Abstract
The Lim-kinase (LIMK) proteins are important for the regulation of the actin cytoskeleton, in particular the control of actin nucleation and depolymerisation via regulation of cofilin, and hence may control a large number of processes during development, including cell tensegrity, migration, cell cycling, and axon guidance. LIMK1/LIMK2 knockouts disrupt spinal cord morphogenesis and synapse formation but other tissues and developmental processes that require LIMK are yet to be fully determined. To identify tissues and cell-types that may require LIMK, we characterised the pattern of LIMK1 protein during mouse embryogenesis. We showed that LIMK1 displays an expression pattern that is temporally dynamic and tissue-specific. In several tissues LIMK1 is detected in cell-types that also express Wilms’ tumour protein 1 and that undergo transitions between epithelial and mesenchymal states, including the pleura, epicardium, kidney nephrons, and gonads. LIMK1 was also found in a subset of cells in the dorsal retina, and in mesenchymal cells surrounding the peripheral nerves. This detailed study of the spatial and temporal expression of LIMK1 shows that LIMK1 expression is more dynamic than previously reported, in particular at sites of tissue–tissue interactions guiding multiple developmental processes.
Keywords: Limk, Kidney, Heart, Epithelia-to-mesenchyme transition, Mesenchyme-to-epithelia transition, Eye, Testes
The regulation of the actin cytoskeleton is essential for a very large number of cellular process including cellular migration, axon guidance, cell cycling, and polarity acquisition and maintenance. LIMK1 and LIMK2 control one of the fundamental aspects of actin dynamics, the depolymerisation of filamentous actin, by phosphorylation of ADF/cofilin (Arber et al., 1998). LIMK1 is primarily phosphorylated by the family of small Rho-GTPases. When phosophorylated, LIMK1 in turn phosphorylates cofilin and blocks its ability to sever and de-nucleate actin filaments. Phosphorylation also inhibits the actin de-branching properties that are normally exerted by cofilin competing with the actin-branching Arp2/3 complex (Arber et al., 1998; Bernard, 2007; Bernstein and Bamburg, 2010). Recently it has been shown that LIMK1 can act downstream of BMP signalling, modulate synapse stability by binding to BMP type II receptors, and control cell cycling by shuttling to the nucleus (Bernstein and Bamburg, 2010; Davila et al., 2007; Eaton and Davis, 2005; Hocking et al., 2009; Wen et al., 2007). In vivo and in vitro experiments on LIMK1 knockout mice have shown that the loss of LIMK1 results in neuronal growth cone, dendritic spine, and actin defects that are associated with abnormal hippocampal long term potentiation and synaptic changes (Meng et al., 2002). LIMK1 knockout mice also display elevated fear response and weakened learning abilities (Meng et al., 2002). Interestingly, the role of LIMK1 in controlling dendritic spine morphology is at least in part modulated by inhibition of LIMK1 through post-transcriptional microRNA-mediated regulation (Schratt et al., 2006). In human patients, chromosomal deletions encompassing LIMK1 and other genes, have been associated with Williams–Beuren syndrome which is characterised by mental retardation, and craniofacial and behavioural defects (Hoogenraad et al., 2004; Scott and Olson, 2007). Many of these symptoms are mimicked in mice carrying targeted deletions of syntenic chromosomal regions (Hua Li et al., 2009). Whilst LIMK1 and LIMK2 double knockouts only cause a moderately more severe phenotype compared to the LIMK1 knockout (Meng et al., 2004), knocking out the LIMK1 effector, n-cofilin/cofilin-1 results in abnormal neural cell migration, neuronal defects and embryonic lethality at E10.5 (Gurniak et al., 2005). This discrepancy in severity between LIMK and n-cofilin mutants may be partly explained by the redundancy provided by relatives of the LIMK family, the testes specific kinases 1 and 2, which also phosphorylate cofilin (Toshima et al., 2001). This notion is supported by recent targeted tissue-specific knockouts of cofilin and its relative destrin in the collecting-duct epithelium of the metanephros where the loss of cofilin and destrin has profound effects on morphology and growth (Kuure et al., 2010).
LIMK1 expression has previously been partially described (Mori et al., 1997; Foletta et al., 2004; Acevedo et al., 2006) but in spite of the emerging importance of LIMK1 for both BMP-receptor signalling and axon guidance, LIMK1 expression is yet to be characterised in the context of specific embryological processes, tissues and cell types. In this work we show that LIMK1 is highly expressed in many neuronal and epithelial tissues where its spatially restricted and dynamic expression pattern correlates with important embryological events. Significantly, LIMK1 is strongly detected in tissues that form by both epithelia-to-mesenchyme (EMT) and mesenchyme-to-epithelia (MET) transitions. However, whilst LIMK1 is persistently co-expressed with the EMT regulator Wilms’ tumour protein 1 (WT1) (Martínez-Estrada et al., 2010) in cells that undergo EMT, LIMK1 is only transiently co-expressed with WT1 during the actual process of MET and is otherwise mutually exclusive with WT1 in these cells.
1. Results
We determined a time-course of localisation of LIMK1 protein in mice during embryogenesis using mouse embryos at stages E10.5, E12.5, E13.5, E14.5, E16.5 and E18.5. LIMK1 plays a central role in actin cytoskeleton regulation (Arber et al., 1998; Bernstein and Bamburg, 2010), and as such would be expected to be widely expressed. We describe a spatially restricted and cell-type specific expression pattern for LIMK1 during organogenesis. Negative controls were performed for each antibody. Non-specific signal from auto-fluorescent erythrocytes were detected and distinguished from real immunofluorescent stains as specified in Experimental Procedures. Erythrocytes are indicated in figures. Where appropriate single-channel LIMK1 immunostaining patterns are presented alongside panels with merged signals from different antibodies.
1.1. Detection of LIMK1 in ectoderm, mesoderm, and endoderm derived tissues
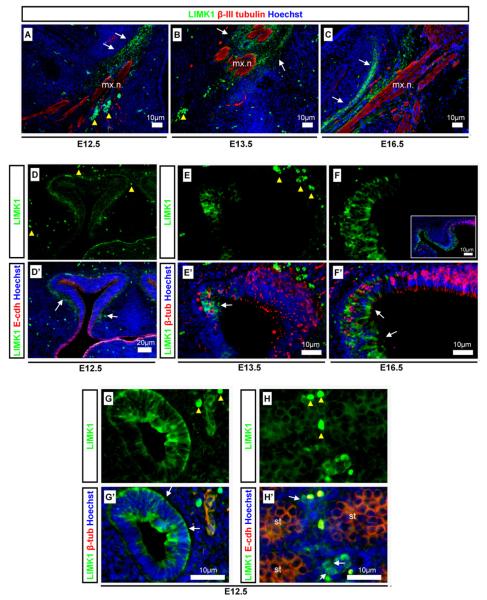
LIMK1 was detected using a monoclonal LIMK1 antibody designed against the LIMK1 PDZ domain with affinity for LIMK1, as previously verified (Lee-Hoeflich et al., 2004; Nadella et al., 2009). LIMK1 was detected in the mesenchymal cells immediately surrounding but not within the β-III tubulin positive maxillary nerve branch of the trigeminal ganglion (Fig. 1A–C), adjacent and within the nasal progressively maturing neuroepithelium (Fig. 1D–F), in the gut epithelium but not the adjacent mesenchyme (Fig. 1G), and in the testes (Fig. 1H). The embryonic testis consists of seminiferous tubules containing germ cells, and supporting interstitial cells such as the androgen producing embryonic Leydig cells (Scott et al., 2009). LIMK1 was strongly detected in cells that were identified histologically as embryonic Leydig cells (Fig. 1H) and at low uniform levels in the seminiferous tubules. LIMK1 was not detected in other interstitial cells. Our results differ from those of Foletta et al., 2004, in that they found LIMK1 to also be strongly expressed in the cytoplasm and nuclei of cells in the seminiferous tubules. A possible explanation for this lies in the number of LIMK1 splice variants, because different antibodies may display different affinities for different LIMK1 isoforms.
Fig. 1.
LIMK1 expression in mesenchymal and epithelial cell types. LIMK1 detection was analysed using neuronal β-III tubulin and epithelial E-cadherin as differentiation and tissue markers. LIMK1 (green) was found in the: (A,C) mesenchyme surrounding the maxillary nerve labeled with β-tubulin (red) from E12.5-E16.5; (D and D′-F and F′) maturing nasal neuroepithelium from E12.5–E16.5 (F-insert showing (F) at lower magnification); (G and G′) intestinal epithelium; (H and H′) developing testes. Scale bars, antibody stains, and embryonic stages are as indicated on images. Yellow arrowheads indicate auto-fluorescent erythrocytes. White arrows indicated detection of LIMK1. mx.n: Maxillary nerve, st: seminiferous tubule.
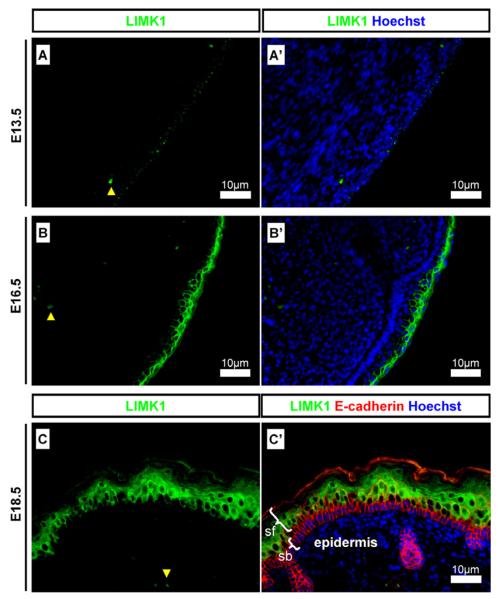
LIMK1 was detected in the skin epidermis. At E13.5 LIMK1 was detected in a punctuated pattern (Fig. 2A) which became elevated and organised as the epidermis developed at E16.5 (Fig. 2B) and E18.5 (Fig. 2C), where it was excluded from the stratum basale but present at high concentrations in the more superficial layers (Fig. 2C).
Fig. 2.
LIMK1 expression in the developing epidermis. Detection of LIMK1 in the epidermis of developing skin. (A and A′) at E13.5 LIMK1 was detected in a punctuated expression pattern in the developing epidermis; (B and B′) at E16.5 LIMK1 was strongly detected in the immature epidermis; (C and C′) at E18.5 LIMK1 was strongly detected in the superficial epidermis but not in the stratum basale. Scale bars and antibody stains are as indicated on images. Yellow arrowheads indicate auto-fluorescent erythrocytes. White brackets indicated epidermal layers. sb: Stratum basale; sf superficial layers.
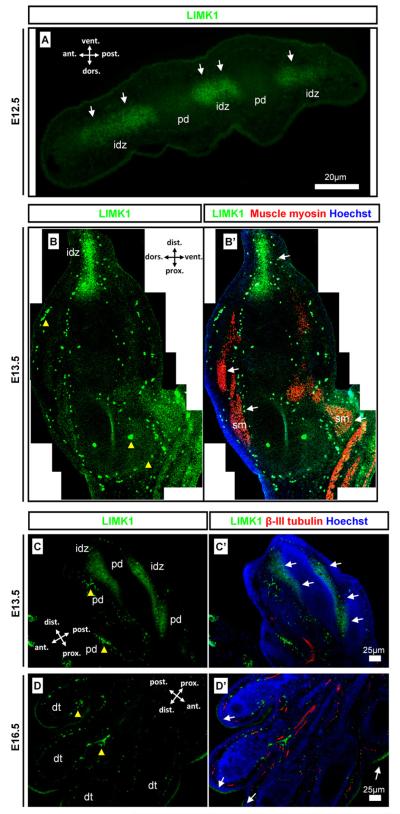
In the limb, LIMK1 was detected interdigitally at E12.5 (Fig. 3A). LIMK1 was weakly detected along the dorso-ventral axis of the limb in mesenchyme and in developing muscle (Fig. 3B). Along the proximo-distal axis LIMK1 was strongly detected in the distal developing hand plate in the interdigital necrotic zones (Fig. 3B). Anterior-posterior sections across the hand plate showed that the interdigital LIMK1 (Fig. 3C) was lost by E16.5 (Fig. 3D) when the digits are separated. Epidermal LIMK1 expression was then detected surrounding the hand plate (Fig. 3D) consistent with the previously described detection of LIMK1 in the epidermis (Fig. 2B).
Fig. 3.
LIMK1 expression in the developing limb. Detection of LIMK1 in the limb in comparison with neuronal β-III tubulin and muscle myosin tissue markers. (A) cross-section of hand plate with LIMK1 signal in the interdigital zones and not in digits; (B and B′) section along the hind limb proximo-distal axis with uniform dorsal and ventral detection, signal in skeletal muscle and high distal interdigital expression; (C and C′) anterior-posterior hand plate section showing strong detection of LIMK1 in interdigital apoptotic zones; (D and D′) Anterior-posterior hand plate section showing formed digits with interdigital epithelium and weak detection of LIMK1 only in the skin epidermis as described in (Fig. 2). Scale bars, antibody stains, orientation, and embryonic stages are as indicated on images. Yellow arrowheads indicate auto-fluorescent erythrocytes. idz: Interdigital zone; pd: presumptive digit; dt: digit; sm: skeletal muscle.
1.2. Localisation of LIMK1 in the eye
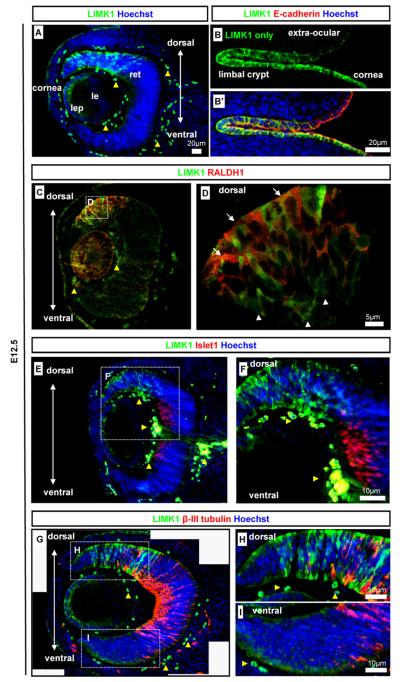
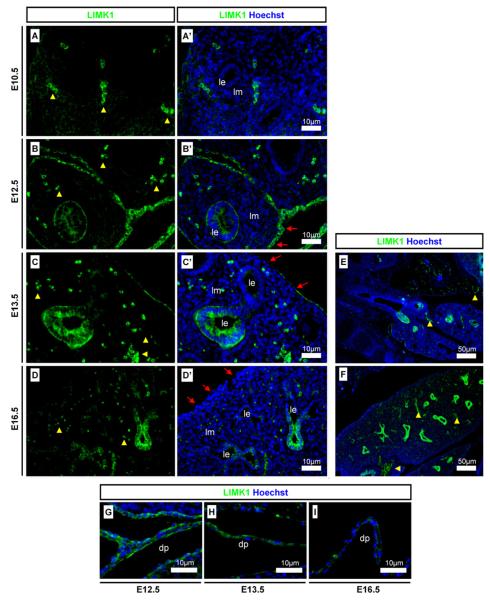
In the embryonic eye, LIMK1 was detected in the developing corneal epithelium, in the lens epithelium, and in the retina ages (Fig. 4A). LIMK1 was strongly detected in the lens epithelium but not in the maturing lens fibres (Fig. 4A). To investigate further the pattern of LIMK1 in the epithelial structures we co-stained with epithelial marker E-cadherin. In the ocular surface from E12.5 LIMK1 displayed a dynamic expression pattern in the epithelial, E-cadherin-positive cornea, limbal crypt, and neighbouring epithelium (Fig. 4B). The epithelium progresses from a columnar type at positions distal to the cornea, through the columnar epithelium of the limbal crypt, and to the proximal corneal position which are of a cuboidal epithelial type at embryonic stages (Collinson et al., 2002). In the extra-ocular epithelium, it was evident that LIMK1 was weakly detected in a punctuated pattern (Fig. 4B) similar to that of early epidermis. In the corneal limbal crypts and the corneal epithelium, LIMK1 was strongly detected in an evenly distributed pattern along the whole apical-basal axis (Fig. 4B).
Fig. 4.
Detection of LIMK1 in the eye. LIMK1 localisation was analysed in the eye using the following markers to detect optic structures and differentiation states: epithelia (E-cadherin), dorsal retina (RALDH1), neuroblast transcription factor (Islet1), and retinal ganglion cells (β-III tubulin). (A) LIMK1 expression in the, lens and corneal epithelia, and in a mosaic pattern in the dorsal retina at E12.5; (B and B′) LIMK1 localisation in the developing corneal, limbal and conjunctival epithelia at E12.5; (C and D) LIMK1 was detected in the dorsal retina but not always in the same cells as another dorsal marker, RALDH1 (co-detection: arrows, no co-detection: arrowheads); (E and F) LIMK1 was not detected in cells positive for neurogenic marker Islet1; (G–I) The expression of LIMK1 and retinal ganglion cell marker β-III tubulin were almost mutually exclusive in developing neurones. Antibody stains, embryonic stages, and scale bars are as specified on images. Dashed-line boxes indicate magnified regions. Yellow arrowheads indicate auto-fluorescent erythrocytes. lep: Lens epithelium; le: lens, ret: retina.
Retinal expression was spatially highly localised and LIMK1 appeared at elevated levels in the cytoplasm of a subset of cells in the dorsal retina and at lower levels in the ventral retina ages (Fig. 4A). We performed co-immunohistochemistry against known retinal markers and LIMK1 to further define the LIMK1 positive cells/domain. Immunofluorescent stains against dorsal retina marker RALDH1 (Fan et al., 2003; Murali et al., 2005), confirmed the dorsal localisation of LIMK1 (Fig. 4C,D). However the cellular staining was not totally coincident – high LIMK1 levels were only detected in a subset of RALDH1 positive cells (Fig. 4D). Transcription factor Islet1 and cytoskeletal protein β-III tubulin are markers for the differentiating retinal neuroblast layer and retinal ganglion cells (RGC) (Pan et al., 2008; Sharma and Netland, 2007). RGC formation occurs in a central to marginal retina direction (Pan et al., 2008; Sharma and Netland, 2007). The dorsal LIMK1 positive cells were not Islet1 positive at E12.5 (Fig. 4E and F). Co-staining analysis of LIMK1 with β-III tubulin showed that LIMK1 positive cells were almost exclusively β-III tubulin-negative (Fig. 4G–I). At later time-points (E13.5 and E16.5) LIMK1 was not detected in the retina (data not shown). Collectively, this data suggest a role for LIMK1 during early retinal neuroblast differentiation prior to the expression of definitive neuronal markers.
1.3. Detection of LIMK1 in the heart, lung and kidney
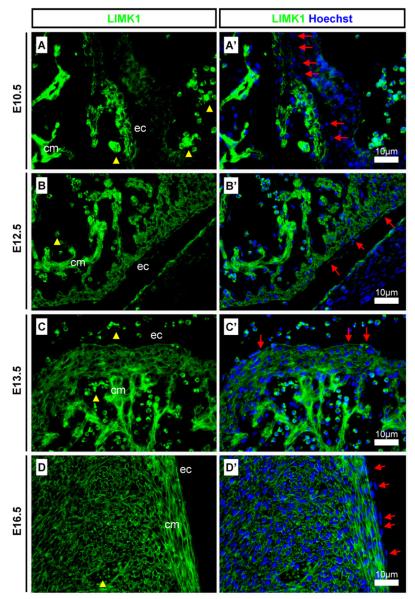
LIMK1 was detected from an early stage in the heart. At E10.5 LIMK1 was found in the cardiomyocytes but not in the epicardial cells (Fig. 5A), however at E12.5 and E13.5 LIMK1 was detected in both cell types (Fig. 5B and C). At E16.5 LIMK1 expression remained strong in the cardiomyocytes and cardiac muscle cells but was lower in the epicardium (Fig. 5D).
Fig. 5.
Detection of LIMK1 in the developing heart. LIMK1 was found in the developing heart. (A and A′) LIMK1 was found in the cardiomyocytes from E10.5-E16.5 and although not detected in the epicardial cells at E10.5, protein was present from E12.5; (B and B′) LIMK1 was detected in cardiomyocytes and in epicardial cells at E12.5 and (C and C′) E13.5; (D and D′) LIMK1 was strongly detected in cardiomyocytes and weakly in epicardial cells at E16.5. Yellow arrowheads indicate auto-fluorescent erythrocytes. Red arrows indicate epicardial cells. Scale bars, antibody stains, and embryonic stages are as indicated on images. ec: Epicardium; cm cardiac muscle.
LIMK1 was not detected in lung epithelium at E10.5 (Fig. 6A) but was strongly detected from E12.5 to E16.5 (Fig. 6B–D). LIMK1 was found to be detected strongly in some branches of the lung epithelium but at very low levels in others (Fig. 6C and D). Lower magnification images highlight the variable detection pattern in lung epithelia at E13.5 and E16.5 (Fig. 6E and F). LIMK1 was strongly detected in the pleura at E12.5 and E13.5 (Fig. 6B and C) but the detection levels were diminished at E16.5 (Fig. 6D). LIMK1 was strongly detected in the diaphragm at E12.5 and E13.5 (Fig. 6G and H) but was found a lower levels at E16.5 (Fig. 6I).
Fig. 6.
Detection of LIMK1 in developing lung tissues. (A and A′) LIMK1 was not detected in the E10.5 lung epithelium; (B and B′) LIMK1 was detected in lung epithelium and in cells of the pleura at E12.5 and (C and C′) E13.5; (D and D′) LIMK1 was present in lung epithelium but only weakly in the pleura at E16.5. (E and F) Low magnification images show a dynamic detection pattern of LIMK1 in branching lung epithelium. LIMK1 was strongly detected in the diaphragm at (G) E12.5 and at (H) E13.5, but weakly at (I) E16.5. Yellow arrowheads indicate auto-fluorescent erythrocytes. Red arrows indicate pleura cells. Scale bars, antibody stains, and embryonic stages are as indicated on images. le: Lung epithelium; lm: lung mesenchyme; dp: diaphragm.
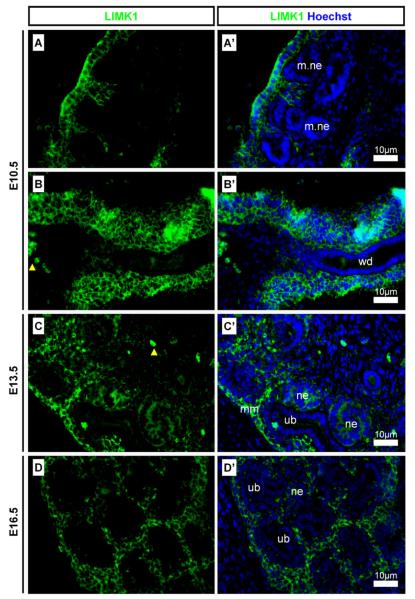
LIMK1 was strongly detected in the temporary mesonephros (Fig. 7A) and in the mesenchyme surrounding the Wolffian duct (Fig. 7B) at E10.5. In the permanent metanephric kidney LIMK1 was strongly detected in the mesenchyme surrounding the ureteric buds, in the stroma, and also in the nephrons (Fig. 7C and D).
Fig. 7.
Detection of LIMK1 in the developing kidney. LIMK1 was detected in mesonephrogenic and metanephrogenic tissues. (A and A′) LIMK1 was found in the mesenchyme surrounding mesonephric nephrons and (B and B′) in the mesenchyme surrounding the Wolffian duct at E10.5; (C and C′) LIMK1 was detected in the metanephric mesenchyme and in nephrons but not in the ureteric bud at E13.5 and at (D and D′) E16.5. Yellow arrowheads indicate auto-fluorescent erythrocytes. Scale bars, antibody stains, and embryonic stages are as indicated on images. wd: Wolffian duct; ub: ureteric bud; mm: metanephric mesenchyme; ne: nephron; m.ne: mesonephric nephron.
1.4. Dynamic localisation of LIMK1 in EMT mesothelial tissues
LIMK1 was detected in a dynamic manner in the mesothelial tissues of the pleura, epicardium, and diaphragm, tissues that all undergo Epithelial-to-Mesenchymal Transition (EMT). EMT is a process that requires the loss of epithelial-specific cadherins, gene expression changes, and loss of apico-basal polarity, in favour of mesenchyme-specific genes and cytoskeletal components (Davies and Garrod, 1997). WT1 has recently been shown to regulate this process in the epicardium via the activation of snail and inhibition of e-cadherin expression (Martínez-Estrada et al., 2010).
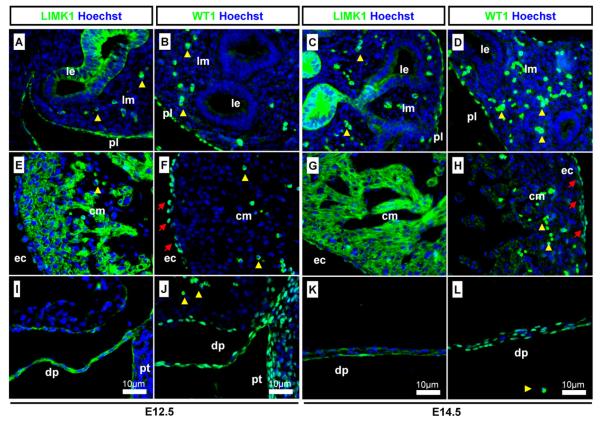
Immunostaining against LIMK1 and WT1 showed that the strong detection of LIMK1 were found in similar patterns to that of WT1 in the pleura (Fig. 8A–D), in the epicardium (but not myocardium) (Fig. 8E–H), and in the diaphragm (Fig. 8I–L). LIMK1 and WT1 were also colocalised in the peritoneal layers (Fig. 8I and J).
Fig. 8.
Detection of LIMK1 in WT1-expressing EMT tissues. Detection of LIMK1 in tissues with cells undergoing EMT using EMT regulating protein WT1 as a differentiation marker. (A and C) LIMK1 in lung epithelium and pleura; (B and D) WT1 in the pleura but not lung epithelium; (E and G) LIMK1 in epicardial and myocardiocyte cells; (F and H) WT1 in epicardial cells (red arrows); (I and K) LIMK1 in the diaphragm and peritoneum; (J and L) WT1 in the diaphragm and peritoneum. Yellow arrowheads indicate auto-fluorescent erythrocytes. Antibody stains, embryonic stages, and scale bars are as specified on images. le: Lung epithelium; lm: lung mesenchyme; ec: epicardium; cm: cardiac muscle; dp: diaphragm; pt: peritoneum.
1.5. Expression of LIMK1 in MET cells in the metanephric kidney
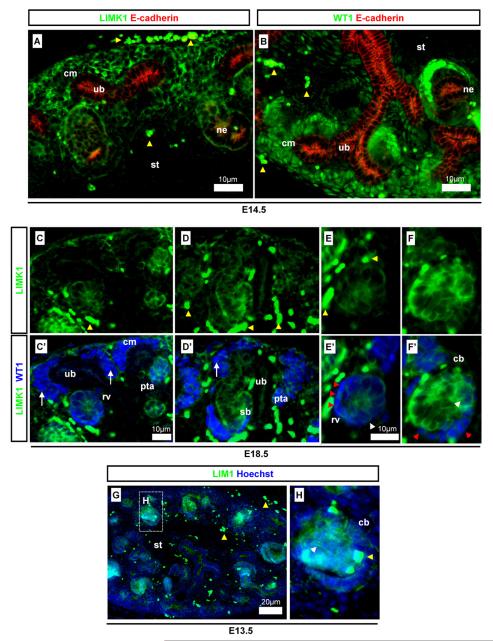
LIMK1 was shown to be detected in EMT tissues and we (Fig. 7) and others (Foletta et al., 2004) have shown LIMK1 to be present in the metanephric kidney, where extensive Mesenchymal-to-Epithelial Transition (MET) occurs (Saxen, 1987). We carefully investigated the specific developmental expression pattern of LIMK1 in this tissue. LIMK1 was detected in the kidney mesenchyme, stroma, and nephrons but not in the ureteric bud/collecting duct (Fig. 9A), as shown using markers WT1 and epithelial E-cadherin. In the kidney, WT1 is a marker for the cap mesenchyme, early differentiating nephrons, and in the more mature nephron, also the cells of the Bowman’s capsule (Armstrong et al., 1993; Huber et al., 2000). E-cadherin is found within the ureteric bud and in the nephron after the MET (Georgas et al., 2009). WT1, which is mainly localised to the nuclei, was detected in an expression pattern similar but not identical to LIMK1 (Fig. 9B). Specifically, LIMK1 was found to be excluded or strongly down-regulated from the cap mesenchyme surrounding the ureteric bud tips; the mesenchyme which contains WT1-positive nephron progenitor cells that will undergo MET and form epithelial nephrons (Fig. 9C and D) (Davies et al., 2004; Saxen, 1987). The process of MET, the opposite of EMT, requires the gain of epithelial characteristics such as epithelial cell junctions, apico-basal polarity, and often planar polarity (Davies and Garrod, 1997). WT1 is essential for nephron formation (Davies et al., 2004). In the nephrons, LIMK1 was detected together with WT1 at the earliest stage of nephron formation in the pretubular aggregate, when mesenchymal nephron progenitor cells tightly cluster together and undergo MET (Fig. 9C). WT1 and LIMK1 were also found together in the subsequent stage of nephron development, the newly formed epithelial renal vesicle (Fig. 9C and E). LIMK1 and WT1 co-localisation decreased after the renal vesicle stage when the MET was complete. In the following comma-shaped and s-shaped body stages of nephron development (Fig. 9D and F), LIMK1 was weakly detected in the strongly WT1-positive cells (those in the visceral and parietal epithelia of the presumptive glomerulus) but remained strongly positive in the tubular compartment where WT1 expression was low. The WT1-positive cells give rise to the glomerular podocytes, a cell-type expressing both epithelial and mesenchymal markers (Armstrong et al., 1993; Yaoita et al., 1999). Lim1 homeobox gene is required for nephron formation and primarily for the progression from the renal vesicle form to later stages of nephron development (Kobayashi et al., 2005). Lim1 is expressed in the renal vesicle and the proximal and distal portions of s-shaped nephrons (Kobayashi et al., 2005). Comparing Lim1 and LIMK1 detection patterns we found that LIMK1 was detected in a pattern highly similar Lim1 in the nephrons (Fig. 9G and H).
Fig. 9.
Detection of LIMK1 in WT1-expressing MET cells of the metanephros. Detection of LIMK1 in cells undergoing MET using WT1 and E-cadherin as a differentiation and structural markers. (A and B) LIMK1 was detected in a pattern that is similar but not identical to WT1 at E14.5; (C, C′, D and D′) weak LIMK1 detection in cap-mesenchyme (arrows) contrasting with strong detection of WT1; (E, E′, F and F′) co-detection of LIMK1 and WT1 in renal vesicles and proximal comma-shaped body nephrons (white arrowheads) and exclusion/down-regulation of LIMK1 from presumptive podocytes (red arrowheads); (G–H) LIM1 expression in tubular comma- and s-shaped nephrons (white arrowheads). Yellow arrowheads indicate auto-fluorescent erythrocytes. Antibody stains, embryonic stages, and scale bars are as specified on images. cm: Cap mesenchyme; ub: ureteric bud; sb: s-shaped body; cb: comma-shaped body; ne: nephron; st: stroma; rv: renal vesicle; pta: pretubular aggregate.
2. Discussion
In this work we investigated LIMK1 protein localisation patterns during embryonic mouse development. It has previously been demonstrated that LIMK1 is present in axon growth cones (Wen et al., 2007) and in the spinal cord (Meng et al., 2002). It is also known to phosphorylate cofilin (Arber et al., 1998), interact with BMP receptors (Eaton and Davis, 2005), and act during cell cycling regulation (Davila et al., 2007). In spite of the apparent significance of LIMK1 little is known about the expression of LIMK1 at the level of tissue formation and patterning. We showed a variable and dynamic expression pattern for LIMK1 in neuronal, epithelial, and mesenchymal cell types. We found the LIMK1 protein localisation pattern particularly interesting in those developing tissues undergoing state transitions and in the retina.
2.1. LIMK1 expression during MET/EMT
Nephrons form when nephron progenitor cells aggregate and progress through MET; a process dependent on WT1 (Davies et al., 2004). Prior to MET, LIMK1 was not detected in the mesenchymal WT1-positive nephron progenitor cells but was immediately detected once cell-aggregation and epithelialisation steps began. However, LIMK1 was quickly lost from the glomerular progenitor cells subsequent to nephron formation when WT1 was further upregulated in these cells. The diaphragm, pleura, and epicardium, on the other hand, form by EMT where at least the normal development of the epicardium is dependent on WT1 regulating snail and e-cadherin expression (Martínez-Estrada et al., 2010). In these EMT tissues, LIMK1 was strongly and stably co-expressed with WT1. As WT1 has recently been shown to act on EMT regulating genes and is essential for nephron formation (Davies et al., 2004), it would be of interest to determine whether LIMK1 may be downstream of WT1 signalling. This could explain the dynamics of the co-expression in the nephron and the differences in MET and EMT tissues. Ureteric bud-specific knockouts of cofilin1 and destrin have demonstrated a necessity for cofilin/destrin in the morphogenesis of the ureteric bud (Kuure et al., 2010) and similarly, it is important to determine if LIMK1 and cofilin1/destrin might have a role in the metanephric mesenchyme. Our data suggests that further research is needed to determine what, if any, role LIMK1 may have during EMT/MET processes as well as elucidating any LIMK1-WT1 connection.
2.2. LIMK1 expression in the eye and retina
LIMK1 is known to act in BMP-regulated axon growth cones in both retinal ganglion and spinal cord cells (Hocking et al., 2009; Wen et al., 2007). Dorsal retinal expression of BMP4 and BMPRI has previously been implicated in retinal patterning during development (Murali et al., 2005). It is interesting to note that the dorsal expression pattern of LIMK1 is similar to that of the retinal BMP signalling domain (Murali et al., 2005). We speculate that since the LIMK1 expression preceded the neurogenesis, as indicated by its Islet1- and β-III tubulin-negative protein localisation domain, LIMK1 may also be an important factor involved in retinal ganglion cell differentiation and possibly apical-basal migration.
The surprisingly dynamic expression pattern of LIMK1 in MET/EMT tissues and in the retina highlights the lack of understanding of what roles LIMK1 play during development. Our data emphasises the need to better understand any redundancy that can resolve the paradox of the relatively mild phenotypes of LIMK1/LIMK2 knockouts compared to cofilin knockouts. Further research is also required to determine whether LIMK1 is involved in mesenchyme and epithelium state transitions as well as for retinal differentiation. Phosphorylated LIMK1 modulates cofilin activity and it would be of importance to in the future also determine the exact pattern of phospho-LIMK1 activity when a LIMK1-positive tissue is investigated in more depth. Likewise, as LIMK1 is present in multiple isoforms, a similar isoform-specific study would be necessary to determine the activity of LIMK1.
3. Experimental procedures
3.1. Immunohistochemistry and embryo preparation
Timed matings were set up for C57 mice overnight and when a vaginal plug was identified in the morning this was considered as E0.5. All animal handling was carried according to UK Home Office protocol. Whole embryos or tissues were dissected from E10.5, E12.5, E13.5, E14.5, E16.5 and E18.5 embryos and were fixed in 4% PFA in 1 × PBS overnight at 4 °C. Tissues were thoroughly rinsed in 1 × PBS, fully dehydrated using sequential dilutions (70%, 85%, 95%) to 100% ethanol, and prepared in xylene overnight at room temperature. Tissues were placed in paraffin wax and embedded for a minimum of 6 h. Embryos and tissues were sectioned at 7 μm intervals, fixed onto poly-l-lysine coated glass slides, and dried at 37 °C overnight. For immunofluorescent stains de-waxing was performed in Histoclear and 100% ethanol. Tissue rehydration was carried out using successively decreased ethanol concentrations (100%, 95%, 85%, 70%, 50%) and finally placed in 1 × PBS. Antigen-retrieval was carried out using 0.1 M citrate buffer and microwave heating (4 × 5 min at 800 W). Samples were thoroughly washed in 1 × PBS prior to antibody blocking in 4% BSA in 1 × PBS for 1–2 h. Primary antibody incubation was carried over night at 4 °C in 4% BSA in 1 × PBS. Where appropriate, primary antibodies were incubated simultaneously. Primary antibodies were used at: 1:100–150 anti-LIMK1 mouse monoclonal IgG1 (BD transduction laboratories), 1:800 anti-β-III tubulin polyclonal rabbit IgG (SIGMA), 1:200 anti-WT1 monoclonal mouse IgG1 (Genetex), 1:100 anti-WT1 polyclonal rabbit IgG C19 (SantaCruz), 1:100 anti-E-cadherin monoclonal mouse IgG2a (BD transduction laboratories), 1:20 anti-Islet1 monoclonal mouse IgG2b (DSHB), 1:20 anti-heavy-chain myosin monoclonal mouse IgG2b MF20 (DSHB), 1:100 anti-RALDH1 polyclonal (Abcam). Fluorescent secondary antibody incubation was carried out for 1–2 h in 4% BSA in 1 × PBS at room temperature. Fluorescent secondary antibodies were purchased from Invitrogen Molecular Probes. Invitrogen Molecular Probes antibodies used were: goat-anti-mouse IgG1 (A21121), donkey-anti-rabbit IgG (A21207), donkey anti-mouse IgG (A10037), goat anti-mouse IgG2a (A21135), and goat anti-mouse IgG2b (A21145). All secondary antibodies were used at 1:200. Samples were mounted in Mowviol with 1:5000 Hoechst under cover slips. Negative controls were performed as above except that slides were incubated in blocking buffer without primary antibody. Auto-fluorescence from erythrocytes was detected most slides. These were readily identified and distinguished from real immunofluorescent signals based on cell morphology (very round cells as single or clusters of cells), detection in multiple channels (both red and green channels), and the off-colour quality of the signal (auto-fluorescent signals are visibly of a different colour to bona fide signal). Fluorescent labelling was viewed using a Nikon Eclipse E400 and each of the fluorophore images were sequentially captured using a Qimagin Qcam camera and Volocity v.5.3.1 software. Images were compiled and merged using public domain software ImageJ v.1.41o (NIH). At least ten stains were performed on sections from throughout embryos for each stage in order to capture all major embryological tissues. The number of embryos used at each stage was: n = 2 at E10.5, n = 2 at E12.5, n > 12 at E12.5, n = 6 at E13.5, n = 3 at E14.5, n = 2 at E16.5, n = 2 at E18.5.
Acknowledgements
We thank Jamie Davies, Peter Hohenstein, Rachel Berry and Mathieu Unbekandt for stimulating discussions, for antibodies, and advice on EMT/MET processes.
Source of funding: This work was funded by the Medical Research Council (UK) (Grant award: G0800901).
Footnotes
Conflict of interest: The authors declare no conflict of interest.
References
- Acevedo K, Moussi N, Li R, Soo P, Bernard O. LIM kinase 2 is widely expressed in all tissues. Journal of Histochemistry and Cytochemistry. 2006;54:487–501. doi: 10.1369/jhc.5C6813.2006. [DOI] [PubMed] [Google Scholar]
- Arber S, Barbayannis FA, Hanser H, Schneider C, S CA, Bernard O, Caroni P. Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase. Nature. 1998;393:805–809. doi: 10.1038/31729. [DOI] [PubMed] [Google Scholar]
- Armstrong JF, Pritchard-Jones K, Bickmore WA, Hastie ND, Bard JBL. The expression of the Wilms-tumor gene, Wt1, in the developing mammalian embryo. Mechanisms of Development. 1993;40(1993):85–97. doi: 10.1016/0925-4773(93)90090-k. [DOI] [PubMed] [Google Scholar]
- Bernard O. Lim kinases, regulators of actin dynamics. International Journal of Biochemistry and Cell Biology. 2007;39:1071–1076. doi: 10.1016/j.biocel.2006.11.011. [DOI] [PubMed] [Google Scholar]
- Bernstein BW, Bamburg JR. ADF/cofilin: a functional node in cell biology. Trends in Cell Biology. 2010;20:187–195. doi: 10.1016/j.tcb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinson JM, Morris L, Reid AI, Ranaesh T, Keighren MA, Flockhart JH, Hill RE, Tan S, Ramaesh K, Dhillon B, West JD. Clonal analysis of patterns of growth, stem cell activity, and cell movement during the development and maintenance of the murine corneal epithelium. Developmental Dynamics. 2002;224:432–440. doi: 10.1002/dvdy.10124. [DOI] [PubMed] [Google Scholar]
- Davies JA, Garrod DR. Molecular aspects of the epithelial phenotype. BioEssays. 1997;19:699–704. doi: 10.1002/bies.950190810. [DOI] [PubMed] [Google Scholar]
- Davies JA, Ladomery M, Hohenstein P, Michael L, Shafe A, Spraggon L, Hastie N. Development of an siRNA-based method for repressing specific genes in renal organ culture and its use to show that the Wt1 tumour suppressor is required for nephron differentiation. Human Molecular Genetics. 2004;13:235–246. doi: 10.1093/hmg/ddh015. [DOI] [PubMed] [Google Scholar]
- Davila M, Jhala D, Ghosh D, Grizzle WE, Chakrabarti R. Expression of LIM kinase 1 is associated with reversible G1/S phase arrest, chromosomal instability and prostate cancer. Molecular Cancer. 2007;6:40. doi: 10.1186/1476-4598-6-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton BA, Davis GW. LIM kinase1 controls synaptic stability downstream of the type II BMP receptor. Neuron. 2005;47:695–708. doi: 10.1016/j.neuron.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Fan X, Molotkov A, Manabe S, Donmoyer CM, Deltour L, Foglio MH, Cuenca AE, Blaner WS, Lipton SA, Duester G. Targeted disruption of Aldh1a1 (Raldh1) provides evidence for a complex mechanism of retinoic acid synthesis in the developing retina. Molecular and Cellular Biology. 2003;23:4637–4648. doi: 10.1128/MCB.23.13.4637-4648.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foletta VC, Moussi N, Sarmiere PD, Bamburg JR, Bernard O. LIM kinase 1, a key regulator of actin dynamics, is widely expressed in embryonic and adult tissues. Experimental Cell Research. 2004;294:392–405. doi: 10.1016/j.yexcr.2003.11.024. [DOI] [PubMed] [Google Scholar]
- Georgas K, Rumballe B, Valerius MT, Chiu HS, Thiagarajan RD, Lesieur E, Aronow BJ, Brunskill EW, Combes AN, Tang D, Taylor D, Grimmond SM, Potter SS, McMahon AP, Little MH. Analysis of early nephron patterning reveals a role for distal RV proliferation in fusion to the ureteric tip via a cap mesenchyme-derived connecting segment. Developmental Biology. 2009;332:273–286. doi: 10.1016/j.ydbio.2009.05.578. [DOI] [PubMed] [Google Scholar]
- Gurniak CB, Perlas E, Witke W. The actin depolymerizing factor n-cofilin is essential for neural tube morphogenesis and neural crest cell migration. Developmental Biology. 2005;278:231–241. doi: 10.1016/j.ydbio.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Hocking JC, Hehr CL, Bertolesi G, Funakoshi H, Nakamura T, McFarlane S. LIMK1 acts downstream of BMP signaling in developing retinal ganglion cell axons but not dendrites. Developmental Biology. 2009;330:273–285. doi: 10.1016/j.ydbio.2009.03.027. [DOI] [PubMed] [Google Scholar]
- Hoogenraad CC, Akhmanova A, Galjart N, de Zeeuw CI. LIMK1 and CLIP-115: linking cytoskeletal defects to Williams syndrome. BioEssays. 2004;26:141–150. doi: 10.1002/bies.10402. [DOI] [PubMed] [Google Scholar]
- Hua Li H, Roy M, Kuscuoglu U, Spencer CM, Halm B, Harrison KC, Bayle JH, Splendore A, Ding F, Meltzer LA, Wright E, Paylor R, Deisseroth K, Francke U. Induced chromosome deletions cause hypersociability and other features of Williams–Beuren syndrome in mice. EMBO Molecular Medicine. 2009;1:50–65. doi: 10.1002/emmm.200900003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber SM, Braun GS, Segerer S, Veh RW, Horster MF. Metanephrogenic mesenchyme-to-epithelium transition induces profound expression changes of ion channels. American Journal of Physiology-Renal Physiology. 2000;279:65–76. doi: 10.1152/ajprenal.2000.279.1.F65. [DOI] [PubMed] [Google Scholar]
- Kobayashi A, Kwan K, Carroll TJ, McMahon AP, Mendelsohn CL, Behringer RR. Distinct and sequential tissue-specific activities of the LIM-class homeobox gene Lim1 for tubular morphogenesis during kidney development. Development. 2005;132:2809–2823. doi: 10.1242/dev.01858. [DOI] [PubMed] [Google Scholar]
- Kuure S, Cebrian C, Machingo Q, Lu BC, Chi X, Hyink D, D’Agati V, Gurniak C, Witke W, Costantini F. Actin depolymerizing factors cofilin1 and destrin are required for ureteric bud branching. Morphogenesis PLoS Genet. 2010;6:1001176. doi: 10.1371/journal.pgen.1001176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee-Hoeflich ST, Causing CG, Podkowa M, Zhao X, Wrana JL, Attisano L. Activation of LIMK1 by binding to the BMP receptor, BMPRII, regulates BMP-dependent dendritogenesis. EMBO. 2004;23:4792–4801. doi: 10.1038/sj.emboj.7600418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Estrada OM, Lettice LA, Essafi A, Guadix JA, Slight J, Velecela V, Hall E, Reichmann J, Devenney PS, Hohenstein P, Hosen N, Hill RE, Muñoz-Chapuli R, Hastie ND. Wt1 is required for cardiovascular progenitor cell formation through transcriptional control of Snail and E-cadherin. Nature Genetics. 2010;42:89–95. doi: 10.1038/ng.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Y, Zhang Y, Tregoubov V, Janus C, Cruz L, Jackson M, Lu W, MacDonald JF, Wang JY, Falls DL, Jia1 Z. Abnormal spine morphology and enhanced LTP in LIMK-1 knockout mice. Neuron. 2002;35:121–133. doi: 10.1016/s0896-6273(02)00758-4. [DOI] [PubMed] [Google Scholar]
- Meng Y, Takahashi H, Meng J, Zhang Y, Lua G, Asrar S, Nakamura T, Jia Z. Regulation of ADF/cofilin phosphorylation and synaptic function by LIM-kinase neuropharmacology. 2004;47:746–754. doi: 10.1016/j.neuropharm.2004.06.030. [DOI] [PubMed] [Google Scholar]
- Mori T, Okano I, Mizuno K, Tohyama M, Wanaka A. Comparison of tissue distribution of two novel serine/threonine kinase genes containing the LIM motif (LIMK-1 and LIMK-2) in the developing rat. Molecular Brain Research. 1997;45:247–254. doi: 10.1016/s0169-328x(96)00257-4. [DOI] [PubMed] [Google Scholar]
- Murali D, Yoshikawa S, Corrigan RR, Plas DJ, Crair MC, Oliver G, Lyons KM, Mishina Y, Furuta Y. Distinct developmental programs require different levels of Bmp signaling during mouse retinal development. Development. 2005;132:913–923. doi: 10.1242/dev.01673. [DOI] [PubMed] [Google Scholar]
- Nadella KS, Saji M, Jacob NK, Pavel E, Ringel MD, Kirschner LS. Regulation of actin function by protein kinase A-mediated phosphorylation of Limk. EMBO Reports. 2009;10:599–605. doi: 10.1038/embor.2009.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan L, Deng M, Xie X, Gan L. ISL1 and BRN3B co-regulate the differentiation of murine retinal ganglion cells. Development. 2008;135:1981–1990. doi: 10.1242/dev.010751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxen L. Organogenesis of the Kidney. Cambridge University Press; 1987. [Google Scholar]
- Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- Scott RW, Olson MF. LIM kinases: function, regulation and association with human disease. Journal of Molecular Medicine. 2007;85:555–568. doi: 10.1007/s00109-007-0165-6. [DOI] [PubMed] [Google Scholar]
- Scott HM, Mason JI. Steroidogenesis in the fetal testis and its susceptibility to disruption by exogenous compounds. Endocrine Reviews. 2009;30:883–925. doi: 10.1210/er.2009-0016. [DOI] [PubMed] [Google Scholar]
- Sharma RK, Netland PA. Early born lineage of retinal neurons express class III β-tubulin isotype. Brain Research. 2007;1176:11–17. doi: 10.1016/j.brainres.2007.07.090. [DOI] [PubMed] [Google Scholar]
- Toshima J, Toshima JY, Amano T, Yang N, Narumiya S, Mizuno K. Cofilin phosphorylation by protein kinase testicular protein kinase 1 and its role in integrin-mediated actin reorganization and focal adhesion formation. Molecular Biology of the Cell. 2001;12:1131–1145. doi: 10.1091/mbc.12.4.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Z, Han L, Bamburg JR, Shim S, Ming G, Zheng JQ. BMP gradients steer nerve growth cones by a balancing act of LIM kinase and Slingshot phosphatase on ADF/cofilin. Journal of Cell Biology. 2007;178:107–119. doi: 10.1083/jcb.200703055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaoita E, Franke WW, Yamamoto T, Kawasaki K, Kihara I. Identification of renal podocytes in multiple species: higher vertebrates are vimentin positive/lower vertebrates are desmin positive. Histochemistry and Cell Biology. 1999;111:107–115. doi: 10.1007/s004180050340. [DOI] [PubMed] [Google Scholar]