Abstract
An increase in cardiac workload, ultimately resulting in hypertrophy, generates oxidative stress and therefore requires the activation of both survival and growth signal pathways. Here, we wanted to characterize the regulators, targets and mechanistic roles of miR-142, a microRNA (miRNA) negatively regulated during hypertrophy. We show that both miRNA-142-3p and -5p are repressed by serum-derived growth factors in cultured cardiac myocytes, in models of cardiac hypertrophy in vivo and in human cardiomyopathic hearts. Levels of miR-142 are inversely related to levels of acetyltransferase p300 and MAPK activity. When present, miR-142 inhibits both survival and growth pathways by directly targeting nodal regulators p300 and gp130. MiR-142 also potently represses multiple components of the NF-κB pathway, preventing cytokine-mediated NO production and blocks translation of α-actinin. Forced expression of miR-142 during hypertrophic growth induced extensive apoptosis and cardiac dysfunction; conversely, loss of miR-142 fully rescued cardiac function in a murine heart failure model. Downregulation of miR-142 is required to enable cytokine-mediated survival signalling during cardiac growth in response to haemodynamic stress and is a critical element of adaptive hypertrophy.
Keywords: apoptosis, epigenetics, hypertrophy, IL6st, nitric oxide
INTRODUCTION
Postnatal growth of the heart, as during childhood and athletic training, requires an increase in myocyte size, or hypertrophy (Grossman, 1980; Moore et al, 1980). Diseases such as hypertension and aortic valvular stenosis that increase cardiac workload also induce cardiac myocyte hypertrophy, often accompanied by alterations in the shape and structure of the heart and by varying degrees of fibrosis, attenuation of blood supply, metabolic alterations, changes in calcium handling and ultimately the activation of harmful signal transduction pathways (Anderson, 2009; Cohn et al, 2000; Ling et al, 2009; Olivetti et al, 1994). Although not established as a direct cause of heart failure, hypertrophy is a frequent precursor and companion of disease- and age-related cardiac dysfunction (Desai et al, in press; Howell, 1981; Lam et al, 2007; Olivetti et al, 1991). A better understanding of the mechanisms by which growth of the heart becomes dysfunctional is needed to improve treatment options for this highly prevalent disorder.
A coordinated reprogramming of gene expression is required for hypertrophy (Bishopric et al, 1987; Dorn et al, 2003) involving activation of multiple signal transduction molecules, including the mitogen-activated protein kinases (MAPK; Dorn & Force, 2005; Sanna et al, 2005), calcium-activated kinases and phosphatases (Backs et al, 2006; Passier et al, 2000; Song et al, 2006; Zhang et al, 2002) and transcription factors such as NF-κB (Craig et al, 2000; Frantz et al, 2003; Li et al, 2004; Mann, 2003; Morishita et al, 1997; Purcell et al, 2001). Activation of the survival pathway transduced via the IL6 receptor IL6st (also known as gp130) to the transcription factor STAT3 is essential for myocyte survival in the face of acute oxidative stress, which typically accompanies mechanical loading (Hilfiker-Kleiner et al, 2004; Hirota et al, 1999; van Empel & De Windt, 2004).
We have previously provided evidence that the coordinated activation of these multiple transcription programs during hypertrophy requires the acetyltransferase p300. p300 and the closely related CREB-binding protein (CBP) have many shared functions (Kalkhoven, 2004; Roth et al, 2003; Shikama et al, 2003; Vo & Goodman, 2001; Yao et al, 1998); haploinsufficiency of either protein leads to Rubinstein-Taybi Syndrome (Bartholdi et al, 2007; Roelfsema et al, 2005). However, the heart appears to have a specific requirement for p300: mice with loss of p300, but not CBP, have impaired expression of sarcomeric genes and die of heart failure between E9.5-11 (Shikama et al, 2003; Yao et al, 1998). Loss of p300 also impairs postnatal myocardial gene expression and growth (Bishopric et al, 1997; Slepak et al, 2001; Wei et al, 2008). p300 levels increase sharply during haemodynamic stress and heart failure, and even relatively small increases or decreases in p300 quantitatively affect the extent of adaptive hypertrophy (Wei et al, 2008), as well as the risk of heart failure (Morimoto et al, 2008; Wei et al, 2008). Hence, the elucidation of downstream effectors of p300 could provide insight into mechanisms of adaptive versus maladaptive cardiac growth.
MicroRNAs (miRNAs) are short ∼18–25 nucleotide non-coding RNAs (Lagos-Quintana et al, 2001; Lau et al, 2001; Lee et al, 1993; Lim et al, 2003; Reinhart et al, 2002) that bind to 7–8 bp complementary sequences in the 3′ untranslated region of target messenger RNAs (mRNAs), inducing their cleavage, or, less often, blocking their translation (Bartel, 2009; Guo et al, 2010). More than 60% of human protein-coding genes may be under the control of miRNAs (Friedman et al, 2009). miRNAs are generated from longer RNA transcripts through several steps of post-transcriptional processing to produce a short double-stranded miRNA; both strands may produce functional miRNAs, designated −5′ and −3′ (Obernosterer et al, 2006). A single miRNA can theoretically target many hundreds of genes (Lewis et al, 2005); in turn, a single mRNA transcript can be targeted by multiple miRNAs. Genes with related functions often have conserved binding sites for the same miRNAs, suggesting that some miRNAs have evolved to influence entire biological pathways (Bartel, 2009; Friedman et al, 2009). miRNAs have been implicated in normal cardiac development and function, as well as in cardiac disorders in humans and animal models (Callis et al, 2009; Care et al, 2007; Cheng et al, 2007; Dong et al, 2010; Landthaler et al, 2004; Sayed et al, 2007; Tatsuguchi et al, 2007; Thum et al, 2007, 2008; van Rooij et al, 2006, 2007; Zhao et al, 2007). Whether miRNAs act as downstream effectors or modulators of p300 during hypertrophy has not been established.
Here, we show that miR-142-5p and -3p, products of the same primary transcript, are downregulated during cardiac hypertrophy by mechanisms requiring p300 and MAP kinase activity. We show that miR-142 directly targets p300 as well as α-actinin, an essential component of the cardiac cytoskeleton. We demonstrate that miR-142 is a global inhibitor of cytokine signalling and function in the myocardium, at least in part through its ability to target gp130. Most strikingly, we show that preventing the physiological downregulation of miR-142 leads to depletion of cytokine-mediated survival signals, induction of apoptosis and development of heart failure during postnatal growth of the heart. We conclude that miR-142 repression is essential for successful cardiac adaptation to changing haemodynamic demand in vivo.
RESULTS
miR-142-5p and -3p are downregulated during cardiac hypertrophy
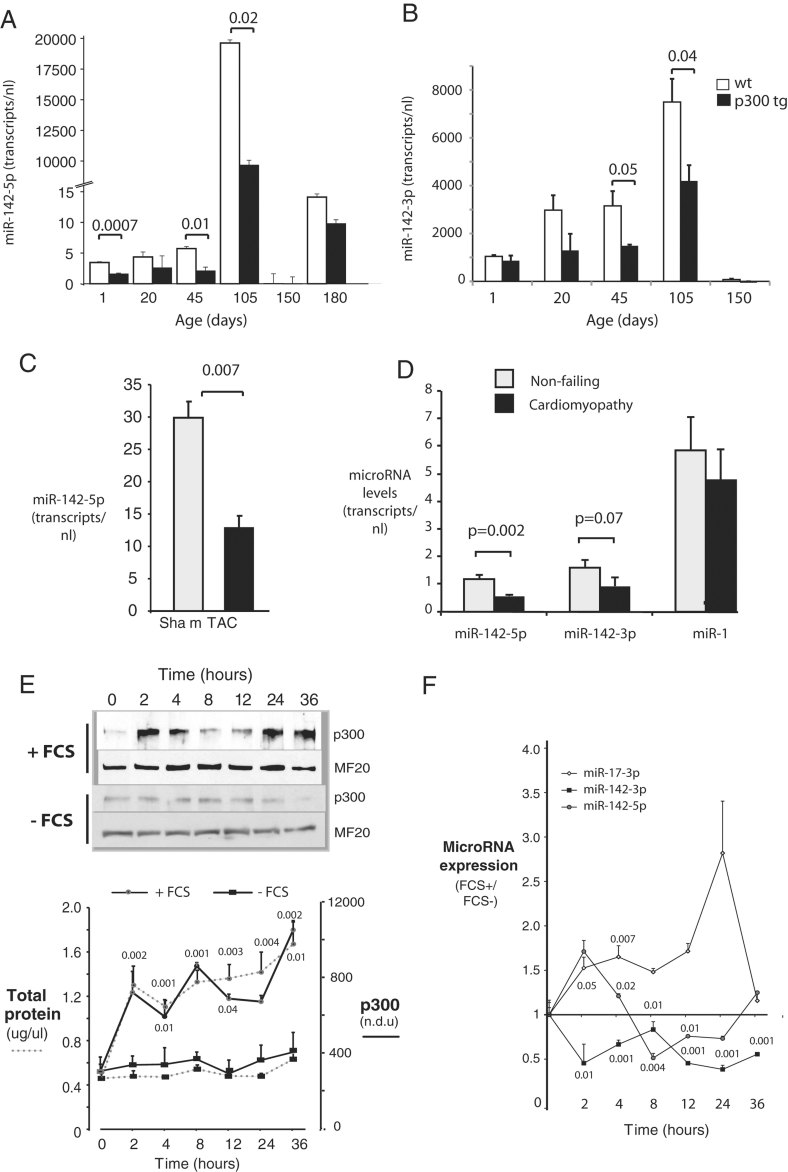
In both p300 transgenic (p300tg) and wild-type (wt) mice, miR-142-5p and -3p levels were very low in the period of adaptive cardiac growth between birth and adulthood at 2 months, relative to levels achieved after 3 months (Fig 1A and B). Both miR-142-5p and -3p were markedly reduced in left ventricular myocardium of p300tg mice compared to their wt littermates at every age examined between birth and 5 months (Fig 1A and B). Relative levels of miR-142-5p and -3p were also age-dependent: before 3 months, miR-142-5p was approximately 5% as abundant as miR-142-3P, which in turn was expressed at about 60% of levels of the highly abundant species miR-1 and miR-let-7c (Fig 1 of Supporting Information). However, miR-142-5p increased relative to miR-142-3p after 105 days of age (Fig 1A and B). Expression of both miR-142 transcripts correlated inversely with hypertrophy in several other systems: miR-142-5p was significantly lower in wt mouse hearts following acute surgically induced pressure overload (Fig 1C) and in human hearts with various types of cardiomyopathy and cardiac enlargement (Table 1 and Fig 1D).
Figure 1. MiR-142-5p and -3p are downregulated during cardiac hypertrophic growth.
- A,B. Repression of miR-142-5p and -3p during early postnatal cardiac growth. Hearts of wt and p300tg mouse littermates were harvested from mice at the indicated ages and miR-142-5p and -3p were quantitated in LV myocardium as described in Methods Section. n = 3 per group. p-Values comparing wt and p300tg hearts are shown where significant. (A) MiR-142-5p. (B) MiR-142-3p.
- C. Repression of miR-142-5p during pressure-overload hypertrophy. Transverse aortic coarctation (TAC) or a sham procedure was performed as descried in Methods Section and miR-142-5p was assayed in LV 3 days later. n = 3 per group.
- D. Repression of miR-142 in failing human heart. MicroRNAs were assayed in LV free wall from cardiomyopathic and non-failing hearts. n = 6 per group. For (A–D), microRNAs are expressed as transcripts per 10 ng total RNA.
- E-F. Repression of miR-142 during cardiac myocyte hypertrophy in culture. NRVM in serum-free culture were placed in fresh media with or without 5% FCS and assayed between 0 and 36 h. Myocyte protein content and p300 levels increase in response to FCS. (Above) Representative immunoblots for p300 and myosin heavy chain (MF20). (Below) Quantitation of myocyte total protein (dotted lines) and p300 content (solid lines) at indicated time points. + FCS, closed red circles; − FCS, closed black rectangles (E). Repression of miR-142 by serum stimulation. MicroRNAs 142-3p and -5p were quantitated by RT-PCR in the same cells as in E. MicroRNA expression levels are normalized to basal levels in unstimulated cells at time 0 (F). For (E–F), n = 3, and p-values are given for comparison between serum-stimulated and non-stimulated cells at the same time point.
Table 1.
Human subjects
| # | ID | Diagnosis | Age | Race | Gender |
|---|---|---|---|---|---|
| 1 | 1030814A1 | Cardiomyopathy | 29 | B | F |
| 2 | 1041601A3 | Cardiomyopathy | 53 | W | M |
| 3 | 1040221A2 | Cardiomyopathy and congestive heart failure | 33 | W | F |
| 4 | 1041487A3 | Ischemic cardiomyopathy | 64 | W | M |
| 5 | 1062110A2 | Ischemic cardiomyopathy | 62 | B | F |
| 6 | 45411A1A | Cardiomyopathy | 58 | W | M |
| 7 | 40695A1B@ | Normal | 70 | U | M |
| 8 | 40696A1B@ | Normal | 64 | U | M |
| 9 | 41001A1B | Normal | 59 | W | F |
| 10 | 41939A1A | Normal | 65 | U | F |
| 11 | 42946A1A | Normal | 71 | W | M |
| 12 | 44376A1B | Normal | 44 | B | M |
Samples were obtained using protocols approved by the University of Miami Institutional Review Board for Human Subjects Research. Left ventricular free wall myocardium was obtained from subjects within 4 h after death or cardiac explantation. Normal hearts were obtained through the Miami Organ and Tissue Procurement Program and the Cooperative Human Tissue Network.
Using a well-characterized cell culture model (Bishopric & Kedes, 1991), we induced hypertrophy of neonatal rat ventricular myocytes (NRVM) in serum-free culture by addition of 5% foetal calf serum (FCS; Fig 1E); as previously reported (Wei et al, 2008), hypertrophy was accompanied by a rapid rise in p300 levels that persisted for at least 36 h (Fig 1E). Over the same interval, both miR-142-5p and -3p fell to ∼50% of basal levels and remained suppressed for at least 24 h (Fig 1F). This suppression did not reflect a global loss of miRNA species, as a genetically unrelated miRNA, miR17-3p, was upregulated under the same conditions (Fig 1F).
Growth factor-induced repression of miR-142 involves MAPK activity
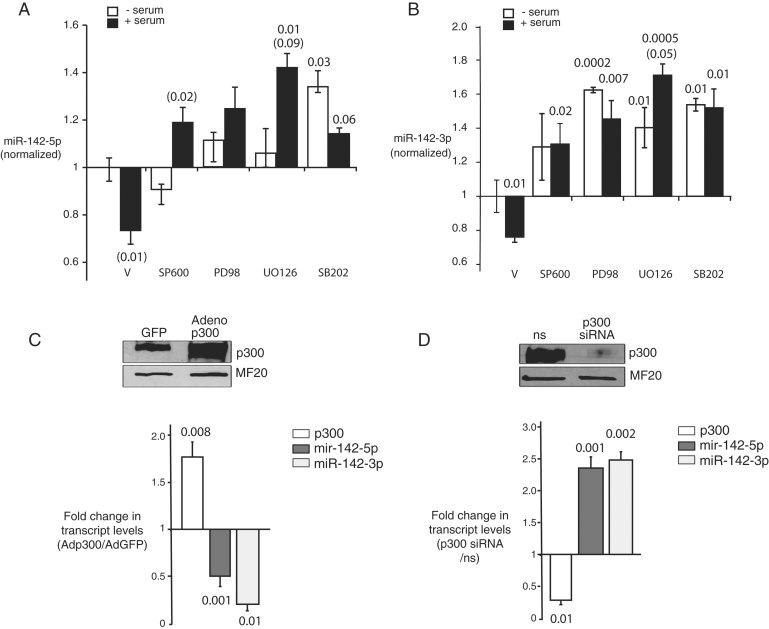
Serum contains a mixture of growth factors that signal through one or more MAPK cascades, including p38 MAPK, c-Jun N-terminal kinases (JNKs) and p42/44 extracellular signal-regulated kinases (ERKs; Sugden & Clerk, 1998). To interrogate the role of MAPK signalling in repression of miR-142, we stimulated cardiac myocytes with serum in the presence of inhibitors of MEK1 (UO126), p42/p44 MAPK/ERK (PD98059), p38MAPK (SB202190) or JNK (SP600125) using optimally selective concentrations (Andreka et al, 2001; Tran et al, 2007). As expected, serum stimulation resulted in the robust induction of p300 and activation of p42/p44 MAPK/ERK, followed by the previously demonstrated repression of miR-142-5p and -3p and hypertrophic growth of myocytes (Figs 2A–C and 1D of Supporting Information). Treatment with any of the MAPK inhibitor compounds blocked the downregulation of both miR-142 species (Fig 2A and B). In addition, all inhibitors increased basal levels of miR-142-3p (Fig 2B). These results indicate that miR-142-5p and 3p are negatively regulated by serum growth factors, likely acting through a combination of MAP kinases.
Figure 2. Inverse regulation of miR-142 by growth signals via MAPK and p300.
- A,B. Repression of miR-142-5p and -3p by serum is reversed by MAPK inhibition. NRVM hypertrophy was induced as in 1D in the presence of MAP kinase inhibitors, or their vehicle (DMSO, V), and miR-142 levels were quantitated at 4 h. SB600 = SB600125 (JNK); PD98 = PD98059 (p42/44 MAPK); UO126 (MEKK1); SB202 = SB202190 (p38MAPK). All values are normalized to basal levels in serum-starved, vehicle-treated cells (V); p-values relative to V are supplied over each column where significant (p-Values in parentheses compare serum-treated to serum-starved cells in each inhibitor treatment group). (A) MiR-142-5p. (B) MiR-142-3p.
- C. p300 represses expression of miR-142(-5p and -3p). NRVM were infected with adenoviral vectors expressing p300 (Ad-p300) or green fluorescent protein (Ad-GFP) and p300, miR-142-5p and -3p were quantitated at 48 h. Values are expressed as the ratio of expression in Ad-p300- versus Ad-GFP-transfected cells. A representative p300 immunoblot is shown.
- D. Loss of p300 is sufficient to induce miR-142-5p and miR-142-3p. NRVM were transfected with anti-p300 siRNA or a ns and p300, miR-142-5p and miR-142-3p were quantitated at 48 h. Values are expressed as the ratio of expression in p300-targeting versus non-targeting siRNA-treated cells. n = at least 3 for all data points, A–D.
Repression of miR-142 by p300
Both ERK/MAPK and p38MAPK phosphorylate and modulate the activity of p300 (Chen et al, 2007; Darieva et al, 2004; Gusterson et al, 2002; Poizat et al, 2005). To determine whether p300 also regulates miR-142, we transduced cardiac myocytes with an adenovirus expressing full-length human p300 (Ad-p300) or with an anti-p300 silencing RNA (siRNA). Ad-p300, but not Ad-GFP virus, significantly increased p300 content at 48 h (Fig 2C), accompanied by a reduction in both miR-142-5p and -3p (Fig 2C, below). Conversely, cardiac myocytes transfected with anti-p300 siRNA but not with a non-silencing sequence (ns) had >70% reduction in p300 levels at 48 h, accompanied by more than doubling of both miR-142-5p and -3p expression (Fig 2D). Thus, p300 is both necessary and sufficient to drive the repression of miR-142 in the absence of hypertrophic extracellular signals or MAPK activation.
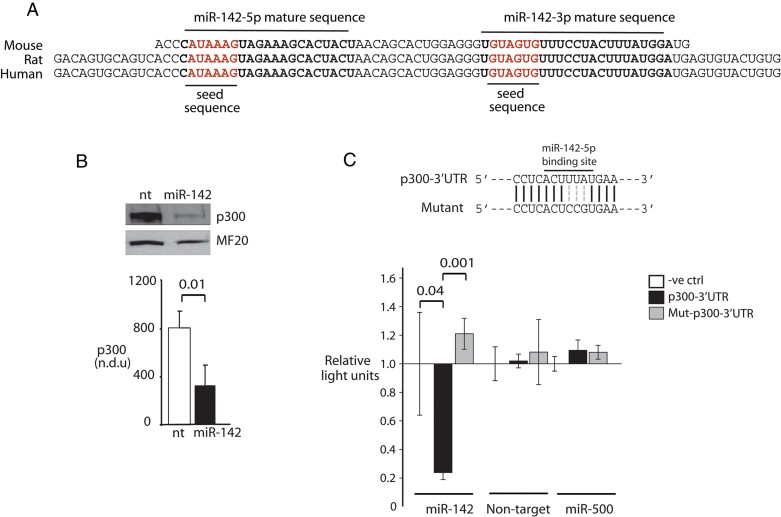
MiR-142 directly targets p300
A query using the TargetScan miRNA target prediction algorithm (http://www.targetscan.org/) revealed a predicted binding site for miR-142-5p in the p300 3′UTR, suggesting that miR-142 might reciprocally inhibit p300 (Fig 3A). To investigate this, we transfected cardiac myocytes with lentiviral vectors encoding both miR-142-5p and -3p (miR-142), or a non-targeting scrambled sequence (NT). Both miRs were expressed from the miR-142 lentivirus, accompanied by a marked decrease in p300 protein levels (Fig 3B). A luciferase expression vector containing the wt p300 3′UTR was efficiently repressed by miR-142 (Fig 3C). Mutation of the predicted miR-142-5p binding site eliminated this repression (Fig 3C), confirming a direct inhibition of p300 by miR-142-5p.
Figure 3. p300 is a direct target of miR-142-5p.
- Conserved p300 3′UTR binding site for miR-142-5p.
- MiR-142 over-expression (OE) represses p300. NRVM were infected with lentivirus encoding the miR-142 hairpin or a NT(see text). p300 levels were quantitated by immunoblot. NDU = normalized densitometry units. n = 3.
- p300 repression is mediated by the 3′UTR miR-142-5p binding site. (Above) MiR-142 binding site and mutated sequence. (Below) Luciferase activity is sensitive to miR-142 binding. Luciferase constructs containing wt or mutant p300 3′UTR sequences were assayed in Cos7 cells stably expressing either RFP alone, miR-142-RFP, or an unrelated microRNA, miR-500-RFP. n = 3.
MiR-142 targets α-actinin
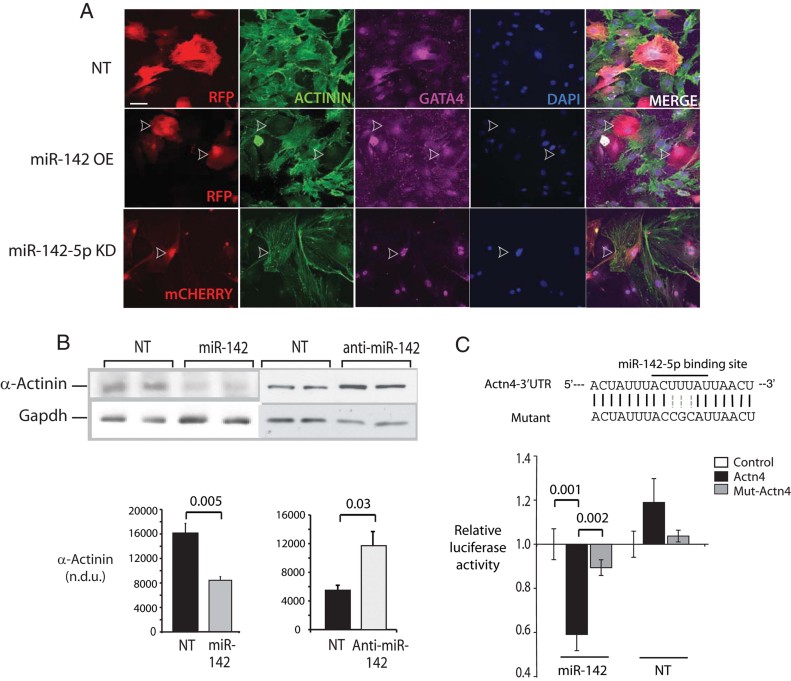
The majority of genes targeted by miRNAs are thought to be regulated through mRNA transcript destabilization (Baek et al, 2008; Bagga et al, 2005). To get a picture of the functional role of miR-142 in the heart as well as to identify potential direct targets, we examined the gene expression profiles of cardiac myocytes with and without miR-142 gain. Significantly regulated genes were identified and selected as potential targets if they underwent reciprocal regulation with miR-142 loss induced by miR-142-5p and -3p antisense inhibitors (Krutzfeldt et al, 2005). Several sarcomeric genes emerged as potential targets (Fig 3 of Supporting Information). To explore the effect of miR-142 on myocyte structure, NRVM were transduced as above to achieve gain or loss of both miR-142-5p and 3p, followed by immunostaining with antibodies directed against pan-α-Actinin, GATA4, α-Myosin heavy chain and F-Actin (Fig 4A and Fig 4 of Supporting Information). MiR-142 modulation had no effect on cell size or on the apparent amount or distribution of myocyte GATA4, Myosin or sarcomeric Actin (Fig 4 of Supporting Information). However, α-Actinin was strikingly depleted in cells overexpressing miR-142 lentivirus (Fig 4A, centre row), compared with cells expressing either a scrambled sequence or anti-miR-142 (Fig 4A, upper and lower rows, respectively). Immunoblots of total cell lysates confirmed the loss of Actinin in miR-142-overexpressing cells, as well as a significant increase of actinin levels in cells expressing anti-miR-142, relative to their scrambled sequence controls (Fig 4B).
Figure 4. α-Actinin is a direct target of miR-142-5p.
- Regulation of α-actinin by miR-142. NRVM were transduced with viral vectors to achieve miR-142 OE or with antisense oligonucleotide targeting miR-142-5p (KD) then fixed and imaged by immunofluorescence 48 h later. Non-targeting sequence, NT sequence. Shown are stains using DAPI (blue) and antibodies directed against GATA4 (purple) and pan-α-Actinin (green). Transduced cells in each field are red due to RFP (top and centre) or mCherry (bottom) fluorescence.
- Regulation of α-Actinin protein content by miR-142. NRVM were transduced with lentiviruses encoding the miR-142 hairpin, an antisense sequence against miR-142, or scrambled sequence controls (NT), for 48 h. Total cell lysates were analysed for α-Actinin expression by immunoblot. n = 3.
- Targeting of the Actn4 3′UTR by miR-142-5p. (Above) A predicted miR-142-5p binding site in the Actn4 3′UTR was mutated. (Below) Luciferase constructs containing either the wt (Actn4) or mutant (mut-Actn4) binding sites, or an unrelated sequence (control) were expressed in Cos7 cells expressing miR-142-5p or a scrambled sequence. Luciferase activity was quantitated at 48 h and expressed relative to activity in the control vector in each of three experiments.
TargetScan and PITA algorithms predicted miR-142-5p-binding sites in both α-Actinin 2 (Actn2) and α-Actinin 4 (Actn4) isoforms; the ACTN4 site is conserved in mouse, rat and humans (Fig 4C). Neither ACTN2 nor ACTN4 mRNAs were differentially expressed in the microarray assays, suggesting that miR-142 might target protein translation rather than transcript stability. Mutation of the miR-142 binding site in the α-Actinin 4 3′UTR and luciferase constructs containing either the wt or mutant 3′UTR were expressed in the presence of miR-142-5p or a scrambled sequence (Fig 4C). MiR-142-5p significantly repressed the wt but not the mutant 3′UTR-containing vector. No repression of either vector was seen with the scrambled sequence, indicating that this α-Actinin isoform is a direct, specific target of miR-142-5p.
MiR-142 globally represses cytokine signalling
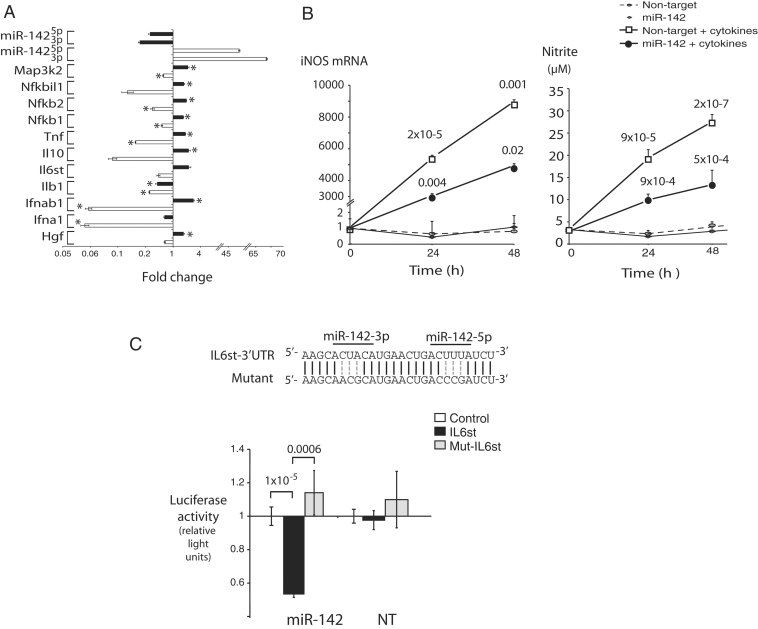
A striking effect of miR-142 overexpression (OE) was the apparent suppression of multiple components of immune signalling in chemokine and cytokine signal transduction pathways, as determined by Gene Ontology (GO) analysis (http://www.geneontology.org). Ninety one genes within the cytokine-related GO functions were differentially regulated, of which 69 were strongly repressed by miR-142. 26 of these genes were examined in cells with miR-142 loss; almost all were correspondingly upregulated (Fig 5A and Table I of Supporting Information). This global repression of cytokine genes was accompanied by a marked functional loss of signalling through the TNFα-NF-κB pathway. Macrophage-derived cytokines, including interleukin 1-β (IL1β), tumour necrosis factor α (TNFα) and interferon gamma (IFNγ), are powerful activators of inducible nitric oxide synthase (iNOS) transcription in cardiac myocytes (Ing et al, 1999). We exposed NRVM to these cytokines after transduction with either miR-142 or non-targeting lentiviruses and monitored iNOS mRNA levels and NO production. As expected, cardiac myocytes expressing the NT produced high levels of both iNOS and NO within 48 h after cytokine exposure, while NRVM expressing miR-142 induced only half as much iNOS and NO under the same conditions (Fig 5B). Since lentiviral transduction was only ∼50% efficient, this result is effectively consistent with elimination of the cytokine response in miR-142-transduced myocytes.
Figure 5. MiR-142 suppresses multiple cytokine signals in cardiac myocytes.
- Cytokine signalling transcripts regulated during miR-142 gain and loss. Relative expression levels of the indicated genes were determined in NRVM following miR-142 OE and knockdown exactly as described in Fig 3A. n = 3, *p < 0.05. Actual p-values along with a complete list of differentially expressed cytokine pathway genes are provided in Table I of Supporting Information.
- MiR-142 blocks cytokine-mediated induction of iNOS. NRVM infected with miR-142- or NT-lentivirus were treated with a mixture of IL1β, TNFα and IFNγ or their vehicle as described in Methods Section and assayed at the indicated time points. (Left) iNOS mRNA was quantitated by qPCR. (Right) Nitrite levels were measured in cell media as a readout of nitric oxide production. n = 8–12 determinations per condition; p-values on top curve compare NT cells with and without cytokines; p-values on miR-142 + cytokines curve are relative to NT cells + cytokines at the same time points.
- gp130 (IL6st) is a direct target of miR-142-5p and 3p. (Above) The gp130 3′UTR paired binding sites for miR-142-5p and miR-142-3p, and a mutant sequence destroying both. (Below) Luciferase vectors incorporating the wt, mutant or a control unrelated 3′UTR sequence were tested for repression by miR-142 in Cos7 cells stably overexpressing miR-142 or a NT sequence.
gp130 is a direct target of miR-142
One potential target identified in the microarray was the cytokine receptor gp130 (encoded by IL6st), previously shown to be critical for survival signalling in the myocardium (Hirota et al, 1999). The 3′UTR of this transcript had predicted binding sites for both miR-142-5p and -3p (Fig 5C). A luciferase construct containing the IL6st 3′UTR, but not a mutant 3′UTR lacking both sites, was strongly repressed by miR-142 but not by the NT (Fig 5C), confirming that gp130 is a direct target of either or both miR-142-5p and -3p.
Forced expression of miR-142 suppresses p300 and reverses p300-driven cytokine gene expression
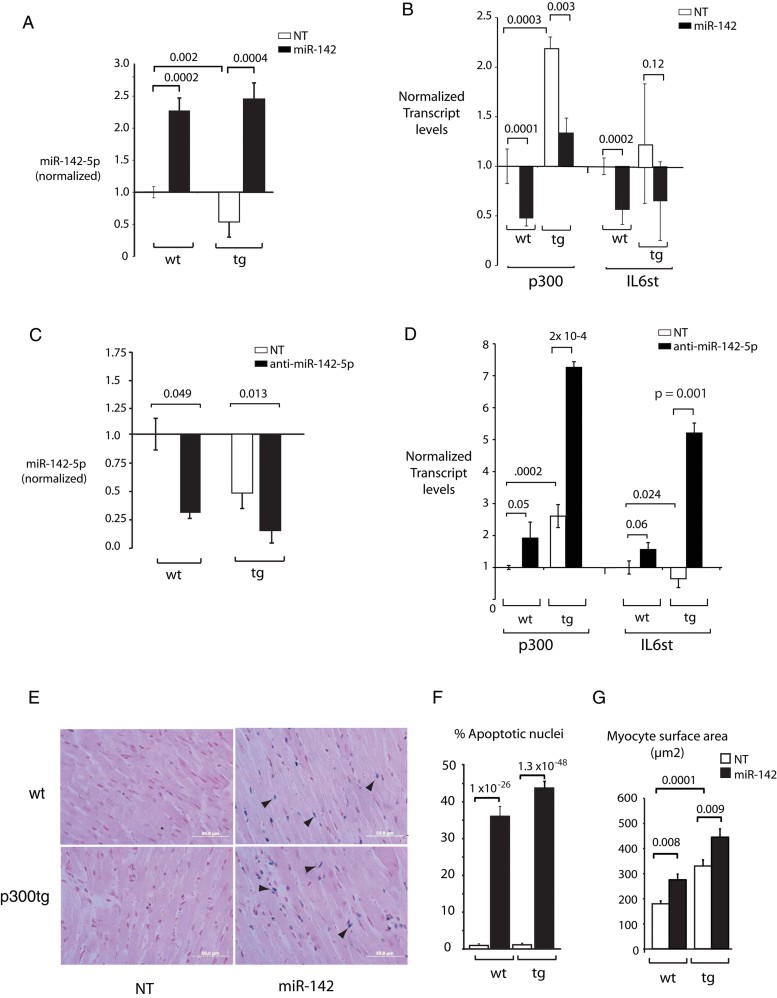
To validate these findings in vivo, we transduced the miR-142- and non-targeting lentiviral vectors into newborn wt mice and p300tg littermates. As we have previously reported, these mice develop hypertrophy that progresses to heart failure over 6–8 months in the majority of individuals (Wei et al, 2008). Consistent with the results in Fig 1A, expression of miR-142-5p was nearly 50% lower in p300tg mice compared to their wt littermates, 2 months after transduction of the non-targeting vector (Fig 6A). Both wt and p300tg animals transduced with miR-142 lentivirus had consistent 2.3–2.5× increases above wt miR-142-5p levels (Fig 6A), indicating successful, sustained expression from the lentiviral vector. Interestingly, miR-142-3p (transcribed from the same transduced sequence) was not significantly upregulated by the lentiviral vector in wt mice, but there was a trend in favour of normalization of miR-142-3p from the depressed levels found in p300tg mice (Fig 6 of Supporting Information). Pathway expression analysis using a custom high-density quantitative polymerase chain reaction (qPCR) array confirmed suppression of the vast majority of cytokine genes previously identified as differentially regulated in cell culture, as well as other predicted targets of miR-142 (see Figs 4, 5 and Table II of Supporting Information). Importantly, polymerase chain reaction (after reverse transcription) (RT-PCR) and Western blotting confirmed that both p300 and gp130 were suppressed in myocardium of all miR-142-transduced mice (Fig 6B and Fig 7 of Supporting Information).
Figure 6. MiR-142 represses growth-associated survival signals in vivo.
- Sustained in vivo transduction of miR-142-5p. Neonatal wt and p300 transgenic littermates (tg) hearts were analysed for miR-142-5p expression at 2 months after transduction with lenti-miR-142 or –NT vectors.
- Repression of p300 and gp130 by miR-142-5p in vivo. Transcripts were quantitated by qPCR as in (A); primers for p300 recognize both endogenous murine and human transgene-derived transcripts. Additional genes and gp130 and p300 protein levels are shown in Figs 5 and 6 of Supporting Information.
- Sustained in vivo silencing of miR-142-5p by transduction of antimiR-142-5p. Neonatal mice hearts transduced with lentivirus expressing anti-miR-142-5p sequences or a scrambled control were analysed for miR-142-5p expression at 2 months.
- Increased expression of p300 and gp130 following miR-142-5p silencing in vivo. Transcripts were quantitated in the same hearts measured in C.
- MiR-142 induces growth-associated apoptosis. Representative LV myocardium images are shown. Apoptotic cells are stained blue (arrowheads). Original magnification: ×60.
- Quantitation of apoptotic cells. n = 10 randomly chosen fields per section in each of five hearts per condition.
- Myocyte enlargement. Cross-sectional surface pixels were measured in 25–30 cells per section in five hearts per condition using Lasso and Histogram tools in Adobe Photoshop and converted to square µM. Note that p300tg myocytes are significantly larger than wt myocytes in both treatment groups.
Transduction of anti-miR-142 induces p300 and gp130 expression in vivo
We used a similar lentiviral vector to deliver an antisense miR (antagomir) to deplete the already low levels of endogenous miR-142-5p in neonatal wt and tg mice, and measured p300 and gp130 levels in left ventricular myocardium at 2 months. This approach resulted in >70% miR-142 knockdown in both genetic backgrounds (Fig 6C). There was a corresponding highly significant increase in both p300 and gp130 expression in anti-miR-142-treated versus non-targeting-vector-treated mice, providing further validation of these targets in vivo (Fig 6D).
Restoration of miR-142 during postnatal and p300-induced growth induces myocyte apoptosis
Macroscopic assessment of wt animals did not reveal significant differences in myocardial thickness, heart weight or heart weight indices between treatment groups (Fig 7A and Table 2). However, the heart weight/tibia length ratio was significantly increased in p300tg mice receiving the miR-142 virus relative to the other three groups (n = 5, p = 0.02). In both genotypes, microscopic evaluation revealed abundant myocyte apoptosis (Fig 6E and F) together with significant compensatory enlargement of the remaining myocytes (Fig 6G) in response to miR-142 transduction. Apoptosis was essentially absent in non-targeting-transduced or anti-miR-142-transduced hearts of either genotype (Fig 6E and F and data not shown). These findings are consistent with a functional suppression of survival signalling by miR-142.
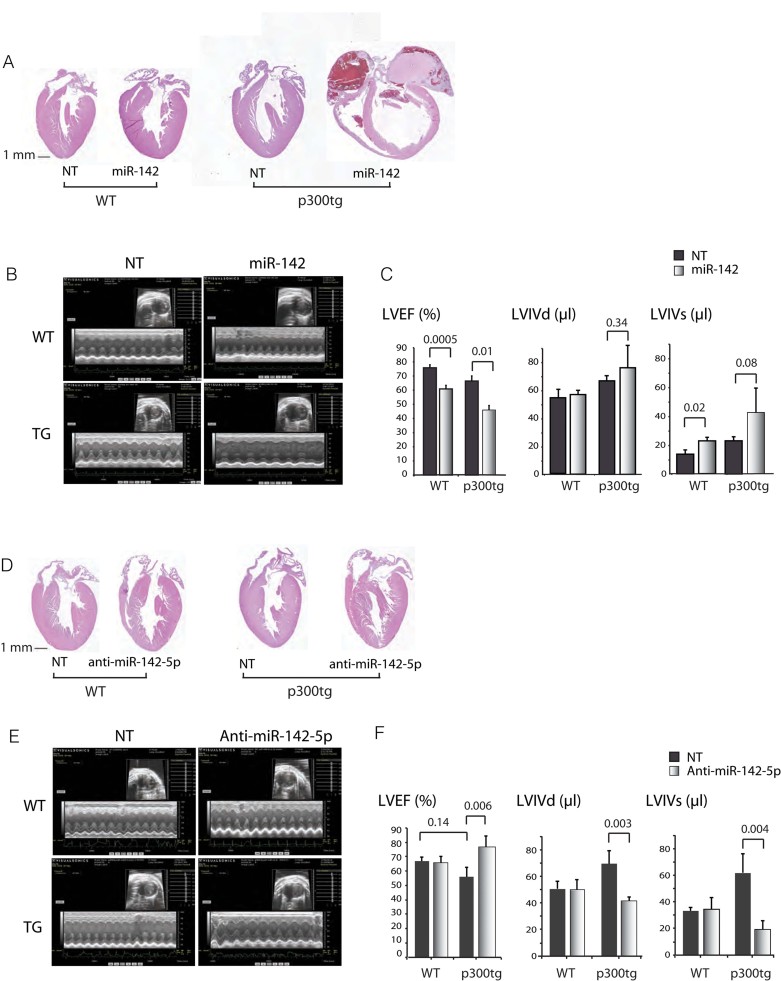
Figure 7. Inverse effects of gain and loss of miR-142 on cardiac function.
5-day old wt and p300tg pups were transduced with the indicated lentiviruses (total dose 108 viral particles/pup), and cardiac contractile function was assessed at 2 months of age.
- A-C. Restoration of miR-142 impairs cardiac function during postnatal growth.
- A. Representative 4 chamber sections of wt and p300tg hearts treated with miR-142 or NT vectors. Haematoxylin and eosin staining of LV tissue from 2.5-month-old mice. Original magnification: 1×.
- B. Representative echocardiographic images showing M-mode and 2D sectors for each genotype and condition.
- C. LV systolic function. Left ventricular ejection fraction (LVEF) and internal dimensions in systole and diastole (LVIDs and LVIDd) are shown. n = 5–8 per group. See also Table III of Supporting Information.
- D-F. Inhibition of miR-142-5p/-3p rescues systolic function in p300tg mice.
- D. Representative four chamber sections of wt and p300tg hearts treated with anti-miR-142 or NT vectors, as in 6A.
- E. Representative 2D and M-mode images.
- F. Quantitation of LVEF, LVIDs and LVIDd. n = 3–6 animals per group. See Table IV of Supporting Information for summary of all parameters.
Table 2.
Summary of organ weights in mice with miR-142 loss and gain
| Genotype | NT | n | miR-142 | n | p-Value | NT | n | Anti-miR-142 | n | p-Value | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Hw/TL | wt | 6.85 ± 0.33 | 19 | 6.79 ± 0.32 | 17 | 0.38 | 6.04 ± 0.37 | 10 | 5.72 ± 0.45 | 8 | 0.57 |
| p300tg | 7.14 ± 0.24 | 13 | 9.40 ± 1.37 | 5 | 0.02 | 6.28 ± 0.10 | 8 | 7.01 ± 0.37 | 11 | 0.10 | |
| Liver (g) | wt | 1.30 ± 0.05 | 19 | 1.08 ± 0.07 | 17 | 0.01 | 0.98 ± 0.09 | 10 | 1.15 ± 0.05 | 8 | 0.13 |
| p300tg | 1.28 ± 0.04 | 13 | 1.32 ± 0.10 | 5 | 0.63 | 1.02 ± 0.06 | 8 | 1.23 ± 0.08 | 11 | 0.05 | |
| Lung (g) | wt | 0.16 ± 0.01 | 19 | 0.18 ± 0.01 | 17 | 0.28 | 0.17 ± 0.01 | 10 | 0.17 ± 0.02 | 8 | 0.99 |
| p300tg | 0.15 ± 0.02 | 13 | 0.19 ± 0.02 | 5 | 0.26 | 0.16 ± 0.01 | 8 | 0.18 ± 0.01 | 11 | 0.28 | |
| Spleen (g) | wt | 0.12 ± 0.01 | 19 | 0.21 ± 0.06 | 17 | 0.18 | 0.11 ± 0.004 | 10 | 0.11 ± 0.01 | 8 | 0.82 |
| p300tg | 0.11 ± 0.01 | 13 | 0.14 ± 0.01 | 5 | 0.02 | 0.09 ± 0.01 | 8 | 0.09 ± 0.01 | 11 | 0.32 |
Gain and loss of miR-142 exert opposing effects on heart function in p300 transgenic mice
Forced expression of miR-142 was associated with significant impact on heart function in both wt and p300tg mice. All miR-142 recipients had reduced LV ejection fraction, fractional shortening and increased systolic and diastolic dimensions relative to their non-targeting-injected, genotype-matched controls (Fig 7A–C and Table III of Supporting Information), indicative of development or worsening of heart failure. In contrast, no ill effect was seen in wt or transgenic mice receiving the anti-miR-142 vector. WT mice receiving non-targeting and antisense vectors were functionally and anatomically indistinguishable (Fig 7D–F and Table 2). Remarkably, in the majority of p300tg mice, depletion of residual miR-142-5p prevented the mild systolic dysfunction seen in their non-targeting-vector-transduced peers (Fig 7F; see also Fig 7C); these mice actually had enhanced function relative to wt mice by multiple measures (chamber volumes, fractional shortening and ejection fraction; Fig 7F and Table IV of Supporting Information). Thus, downregulation of miR-142-5p was able to prevent deterioration and possibly enhance function induced by sustained p300 signalling.
DISCUSSION
Downregulation of miR-142 during cardiac growth is critical to cell survival and cardiac performance
In this study, we demonstrate an inverse relationship between levels of miR-142 and cardiac hypertrophy in multiple physiologic and pathologic settings. We further show that growth signals transduced by MAPK and p300 lead to physiologic repression of miR-142, and that this repression is required to prevent myocyte loss and to permit adaptive hypertrophic growth in vivo. The myocardium is vulnerable to apoptosis during acute increases in workload (Condorelli et al, 1999; Teiger et al, 1996). Consequently, successful adaptation to changing haemodynamic demand requires both myocyte growth and activation of myocyte survival signals. MiR-142-5p directly targets two nodal regulators of these pathways: acetyltransferase p300, a powerful regulator of multiple transcription factors involved in cardiac growth (Wei et al, 2008), and gp130, the cell surface receptor for IL-6, cardiotrophin-1 and related cytokines providing survival signals through JAK/STAT (Fujio et al, 1997; Wollert & Chien, 1997). The critical importance of the gp130 receptor for myocyte survival during load-induced growth is well established (Burchfield et al, 2008; Fujio et al, 1997; Sheng et al, 1997). A key finding is that mice lacking gp130 undergo massive myocardial apoptosis, develop dilated cardiomyopathy and die of heart failure within days after experimental pressure overload (Hirota et al, 1999). Here, we show that by targeting gp130, miR-142 OE results in a similar phenotype during physiologic cardiac growth. Thus, while miR-142 targets both p300 and gp130, its overriding effect in vivo is not repression of hypertrophy, but rather repression of a cytokine-mediated survival signal pathway. While the repression of cytokine signalling under basal conditions may be desirable, this pathway must be activated to permit adaptive growth, requiring repression of miR-142.
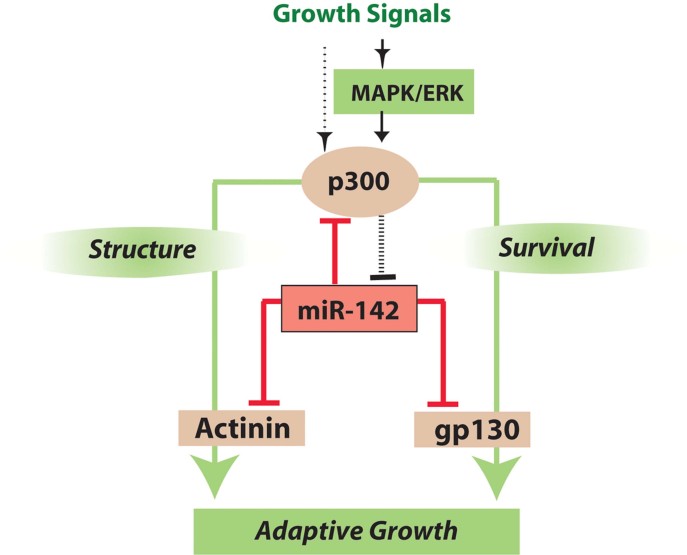
A mutual inhibitory loop between p300 and miR-142
We envision that under basal conditions, miR-142-5p plays a homeostatic role, maintaining p300 at low levels through direct targeting of its mRNA (Fig 8). Under conditions of increased demand, p300 and MAP kinases activation transiently repress miR-142. Regulatory feedback loops involving miRNAs represent a recurring motif in gene regulation (Martinez et al, 2008). Although the precise mechanism of repression remains to be established, ERK/MAPK signalling is known to activate p300 (Gusterson et al, 2002), and ERK signalling could repress miR-142 through this mechanism. Other miRNAs have been shown to be positively regulated by ERK activation (Paroo et al, 2009; Terasawa et al, 2009). ERK1,2 phosphorylates and stabilizes trans-activated RNA binding protein (TRBP), a key member of the miRNA-processing complex, increasing production of the mature forms of many miRNAs; however, let-7 appears to be repressed by ERK through an undetermined mechanism (Paroo et al, 2009). Yuan et al found that expression of the miR-142 precursor in lymphoblastoid cells was inhibited by a complex of transcription factors including LMO2, GATA1 and E47 (Yuan et al, 2008). The logarithmic increase in miR-142 in both wt and p300tg mice between 2 and 3 months suggests that there may be additional positive regulators of miR-142 that act during later stages of myocardial maturation associated with reproductive maturity.
Figure 8. A mutual inhibitory loop involving p300 and miR-142 controls survival signalling during cardiac myocyte growth.
MicroRNAs encoded by the miR-142 hairpin play a homeostatic role by inhibiting the expression of genes that are not required during basal conditions, but which have nodal roles in regulating cardiac survival signalling and growth during periods of increased demand. Under stress, mechanical and other growth signals induce p300 accumulation and activate MAP kinases, which increase p300 HAT activity. MiR-142-5p and 3p are repressed by activation of MAPK and p300. The loss of miR-142 removes a brake on the induction of genes required for myocyte function and survival, permitting adaptive growth of the working myocardium.
miR-142 regulates the myocyte cytoskeleton
Another important finding of this study is that expression of α-Actinin is profoundly negatively regulated by miR-142-5p. The α-Actinins belong to a superfamily of cell structural proteins that includes the Dystrophins, loss of which can compromise sarcolemmal integrity leading to myocyte death (Lapidos et al, 2004). Actinins 1 and 2 have been implicated as disease genes in hypertrophic and dilated cardiomyopathy (D'Amico et al, 2006; Mohapatra et al, 2003). Actinin-4 has been identified in both cytosolic and nuclear compartments, and functionally interacts with nuclear proteins that regulate cell growth, transcription and apoptosis (Chakraborty et al, 2006; Khotin et al, 2010; Khurana et al, 1850; Liu et al, 2004). Inappropriately high levels of miR-142 could thus impair Actinin-mediated structural integrity, leading to apoptosis under conditions of stress.
Global repression of immune response genes by miR-142
One of the most striking findings of our study is the potent repressive effect of miR-142-5p/-3p on genes controlling every aspect of immune response, and the corresponding suppression of the response to inflammatory cytokines. Expression of iNOS is extremely sensitive to macrophage-derived cytokines via multiple promoter binding sites for NF-kB, and this response was strongly blunted by miR-142. We have identified at least 26 components of immune signalling pathways, including both NF-κB1 and -2 and isoforms, that are directly or indirectly regulated by miR-142 in vivo. By targeting an entire cytokine response program, miR-142 acts as a critical regulator of immune responses that are likely to be important in multiple tissues. MiR-142 is abundantly expressed in lymphocytes and cells of haematopoietic lineage (Landgraf et al, 2007), although T regulatory cells appear to have relatively low levels of miR-142-3p (Zhao et al, in press). MiR-142-3p is differentially regulated in various models of inflammation (Moschos et al, 2007; Recchiuti et al, in press) and regulates inflammatory mediators in cells of the immune system, including IL-6 (Huang et al, 2009; Sun et al, in press). Other miRNAs implicated in immune signalling include miR-181a, miR-146, miR-155 and miR-125b (Li et al, 2007; Taganov et al, 2006; Tili et al, 2007). p300 itself has previously been identified as a critical regulator of immune gene transcription (Merika et al, 1998) and is targeted in monocytes by miR-132 (Lagos et al, 2010). The extent of functional overlap among these miRNAs remains to be determined.
To summarize, we have shown that miR-142 is a potent repressor of cytokine signalling in the myocardium, and this miRNA must itself be repressed in order for adaptive cardiac hypertrophy to take place. Although the mechanism of repression involves ERK signalling and p300, it seems likely that unidentified factors downstream are directly responsible. Our observations open up the interesting concept that certain growth and survival pathways are under strong tonic inhibition in the heart at equilibrium. It seems likely that additional miRNAs are involved in maintaining this cardiac homeostasis. The identity and function of these miRNAs and homeostatic mechanisms represent important areas for future investigation.
MATERIALS AND METHODS
Materials and reagents
A comprehensive list of Materials and Reagents can be found in the Supporting Information. For over-expression and knockdown of p300 and miR-142 in primary cardiac myocytes and in vivo, we used Turbo-RFP-tagged pLemir lentivirus constructs for miRNA-142 transduction and expression from Open Biosystems Products Inc., Huntsville AL. The mCherry-tagged pEZX-AM03 lentiviral vectors expressing the miR-142-5p inhibitor (antagomir) and scrambled sequences, as well as luciferase vectors containing selected 3′UTRs, were obtained from GeneCopoeia, Rockville MD. Each of these lentiviral expression constructs utilizes the cytomegalovirus (CMV) promoter. Locked nucleic acid (LNA) oligonucleotides for knockdown of miR-142 (-5p and -3p) were obtained from Exiqon, Woburn, MA.
The paper explained
PROBLEM:
Cardiac hypertrophy accompanies the majority of disease- and age-related cardiac pathologies. However, a direct cause-and-effect relationship between hypertrophy and heart failure is so far lacking, and it is clear that hypertrophy can be an important adaptive response in certain settings, particularly during normal postnatal growth. Cardiac hypertrophy is the end result of complex signals that are activated when the haemodynamic workload is acutely or chronically increased. Increased work entails significant oxidative stress, hence, successful adaptation to loading requires both increased cell mass and the presence of robust anti-apoptotic signals, such as those mediated by STAT3. Elucidating the downstream targets of stress-induced signals is key to understanding the difference between adaptive and pathological hypertrophy.
RESULTS:
In this paper, we describe two miRNAs encoded by the same primary transcript (miR-142-5p and miR-142-3p) that target p300 and gp130, nodal mediators of growth and STAT3-mediated survival signals, respectively, ensuring that these signals remain quiescent in the non-stressed heart. We also demonstrate that these miRNAs are downregulated in various models of hypertrophic stress, allowing for an effective adaptive response. Repression of these miRNAs requires MAPK and p300, establishing a mutually inhibitory circuit between these growth activators and miR-142.
IMPACT:
Our findings point to a novel, biologically important mechanism that mutes critical components of the stress responsive gene program in the heart under normal conditions; the removal of this standing inhibition represents a novel mechanism in cardiac hypertrophy that may be amenable to therapeutic exploitation in heart failure.
Methods
Methods for primary culture of neonatal rat cardiac myocytes have been previously described (Bishopric & Kedes, 1991).
Methods for realtime PCR, Western blotting, measurement of nitrite release, cell imaging techniques including morphometry apoptosis detection immunofluorescence and image acquisition and processing are described in the Supporting Information.
Reporter assays
All luciferase assays were performed on Cos7 cells stably expressing miRNAs or control scrambled sequences. Fourty eight hours post-transfection, cell extracts were assayed for luciferase activity using the Luc-Pair miR luciferase assay kit (Genecoepia). Relative reporter activities were expressed as luminescence units normalized to Renilla luciferase activity. Luminescence was quantitated using a multimode microplate reader (BMG Labtech).
In vivo transduction of miR-142
All experiments were conducted under University of Miami Animal Care and Use Committee-approved protocols. Newborn (5 day) wt and p300tg pups received 8 × 108 viral particles of lentivirus expressing miR-142 or a control sequence via external jugular vein injection using a previously described method, with minor modifications (Kienstra et al, 2007). In brief, 5-day-old wt mouse pups and littermates with myocyte-specific OE of p300 (BS line; Wei et al, 2008) were placed on a transilluminator and the external jugular vein was injected with 8 × 108 viral particles of miR-142 or control. Injected pups were returned to their mothers until weaning at 3 weeks of age.
Microarrays
Rat Gene Expression microarray analysis was performed using non-Affymetrix single channel arrays (Ocean Ridge Biosciences). Microarray data is available at GEO (http://www.ncbi.nlm.nih.gov/gds) under accession numbers GSE31121 and GPL14011.
Statistical analysis
Microarray and low-density TaqMan array expression ratios were calculated as the power-2 exponential of the log2 differences. The acceptance criteria for gene array expression changes was a minimum 1.7-fold change and a one-way Analysis of Variance (ANOVA) t-test p-value of <0.05. The direction of change was validated in each condition by qPCR in a randomly chosen subset of 26 genes. Data analysis and hierarchical clustering were performed using XLSTAT (http://www.xlstat.com). A custom heat map display program was created in the R software environment (http://www.r-project.org/). For all other studies, analysis of variance (ANOVA) was used for comparison of multiple results within a single experiment, followed by Student's t-test using one- or two-tailed distributions as appropriate.
Acknowledgments
We thank Dr. David Willoughby (Ocean Ridge Biosciences) for his generous assistance in performance and analysis of the microarray data and Drs. Jochen Reiser and Marc Lippman for critical reviews of the manuscript. This work was supported by grants from the NIH (R-01-HL71094 to NHB), the Florida Heart Research Institute (NHB), the Fondation Leducq Transatlantic Network of Excellence 05-CVD-02 (NHB) and an American Heart Association Predoctoral Fellowship Award (SS).
Supporting Information is available at EMBO Molecular Medicine online.
Conflict of interest statement: Dr. Bishopric has collaborated with Regulus Pharma, Inc. on an unrelated project. The other authors declare that they have no conflict of interest.
Author contributions
SS performed the experiments, obtained funding for the work, analysed the data and wrote the manuscript; JL performed the echocardiography and conducted the functional analysis of the mice; JW performed the in vivo injections and surgical manipulations of the mice; HY performed miRNA analyses that identified miR-142; TZ collected and analysed the human myocardial tissue for levels of miR-142. NHB designed experiments, obtained funding for the work, analyzed the data, and wrote the manuscript.
Supplementary material
Detailed facts of importance to specialist readers are published as ”Supporting Information”. Such documents are peer-reviewed, but not copy-edited or typeset. They are made available as submitted by the authors.
References
- Anderson ME. CaMKII and a failing strategy for growth in heart. J Clin Invest. 2009;119:1082–1085. doi: 10.1172/JCI39262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreka P, Dougherty C, Slepak TI, Webster KA, Bishopric NH. Cytoprotection by Jun kinase during nitric oxide-induced cardiac myocyte apoptosis. Circ Res. 2001;88:305–312. doi: 10.1161/01.res.88.3.305. [DOI] [PubMed] [Google Scholar]
- Backs J, Song K, Bezprozvannaya S, Chang S, Olson EN. CaM kinase II selectively signals to histone deacetylase 4 during cardiomyocyte hypertrophy. J Clin Invest. 2006;116:1853–1864. doi: 10.1172/JCI27438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholdi D, Roelfsema JH, Papadia F, Breuning MH, Niedrist D, Hennekam RC, Schinzel A, Peters DJ. Genetic heterogeneity in Rubinstein-Taybi syndrome: delineation of the phenotype of the first patients carrying mutations in EP300. J Med Genet. 2007;44:327–333. doi: 10.1136/jmg.2006.046698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishopric NH, Kedes L. Adrenergic regulation of the skeletal alpha-actin gene promoter during myocardial cell hypertrophy. Proc Natl Acad Sci USA. 1991;88:2132–2136. doi: 10.1073/pnas.88.6.2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishopric NH, Simpson PC, Ordahl CP. Induction of the skeletal actin gene in alpha1-adrenoceptor mediated hypertrophy of rat cardiac myocytes. J Clin Invest. 1987;80:1194–1199. doi: 10.1172/JCI113179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishopric NH, Zeng G-Q, Sato B, Webster KA. Adenovirus E1A inhibits cardiac myocyte-specific gene expression through its amino terminus. J Biol Chem. 1997;272:20584–20594. doi: 10.1074/jbc.272.33.20584. [DOI] [PubMed] [Google Scholar]
- Burchfield J, Iwasaki M, Koyanagi M, Urbich C, Rosenthal N, Zeiher AM, Dimmeler S. Interleukin-10 from transplanted bone marrow mononuclear cells contributes to cardiac protection following myocardial infarction. Circ Res. 2008;103:203–211. doi: 10.1161/CIRCRESAHA.108.178475. [DOI] [PubMed] [Google Scholar]
- Callis TE, Pandya K, Seok HY, Tang RH, Tatsuguchi M, Huang ZP, Chen JF, Deng Z, Gunn B, Shumate J, et al. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J Clin Invest. 2009;119:2772–2786. doi: 10.1172/JCI36154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Care A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, Bang ML, Segnalini P, Gu Y, Dalton ND, et al. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007;13:613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- Chakraborty S, Reineke EL, Lam M, Li X, Liu Y, Gao C, Khurana S, Kao HY. Alpha-actinin 4 potentiates myocyte enhancer factor-2 transcription activity by antagonizing histone deacetylase 7. J Biol Chem. 2006;281:35070–35080. doi: 10.1074/jbc.M602474200. [DOI] [PubMed] [Google Scholar]
- Chen YJ, Wang YN, Chang WC. ERK2-mediated C-terminal serine phosphorylation of p300 is vital to the regulation of epidermal growth factor-induced keratin 16 gene expression. J Biol Chem. 2007;282:27215–27228. doi: 10.1074/jbc.M700264200. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Ji R, Yue J, Yang J, Liu X, Chen H, Dean DB, Zhang C. MicroRNAs are aberrantly expressed in hypertrophic heart: Do they play a role in cardiac hypertrophy. Am J Pathol. 2007;170:1831–1840. doi: 10.2353/ajpath.2007.061170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn JN, Ferrari R, Sharpe N. Cardiac remodeling–concepts and clinical implications: a consensus paper from an international forum on cardiac remodeling. J Am Coll Cardiol. 2000;35:569–582. doi: 10.1016/s0735-1097(99)00630-0. [DOI] [PubMed] [Google Scholar]
- Condorelli G, Morisco C, Stassi G, Notte A, Farina F, Sgaramella G, de Rienzo A, Roncarati R, Trimarco B, Lembo G. Increased cardiomyocyte apoptosis and changes in proapoptotic and antiapoptotic genes bax and bcl-2 during left ventricular adaptations to chronic pressure overload in the rat. Circulation. 1999;99:3071–3078. doi: 10.1161/01.cir.99.23.3071. [DOI] [PubMed] [Google Scholar]
- Craig R, Larkin A, Mingo AM, Thuerauf DJ, Andrews C, McDonough PM, Glembotski CC. p38 MAPK and NF-kappa B collaborate to induce interleukin-6 gene expression and release. Evidence for a cytoprotective autocrine signaling pathway in a cardiac myocyte model system. J Biol Chem. 2000;275:23814–23824. doi: 10.1074/jbc.M909695199. [DOI] [PubMed] [Google Scholar]
- D'Amico A, Graziano C, Pacileo G, Petrini S, Nowak KJ, Boldrini R, Jacques A, Feng JJ, Porfirio B, Sewry CA, et al. Fatal hypertrophic cardiomyopathy and nemaline myopathy associated with ACTA1 K336E mutation. Neuromuscul Disord. 2006;16:548–552. doi: 10.1016/j.nmd.2006.07.005. [DOI] [PubMed] [Google Scholar]
- Darieva Z, Lasunskaia EB, Campos MN, Kipnis TL, Da Silva WD. Activation of phosphatidylinositol 3-kinase and c-Jun-N-terminal kinase cascades enhances NF-kappaB-dependent gene transcription in BCG-stimulated macrophages through promotion of p65/p300 binding. J Leukoc Biol. 2004;75:689–697. doi: 10.1189/jlb.0603280. [DOI] [PubMed] [Google Scholar]
- Desai RV, Ahmed MI, Mujib M, Aban IB, Zile MR, Ahmed A. Natural history of concentric left ventricular geometry in community-dwelling older adults without heart failure during seven years of follow-up. Am J Cardiol. 107:321–324. doi: 10.1016/j.amjcard.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong DL, Chen C, Huo R, Wang N, Li Z, Tu YJ, Hu JT, Chu X, Huang W, Yang BF. Reciprocal repression between microRNA-133 and calcineurin regulates cardiac hypertrophy: a novel mechanism for progressive cardiac hypertrophy. Hypertension. 2010;55:946–952. doi: 10.1161/HYPERTENSIONAHA.109.139519. [DOI] [PubMed] [Google Scholar]
- Dorn GW, 2nd, Force T. Protein kinase cascades in the regulation of cardiac hypertrophy. J Clin Invest. 2005;115:527–537. doi: 10.1172/JCI24178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn GW, 2nd, Robbins J, Sugden PH. Phenotyping hypertrophy: eschew obfuscation. Circ Res. 2003;92:1171–1175. doi: 10.1161/01.RES.0000077012.11088.BC. [DOI] [PubMed] [Google Scholar]
- Frantz S, Fraccarollo D, Wagner H, Behr TM, Jung P, Angermann CE, Ertl G, Bauersachs J. Sustained activation of nuclear factor kappa B and activator protein 1 in chronic heart failure. Cardiovasc Res. 2003;57:749–756. doi: 10.1016/s0008-6363(02)00723-x. [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujio Y, Kunisada K, Hirota H, Yamauchi-Takihara K, Kishimoto T. Signals through gp130 upregulate bcl-x gene expression via STAT1- binding cis-element in cardiac myocytes. JClinInvest. 1997;99:2898–2905. doi: 10.1172/JCI119484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman W. Cardiac hypertrophy: Useful adaptation or pathologic process. Am J Med. 1980;69:576–584. doi: 10.1016/0002-9343(80)90471-4. [DOI] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusterson R, Brar B, Faulkes D, Giordano A, Chrivia J, Latchman D. The transcriptional co-activators CBP and p300 are activated via phenylephrine through the p42/p44 MAPK cascade. J Biol Chem. 2002;277:2517–2524. doi: 10.1074/jbc.M104626200. [DOI] [PubMed] [Google Scholar]
- Hilfiker-Kleiner D, Hilfiker A, Fuchs M, Kaminski K, Schaefer A, Schieffer B, Hillmer A, Schmiedl A, Ding Z, Podewski E, et al. Signal transducer and activator of transcription 3 is required for myocardial capillary growth, control of interstitial matrix deposition, and heart protection from ischemic injury. Circ Res. 2004;95:187–195. doi: 10.1161/01.RES.0000134921.50377.61. [DOI] [PubMed] [Google Scholar]
- Hirota H, Chen J, Betz UA, Rajewsky K, Gu Y, Ross J, Jr, Muller W, Chien KR. Loss of a gp130 cardiac muscle cell survival pathway is a critical event in the onset of heart failure during biomechanical stress. Cell. 1999;97:189–198. doi: 10.1016/s0092-8674(00)80729-1. [DOI] [PubMed] [Google Scholar]
- Howell TH. Heart weights among octogenarians. J Am Geriatr Soc. 1981;29:572–575. doi: 10.1111/j.1532-5415.1981.tb01262.x. [DOI] [PubMed] [Google Scholar]
- Huang B, Zhao J, Lei Z, Shen S, Li D, Shen GX, Zhang GM, Feng ZH. miR-142-3p restricts cAMP production in CD4+CD25− T cells and CD4+CD25+ TREG cells by targeting AC9 mRNA. EMBO Rep. 2009;10:180–185. doi: 10.1038/embor.2008.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ing DJ, Zang J, Dzau VJ, Webster KA, Bishopric NH. Modulation of cytokine-induced cardiac myocyte apoptosis by nitric oxide, Bak and Bcl-x. CircRes. 1999;84:21–33. doi: 10.1161/01.res.84.1.21. [DOI] [PubMed] [Google Scholar]
- Kalkhoven E. CBP and p300: HATs for different occasions. Biochem Pharmacol. 2004;68:1145–1155. doi: 10.1016/j.bcp.2004.03.045. [DOI] [PubMed] [Google Scholar]
- Khotin M, Turoverova L, Aksenova V, Barlev N, Borutinskaite VV, Vener A, Bajenova O, Magnusson KE, Pinaev GP, Tentler D. Proteomic analysis of ACTN4-interacting proteins reveals its putative involvement in mRNA metabolism. Biochem Biophys Res Commun. 2010;397:192–196. doi: 10.1016/j.bbrc.2010.05.079. [DOI] [PubMed] [Google Scholar]
- Khurana S, Chakraborty S, Cheng X, Su YT, Kao HY. The actin-binding protein, actinin alpha 4 (ACTN4), is a nuclear receptor coactivator that promotes proliferation of MCF-7 breast cancer cells. J Biol Chem. 286:1850–1859. doi: 10.1074/jbc.M110.162107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kienstra KA, Freysdottir D, Gonzales NM, Hirschi KK. Murine neonatal intravascular injections: modeling newborn disease. J Am Assoc Lab Anim Sci. 2007;46:50–54. [PubMed] [Google Scholar]
- Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- Lagos D, Pollara G, Henderson S, Gratrix F, Fabani M, Milne RS, Gotch F, Boshoff C. miR-132 regulates antiviral innate immunity through suppression of the p300 transcriptional co-activator. Nat Cell Biol. 2010;12:513–519. doi: 10.1038/ncb2054. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Lam CS, Roger VL, Rodeheffer RJ, Bursi F, Borlaug BA, Ommen SR, Kass DA, Redfield MM. Cardiac structure and ventricular–vascular function in persons with heart failure and preserved ejection fraction from Olmsted County, Minnesota. Circulation. 2007;115:1982–1990. doi: 10.1161/CIRCULATIONAHA.106.659763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol. 2004;14:2162–2167. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Lapidos KA, Kakkar R, McNally EM. The dystrophin glycoprotein complex: signaling strength and integrity for the sarcolemma. Circ Res. 2004;94:1023–1031. doi: 10.1161/01.RES.0000126574.61061.25. [DOI] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li Y, Ha T, Gao X, Kelley J, Williams DL, Browder IW, Kao RL, Li C. NF-kappaB activation is required for the development of cardiac hypertrophy in vivo. Am J Physiol Heart Circ Physiol. 2004;287:H1712–1720. doi: 10.1152/ajpheart.00124.2004. [DOI] [PubMed] [Google Scholar]
- Li QJ, Chau J, Ebert PJ, Sylvester G, Min H, Liu G, Braich R, Manoharan M, Soutschek J, Skare P, et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129:147–161. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP. Vertebrate microRNA genes. Science. 2003;299:1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- Ling H, Zhang T, Pereira L, Means CK, Cheng H, Gu Y, Dalton ND, Peterson KL, Chen J, Bers D, et al. Requirement for Ca2+/calmodulin-dependent kinase II in the transition from pressure overload-induced cardiac hypertrophy to heart failure in mice. J Clin Invest. 2009;119:1230–1240. doi: 10.1172/JCI38022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu QY, Lei JX, LeBlanc J, Sodja C, Ly D, Charlebois C, Walker PR, Yamada T, Hirohashi S, Sikorska M. Regulation of DNaseY activity by actinin-alpha4 during apoptosis. Cell Death Differ. 2004;11:645–654. doi: 10.1038/sj.cdd.4401401. [DOI] [PubMed] [Google Scholar]
- Mann DL. Stress-activated cytokines and the heart: from adaptation to maladaptation. Annu Rev Physiol. 2003;65:81–101. doi: 10.1146/annurev.physiol.65.092101.142249. [DOI] [PubMed] [Google Scholar]
- Martinez NJ, Ow MC, Barrasa MI, Hammell M, Sequerra R, Doucette-Stamm L, Roth FP, Ambros VR, Walhout AJ. A C. elegans genome-scale microRNA network contains composite feedback motifs with high flux capacity. Genes Dev. 2008;22:2535–2549. doi: 10.1101/gad.1678608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merika M, Williams AJ, Chen G, Collins T, Thanos D. Recruitment of CBP/p300 by the IFN beta enhanceosome is required for synergistic activation of transcription. Mol Cell. 1998;1:277–287. doi: 10.1016/s1097-2765(00)80028-3. [DOI] [PubMed] [Google Scholar]
- Mohapatra B, Jimenez S, Lin JH, Bowles KR, Coveler KJ, Marx JG, Chrisco MA, Murphy RT, Lurie PR, Schwartz RJ, et al. Mutations in the muscle LIM protein and alpha-actinin-2 genes in dilated cardiomyopathy and endocardial fibroelastosis. Mol Genet Metab. 2003;80:207–215. doi: 10.1016/s1096-7192(03)00142-2. [DOI] [PubMed] [Google Scholar]
- Moore GW, Hutchins GM, Bulkley BH, Tseng JS, Ki PF. Constituents of the human ventricular myocardium: connective tissue hyperplasia accompanying muscular hypertrophy. Am Heart J. 1980;100:610–616. doi: 10.1016/0002-8703(80)90224-0. [DOI] [PubMed] [Google Scholar]
- Morimoto T, Sunagawa Y, Kawamura T, Takaya T, Wada H, Nagasawa A, Komeda M, Fujita M, Shimatsu A, Kita T, et al. The dietary compound curcumin inhibits p300 histone acetyltransferase activity and prevents heart failure in rats. J Clin Invest. 2008;118:868–878. doi: 10.1172/JCI33160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishita R, Sugimoto T, Aoki M, Kida I, Tomita N, Moriguchi A, Maeda K, Sawa Y, Kaneda Y, Higaki J, et al. In vivo transfection of cis element “decoy” against nuclear factor-kappaB binding site prevents myocardial infarction. Nat Med. 1997;3:894–899. doi: 10.1038/nm0897-894. [DOI] [PubMed] [Google Scholar]
- Moschos SA, Williams AE, Perry MM, Birrell MA, Belvisi MG, Lindsay MA. Expression profiling in vivo demonstrates rapid changes in lung microRNA levels following lipopolysaccharide-induced inflammation but not in the anti-inflammatory action of glucocorticoids. BMC Genomics. 2007;8:240. doi: 10.1186/1471-2164-8-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obernosterer G, Leuschner PJ, Alenius M, Martinez J. Post-transcriptional regulation of microRNA expression. RNA. 2006;12:1161–1167. doi: 10.1261/rna.2322506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivetti G, Melissari M, Capasso JM, Anversa P. Cardiomyopathy of the aging human heart. Myocyte loss and reactive cellular hypertrophy. Circ Res. 1991;68:1560–1568. doi: 10.1161/01.res.68.6.1560. [DOI] [PubMed] [Google Scholar]
- Olivetti G, Melissari M, Balbi T, Quaini F, Cigola E, Sonnenblick EH, Anversa P. Myocyte cellular hypertrophy is responsible for ventricular remodelling in the hypertrophied heart of middle aged individuals in the absence of cardiac failure. Cardiovasc Res. 1994;28:1199–1208. doi: 10.1093/cvr/28.8.1199. [DOI] [PubMed] [Google Scholar]
- Paroo Z, Ye X, Chen S, Liu Q. Phosphorylation of the human microRNA-generating complex mediates MAPK/Erk signaling. Cell. 2009;139:112–122. doi: 10.1016/j.cell.2009.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passier R, Zeng H, Frey N, Naya FJ, Nicol RL, McKinsey TA, Overbeek P, Richardson JA, Grant SR, Olson EN. CaM kinase signaling induces cardiac hypertrophy and activates the MEF2 transcription factor in vivo. J Clin Invest. 2000;105:1395–1406. doi: 10.1172/JCI8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poizat C, Puri PL, Bai Y, Kedes L. Phosphorylation-dependent degradation of p300 by doxorubicin-activated p38 mitogen-activated protein kinase in cardiac cells. Mol Cell Biol. 2005;25:2673–2687. doi: 10.1128/MCB.25.7.2673-2687.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell NH, Tang G, Yu C, Mercurio F, DiDonato JA, Lin A. Activation of NF-kappa B is required for hypertrophic growth of primary rat neonatal ventricular cardiomyocytes. Proc Natl Acad Sci USA. 2001;98:6668–6673. doi: 10.1073/pnas.111155798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recchiuti A, Krishnamoorthy S, Fredman G, Chiang N, Serhan CN. MicroRNAs in resolution of acute inflammation: identification of novel resolvin D1-miRNA circuits. FASEB J. 25:544–560. doi: 10.1096/fj.10-169599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Genes Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelfsema JH, White SJ, Ariyurek Y, Bartholdi D, Niedrist D, Papadia F, Bacino CA, den Dunnen JT, van Ommen GJ, Breuning MH, et al. Genetic heterogeneity in Rubinstein-Taybi syndrome: mutations in both the CBP and EP300 genes cause disease. Am J Hum Genet. 2005;76:572–580. doi: 10.1086/429130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth JF, Shikama N, Henzen C, Desbaillets I, Lutz W, Marino S, Wittwer J, Schorle H, Gassmann M, Eckner R. Differential role of p300 and CBP acetyltransferase during myogenesis: p300 acts upstream of MyoD and Myf5. EMBO J. 2003;22:5186–5196. doi: 10.1093/emboj/cdg473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanna B, Bueno OF, Dai YS, Wilkins BJ, Molkentin JD. Direct and indirect interactions between calcineurin-NFAT and MEK1-extracellular signal-regulated kinase 1/2 signaling pathways regulate cardiac gene expression and cellular growth. Mol Cell Biol. 2005;25:865–878. doi: 10.1128/MCB.25.3.865-878.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayed D, Hong C, Chen IY, Lypowy J, Abdellatif M. MicroRNAs play an essential role in the development of cardiac hypertrophy. Circ Res. 2007;100:416–424. doi: 10.1161/01.RES.0000257913.42552.23. [DOI] [PubMed] [Google Scholar]
- Sheng Z, Knowlton K, Chen J, Hoshijima M, Brown JH, Chien KR. Cardiotrophin 1 (CT-1) inhibition of cardiac myocyte apoptosis via a mitogen-activated protein kinase-dependent pathway. Divergence from downstream CT-1 signals for myocardial cell hypertrophy. J Biol Chem. 1997;272:5783–5791. doi: 10.1074/jbc.272.9.5783. [DOI] [PubMed] [Google Scholar]
- Shikama N, Lutz W, Kretzschmar R, Sauter N, Roth JF, Marino S, Wittwer J, Scheidweiler A, Eckner R. Essential function of p300 acetyltransferase activity in heart, lung and small intestine formation. EMBO J. 2003;22:5175–5185. doi: 10.1093/emboj/cdg502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slepak TI, Webster KA, Zang J, Prentice H, O'Dowd A, Hicks MN, Bishopric NH. Control of cardiac-specific transcription by p300 through myocyte enhancer factor-2D. J Biol Chem. 2001;276:7575–7585. doi: 10.1074/jbc.M004625200. [DOI] [PubMed] [Google Scholar]
- Song K, Backs J, McAnally J, Qi X, Gerard RD, Richardson JA, Hill JA, Bassel-Duby R, Olson EN. The transcriptional coactivator CAMTA2 stimulates cardiac growth by opposing class II histone deacetylases. Cell. 2006;125:453–466. doi: 10.1016/j.cell.2006.02.048. [DOI] [PubMed] [Google Scholar]
- Sugden PH, Clerk A. Cellular mechanisms of cardiac hypertrophy. J Mol Med. 1998;76:725–746. doi: 10.1007/s001090050275. [DOI] [PubMed] [Google Scholar]
- Sun Y, Varambally S, Maher CA, Cao Q, Chockley P, Toubai T, Malter C, Nieves E, Tawara I, Wang Y, et al. Targeting of microRNA-142-3p in dendritic cells regulates endotoxin-induced mortality. Blood. 117:6172–6183. doi: 10.1182/blood-2010-12-325647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsuguchi M, Seok HY, Callis TE, Thomson JM, Chen JF, Newman M, Rojas M, Hammond SM, Wang DZ. Expression of microRNAs is dynamically regulated during cardiomyocyte hypertrophy. J Mol Cell Cardiol. 2007;42:1137–1141. doi: 10.1016/j.yjmcc.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teiger E, Than VD, Richard L, Wisnewsky C, Tea BS, Gaboury L, Tremblay J, Schwartz K, Hamet P. Apoptosis in pressure overload-induced heart hypertrophy in the rat. J Clin Invest. 1996;97:2891–2897. doi: 10.1172/JCI118747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terasawa K, Ichimura A, Sato F, Shimizu K, Tsujimoto G. Sustained activation of ERK1/2 by NGF induces microRNA-221 and 222 in PC12 cells. FEBS J. 2009;276:3269–3276. doi: 10.1111/j.1742-4658.2009.07041.x. [DOI] [PubMed] [Google Scholar]
- Thum T, Galuppo P, Wolf C, Fiedler J, Kneitz S, van Laake LW, Doevendans PA, Mummery CL, Borlak J, Haverich A, et al. MicroRNAs in the human heart. A clue to fetal gene reprogramming in heart failure. Circulation. 2007;116:258–267. doi: 10.1161/CIRCULATIONAHA.107.687947. [DOI] [PubMed] [Google Scholar]
- Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, Galuppo P, Just S, Rottbauer W, Frantz S, et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456:980–984. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- Tili E, Michaille JJ, Cimino A, Costinean S, Dumitru CD, Adair B, Fabbri M, Alder H, Liu CG, Calin GA, et al. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol. 2007;179:5082–5089. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- Tran TH, Andreka P, Rodrigues CO, Webster KA, Bishopric NH. Jun kinase delays caspase-9 activation by interaction with the apoptosome. J Biol Chem. 2007;282:20340–20350. doi: 10.1074/jbc.M702210200. [DOI] [PubMed] [Google Scholar]
- van Empel VP, De Windt LJ. Myocyte hypertrophy and apoptosis: a balancing act. Cardiovasc Res. 2004;63:487–499. doi: 10.1016/j.cardiores.2004.02.013. [DOI] [PubMed] [Google Scholar]
- van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, Richardson JA, Olson EN. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci USA. 2006;103:18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- Vo NK, Goodman RH. CBP and p300 in transcriptional regulation. J Biol Chem. 2001;276:13505–13508. doi: 10.1074/jbc.R000025200. [DOI] [PubMed] [Google Scholar]
- Wei JQ, Shehadeh LA, Mitrani JM, Pessanha M, Slepak TI, Webster KA, Bishopric NH. Quantitative control of adaptive cardiac hypertrophy by acetyltransferase p300. Circulation. 2008;118:934–946. doi: 10.1161/CIRCULATIONAHA.107.760488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollert KC, Chien KR. Cardiotrophin-1 and the role of gp130-dependent signaling pathways in cardiac growth and development. J Mol Med. 1997;75:492–501. doi: 10.1007/s001090050134. [DOI] [PubMed] [Google Scholar]
- Yao TP, Oh SP, Fuchs M, Zhou ND, Ch'ng LE, Newsome D, Bronson RT, Li E, Livingston DM, Eckner R. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93:361–372. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- Yuan W, Sun W, Yang S, Du J, Zhai CL, Wang ZQ, Zhang J, Zhu TH. Downregulation of microRNA-142 by proto-oncogene LMO2 and its co-factors. Leukemia. 2008;22:1067–1071. doi: 10.1038/sj.leu.2405001. [DOI] [PubMed] [Google Scholar]
- Zhang CL, McKinsey TA, Chang S, Antos CL, Hill JA, Olson EN. Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell. 2002;110:479–488. doi: 10.1016/s0092-8674(02)00861-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- Zhao J, Cao Y, Lei Z, Yang Z, Zhang B. Huang B selective depletion of CD4+CD25+Foxp3+ regulatory T cells by low-dose cyclophosphamide is explained by reduced intracellular ATP levels. Cancer Res. 70:4850–4858. doi: 10.1158/0008-5472.CAN-10-0283. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.