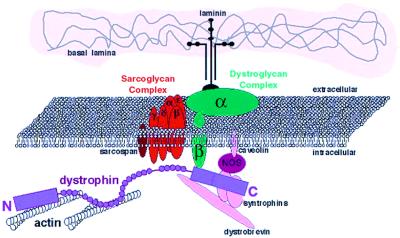
Muscle cells contain a membrane-spanning complex of proteins that are associated with dystrophin, which is a spectrin-related protein of the muscle membrane cytoskeleton (1). The members of this complex include membrane-spanning subunits, such as β-dystroglycan and the sarcoglycans, as well as strictly intracellular and extracellular components. As diagrammed in Fig. 1, the complex connects the cortical actin cytoskeleton (via dystrophin interactions) to the extracellular matrix (via interactions with laminin). Considerable work has focused on identifying the members of this complex and delineating the functions of the individual parts and the complex as a whole. Attention directed at this complex, and indeed the discovery of dystrophin (2), largely stems from the fact that the loss of dystrophin in humans results in loss of the entire protein complex shown in Fig. 1 and causes a progressive, lethal muscle-wasting disease, known as Duchenne muscular dystrophy (DMD).
Figure 1.
Schematic representation of the dystrophin-associated glycoprotein complex. The N-terminal, actin-binding domain of dystrophin in purple is associated with the cortical actin. The C-terminal domain associates with β-dystroglycan and with α- and β-syntrophin and dystrobrevin. nNOS is known to interact with the syntrophins as well as with caveolin.
Two papers in this issue of PNAS address unresolved issues concerning the function of dystrophin and the dystrophin-associated proteins. First, the paper by Wang et al. (3) evaluates the ability of dystrophin molecules that have large segments deleted to rescue skeletal muscles of the mouse model of DMD (mdx mouse). The motivation for this study is to design a functional dystrophin molecule that can fit into the most promising vector for skeletal muscle-directed gene therapy, recombinant adeno-associated virus. However, this and other such studies also will help to elucidate the functional roles of the dystrophin domains. The paper by Sander et al. (4) focuses on another member of the complex, neuronal nitric oxide synthase (nNOS). Although nNOS is not entirely missing in the skeletal muscles of DMD patients or mdx mice, its membrane localization is lost because of the absence of dystrophin and the dystrophin-associated glycoprotein complex (5). The question that Sander et al. pose, based on previous studies in mice (6), is whether or not the proper localization of nNOS in patients is necessary to regulate blood flow to skeletal muscle during exercise.
Although elements of the dystrophin gene and protein structure have been determined, and multiple components of the dystrophin-associated glycoprotein complex have been identified (see Figs. 1 and 2; refs. 1 and 2), the functional roles of dystrophin and the complex have proven more difficult to ascertain. Most investigators now believe that the functional defect in dystrophin-deficient muscle is caused by primary structural weakness in connections spanning the sarcolemma between the cortical actin and the extracellular matrix (7–10). This connection forms the major pathway for transmitting the forces generated in the muscle sarcomeres to the extracellular connective tissues. In the absence of the dystrophin complex, force transmission may occur solely via integrin complexes. Although integrin-containing focal adhesions play this role in nonmuscle cells, they may not be able to physically withstand and transmit the high forces generated within muscle cells. A number of investigators (11–13) have proposed that the dystrophin-associated glycoprotein complex plays important signaling roles in addition to its structural role. Certainly there have been demonstrations of altered NO production (6), as well as possible alterations in ion channel function (14, 15) caused by the loss of the complex.
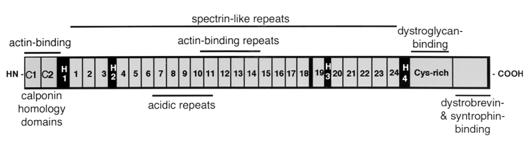
Figure 2.
Domains of the dystrophin molecule. The N terminus contains the primary actin-binding site, whereas the C terminus contains the β-dystroglycan, dystrobrevin, and syntrophin-binding sites. The N- and C-terminal domains are connected by 24 spectrin-like repeats, some of which have been shown to bind actin (9). The four “hinge” regions are denoted H1–H4.
As diagrammed in Fig. 2, the dystrophin molecule has a N-terminal actin-binding domain (two calponin homology domains) followed by 24 spectrin-like triple helical repeats, with four intervening nonhelical segments (commonly referred to as “hinge” regions). The C terminus of dystrophin contains a cysteine-rich domain that binds to dystroglycan (16, 17) followed by a region that associates with dystrobrevin, syntrophin, and indirectly, with nNOS (18, 19). The loss of the C-terminal domain of dystrophin in the mdx mouse (and in DMD) is responsible for the loss of the complex as a whole. This was demonstrated by the expression of a dystrophin-related protein, Dp71, in the skeletal muscles of the mdx mouse (20). Dp71 contains the dystrophin C-terminal domain, but is missing the N-terminal actin-binding domain and all of the spectrin-like repeats. Interestingly, even though expression of Dp71 in the mdx mouse restored the dystrophin-associated proteins to the membrane, the expression resulted in more severe muscle degeneration as compared with the complete absence of dystrophin and the complex. This finding implies that although localization of the complex may be necessary for its function, it is not sufficient to restore dystrophic muscle to a healthy state.
Conceptually, the simplest approach to gene therapy for DMD would be to express full-length dystrophin in muscle. Daunting obstacles, including generation of an efficient gene delivery vector, a means to deliver it to all of the muscles of the body, and possible immunological complications must be overcome before this can be a reality. Of all of the gene delivery vectors evaluated in skeletal muscle, recombinant adeno-associated virus (rAAV) is the most efficient at delivery to adult skeletal muscle without generating an immune response (21, 22). However, it can accommodate only a total genome of approximately 5 kb, whereas the dystrophin coding sequence is almost 14 kb. Although advances that enable the trans-splicing of independent rAAV vectors have been demonstrated (23), the size of dystrophin would require the efficient delivery of at least three different AAV vectors to express a fully intact molecule. Therefore, the question remains as to which parts of dystrophin can be removed and still provide functional protection to the muscle.
Perhaps the more important question is whether or not any dystrophin molecule that is missing most of its domains can prevent muscle degeneration. The question is partly answered in nature, by the observation that a less severe muscle degenerative disease, Becker muscular dystrophy, is caused by in-frame deletions in the dystrophin coding sequence that results in production of truncated dystrophin molecules. The severity of the resulting disease varies greatly, depending on the region that is deleted. Indeed, a truncated dystrophin that has been investigated by a number of groups was first detected in a patient that presented with late-onset, mild dystrophy (24). However, if a major deletion that retains most of the function of full-length dystrophin has been created by accidents of nature, then humans carrying such deletions would not present with disease and their mutations would go undetected.
The paper of Wang et al. (3) provides an important demonstration that considerable protection of muscle can be achieved by a dystrophin molecule that is missing most of its spectrin-like repeats and the portion of the C terminus containing the binding sites for dystrobrevin and syntrophin. However, only the diaphragm of the mdx shows severe muscle degeneration analogous to human DMD (25). Thus it is unclear whether the truncated dystrophin constructs introduced into the limb muscles by Wang et al. can withstand the repeated workloads experienced by the mouse diaphragm, which would be more predictive of efficacy in humans. Although the truncated dystrophins did provide protection against the forces generating during swimming, evaluation of the ability of the transduced muscles to withstand the high forces generated during lengthening contractions (8) might give additional insights into the degree of protection that is afforded. Nevertheless, the lack of ongoing degeneration/regeneration in the muscles expressing some of the truncated constructs provides evidence of significant protection and benefit.
A rational design of a truncated dystrophin for gene therapy would require knowing the function of all of the domains, including the individual spectrin-like repeats. A number of truncation studies in transgenic mdx mice (26) have delineated the minimal C-terminal domain that allows for localization of the complex members, including nNOS (27). What is less clear is what is the role or roles of the many spectrin-like repeats. Work from the Ervasti lab (9) has revealed that some of the repeats display actin binding and may contribute to a large actin-binding interface that includes the N-terminal domain. It is possible that the multiple actin-interactions along the molecule lead to cross-linking and organization of the cortical actin that is necessary for the transmission of force to the extracellular matrix. It is also possible that all of the spectrin-like repeats are necessary to generate a dystrophin that is physically capable of withstanding and transmitting the high forces developed in skeletal muscle. Evolution provides no help in deciding which dystrophin domains are the most important, as the Caenorhabditis elegans homologue of dystrophin, dys-1, contains all of the domains of mammalian dystrophin, including the same number of spectrin-like repeats (28). This level of conservation gives credence to the notion that any truncated dystrophin will likely lose some of its function.
Any truncated dystrophin molecule that is likely to maintain the majority of its function will require the actin-binding domain of the N terminus and a sufficient portion of the C terminus to allow localization of the critical complex members. This obviously includes the dystro- and sarcoglycans, which are necessary for complex formation and function. But is there a requirement for the components that bind to the extreme C terminus of dystrophin, with include the syntrophins and, indirectly, nNOS? Syntrophin knockout mice have been generated and the animals do not have a pronounced dystrophic phenotype (13, 29). However, the loss of the syntrophins does lead to the loss of nNOS localization. In contrast, the loss of the extreme C terminus of dystrophin, which has been shown to bind the syntrophins and dystrobrevin, does not result in the loss of nNOS, syntrophin, or dystrobrevin (25). Perhaps this localization is mediated through other members of the dystrophin complex, or, as in the case of actin, via alternative sites on the dystrophin protein.
The paper of Sander et al. (4) demonstrates that proper nNOS localization is necessary to increase blood flow to skeletal muscle in humans during exercise. The loss of local control of blood perfusion could render dystrophin-deficient fibers ischemic during contraction, which would exacerbate their functional deficits. This could in part explain why one often observed clusters of degenerating fibers in dystrophic muscles and why incomplete gene transfer of dystrophin seems to provide benefit to the fibers that surround a dystrophin-expressing fiber. The importance of the nNOS localization may not be apparent in animal models in which the muscles are not exposed to rigorous exercise. Clearly, this localization and potential coordination of nNOS signaling is an important function of the dystrophin-associated glycoprotein complex. Future attempts at generating a truncated dystrophin for therapeutic purposes should evaluate the localization of nNOS.
The papers of Wang et al. and Sander et al. represent important contributions to the understanding of dystrophin and the dystrophin-associated glycoprotein complex. Wang et al. provide a surprising demonstration that a small piece of the dystrophin molecule can provide much of the protective function of the full-length molecule. The data of Sander et al. underscore the importance of the role of the complex in localizing other proteins. In particular, the localization of nNOS may be critical for normal muscle function. Loss of the localization may play a major role in the development of the pathology of DMD.
Dystrophin is a protein of many domains, connecting to and organizing a complex of many members. All of its parts likely play important roles. Delineating which are the most important may be of immediate interest to gene therapists, but in the long term we need to understand all of the roles of this truly complex complex.
Acknowledgments
We thank Elizabeth McNally for providing the diagram in Fig. 1. H.L.S. acknowledges grant support from the National Institutes of Health and the Muscular Dystrophy Association.
Footnotes
References
- 1.Campbell K P. Cell. 1995;80:675–679. doi: 10.1016/0092-8674(95)90344-5. [DOI] [PubMed] [Google Scholar]
- 2.Hoffman E P, Brown R H, Kunkel L M. Cell. 1987;51:919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- 3.Wang B, Li J, Xiao X. Proc Natl Acad Sci USA. 2000;97:13714–13719. doi: 10.1073/pnas.240335297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sander M, Chavoshan B, Harris S A, Iannacconne S T, Stull J T, Thomas G D, Victor R G. Proc Natl Acad Sci USA. 2000;97:13818–13823. doi: 10.1073/pnas.250379497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brenman J E, Chao D S, Xia H, Aldape K, Bredt D S. Cell. 1995;82:743–752. doi: 10.1016/0092-8674(95)90471-9. [DOI] [PubMed] [Google Scholar]
- 6.Thomas G D, Sander M, Lau K S, Huang P L, Stull J T, Victor R G. Proc Natl Acad Sci USA. 1998;95:15090–15095. doi: 10.1073/pnas.95.25.15090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ervasti J R, Campbell K P. J Cell Biol. 1993;122:809–823. doi: 10.1083/jcb.122.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petrof B J, Shrager J B, Stedman H H, Kelly A M, Sweeney H L. Proc Natl Acad Sci USA. 1993;90:3710–3714. doi: 10.1073/pnas.90.8.3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amann K J, Renley B A, Ervasti J M. J Biol Chem. 1998;273:28419–28423. doi: 10.1074/jbc.273.43.28419. [DOI] [PubMed] [Google Scholar]
- 10.Straub V, Rafael J A, Chamberlain J S, Campbell K P. J Cell Biol. 1997;139:375–385. doi: 10.1083/jcb.139.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hack A, Cordier L, Shoturma D, Lam M, Sweeney H, McNally E. Proc Natl Acad Sci USA. 1999;96:10723–10728. doi: 10.1073/pnas.96.19.10723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rafael J A, Townsend E R, Squire S E, Potter A C, Chamberlain J S, Davies K E. Hum Mol Genet. 2000;9:1357–1367. doi: 10.1093/hmg/9.9.1357. [DOI] [PubMed] [Google Scholar]
- 13.Adams M E, Kramarcy N, Krall S P, Rossi S G, Rotundo R L, Sealock R, Froehner S C. J Cell Biol. 2000;150:1385–1398. doi: 10.1083/jcb.150.6.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fong P, Turner P R, Denetclaw W F, Steinhardt R A. Science. 1990;250:673–676. doi: 10.1126/science.2173137. [DOI] [PubMed] [Google Scholar]
- 15.Franco A, Lansman J B. Nature (London) 1990;344:670–673. doi: 10.1038/344670a0. [DOI] [PubMed] [Google Scholar]
- 16.Jung D, Yang B, Meyer J, Chamberlain J S, Campbell K P. J Biol Chem. 1995;270:27305–27310. doi: 10.1074/jbc.270.45.27305. [DOI] [PubMed] [Google Scholar]
- 17.Suzuki A, Yoshida M, Yamamoto H, Ozawa E. FEBS Lett. 1992;308:154–160. doi: 10.1016/0014-5793(92)81265-n. [DOI] [PubMed] [Google Scholar]
- 18.Sadoulet-Puccio H M, Rajala M, Kunkel L M. Proc Natl Acad Sci USA. 1997;94:12413–12418. doi: 10.1073/pnas.94.23.12413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suzuki A, Yoshida M, Ozawa E. J Cell Biol. 1995;128:373–381. doi: 10.1083/jcb.128.3.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greenberg D S, Sunada Y, Campbell K P, Yaffe D, Nudel U. Nat Genet. 1994;8:340–344. doi: 10.1038/ng1294-340. [DOI] [PubMed] [Google Scholar]
- 21.Cordier L, Hack A A, Scot M O, Barton-Davis E R, Gao G, Wilson J M, McNally E M, Sweeney H L. Mol Ther. 2000;1:119–129. doi: 10.1006/mthe.1999.0019. [DOI] [PubMed] [Google Scholar]
- 22.Xiao X, Li J, Samulski R J. J Virol. 1996;70:8098–8108. doi: 10.1128/jvi.70.11.8098-8108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan Z, Zhang Y, Duan D, Engelhardt J F. Proc Natl Acad Sci USA. 2000;97:6716–6721. doi: 10.1073/pnas.97.12.6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.England S B, Nicholson L V, Johnson M A, Forrest S M, Love D R, Zubrzycka-Gaarn E E, Bulman D E, Harris J B, Davies K E. Nature (London) 1990;343:180–182. doi: 10.1038/343180a0. [DOI] [PubMed] [Google Scholar]
- 25.Stedman H H, Sweeney H L, Shrager J B, Maguire H C, Panettieri R A, Petrof B, Narusawa M, Leferovich J M, Sladky J T, Kelly A M. Nature (London) 1991;352:536–539. doi: 10.1038/352536a0. [DOI] [PubMed] [Google Scholar]
- 26.Rafael J A, Cox G A, Corrado K, Jung D, Campbell K P, Chamberlain J S. J Cell Biol. 1996;134:93–102. doi: 10.1083/jcb.134.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crawford G E, Faulkner J A, Crosbie R H, Campbell K P, Froehner S C, Chamberlain J S. J Cell Biol. 2000;150:1399–1410. doi: 10.1083/jcb.150.6.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bessou C, Giugia J B, Franks C J, Holden-Dye L, Segalat L. Neurogenetics. 1998;2:61–72. doi: 10.1007/s100480050053. [DOI] [PubMed] [Google Scholar]
- 29.Kameya S, Miyagoe Y, Nonaka I, Ikemoto T, Endo M, Hanaoka K, Nabeshim Y, Takeda S. J Biol Chem. 1999;274:2193–2200. doi: 10.1074/jbc.274.4.2193. [DOI] [PubMed] [Google Scholar]