This work examines two types of histone chaperones and shows that depletion of NAP1 leads to decreased somatic homologous recombination (HR), but depletion of CAF-1 leads to increased HR and telomere shortening, which are NAP1 dependent and NAP1 independent, respectively. These results highlight distinct nucleosome assembly/disassembly pathways in regulation of genome stability and variability.
Abstract
Homologous recombination (HR) is essential for maintaining genome integrity and variability. To orchestrate HR in the context of chromatin is a challenge, both in terms of DNA accessibility and restoration of chromatin organization after DNA repair. Histone chaperones function in nucleosome assembly/disassembly and could play a role in HR. Here, we show that the NUCLEOSOME ASSEMBLY PROTEIN1 (NAP1) family histone chaperones are required for somatic HR in Arabidopsis thaliana. Depletion of either the NAP1 group or NAP1-RELATED PROTEIN (NRP) group proteins caused a reduction in HR in plants under normal growth conditions as well as under a wide range of genotoxic or abiotic stresses. This contrasts with the hyperrecombinogenic phenotype caused by the depletion of the CHROMATIN ASSEMBLY FACTOR-1 (CAF-1) histone chaperone. Furthermore, we show that the hyperrecombinogenic phenotype caused by CAF-1 depletion relies on NRP1 and NRP2, but the telomere shortening phenotype does not. Our analysis of DNA lesions, H3K56 acetylation, and expression of DNA repair genes argues for a role of NAP1 family histone chaperones in nucleosome disassembly/reassembly during HR. Our study highlights distinct functions for different families of histone chaperones in the maintenance of genome stability and establishes a crucial function for NAP1 family histone chaperones in somatic HR.
INTRODUCTION
Homologous recombination (HR) has two key functions in life processes (Schuermann et al., 2005; Heyer et al., 2010). In meiosis, HR is required for accurate chromosome segregation and forms the basis of genetic mixing between maternal and paternal origin chromosomes, thus contributing to novel combinations of genetic traits that enable both natural organism evolution and agricultural breeding programs. In somatic tissues, HR is also important for repairing damaged DNA to maintain genome integrity, hence contributing to the normal growth and development of an organism. The molecular mechanisms that regulate HR at the DNA level for double-strand break (DSB) signaling and repair have been characterized in great detail (Schuermann et al., 2005; Heyer et al., 2010; Knoll and Puchta, 2011), but little is known about HR regulation in the context of chromatin.
The basic repeating unit of chromatin is the nucleosome, which consists of 146 bp of DNA wrapped around a histone octamer composed of an (H3-H4)2 tetramer at the center and two H2A-H2B dimers attached symmetrically on either side (Luger et al., 1997). Nucleosome compaction limits DNA accessibility to DNA repair proteins, including those involved in HR. Also, proper chromatin structure needs to be restored after DNA repair. Dynamic nucleosome disassembly/reassembly mediated by histone chaperones is thought to play an important role in DNA repair (Ransom et al., 2010; Avvakumov et al., 2011). However, depletion of CHROMATIN ASSEMBLY FACTOR-1 (CAF-1), an evolutionarily conserved histone H3/H4-type chaperone, does not reduce but rather elevates HR in Arabidopsis thaliana plants (Endo et al., 2006; Kirik et al., 2006) as well as in yeast cells (Endo et al., 2010). Although another histone H3/H4-type chaperone ANTI-SILENCING FUNCTION1 (ASF1) is required for HR in yeast, this is associated with ASF1 function in histone H3 Lys 56 acetylation (H3K56ac) rather than in physical deposition of histones in nucleosome assembly (Endo et al., 2010; Costelloe and Lowndes, 2010). Therefore, an active role of histone chaperone–mediated nucleosome disassembly/reassembly in HR remains elusive.
NUCLEOSOME ASSEMBLY PROTEIN1 (NAP1) belongs to a family of evolutionarily conserved histone chaperones. NAP1 binds all types of histones in vitro and is found primarily associated with H2A/H2B in vivo and thus is considered as an H2A/H2B-type histone chaperone (Dong et al., 2003, 2005; Park and Luger, 2006; Zhu et al., 2006). NAP1 proteins are implicated in histone trafficking (Mosammaparast et al., 2002; Miyaji-Yamaguchi et al., 2003; Dong et al., 2005), nucleosome assembly (Ito et al., 1996; Andrews et al., 2010), and disassembly (Lorch et al., 2006; Walfridsson et al., 2007). In Arabidopsis, the NAP1 family comprises four NAP1 genes (NAP1;1, NAP1;2, NAP1;3, and NAP1;4) and two NAP1-RELATED PROTEIN genes (NRP1 and NRP2) (Galichet and Gruissem, 2006; Zhu et al., 2006). Remarkably, loss-of-function allelic triple mutants of the three ubiquitously expressed NAP1 genes, nap1;1-1 nap1;2-1 nap1;3-1 and nap1;1-1 nap1;2-1 nap1;3-2, show a normal growth phenotype (Liu et al., 2009a, 2009b), and the double mutant nrp1-1 nrp2-1 displays only a mild short-root phenotype (Zhu et al., 2006). Only under stress conditions do the nap1;1-1 nap1;2-1 nap1;3-1 and nap1;1-1 nap1;2-1 nap1;3-2 triple mutants exhibit detectable defects in nucleotide excision repair of DNA (Liu et al., 2009a), and the nrp1-1 nrp2-1 double mutant shows an increased level of DNA damage (Zhu et al., 2006).
Here, we report that somatic HR is impaired in the nap1;1-1 nap1;2-1 nap1;3-1 and nap1;1-1 nap1;2-1 nap1;3-2 triple mutants and in the nrp1-1 nrp2-1 double mutant plants under standard growth conditions as well as under a diversity of stresses. In addition, we show that nrp1-1 nrp2-1 suppresses the hyperrecombinogenic phenotype of the CAF-1–deficient mutant fas2-4 but not the telomere shortening and developmental-defective phenotypes. Elevated levels of DSBs and expression of DNA repair genes observed in fas2-4 were not suppressed by nrp1-1 nrp2-1, indicating that NRP1 and NRP2 are required for HR downstream of DSBs caused by CAF-1 depletion. Our results clarify distinct functions of the CAF-1 and NAP1 family histone chaperones in the maintenance of genome stability and unravel a crucial role of NAP1 but not CAF-1 in nucleosome disassembly/reassembly during the HR process.
RESULTS
Depletion of NAP1s or NRPs Reduces Somatic HR
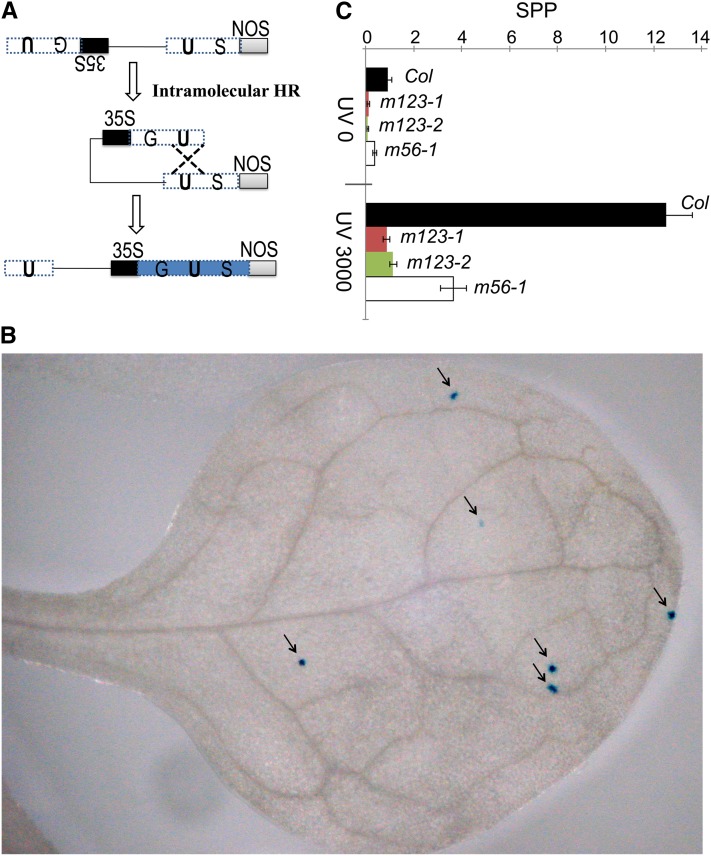
To analyze the role of NAP1s and NRPs in somatic HR, we used the recombination substrate line 1406, which contains a nonfunctional reporter transgene consisting of two separated fragments of the β-glucuronidase (GUS) gene (Gherbi et al., 2001; Schuermann et al., 2009). The two fragments contain an overlapping region and are spatially arranged as inverted repeats that restore a functional GUS after an HR event (Figure 1A; Schuermann et al., 2009). The restored GUS activity can be visualized in planta as a blue spot/sector by histochemical staining (Figure 1B). We introgressed the 1406 recombination substrate into nap1;1-1 nap1;2-1 nap1;3-1 (in short, m123-1), nap1;1-1 nap1;2-1 nap1;3-2 (m123-2), or nrp1-1 nrp2-1 (m56-1) mutants by genetic crosses and obtained the corresponding homozygous lines. The number of somatic HR events per plant was determined, and statistical analysis was performed based on a total of 60 to 90 individual plants in each experiment for a given genotype. Under standard growth conditions, we found that HR capacity was reduced in the mutants by more than eightfold in nap1;1-1 nap1;2-1 nap1;3-1 or nap1;1-1 nap1;2-1 nap1;3-2 and over twofold in nrp1-1 nrp2-1 compared with wild-type Columbia (Col) (Figure 1C). Exposure to UV-C irradiation increased HR frequency in Col as well as in nap1;1-1 nap1;2-1 nap1;3-1, nap1;1-1 nap1;2-1 nap1;3-2, and nrp1-1 nrp2-1. In these conditions, a reduction in HR frequency in the mutants, compared with Col, becomes increasingly obvious (Figure 1C). The effects of the mutant backgrounds and UV-C treatment on HR were consistently reproducible and statistically significant (see Supplemental Table 1 online).
Figure 1.
Somatic HR Is Reduced in the nap1;1-1 nap1;2-1 nap1;3-1, nap1;1-1 nap1;2-1 nap1;3-2, and nrp1-1 nrp2-1 Mutants Compared with Wild-Type Col.
(A) Scheme of GUS-based recombination substrate and intramolecular HR in line 1406. Intramolecular interaction between inverted repeats (U) gives rise to the restoration of a functional GUS gene through HR.
(B) Example showing blue spots/sectors as a result of assembly of functional GUS through HR events in an Arabidopsis leaf. Arrows indicate independent events.
(C) HR capacity of the nap1;1-1 nap1;2-1 nap1;3-1 (m123-1), nap1;1-1 nap1;2-1 nap1;3-2 (m123-2), and nrp1-1 nrp2-1 (m56-1) mutants compared with wild-type Col. Spots/sectors per plant (SPP) were assessed for each genotype on a population of 60 to 90 individual plants untreated (UV 0) or treated with UV-C irradiation at a dose of 3000 J m−2 (UV 3000). Average number of SPP from three independent experiments is shown with se bars (refer to Supplemental Table 1 online for statistical analysis).
NRP1 and NRP2 Are Required for Both Intramolecular and Intermolecular HR
The nrp1-1 nrp2-1, nap1;1-1 nap1;2-1 nap1;3-1, and nap1;1-1 nap1;2-1 nap1;3-2 mutants show similar HR defects. We next focused on nrp1-1 nrp2-1 for more detailed analyses. To address whether the observed somatic HR defects are reproducible with different recombination substrates, the line 1445 containing a similar GUS reporter construct as in 1406 but at a different genome location (Fritsch et al., 2004; Schuermann et al., 2009) was introgressed into nrp1-1 nrp2-1 by crossing. Similar to 1406, 1445 also showed a reduction in somatic HR in the nrp1-1 nrp2-1 mutant background compared with Col under either standard growth conditions or UV treatment (Figure 2A). In both 1406 and 1445, HR involves DNA sequences within the same molecule, thus representing intramolecular recombination events (Figure 1A; Schuermann et al., 2009). To investigate HR between homologous chromosomes and/or sister chromatids, we introgressed into nrp1-1 nrp2-1 the substrate line IC9C, which harbors two GUS fragments in a spatial orientation that can restore a functional GUS only by intermolecular HR (Figure 2B; Molinier et al., 2004; Schuermann et al., 2009). As shown in Figure 2A, IC9C also displayed reduced HR capacity in nrp1-1 nrp2-1 compared with Col. In summary, with all tested recombination substrate lines, the nrp1-1 nrp2-1 mutant showed reduced somatic HR, indicating that NRP1 and NRP2 are required for HR, independent of the structure of the recombination substrates.
Figure 2.
Reduced HR in nrp1-1 nrp2-1 Is Detected with Various Recombination Reporter Substrates.
(A) HR capacity of 1406, 1445, and IC9C in nrp1-1 nrp2-1 (m56-1), compared with Col, was assessed as described in Figure 1. Line 1445 contains the same GUS-based HR substrate as in 1406 but at a different genome position. Line IC9C contains a distinct GUS-based recombination substrate, which detects intermolecular HR. SPP, spots/sectors per plant.
(B) Scheme of GUS-based recombination substrate and intermolecular HR in line IC9C. The spatial orientation of the two GUS fragments requires intermolecular interaction between homologous chromosomes or sister chromatids to restore a functional GUS gene through HR.
[See online article for color version of this figure.]
NRP1 and NRP2 Are Required for HR in Response to Multiple Types of Stresses
Both Col and nrp1-1 nrp2-1 are responsive to UV-induced HR (Figures 1C and 2) in a dosage-dependent manner (see Supplemental Table 1 online). In addition, we tested the HR response to several different mutagenic chemicals, including mitomycin C (MMC), methyl methanesulfonate (MMS), and bleomycin. These chemicals induced HR drastically, and in all tested conditions, nrp1-1 nrp2-1 exhibited a dramatically reduced HR compared with Col (Figure 3A). By contrast, the single mutant nrp1-1 (m5-1) showed HR levels comparable to Col under either standard or bleomycin-treated plant growth conditions (Figure 3B), indicating a redundant function of NRP1 and NRP2 in HR.
Figure 3.
HR Capacity in nrp1-1 nrp2-1 Is Impaired in Response to Diverse Types of Stresses.
(A) HR capacity of 1406 in nrp1-1 nrp2-1 (m56-1) compared with Col in response to the mutagenic chemical MMC, MMS, or bleomycin. SPP, spots/sectors per plant.
(B) Comparison of HR in the double mutant nrp1-1 nrp2-1 (m56-1), the single mutant nrp1-1 (m5-1), and Col.
(C) HR capacity of 1406 in nrp1-1 nrp2-1 (m56-1) compared with Col in response to the DNA synthesis inhibitor HU.
(D) HR capacity of 1406 in nrp1-1 nrp2-1 (m56-1) compared with Col in response to salt stress and to the phytohormone ABA.
HR capacity was assessed as described in Figure 1. Average number of SPP from three independent experiments is shown with se bars (refer to Supplemental Table 1 online for statistical analysis).
Treatments with physical and chemical mutagenic agents induce various kinds of DNA damage, including DSBs, but spontaneous HR might also be initiated by replication-associated DNA intermediates (such as collapsed replication forks) that differ from classical DSBs. We found that treatment with the DNA replication–blocking compound hydroxyurea (HU) stimulates HR in nrp1-1 nrp2-1 and Col (Figure 3C). Abiotic stresses induce the accumulation of the phytohormone abscisic acid (ABA), which inhibits DNA replication and increases HR (Yin et al., 2009). Consistently, we found that both salt stress and ABA treatments stimulated HR in nrp1-1 nrp2-1 and Col (Figure 3D), albeit to a lower extent compared with induction by HU or mutagenic chemicals. Again, in all tested conditions, nrp1-1 nrp2-1 showed a remarkably reduced HR capacity compared with Col.
The intensity of blue-stained spots/sectors did not show a significant difference between nrp1-1 nrp2-1 and Col, and the 35S-promoter drove similar levels of reporter GUS expression in nrp1-1 nrp2-1 and Col (see Supplemental Figure 1 online). This strongly argues against the assumption that transcriptional repression might lead to a reduced number of blue spots/sectors in nrp1-1 nrp2-1. Distinct from the highly significant differences between untreated and treated plants as well as between nrp1-1 nrp2-1 and Col, HR induction caused by a given treatment is significantly different between nrp1-1 nrp2-1 and Col only in a few cases, while in most cases HR induction is relatively similar between nrp1-1 nrp2-1 and Col (see Supplemental Table 1 online).
Taken together, these data indicate that NRP1 and NRP2 are required for somatic HR in plant responses to diverse types of stresses and that this requirement is positioned at later steps, whereas early signal transduction steps of the different stress pathways in HR initiation are largely unaffected in nrp1-1 nrp2-1.
NRP1 and NRP2 Depletion Suppresses the Hyperrecombinogenic Phenotype of the fas2-4 Mutant
The hyporecombinogenic phenotype of nrp1-1 nrp2-1 contrasts with the previously reported hyperrecombinogenic phenotype of the CAF-1–deficient mutants fas1 and fas2 (Endo et al., 2006; Kirik et al., 2006). To investigate the interaction between CAF-1 and NRP1/NRP2, we crossed nrp1-1 nrp2-1 with the line 1415 (fas2-4), which carries a GUS reporter construct similar to 1406 and 1445 but at a new genome location in the CAF-1–deficient mutant fas2-4 background (Endo et al., 2006), and we obtained homozygous 1415 in the triple mutant nrp1-1 nrp2-1 fas2-4 (m56-1fas2-4) background. The triple mutant plants showed developmental phenotypes largely similar to those of fas2-4, including early shoot branching, enhanced leaf serrations, abnormal phyllotaxy, and perturbed numbers of sepals, petals, and stamens (Figures 4A to 4D). Only very mild phenotypic differences could be observed; for example, the triple mutant plant rosette leaves are flattened and bigger than those of fas2-4 (Figure 4B).
Figure 4.
The Triple Mutant nrp1-1 nrp2-1 fas2-4 Shows a Morphological Phenotype Largely Similar to That of fas2-4.
(A) Phenotype of representative 5-week-old plants of wild-type Col and mutants nrp1-1 nrp2-1 (m56-1), fas2-4, and nrp1-1 nrp2-1 fas2-4 (m56-1fas2-4).
(B) Comparison of rosette leaves from 5-week-old wild-type and mutants plants.
(C) Comparison of inflorescences from 6-week-old wild-type and mutants plants.
(D) Percentage of flowers showing the indicated number of floral organs in wild-type and mutants plants. Analysis was performed on more than 100 flowers for each genotype randomly collected from 6- to 7-week-old plants.
Consistent with previous reports (Endo et al., 2006; Kirik et al., 2006), fas2-4 showed a dramatically increased HR frequency compared with Col under standard growth conditions as well as under UV-C or bleomycin treatments (Figure 5A). Strikingly, the triple mutant nrp1-1 nrp2-1 fas2-4 showed a somatic HR capacity similar to that of the double mutant nrp1-1 nrp2-1 (Figure 5A; see Supplemental Table 1 online), establishing that the hyperrecombinogenic phenotype of fas2-4 relies on NRP1 and NRP2 function.
Figure 5.
The Triple Mutant nrp1-1 nrp2-1 fas2-4 Exhibits HR Capacity Similar to nrp1-1 nrp2-1 but Telomere Length Similar to fas2-4.
(A) HR capacity of the nrp1-1 nrp2-1 (m56-1), fas2-4, and nrp1-1 nrp2-1 fas2-4 (m56-1fas2-4) mutants compared with wild-type Col under diverse plant growth conditions. HR was detected using the reporter line 1415, which contains the same GUS-based recombination substrate as in 1406 and 1445 but at a different genome position. Average number of spots/sectors per plant (SPP) from three independent experiments is shown with se bars (refer to Supplemental Table 1 online for statistical analysis).
(B) Telomere length comparison between wild-type and mutants. Genomic DNA was digested with MseI. DNA gel blot analysis was performed using a digoxigenin-labeled telomere repeat as the probe. Note that telomeres are shortened in fas2-4 as well as in nrp1-1 nrp2-1 fas2-4 (m56-1fas2-4) but not in nrp1-1 nrp2-1 (m56-1) compared with Col.
Next, we investigated the roles of NRPs and CAF-1 in telomere length maintenance. Telomeres protect the chromosome ends from unscheduled DNA repair reactions and degradation. In agreement with a previous report (Mozgová et al., 2010), we found that telomeres were shortened in fas2-4 (Figure 5B). By contrast, telomere length was comparable between nrp1-1 nrp2-1 and Col and between nrp1-1 nrp2-1 fas2-4 and fas2-4 (Figure 5B), indicating that depletion of NRP1 and NRP2 does not affect telomere length maintenance and cannot attenuate the telomere shortening caused by CAF-1 depletion.
Possible Mechanism of Differently Altered HR Frequency in the nrp1-1 nrp2-1 and fas2-4 Mutants
To gain insight into what causes altered somatic HR frequencies in the mutants, we first analyzed the levels of DNA damage by comet assays (Zhu et al., 2006). We found that fas2-4 caused increased DNA damage (Figures 6A and 6B), indicating that CAF-1 depletion renders the genome unstable and that the observed HR increase might result from the high level of DNA lesions. Interestingly, increased levels of DNA lesions were also observed in nrp1-1 nrp2-1 and nrp1-1 nrp2-1 fas2-4 (Figures 6A and 6B), indicating that the HR requirement of NRP1 and NRP2 is located downstream of DNA damage. It is noteworthy that nrp1-1 nrp2-1 fas2-4 exhibits a hyporecombinogenic phenotype but a further enhanced level of DNA lesions. NRP1/NRP2 and CAF-1 thus have independent functions in HR as well as in DNA damage repair in general.
Figure 6.
Effects of nrp1-1 nrp2-1 and fas2-4 on DNA Damage, H3K56ac, and Expression of DNA Repair Genes.
(A) Representative comet images of wild-type Col and mutants nrp1-1 nrp2-1 (m56-1), fas2-4, and nrp1-1 nrp2-1 fas2-4 (m56-1fas2-4) nuclei. Note the intact nucleus at the left and comet tail formed by fragmented nuclear DNA to the right on each panel.
(B) DNA damage level as measured by the percentage of DNA in the tail of nuclei in comet assays. The mean value of more than 200 nuclei is shown with sd bars.
(C) Comparison of H3K56ac levels in wild-type Col and mutants. H3 antibody was used as a loading control.
(D) Expression level of DNA repair genes as measured by quantitative RT-PCR. Values are presented as relative to levels in untreated wild-type Col (set as 1) and represent the mean from three biological repeats with se bars.
In yeast and animals, H3K56ac is involved in the response to DNA damage, in nucleosome reassembly after DNA repair, and in HR (Costelloe and Lowndes, 2010; Endo et al., 2010). Our analysis revealed that nrp1-1 nrp2-1, fas2-4, and nrp1-1 nrp2-1 fas2-4 mutants exhibit an H3K56ac level similar to Col under either standard growth conditions or in bleomycin-treated plants (Figure 6C). This indicates that the HR requirement of NRP1 and NRP2 is H3K56ac independent, which contrasts with the H3K56ac-dependent HR requirement of ASF1 in yeast (Endo et al., 2010).
We next investigated the expression of several genes encoding evolutionarily conserved components implicated in DNA repair (Schuermann et al., 2005; Heyer et al., 2010; Knoll and Puchta, 2011). We found that genes involved in HR, including BRCA1 (for BREAST CANCER SUSCEPTIBILITY1), RAD51 (for RADIATION SENSITIVE51), PARP1 (for POLY ADP-RIBOSE POLYMERASE1), RAD54, and PARP2, were upregulated in fas2-4 compared with Col under standard plant growth conditions (Figure 6D). However, following bleomycin induction, we found that these genes were expressed at lower levels in fas2-4 compared with Col. By contrast, ATM encoding a signaling kinase expressed in response to DNA damage, as well as KU70, KU80, LIG-I, and LIG-IV, which are involved in nonhomologous end joining (NHEJ) of DNA repair, were constantly expressed in fas2-4 compared with Col (only shown for ATM in Figure 6D). In nrp1-1 nrp2-1, apart from PARP2, which showed increased expression under standard plant growth conditions, all other examined genes showed similar expression levels as in Col (Figure 6D). In nrp1-1 nrp2-1 fas2-4, HR genes showed expression levels higher than those observed in fas2-4 (Figure 6D). Taken together, these data do not support the assumption that altered transcription levels of DNA repair genes are responsible for the HR phenotypes observed in fas2-4, nrp1-1 nrp2-1, or nrp1-1 nrp2-1 fas2-4 mutants. We conclude that somatic HR depends on NAP1 family proteins but not CAF-1, likely because of a critical role of NAP1 in nucleosome disassembly/reassembly required in HR.
DISCUSSION
In response to DNA damage, detection of lesions and repair of DNA occur in the context of chromatin structure. Mechanisms to manipulate chromatin structure, including covalent histone modifications, incorporation of histone variants, ATP-dependent chromatin remodeling, and nucleosome assembly/disassembly, play crucial functions in transcription, replication, and DNA repair. Indeed, DSB can cause a loss of nucleosomes within a few kilobases of the break (Tsukuda et al., 2005). HR is one of the two major pathways involved in repair of DSBs, but only a few factors involved in chromatin structure manipulation have been so far identified as playing a role in HR (Ransom et al., 2010; Avvakumov et al., 2011; Ceballos and Heyer, 2011; Lukas et al., 2011). HR events occur spontaneously and broadly throughout the genome; hence, it is difficult to monitor and detect HR products. The GUS reporter system allowing detection of HR in a single cell within the whole plant was developed and has been widely used and proven as a powerful tool in the study of somatic HR in plants (Swoboda et al., 1994; Gherbi et al., 2001; Fritsch et al., 2004; Molinier et al., 2004; Endo et al., 2006; Kirik et al., 2006; Schuermann et al., 2009; Yin et al., 2009; and others). Using this in vivo system, we show that the NAP1 family of histone chaperones is required for somatic HR in Arabidopsis. The NAP1 group and NRP group proteins have sequence similarities and similar structural organization, and both groups of proteins bind H2A and H2B (Zhu et al., 2006; Liu et al., 2009a). The reduced but not null HR capacity observed in the nap1;1-1 nap1;2-1 nap1;3-1, nap1;1-1 nap1;2-1 nap1;3-2, and nrp1-1 nrp2-1 mutant plants implies a redundant function between the NAP1 group and NRP group proteins in HR.
For in planta somatic HR assays, we used both the intramolecular HR GUS reporter lines 1406, 1415, and 1445 (Figure 1A; Gherbi et al., 2001; Schuermann et al., 2009) and the intermolecular HR GUS reporter line IC9C (Figure 2B; Molinier et al., 2004; Schuermann et al., 2009). Independent of the recombination substrate structure, the nrp1-1 nrp2-1 mutant displayed a reduced somatic HR with all reporter lines used. The HR reduction found in this study is in line with previous reported genotoxin-hypersensitive phenotypes of the nap1;1-1 nap1;2-1 nap1;3-1, nap1;1-1 nap1;2-1 nap1;3-2, and nrp1-1 nrp2-1 mutants (Zhu et al., 2006; Liu et al., 2009a). Most genotoxins can cause multiple kinds of DNA damage, but each also exhibits specificity (e.g., UV primarily induces cyclobutane pyrimidine dimers and 6-4 photoproducts, MMS generates methylated bases, MMC mainly forms interstrand cross-links, and the radiomimetic agent bleomycin produces strand breaks). DNA damage can also arise from stalled DNA replication forks and by free radical attack of highly reactive oxygen species that accumulate as a consequence of intrinsic cellular metabolism and/or upon plant exposure to environmental stresses. Multiple repair pathways exist, and HR constitutes a key repair pathway for complex DNA damage, including DSBs, interstrand cross-links, and DNA gaps (Heyer et al., 2010). In agreement with previous reports (Schuermann et al., 2005, 2009; Boyko et al., 2006; Yin et al., 2009), we found that treatment with UV-C, MMS, MMC, bleomycin, HU, salinity, or ABA induced somatic HR in wild-type plants. Under the same treatment, mutant plants showed a lower HR capacity, indicating that NRP1 and NRP2, and possibly also NAP1 genes, are required for HR in plant responses to diverse types of genotoxic and abiotic stresses. Because the nrp1-1 nrp2-1 as well as the nap1;1-1 nap1;2-1 nap1;3-2 and nap1;1-1 nap1;2-1 nap1;3-2 mutants showed largely similar fold induction of HR in response to the treatments as did the wild-type Col, we conclude that NRP/NAP1 proteins are not involved in the initial steps of DNA damage response signaling. This conclusion is also supported by the observation that nrp1-1 nrp2-1 and Col exhibit similar levels of H3K56ac. The high levels of DNA lesions detected in nrp1-1 nrp2-1 indicate that the impaired HR might be located downstream of DSB formation.
Upon damage, the BRCA1 protein is recruited early in DSB repair, and its depletion impairs HR (Heyer et al., 2010; Lukas et al., 2011). The RAD51 protein binds to the ends of the DNA break to form RAD51 single-stranded DNA nucleoprotein filaments and is involved in strand invasion in HR. The RAD54 protein, a member of the SWI2/SNF2 family of ATP-dependent chromatin remodeling factors, plays diverse roles throughout HR processes, including stabilization of nucleoprotein filaments, facilitation of D-loop formation, and heteroduplex extension (Ceballos and Heyer, 2011; Knoll and Puchta, 2011). Our analyses show that expression of BRCA1, RAD51, and RAD54 is unaffected in nrp1-1 nrp2-1, excluding the hypothesis that NRPs regulate HR through transcriptional control of these key genes. Histone chaperones are not known to physically interact and directly recruit any DNA repair proteins, but their function in nucleosome assembly/disassembly might be crucial for DNA end resection, strand invasion, D-loop formation, and heteroduplex extension during HR.
Because of their peripheral location on the nucleosome, H2A and H2B are more dynamic and are removed before H3 and H4 during nucleosome disassembly. NAP1 is considered to be an H2A/H2B-type histone chaperone and is routinely used to chaperone all four types of histones for the purpose of nucleosome assembly or disassembly in vitro. In vitro nucleosome disassembly requires NAP1 together with an ATP-dependent chromatin-remodeling factor (Lorch et al., 2006), and NAP1 together with CHD1 also promotes nucleosome disassembly during transcription in vivo (Walfridsson et al., 2007). Similar to NAP1/NRPs, the ATP-dependent chromatin-remodeling factor INO80 is required for HR in Arabidopsis (Fritsch et al., 2004). It is reasonable to speculate that NAP1 and INO80 may act in nucleosome disassembly to facilitate access of DNA repair complexes in HR. Alternatively or in addition, NAP1 may act together with RAD54 in nucleosome removal for resection of DNA to generate single-stranded overhangs for strand invasion and/or for heteroduplex extension during HR. Thermodynamic analysis revealed that NAP1 has a crucial role in the elimination of competing, non-nucleosomal histone–DNA interactions (Andrews et al., 2010). Such a role might be necessary to avoid aberrant DNA structure formation and atypical histone-DNA associations that block HR.
In sharp contrast with the essential role of NAP1/NRPs in HR, depletion of CAF-1 caused a hyperrecombinogenic phenotype (Endo et al., 2006; Kirik et al., 2006; this study). Interestingly, NRP1 and NRP2 are required for the hyperrecombinogenic phenotype of the fas2-4 mutant as shown by the hyporecombinogenic phenotype of the triple mutant nrp1-1 nrp2-1 fas2-4. CAF-1 chaperones H3-H4 and plays a pivotal role in nucleosome assembly during DNA replication (Ransom et al., 2010). It has also been hypothesized that H3K56ac-H4 dimers are transferred from ASF1 to CAF-1, subsequently ensuring rapid formation of (H3-H4)2 tetramers and, subsequently, of the nucleosome in DNA repair and HR (Costelloe and Lowndes, 2010; Endo et al., 2010). Our data do not exclude a possible role of CAF-1 in nucleosome assembly during DNA repair but clearly establish that CAF-1 is not a determinant factor in somatic HR. Sustaining the previous notion that the moderately elevated expression of some HR genes is unlikely to account for the drastically increased HR frequency in the fas mutants (Kirik et al., 2006), our analysis of bleomycin treatment clearly shows that the HR level is not correlated with the HR gene expression level in fas2-4 compared with Col. In addition, we found that the triple mutant nrp1-1 nrp2-1 fas2-4, compared with fas2-4, exhibits an attenuated HR increase but similar or even higher expression levels of BRCA1, RAD51, PARP1, PARP2, and RAD54. This finding establishes that increased expression of these genes is insufficient to induce HR in the absence of NRP1 and NRP2. Increased HR in fas mutants is likely a consequence or an indirect effect caused by high levels of DNA lesions associated with the replication and chromatin assembly delay in the absence of CAF-1 function. It is also noteworthy that the fas mutants exhibit increased frequency of T-DNA integration into the plant genome (Endo et al., 2006), whereas RNA interference–mediated silencing of NFA2 (NAP1;2) or NFA3 (NAP1;3) reduces T-DNA transformation efficiency (Crane and Gelvin, 2007). T-DNA integration occurs through the NHEJ process; determining the precise role of CAF-1 and NAP1 in NHEJ will require further investigation.
The triple mutant nrp1-1 nrp2-1 fas2-4 shows a morphological phenotype largely similar to fas2-4, indicating that HR frequency is not associated with the plant morphological phenotype. Depletion of Arabidopsis CAF-1 severely affects global genome transcription and replication and subsequently cell division and differentiation during plant growth and development (reviewed in Ramirez-Parra and Gutierrez, 2007). By contrast, depletion of NAP1 or NRPs only affects expression of a small number of genes and does not affect aboveground morphology of plant growth and development (Zhu et al., 2006; Liu et al., 2009a). Our observation corroborates previous studies on the distinct roles of CAF-1 and NRPs in the regulation of plant growth and development. NRPs also contrast with CAF-1 in telomere length control. While telomeres are shortened in the fas mutants (Mozgová et al., 2010; this study), depletion of NRP1 and NRP2 does not affect telomere length. Telomeres contain tandem short DNA sequence repeats, thus representing potential substrates for HR. HR at telomeres is tightly suppressed and is only allowed in the absence of telomerase activity or some repair factors (Eckert-Boulet and Lisby, 2010). HR is unlikely to be responsible for the telomere shortening in the fas mutants (Mozgová et al., 2010). The mechanism leading to telomere shortening in the fas mutants is still unknown, but our analysis of the triple mutant nrp1-1 nrp2-1 fas2-4 demonstrates that NRP1 and NRP2 are not involved.
In conclusion, our discovery of histone chaperones involved in HR and DNA damage repair in general will allow the advancement of knowledge of the mechanisms of chromatin regulation in genome stability and flexibility. Our results establish that CAF-1 and NAP1 family histone chaperones have distinct roles and that NAP1/NRP proteins, but not CAF-1, are required for somatic HR in Arabidopsis. Because of the high level of conservation of histone chaperones and HR machinery, NAP1-mediated chromatin regulation might also prove crucial for HR in other eukaryotes.
METHODS
Plant Materials
All plant lines used in this study were derived from the Col ecotype. The double mutant nrp1-1 nrp2-1 (m56-1) and the triple mutants nap1;1-1 nap1;2-1 nap1;3-1 (m123-1) and nap1;1-1 nap1;2-1 nap1;3-2 (m123-2) have been described previously (Zhu et al., 2006; Liu et al., 2009a, 2009b). The GUS-based HR reporter lines 1406, 1445, IC9C, and 1415 have also been described previously (Gherbi et al., 2001; Fritsch et al., 2004; Molinier et al., 2004; Schuermann et al., 2009). The reporter lines were crossed with the indicated mutant, and homozygous lines in the mutant background were obtained in F3 and F4 populations through PCR-based genotyping of 50 to 80 individual plants per line. The 1415 (fas2-4) line (Endo et al., 2006) was crossed with nrp1-1 nrp2-1, and 1415 in the triple mutant nrp1-1 nrp2-1 fas2-4 background was obtained in the F4 population through phenotyping combined with PCR-based genotyping.
Growth Conditions and Stress Treatments
For seed production, plants were grown on standard potting soil at 21°C under a 16-h-light/8-h-dark photoperiod in a glasshouse. Somatic HR was studied using in vitro–grown plants. Seeds were surface sterilized (70 and 95% ethanol for 10 min) and plated on agar Murashige and Skoog (MS) medium (MS salts, 1% Suc, pH 5.8, and 0.9% bacto-agar). After stratification at 4°C for 2 d in the dark, plates were incubated in a growth chamber at 21°C under a 16-h-light/8-h-dark photoperiod. For UV-C (254 nm) treatment, 14-d-old plants were irradiated with the indicated dosage using a Stratalinker (UV Crosslinker 2400). Plants were immediately returned to the growth chamber after UV-C exposure. Somatic HR events were examined 4 d after UV-C treatment. For MMC, MMS, bleomycin, HU, and NaCl treatments, 14-d-old plants were incubated in liquid MS supplemented with the indicated concentration of the respective chemical for 1 d and then grown in chemical-free MS medium for 3 d and finally assessed for HR events. The ABA treatment was performed according to Yin et al. (2009).
Histochemical Staining and HR Assessment
Restoration of the reporter gene was detected by histochemical GUS staining according to the standard procedure (Jefferson et al., 1987) in a modified buffer containing 0.04% 5-bromo-4-chloro-3-indolyl-β-d-glucuronide, 50 mM NaHPO4, pH 7.0, 2 mM K4Fe(CN)6, 2 mM K3Fe(CN)6, 5 mM EDTA, and 0.1% Triton X-100. The number of HR events per plant (60 to 90 plants) was assessed visually using a stereomicroscope. Average HR of each sample (per genotype for the indicated treatment) was calculated from three independent experiments and statistically analyzed by analysis of variance using OriginPro 7.5 software (OriginLab).
Telomere Length Analysis
Genomic DNA isolated from 5-week-old plants was used for telomere length analysis according to Mozgová et al. (2010). The hybridization probe was prepared using DIG High Prime DNA Labeling and Detection Starter Kit I and hybridization performed according to the manufacturer’s instructions (Roche Applied Science).
Comet Assay and H3K56ac Analysis
Fourteen-day-old plants were incubated in liquid MS supplemented with the indicated concentration of bleomycin for 6 h and then harvested in liquid nitrogen. Comet assay and the following evaluation were performed as described (Zhu et al., 2006). Histones were extracted as previously described (Yu et al., 2004), except that 25 mM nicotinamide (N0636; Sigma-Aldrich), an inhibitor of NAD-dependent histone deacetylase, was included in the extraction buffer. The extracted histones were separated by 15% SDS-PAGE and transferred to a polyvinylidene fluoride membrane (Millipore). Protein gel blotting was performed using antibodies anti-H3K56ac (07-677; Millipore) and anti-H3 (05-499; Millipore).
Gene Expression Analysis
RNA was isolated from untreated and bleomycin-treated plants and gene expression levels analyzed by quantitative real-time RT-PCR, according to the previously described procedures (Liu et al., 2009a). Melting curve analysis was performed to verify the amplification of a single PCR product. The threshold cycle value was set so that the fluorescent signal is above the baseline noise but as low as possible in the exponential amplification phase. The BRCA1 primers used were 5′-TGCTCAGGGCTCACAGTTGAAGA-3′ and 5′-TTGCAGGCTCCGTTTTCATTGATTG-3′. RAD51, RAD54, and ATM primers were described by Liu et al. (2009a) and the remaining primers described by Zhu et al. (2011).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At4g26110 (NAP1;1), At2g19480 (NAP1;2), At5g56950 (NAP1;3), At1g74560 (NRP1), At1g18800 (NRP2), At5g64630 (FAS2), At4g21070 (BRCA1), At5g20850 (RAD51), At3g19210 (RAD54), At4g02390 (PARP1), At2g31320 (PARP2), At3g48190 (ATM), At1g16970 (KU70), At1g48050 (KU80), At1g08130 (LIG-I), and At5g57160 (LIG-IV).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. RT-PCR Analysis Reveals Similar Expression Levels of 35S:GUS in nrp1-1 nrp2-1 (m56-1) and Col under Either Untreated or Bleomycin-Treated Plant Growth Conditions.
Supplemental Table 1. Comparison of HR Capacity of GUS Recombination Substrates between Wild-Type and Mutant Plants upon Different Growth Treatments.
Supplementary Material
Acknowledgments
We thank Babara Hohn, Zhizhong Gong, and Endo Masaki for kindly providing us with 1406 (Col), 1445 (Col), IC9C (Col), 1415 (Col), and 1415 (fas2-4) seeds, Xiaozhen Yao for assistance in telomere length analysis, and Andre Steinmetz and Emily J. McCallum for critical reading of the article. This work was supported by the National Basic Research Program of China (973 Program; Grants 2011CB944600 and 2012CB910500), by the National Natural Science Foundation of China (Grants NSF30800630, NSF30971443, and NSF90919003), and by the French Centre National de la Recherche Scientifique (PICS 4198). J.G. was supported in part by a foreign student fellowship from the French Ministère de l’Education Nationale, de l’Enseignement Supérieur et de la Recherche.
AUTHOR CONTRIBUTIONS
W.-H.S. conceived and directed the project. W.-H.S. and A.D. designed and supervised the research. J.G. and Y.Z. conducted the majority of experiments. W.Z. conducted the telomere length experiment. J.M. contributed the analytic tool for HR. J.G., Y.Z., W.Z., A.D., and W.-H.S. analyzed data. W.-H.S. wrote the article. All authors contributed to writing the article.
Glossary
- HR
homologous recombination
- DSB
double-strand break
- H3K56ac
H3 Lys 56 acetylation
- GUS
β-glucuronidase
- Col
Columbia
- MMC
mitomycin C
- MMS
methyl methanesulfonate
- HU
hydroxyurea
- ABA
abscisic acid
- NHEJ
nonhomologous end joining
References
- Andrews A.J., Chen X., Zevin A., Stargell L.A., Luger K. (2010). The histone chaperone Nap1 promotes nucleosome assembly by eliminating nonnucleosomal histone DNA interactions. Mol. Cell 37: 834–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avvakumov N., Nourani A., Côté J. (2011). Histone chaperones: modulators of chromatin marks. Mol. Cell 41: 502–514 [DOI] [PubMed] [Google Scholar]
- Boyko A., Hudson D., Bhomkar P., Kathiria P., Kovalchuk I. (2006). Increase of homologous recombination frequency in vascular tissue of Arabidopsis plants exposed to salt stress. Plant Cell Physiol. 47: 736–742 [DOI] [PubMed] [Google Scholar]
- Ceballos S.J., Heyer W.D. (2011). Functions of the Snf2/Swi2 family Rad54 motor protein in homologous recombination. Biochim. Biophys. Acta 1809: 509–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costelloe T., Lowndes N.F. (2010). Chromatin assembly and signalling the end of DNA repair requires acetylation of histone H3 on lysine 56. Subcell. Biochem. 50: 43–54 [DOI] [PubMed] [Google Scholar]
- Crane Y.M., Gelvin S.B. (2007). RNAi-mediated gene silencing reveals involvement of Arabidopsis chromatin-related genes in Agrobacterium-mediated root transformation. Proc. Natl. Acad. Sci. USA 104: 15156–15161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong A., Liu Z., Zhu Y., Yu F., Li Z., Cao K., Shen W.H. (2005). Interacting proteins and differences in nuclear transport reveal specific functions for the NAP1 family proteins in plants. Plant Physiol. 138: 1446–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong A., Zhu Y., Yu Y., Cao K., Sun C., Shen W.H. (2003). Regulation of biosynthesis and intracellular localization of rice and tobacco homologues of nucleosome assembly protein 1. Planta 216: 561–570 [DOI] [PubMed] [Google Scholar]
- Eckert-Boulet N., Lisby M. (2010). Regulation of homologous recombination at telomeres in budding yeast. FEBS Lett. 584: 3696–3702 [DOI] [PubMed] [Google Scholar]
- Endo H., Kawashima S., Sato L., Lai M.S., Enomoto T., Seki M., Horikoshi M. (2010). Chromatin dynamics mediated by histone modifiers and histone chaperones in postreplicative recombination. Genes Cells 15: 945–958 [DOI] [PubMed] [Google Scholar]
- Endo M., Ishikawa Y., Osakabe K., Nakayama S., Kaya H., Araki T., Shibahara K., Abe K., Ichikawa H., Valentine L., Hohn B., Toki S. (2006). Increased frequency of homologous recombination and T-DNA integration in Arabidopsis CAF-1 mutants. EMBO J. 25: 5579–5590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsch O., Benvenuto G., Bowler C., Molinier J., Hohn B. (2004). The INO80 protein controls homologous recombination in Arabidopsis thaliana. Mol. Cell 16: 479–485 [DOI] [PubMed] [Google Scholar]
- Galichet A., Gruissem W. (2006). Developmentally controlled farnesylation modulates AtNAP1;1 function in cell proliferation and cell expansion during Arabidopsis leaf development. Plant Physiol. 142: 1412–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gherbi H., Gallego M.E., Jalut N., Lucht J.M., Hohn B., White C.I. (2001). Homologous recombination in planta is stimulated in the absence of Rad50. EMBO Rep. 2: 287–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer W.D., Ehmsen K.T., Liu J. (2010). Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 44: 113–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Bulger M., Kobayashi R., Kadonaga J.T. (1996). Drosophila NAP-1 is a core histone chaperone that functions in ATP-facilitated assembly of regularly spaced nucleosomal arrays. Mol. Cell. Biol. 16: 3112–3124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik A., Pecinka A., Wendeler E., Reiss B. (2006). The chromatin assembly factor subunit FASCIATA1 is involved in homologous recombination in plants. Plant Cell 18: 2431–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoll A., Puchta H. (2011). The role of DNA helicases and their interaction partners in genome stability and meiotic recombination in plants. J. Exp. Bot. 62: 1565–1579 [DOI] [PubMed] [Google Scholar]
- Liu Z., Zhu Y., Gao J., Yu F., Dong A., Shen W.H. (2009a). Molecular and reverse genetic characterization of NUCLEOSOME ASSEMBLY PROTEIN1 (NAP1) genes unravels their function in transcription and nucleotide excision repair in Arabidopsis thaliana. Plant J. 59: 27–38 [DOI] [PubMed] [Google Scholar]
- Liu Z.Q., Gao J., Dong A.W., Shen W.H. (2009b). A truncated Arabidopsis NUCLEOSOME ASSEMBLY PROTEIN 1, AtNAP1;3T, alters plant growth responses to abscisic acid and salt in the Atnap1;3-2 mutant. Mol. Plant 2: 688–699 [DOI] [PubMed] [Google Scholar]
- Lorch Y., Maier-Davis B., Kornberg R.D. (2006). Chromatin remodeling by nucleosome disassembly in vitro. Proc. Natl. Acad. Sci. USA 103: 3090–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K., Mäder A.W., Richmond R.K., Sargent D.F., Richmond T.J. (1997). Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389: 251–260 [DOI] [PubMed] [Google Scholar]
- Lukas J., Lukas C., Bartek J. (2011). More than just a focus: The chromatin response to DNA damage and its role in genome integrity maintenance. Nat. Cell Biol. 13: 1161–1169 [DOI] [PubMed] [Google Scholar]
- Miyaji-Yamaguchi M., Kato K., Nakano R., Akashi T., Kikuchi A., Nagata K. (2003). Involvement of nucleocytoplasmic shuttling of yeast Nap1 in mitotic progression. Mol. Cell. Biol. 23: 6672–6684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinier J., Ries G., Bonhoeffer S., Hohn B. (2004). Interchromatid and interhomolog recombination in Arabidopsis thaliana. Plant Cell 16: 342–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosammaparast N., Ewart C.S., Pemberton L.F. (2002). A role for nucleosome assembly protein 1 in the nuclear transport of histones H2A and H2B. EMBO J. 21: 6527–6538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozgová I., Mokros P., Fajkus J. (2010). Dysfunction of chromatin assembly factor 1 induces shortening of telomeres and loss of 45S rDNA in Arabidopsis thaliana. Plant Cell 22: 2768–2780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y.J., Luger K. (2006). Structure and function of nucleosome assembly proteins. Biochem. Cell Biol. 84: 549–558 [DOI] [PubMed] [Google Scholar]
- Ramirez-Parra E., Gutierrez C. (2007). The many faces of chromatin assembly factor 1. Trends Plant Sci. 12: 570–576 [DOI] [PubMed] [Google Scholar]
- Ransom M., Dennehey B.K., Tyler J.K. (2010). Chaperoning histones during DNA replication and repair. Cell 140: 183–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuermann D., Fritsch O., Lucht J.M., Hohn B. (2009). Replication stress leads to genome instabilities in Arabidopsis DNA polymerase delta mutants. Plant Cell 21: 2700–2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuermann D., Molinier J., Fritsch O., Hohn B. (2005). The dual nature of homologous recombination in plants. Trends Genet. 21: 172–181 [DOI] [PubMed] [Google Scholar]
- Swoboda P., Gal S., Hohn B., Puchta H. (1994). Intrachromosomal homologous recombination in whole plants. EMBO J. 13: 484–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukuda T., Fleming A.B., Nickoloff J.A., Osley M.A. (2005). Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature 438: 379–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walfridsson J., Khorosjutina O., Matikainen P., Gustafsson C.M., Ekwall K. (2007). A genome-wide role for CHD remodelling factors and Nap1 in nucleosome disassembly. EMBO J. 26: 2868–2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H., Zhang X., Liu J., Wang Y., He J., Yang T., Hong X., Yang Q., Gong Z. (2009). Epigenetic regulation, somatic homologous recombination, and abscisic acid signaling are influenced by DNA polymerase epsilon mutation in Arabidopsis. Plant Cell 21: 386–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Dong A., Shen W.H. (2004). Molecular characterization of the tobacco SET domain protein NtSET1 unravels its role in histone methylation, chromatin binding, and segregation. Plant J. 40: 699–711 [DOI] [PubMed] [Google Scholar]
- Zhu Y., Dong A., Meyer D., Pichon O., Renou J.P., Cao K., Shen W.H. (2006). Arabidopsis NRP1 and NRP2 encode histone chaperones and are required for maintaining postembryonic root growth. Plant Cell 18: 2879–2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Weng M., Yang Y., Zhang C., Li Z., Shen W.H., Dong A. (2011). Arabidopsis homologues of the histone chaperone ASF1 are crucial for chromatin replication and cell proliferation in plant development. Plant J. 66: 443–455 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.