their importance for immunity and inflammation. The role of atypical ubiquitin chains and their role in regulating normal physiology is an exciting area of research and unveiling the complex cross talks between ubiquitylation and other PTMs will continue to be a challenging issue, as exemplified by recent findings that phosphorylation controls the ubiquitin-dependent selective autophagy processes (Matsumoto et al, 2011; Wild et al, 2011). Interdisciplinary approaches have substantially enhanced our understanding of the ubiquitin machinery in the last years and there is no doubt that drugs targeting the ubiquitin machinery either alone and in combination with others will be saviours of the future.
Pending issues
The functional interplay between various PTMs.
What is the basis of spatio-temporal regulation in the ubiquitin system?
How is the length and linkage specificity of ubiquitin chains controlled on endogenous substrates in vivo?
What are the (patho)physiological roles of atypical ubiquitin chains?
How is the UPS epigenetically regulated?
Acknowledgments
The authors declare that they have no conflict of interest.
References
- Adams JM, Cory S. The Bcl-2 apoptotic switch in cancer development and therapy. Oncogene. 2007;26:1324–1337. doi: 10.1038/sj.onc.1210220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J, Gordon A, Pritchard-Jones K, Shipley J. Genes, chromosomes, and rhabdomyosarcoma. Genes, Chromosomes, Cancer. 1999;26:275–285. [PubMed] [Google Scholar]
- Annunziata CM, Davis RE, Demchenko Y, Bellamy W, Gabrea A, Zhan F, Lenz G, Hanamura I, Wright G, Xiao W, et al. Frequent engagement of the classical and alternative NF-kappaB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell. 2007;12:115–130. doi: 10.1016/j.ccr.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandi SR, Brandts C, Rensinghoff M, Grundler R, Tickenbrock L, Kohler G, Duyster J, Berdel WE, Muller-Tidow C, Serve H, et al. E3 ligase-defective Cbl mutants lead to a generalized mastocytosis and myeloproliferative disease. Blood. 2009;114:4197–4208. doi: 10.1182/blood-2008-12-190934. [DOI] [PubMed] [Google Scholar]
- Bedford L, Lowe J, Dick LR, Mayer RJ, Brownell JE. Ubiquitin-like protein conjugation and the ubiquitin-proteasome system as drug targets. Nat Rev Drug Discov. 2011;10:29–46. doi: 10.1038/nrd3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergink S, Jentsch S. Principles of ubiquitin and SUMO modifications in DNA repair. Nature. 2009;458:461–467. doi: 10.1038/nature07963. [DOI] [PubMed] [Google Scholar]
- Bertrand MJ, Doiron K, Labbe K, Korneluk RG, Barker PA, Saleh M. Cellular inhibitors of apoptosis cIAP1 and cIAP2 are required for innate immunity signaling by the pattern recognition receptors NOD1 and NOD2. Immunity. 2009;30:789–801. doi: 10.1016/j.immuni.2009.04.011. [DOI] [PubMed] [Google Scholar]
- Bhoj VG, Chen ZJ. Ubiquitylation in innate and adaptive immunity. Nature. 2009;458:430–437. doi: 10.1038/nature07959. [DOI] [PubMed] [Google Scholar]
- Bignell GR, Warren W, Seal S, Takahashi M, Rapley E, Barfoot R, Green H, Brown C, Biggs PJ, Lakhani SR, et al. Identification of the familial cylindromatosis tumour-suppressor gene. Nat Genet. 2000;25:160–165. doi: 10.1038/76006. [DOI] [PubMed] [Google Scholar]
- Bonnet MC, Preukschat D, Welz PS, van Loo G, Ermolaeva MA, Bloch W, Haase I, Pasparakis M. The adaptor protein FADD protects epidermal keratinocytes from necroptosis in vivo and prevents skin inflammation. Immunity. 2011;35:572–582. doi: 10.1016/j.immuni.2011.08.014. [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Nijman SM, Dirac AM, Bernards R. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-kappaB. Nature. 2003;424:797–801. doi: 10.1038/nature01811. [DOI] [PubMed] [Google Scholar]
- Cardozo T, Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Mol Cell Biol. 2004;5:739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- Casas S, Nagy B, Elonen E, Aventin A, Larramendy ML, Sierra J, Ruutu T, Knuutila S. Aberrant expression of HOXA9, DEK, CBL and CSF1R in acute myeloid leukemia. Leuk Lymphoma. 2003;44:1935–1941. doi: 10.1080/1042819031000119299. [DOI] [PubMed] [Google Scholar]
- Chen Q, Xie W, Kuhn DJ, Voorhees PM, Lopez-Girona A, Mendy D, Corral LG, Krenitsky VP, Xu W, Moutouh-de Parseval L, et al. Targeting the p27 E3 ligase SCF(Skp2) results in p27- and Skp2-mediated cell-cycle arrest and activation of autophagy. Blood. 2008;111:4690–4699. doi: 10.1182/blood-2007-09-112904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Shan J, Zhu WG, Qin J, Gu W. Transcription-independent ARF regulation in oncogenic stress-mediated p53 responses. Nature. 2010;464:624–627. doi: 10.1038/nature08820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Frezza M, Schmitt S, Kanwar J, Dou QP. Bortezomib as the first proteasome inhibitor anticancer drug: current status and future perspectives. Curr Cancer Drug Targets. 2011;11:239–253. doi: 10.2174/156800911794519752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clague MJ, Coulson JM, Urbe S. Cellular functions of the DUBs. J Cell Sci. 2012;125:277–286. doi: 10.1242/jcs.090985. [DOI] [PubMed] [Google Scholar]
- Crusio KM, King B, Reavie LB, Aifantis I. The ubiquitous nature of cancer: the role of the SCF(Fbw7) complex in development and transformation. Oncogene. 2010;29:4865–4873. doi: 10.1038/onc.2010.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejardin E. The alternative NF-kappaB pathway from biochemistry to biology: pitfalls and promises for future drug development. Biochem Pharmacol. 2006;72:1161–1179. doi: 10.1016/j.bcp.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Deribe YL, Pawson T, Dikic I. Post-translational modifications in signal integration. Nat Struct Mol Biol. 2010;17:666–672. doi: 10.1038/nsmb.1842. [DOI] [PubMed] [Google Scholar]
- Di J, Zheng Y, Zhang J. Reactivation of p53 by inhibiting Mdm2 E3 ligase: a novel antitumor approach. Curr Cancer Drug Targets. 2011;11:987–994. doi: 10.2174/156800911797264789. [DOI] [PubMed] [Google Scholar]
- Dierlamm J, Baens M, Wlodarska I, Stefanova-Ouzounova M, Hernandez JM, Hossfeld DK, De Wolf-Peeters C, Hagemeijer A, Van den Berghe H, Marynen P. The apoptosis inhibitor gene API2 and a novel 18q gene, MLT, are recurrently rearranged in the t(11;18)(q21;q21) associated with mucosa-associated lymphoid tissue lymphomas. Blood. 1999;93:3601–3609. [PubMed] [Google Scholar]
- Dikic I, Wakatsuki S, Walters KJ. Ubiquitin-binding domains—from structures to functions. Nat Rev Mol Cell Biol. 2009;10:659–671. doi: 10.1038/nrm2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan S, Cermak L, Pagan JK, Rossi M, Martinengo C, di Celle PF, Chapuy B, Shipp M, Chiarle R, Pagano M. FBXO11 targets BCL6 for degradation and is inactivated in diffuse large B-cell lymphomas. Nature. 2012;481:90–93. doi: 10.1038/nature10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duprez L, Takahashi N, Van Hauwermeiren F, Vandendriessche B, Goossens V, Vanden Berghe T, Declercq W, Libert C, Cauwels A, Vandenabeele P. RIP kinase-dependent necrosis drives lethal systemic inflammatory response syndrome. Immunity. 2011;35:908–918. doi: 10.1016/j.immuni.2011.09.020. [DOI] [PubMed] [Google Scholar]
- Feoktistova M, Geserick P, Kellert B, Dimitrova DP, Langlais C, Hupe M, Cain K, MacFarlane M, Hacker G, Leverkus M. cIAPs block Ripoptosome formation, a RIP1/caspase-8 containing intracellular cell death complex differentially regulated by cFLIP isoforms. Mol Cell. 2011;43:449–463. doi: 10.1016/j.molcel.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman CS, O'Donnell MA, Legarda-Addison D, Ng A, Cardenas WB, Yount JS, Moran TM, Basler CF, Komuro A, Horvath CM, et al. The tumour suppressor CYLD is a negative regulator of RIG-I-mediated antiviral response. EMBO Rep. 2008;9:930–936. doi: 10.1038/embor.2008.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulda S, Vucic D. Targeting IAP proteins for therapeutic intervention in cancer. Nat Rev Drug Disc. 2012;11:109–124. doi: 10.1038/nrd3627. [DOI] [PubMed] [Google Scholar]
- Georgescu MM. PTEN tumor suppressor network in PI3K-Akt pathway control. Genes Cancer. 2010;1:1170–1177. doi: 10.1177/1947601911407325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach B, Cordier SM, Schmukle AC, Emmerich CH, Rieser E, Haas TL, Webb AI, Rickard JA, Anderton H, Wong WW, et al. Linear ubiquitination prevents inflammation and regulates immune signalling. Nature. 2011;471:591–596. doi: 10.1038/nature09816. [DOI] [PubMed] [Google Scholar]
- Grabbe C, Husnjak K, Dikic I. The spatial and temporal organization of ubiquitin networks. Nat Rev Mol Cell Biol. 2011;12:295–307. doi: 10.1038/nrm3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grand FH, Hidalgo-Curtis CE, Ernst T, Zoi K, Zoi C, McGuire C, Kreil S, Jones A, Score J, Metzgeroth G, et al. Frequent CBL mutations associated with 11q acquired uniparental disomy in myeloproliferative neoplasms. Blood. 2009;113:6182–6192. doi: 10.1182/blood-2008-12-194548. [DOI] [PubMed] [Google Scholar]
- Gunther C, Martini E, Wittkopf N, Amann K, Weigmann B, Neumann H, Waldner MJ, Hedrick SM, Tenzer S, Neurath MF, et al. Caspase-8 regulates TNF-alpha-induced epithelial necroptosis and terminal ileitis. Nature. 2011;477:335–339. doi: 10.1038/nature10400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haglund K, Dikic I. Ubiquitylation and cell signaling. EMBO J. 2005;24:3353–3359. doi: 10.1038/sj.emboj.7600808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Zhong CQ, Zhang DW. Programmed necrosis: backup to and competitor with apoptosis in the immune system. Nat Immunol. 2011;12:1143–1149. doi: 10.1038/ni.2159. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hirsch C, Gauss R, Horn SC, Neuber O, Sommer T. The ubiquitylation machinery of the endoplasmic reticulum. Nature. 2009;458:453–460. doi: 10.1038/nature07962. [DOI] [PubMed] [Google Scholar]
- Hoeller D, Dikic I. Targeting the ubiquitin system in cancer therapy. Nature. 2009;458:438–444. doi: 10.1038/nature07960. [DOI] [PubMed] [Google Scholar]
- Hovelmeyer N, Wunderlich FT, Massoumi R, Jakobsen CG, Song J, Worns MA, Merkwirth C, Kovalenko A, Aumailley M, Strand D, et al. Regulation of B cell homeostasis and activation by the tumor suppressor gene CYLD. J Exp Med. 2007;204:2615–2627. doi: 10.1084/jem.20070318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda F, Dikic I. Atypical ubiquitin chains: new molecular signals. ‘Protein Modifications: Beyond the Usual Suspects’ review series. EMBO Rep. 2008;9:536–542. doi: 10.1038/embor.2008.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda F, Deribe YL, Skanland SS, Stieglitz B, Grabbe C, Franz-Wachtel M, van Wijk SJ, Goswami P, Nagy V, Terzic J, et al. SHARPIN forms a linear ubiquitin ligase complex regulating NF-kappaB activity and apoptosis. Nature. 2011;471:637–641. doi: 10.1038/nature09814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inuzuka H, Shaik S, Onoyama I, Gao D, Tseng A, Maser RS, Zhai B, Wan L, Gutierrez A, Lau AW, et al. SCF(FBW7) regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature. 2011;471:104–109. doi: 10.1038/nature09732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai K, Tokunaga F. Linear polyubiquitination: a new regulator of NF-kappaB activation. EMBO Rep. 2009;10:706–713. doi: 10.1038/embor.2009.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- Jin W, Reiley WR, Lee AJ, Wright A, Wu X, Zhang M, Sun SC. Deubiquitinating enzyme CYLD regulates the peripheral development and naive phenotype maintenance of B cells. J Biol Chem. 2007;282:15884–15893. doi: 10.1074/jbc.M609952200. [DOI] [PubMed] [Google Scholar]
- Kaiser WJ, Upton JW, Long AB, Livingston-Rosanoff D, Daley-Bauer LP, Hakem R, Caspary T, Mocarski ES. RIP3 mediates the embryonic lethality of caspase-8-deficient mice. Nature. 2011;471:368–372. doi: 10.1038/nature09857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Sanada M, Kato I, Sato Y, Takita J, Takeuchi K, Niwa A, Chen Y, Nakazaki K, Nomoto J, et al. Frequent inactivation of A20 in B-cell lymphomas. Nature. 2009;459:712–716. doi: 10.1038/nature07969. [DOI] [PubMed] [Google Scholar]
- Keats JJ, Fonseca R, Chesi M, Schop R, Baker A, Chng WJ, Van Wier S, Tiedemann R, Shi CX, Sebag M, et al. Promiscuous mutations activate the noncanonical NF-kappaB pathway in multiple myeloma. Cancer Cell. 2007;12:131–144. doi: 10.1016/j.ccr.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- Koblish HK, Zhao S, Franks CF, Donatelli RR, Tominovich RM, LaFrance LV, Leonard KA, Gushue JM, Parks DJ, Calvo RR, et al. Benzodiazepinedione inhibitors of the Hdm2:p53 complex suppress human tumor cell proliferation in vitro and sensitize tumors to doxorubicin in vivo. Mol Cancer Ther. 2006;5:160–169. doi: 10.1158/1535-7163.MCT-05-0199. [DOI] [PubMed] [Google Scholar]
- Kovalenko A, Chable-Bessia C, Cantarella G, Israel A, Wallach D, Courtois G. The tumour suppressor CYLD negatively regulates NF-kappaB signalling by deubiquitination. Nature. 2003;424:801–805. doi: 10.1038/nature01802. [DOI] [PubMed] [Google Scholar]
- Lee DF, Kuo HP, Liu M, Chou CK, Xia W, Du Y, Shen J, Chen CT, Huo L, Hsu MC, et al. KEAP1 E3 ligase-mediated downregulation of NF-kappaB signaling by targeting IKKbeta. Mol Cell. 2009;36:131–140. doi: 10.1016/j.molcel.2009.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JH, Stirling B, Derry J, Koga T, Jono H, Woo CH, Xu H, Bourne P, Ha UH, Ishinaga H, et al. Tumor suppressor CYLD regulates acute lung injury in lethal Streptococcus pneumoniae infections. Immunity. 2007;27:349–360. doi: 10.1016/j.immuni.2007.07.011. [DOI] [PubMed] [Google Scholar]
- Lim JH, Ha UH, Woo CH, Xu H, Li JD. CYLD is a crucial negative regulator of innate immune response in Escherichia coli pneumonia. Cell Microbiol. 2008;10:2247–2256. doi: 10.1111/j.1462-5822.2008.01204.x. [DOI] [PubMed] [Google Scholar]
- Lind H, Zienolddiny S, Ekstrom PO, Skaug V, Haugen A. Association of a functional polymorphism in the promoter of the MDM2 gene with risk of nonsmall cell lung cancer. Int J Cancer. 2006;119:718–721. doi: 10.1002/ijc.21872. [DOI] [PubMed] [Google Scholar]
- Lipkowitz S, Weissman AM. RINGs of good and evil: RING finger ubiquitin ligases at the crossroads of tumour suppression and oncogenesis. Nat Rev Cancer. 2011;11:629–643. doi: 10.1038/nrc3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann M, Jensen ON. Proteomic analysis of post-translational modifications. Nat Biotechnol. 2003;21:255–261. doi: 10.1038/nbt0303-255. [DOI] [PubMed] [Google Scholar]
- Massoumi R, Chmielarska K, Hennecke K, Pfeifer A, Fassler R. Cyld inhibits tumor cell proliferation by blocking Bcl-3-dependent NF-kappaB signaling. Cell. 2006;125:665–677. doi: 10.1016/j.cell.2006.03.041. [DOI] [PubMed] [Google Scholar]
- Masuya D, Huang C, Liu D, Nakashima T, Yokomise H, Ueno M, Nakashima N, Sumitomo S. The HAUSP gene plays an important role in non-small cell lung carcinogenesis through p53-dependent pathways. J Pathol. 2006;208:724–732. doi: 10.1002/path.1931. [DOI] [PubMed] [Google Scholar]
- Mathew R, White E. Why sick cells produce tumors: the protective role of autophagy. Autophagy. 2007;3:502–505. doi: 10.4161/auto.4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matmati M, Jacques P, Maelfait J, Verheugen E, Kool M, Sze M, Geboes L, Louagie E, Guire CM, Vereecke L, et al. A20 (TNFAIP3) deficiency in myeloid cells triggers erosive polyarthritis resembling rheumatoid arthritis. Nat Genet. 2011;43:908–912. doi: 10.1038/ng.874. [DOI] [PubMed] [Google Scholar]
- Matsumoto G, Wada K, Okuno M, Kurosawa M, Nukina N. Serine 403 phosphorylation of p62/SQSTM1 regulates selective autophagic clearance of ubiquitinated proteins. Mol Cell. 2011;44:279–289. doi: 10.1016/j.molcel.2011.07.039. [DOI] [PubMed] [Google Scholar]
- Menin C, Scaini MC, De Salvo GL, Biscuola M, Quaggio M, Esposito G, Belluco C, Montagna M, Agata S, D'Andrea E, et al. Association between MDM2-SNP309 and age at colorectal cancer diagnosis according to p53 mutation status. J Natl Cancer Inst. 2006;98:285–288. doi: 10.1093/jnci/djj054. [DOI] [PubMed] [Google Scholar]
- Miliani de Marval PL, Zhang Y. The RP-Mdm2-p53 pathway and tumorigenesis. Oncotarget. 2011;2:234–238. doi: 10.18632/oncotarget.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina TJ, Kishihara K, Siderovski DP, van Ewijk W, Narendran A, Timms E, Wakeham A, Paige CJ, Hartmann KU, Veillette A, et al. Profound block in thymocyte development in mice lacking p56lck. Nature. 1992;357:161–164. doi: 10.1038/357161a0. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay D, Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007;315:201–205. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- Naumann I, Kappler R, von Schweinitz D, Debatin KM, Fulda S. Bortezomib primes neuroblastoma cells for TRAIL-induced apoptosis by linking the death receptor to the mitochondrial pathway. Clin Cancer Res. 2011;17:3204–3218. doi: 10.1158/1078-0432.CCR-10-2451. [DOI] [PubMed] [Google Scholar]
- Oberoi TK, Dogan T, Hocking JC, Scholz RP, Mooz J, Anderson CL, Karreman C, Meyer Zu Heringdorf D, Schmidt G, Ruonala M, et al. IAPs regulate the plasticity of cell migration by directly targeting Rac1 for degradation. EMBO J. 2012;31:14–28. doi: 10.1038/emboj.2011.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onoyama I, Tsunematsu R, Matsumoto A, Kimura T, de Alboran IM, Nakayama K, Nakayama KI. Conditional inactivation of Fbxw7 impairs cell-cycle exit during T cell differentiation and results in lymphomatogenesis. J Exp Med. 2007;204:2875–2888. doi: 10.1084/jem.20062299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto T, Horn S, Brockmann M, Eilers U, Schuttrumpf L, Popov N, Kenney AM, Schulte JH, Beijersbergen R, Christiansen H, et al. Stabilization of N-Myc is a critical function of Aurora A in human neuroblastoma. Cancer Cell. 2009;15:67–78. doi: 10.1016/j.ccr.2008.12.005. [DOI] [PubMed] [Google Scholar]
- Pasparakis M. Regulation of tissue homeostasis by NF-kappaB signalling: implications for inflammatory diseases. Nat Rev Immunol. 2009;9:778–788. doi: 10.1038/nri2655. [DOI] [PubMed] [Google Scholar]
- Pereg Y, Liu BY, O'Rourke KM, Sagolla M, Dey A, Komuves L, French DM, Dixit VM. Ubiquitin hydrolase Dub3 promotes oncogenic transformation by stabilizing Cdc25A. Nat Cell Biol. 2010;12:400–406. doi: 10.1038/ncb2041. [DOI] [PubMed] [Google Scholar]
- Perkins ND. Integrating cell-signalling pathways with NF-kappaB and IKK function. Nat Rev Mol Cell Biol. 2007;8:49–62. doi: 10.1038/nrm2083. [DOI] [PubMed] [Google Scholar]
- Rahighi S, Ikeda F, Kawasaki M, Akutsu M, Suzuki N, Kato R, Kensche T, Uejima T, Bloor S, Komander D, et al. Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell. 2009;136:1098–1109. doi: 10.1016/j.cell.2009.03.007. [DOI] [PubMed] [Google Scholar]
- Reiley W, Zhang M, Wu X, Granger E, Sun SC. Regulation of the deubiquitinating enzyme CYLD by IkappaB kinase gamma-dependent phosphorylation. Mol Cell Biol. 2005;25:3886–3895. doi: 10.1128/MCB.25.10.3886-3895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiley WW, Zhang M, Jin W, Losiewicz M, Donohue KB, Norbury CC, Sun SC. Regulation of T cell development by the deubiquitinating enzyme CYLD. Nat Immunol. 2006;7:411–417. doi: 10.1038/ni1315. [DOI] [PubMed] [Google Scholar]
- Reiley WW, Jin W, Lee AJ, Wright A, Wu X, Tewalt EF, Leonard TO, Norbury CC, Fitzpatrick L, Zhang M, et al. Deubiquitinating enzyme CYLD negatively regulates the ubiquitin-dependent kinase Tak1 and prevents abnormal T cell responses. J Exp Med. 2007;204:1475–1485. doi: 10.1084/jem.20062694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruschak AM, Slassi M, Kay LE, Schimmer AD. Novel proteasome inhibitors to overcome bortezomib resistance. J Natl Cancer Inst. 2011;103:1007–1017. doi: 10.1093/jnci/djr160. [DOI] [PubMed] [Google Scholar]
- Sargin B, Choudhary C, Crosetto N, Schmidt MH, Grundler R, Rensinghoff M, Thiessen C, Tickenbrock L, Schwable J, Brandts C, et al. Flt3-dependent transformation by inactivating c-Cbl mutations in AML. Blood. 2007;110:1004–1012. doi: 10.1182/blood-2007-01-066076. [DOI] [PubMed] [Google Scholar]
- Sayers TJ, Murphy WJ. Combining proteasome inhibition with TNF-related apoptosis-inducing ligand (Apo2L/TRAIL) for cancer therapy. Cancer Immunol Immunother. 2006;55:76–84. doi: 10.1007/s00262-005-0676-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt MH, Dikic I. The Cbl interactome and its functions. Nat Rev Mol Cell Biol. 2005;6:907–918. doi: 10.1038/nrm1762. [DOI] [PubMed] [Google Scholar]
- Schulman BA, Harper JW. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat Rev Mol Cell Biol. 2009;10:319–331. doi: 10.1038/nrm2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwickart M, Huang X, Lill JR, Liu J, Ferrando R, French DM, Maecker H, O'Rourke K, Bazan F, Eastham-Anderson J, et al. Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature. 2010;463:103–107. doi: 10.1038/nature08646. [DOI] [PubMed] [Google Scholar]
- Seet BT, Dikic I, Zhou MM, Pawson T. Reading protein modifications with interaction domains. Nat Rev Mol Cell Biol. 2006;7:473–483. doi: 10.1038/nrm1960. [DOI] [PubMed] [Google Scholar]
- Seymour RE, Hasham MG, Cox GA, Shultz LD, Hogenesch H, Roopenian DC, Sundberg JP. Spontaneous mutations in the mouse Sharpin gene result in multiorgan inflammation, immune system dysregulation and dermatitis. Genes Immun. 2007;8:416–421. doi: 10.1038/sj.gene.6364403. [DOI] [PubMed] [Google Scholar]
- Song MS, Salmena L, Carracedo A, Egia A, Lo-Coco F, Teruya-Feldstein J, Pandolfi PP. The deubiquitinylation and localization of PTEN are regulated by a HAUSP-PML network. Nature. 2008;455:813–817. doi: 10.1038/nature07290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub CS. Targeting IAPs as an approach to anti-cancer therapy. Curr Top Med Chem. 2011;11:291–316. doi: 10.2174/156802611794072623. [DOI] [PubMed] [Google Scholar]
- Strobel P, Zettl A, Ren Z, Starostik P, Riedmiller H, Storkel S, Muller-Hermelink HK, Marx A. Spiradenocylindroma of the kidney: clinical and genetic findings suggesting a role of somatic mutation of the CYLD1 gene in the oncogenesis of an unusual renal neoplasm. Am J Surg Pathol. 2002;26:119–124. doi: 10.1097/00000478-200201000-00016. [DOI] [PubMed] [Google Scholar]
- Sun SC. Deubiquitylation and regulation of the immune response. Nat Rev Immunol. 2008;8:501–511. doi: 10.1038/nri2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun SC. CYLD: a tumor suppressor deubiquitinase regulating NF-kappaB activation and diverse biological processes. Cell Death Differ. 2010;17:25–34. doi: 10.1038/cdd.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- Tenev T, Bianchi K, Darding M, Broemer M, Langlais C, Wallberg F, Zachariou A, Lopez J, Macfarlane M, Cain K, et al. The Ripoptosome, a signaling platform that assembles in response to genotoxic stress and loss of IAPs. Mol cell. 2011;43:432–448. doi: 10.1016/j.molcel.2011.06.006. [DOI] [PubMed] [Google Scholar]
- Tokunaga F, Nakagawa T, Nakahara M, Saeki Y, Taniguchi M, Sakata S, Tanaka K, Nakano H, Iwai K. SHARPIN is a component of the NF-kappaB-activating linear ubiquitin chain assembly complex. Nature. 2011;471:633–636. doi: 10.1038/nature09815. [DOI] [PubMed] [Google Scholar]
- Trotman LC, Wang X, Alimonti A, Chen Z, Teruya-Feldstein J, Yang H, Pavletich NP, Carver BS, Cordon-Cardo C, Erdjument-Bromage H, et al. Ubiquitination regulates PTEN nuclear import and tumor suppression. Cell. 2007;128:141–156. doi: 10.1016/j.cell.2006.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterkircher T, Cristofanon S, Vellanki SH, Nonnenmacher L, Karpel-Massler G, Wirtz CR, Debatin KM, Fulda S. Bortezomib primes glioblastoma, including glioblastoma stem cells, for TRAIL by increasing tBid stability and mitochondrial apoptosis. Clin Cancer Res. 2011;17:4019–4030. doi: 10.1158/1078-0432.CCR-11-0075. [DOI] [PubMed] [Google Scholar]
- Vallabhapurapu S, Matsuzawa A, Zhang W, Tseng PH, Keats JJ, Wang H, Vignali DA, Bergsagel PL, Karin M. Nonredundant and complementary functions of TRAF2 and TRAF3 in a ubiquitination cascade that activates NIK-dependent alternative NF-kappaB signaling. Nat Immunol. 2008;9:1364–1370. doi: 10.1038/ni.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenabeele P, Galluzzi L, Vanden Berghe T, Kroemer G. Molecular mechanisms of necroptosis: an ordered cellular explosion. Nat Rev Mol Cell Biol. 2010;11:700–714. doi: 10.1038/nrm2970. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. Regulated protein degradation. Trends Biochem Sci. 2005;30:283–286. doi: 10.1016/j.tibs.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- Vereecke L, Beyaert R, van Loo G. The ubiquitin-editing enzyme A20 (TNFAIP3) is a central regulator of immunopathology. Trends Immunol. 2009;30:383–391. doi: 10.1016/j.it.2009.05.007. [DOI] [PubMed] [Google Scholar]
- Verma R, Peters NR, D'Onofrio M, Tochtrop GP, Sakamoto KM, Varadan R, Zhang M, Coffino P, Fushman D, Deshaies RJ, et al. Ubistatins inhibit proteasome-dependent degradation by binding the ubiquitin chain. Science. 2004;306:117–120. doi: 10.1126/science.1100946. [DOI] [PubMed] [Google Scholar]
- Vivanco I, Rohle D, Versele M, Iwanami A, Kuga D, Oldrini B, Tanaka K, Dang J, Kubek S, Palaskas N, et al. The phosphatase and tensin homolog regulates epidermal growth factor receptor (EGFR) inhibitor response by targeting EGFR for degradation. Proc Natl Acad Sci USA. 2010;107:6459–6464. doi: 10.1073/pnas.0911188107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vucic D, Dixit VM, Wertz IE. Ubiquitylation in apoptosis: a post-translational modification at the edge of life and death. Nat Rev Mol Cell Biol. 2011;12:439–452. doi: 10.1038/nrm3143. [DOI] [PubMed] [Google Scholar]
- Walczak H, Iwai K, Dikic I. Generation and physiological roles of linear ubiquitin chains. BMC Biol. 2012;10:23. doi: 10.1186/1741-7007-10-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Trotman LC, Koppie T, Alimonti A, Chen Z, Gao Z, Wang J, Erdjument-Bromage H, Tempst P, Cordon-Cardo C, et al. NEDD4-1 is a proto-oncogenic ubiquitin ligase for PTEN. Cell. 2007;128:129–139. doi: 10.1016/j.cell.2006.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Du F, Wang X. TNF-alpha induces two distinct caspase-8 activation pathways. Cell. 2008;133:693–703. doi: 10.1016/j.cell.2008.03.036. [DOI] [PubMed] [Google Scholar]
- Welz PS, Wullaert A, Vlantis K, Kondylis V, Fernandez-Majada V, Ermolaeva M, Kirsch P, Sterner-Kock A, van Loo G, Pasparakis M. FADD prevents RIP3-mediated epithelial cell necrosis and chronic intestinal inflammation. Nature. 2011;477:330–334. doi: 10.1038/nature10273. [DOI] [PubMed] [Google Scholar]
- Wertz IE, Dixit VM. Signaling to NF-kappaB: regulation by ubiquitination. Cold Spring Harb Perspect Biol. 2010;2:a003350. doi: 10.1101/cshperspect.a003350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz IE, O'Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, Wu P, Wiesmann C, Baker R, Boone DL, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- Wertz IE, Kusam S, Lam C, Okamoto T, Sandoval W, Anderson DJ, Helgason E, Ernst JA, Eby M, Liu J, et al. Sensitivity to antitubulin chemotherapeutics is regulated by MCL1 and FBW7. Nature. 2011;471:110–114. doi: 10.1038/nature09779. [DOI] [PubMed] [Google Scholar]
- Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, Richter B, Korac J, Waidmann O, Choudhary C, et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333:228–233. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Ludwig RL, Jensen JP, Pierre SA, Medaglia MV, Davydov IV, Safiran YJ, Oberoi P, Kenten JH, Phillips AC, et al. Small molecule inhibitors of HDM2 ubiquitin ligase activity stabilize and activate p53 in cells. Cancer Cell. 2005;7:547–559. doi: 10.1016/j.ccr.2005.04.029. [DOI] [PubMed] [Google Scholar]
- Yang Y, Kitagaki J, Dai RM, Tsai YC, Lorick KL, Ludwig RL, Pierre SA, Jensen JP, Davydov IV, Oberoi P, et al. Inhibitors of ubiquitin-activating enzyme (E1), a new class of potential cancer therapeutics. Cancer Res. 2007;67:9472–9481. doi: 10.1158/0008-5472.CAN-07-0568. [DOI] [PubMed] [Google Scholar]
- Yang WL, Wang J, Chan CH, Lee SW, Campos AD, Lamothe B, Hur L, Grabiner BC, Lin X, Darnay BG, et al. The E3 ligase TRAF6 regulates Akt ubiquitination and activation. Science. 2009;325:1134–1138. doi: 10.1126/science.1175065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim EK, Peng G, Dai H, Hu R, Li K, Lu Y, Mills GB, Meric-Bernstam F, Hennessy BT, Craven RJ, et al. Rak functions as a tumor suppressor by regulating PTEN protein stability and function. Cancer Cell. 2009;15:304–314. doi: 10.1016/j.ccr.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Luo K, Zhang L, Cheville JC, Lou Z. USP10 regulates p53 localization and stability by deubiquitinating p53. Cell. 2010;140:384–396. doi: 10.1016/j.cell.2009.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarnegar BJ, Wang Y, Mahoney DJ, Dempsey PW, Cheung HH, He J, Shiba T, Yang X, Yeh WC, Mak TW, et al. Noncanonical NF-kappaB activation requires coordinated assembly of a regulatory complex of the adaptors cIAP1, cIAP2, TRAF2 and TRAF3 and the kinase NIK. Nat Immunol. 2008;9:1371–1378. doi: 10.1038/ni.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zender L, Spector MS, Xue W, Flemming P, Cordon-Cardo C, Silke J, Fan ST, Luk JM, Wigler M, Hannon GJ, et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Stirling B, Temmerman ST, Ma CA, Fuss IJ, Derry JM, Jain A. Impaired regulation of NF-kappaB and increased susceptibility to colitis-associated tumorigenesis in CYLD-deficient mice. J Clin Invest. 2006;116:3042–3049. doi: 10.1172/JCI28746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Wu X, Lee AJ, Jin W, Chang M, Wright A, Imaizumi T, Sun SC. Regulation of IkappaB kinase-related kinases and antiviral responses by tumor suppressor CYLD. J Biol Chem. 2008;283:18621–18626. doi: 10.1074/jbc.M801451200. [DOI] [PMC free article] [PubMed] [Google Scholar]