Abstract
Survivin (SVV) is a protein that belongs to the inhibitor of apoptosis proteins (IAP) family and is involved in the G2/M phase progression of the cell cycle as a spindle‑associated molecule. The biological features of this protein are well documented and its activity appears to be involved in mitochondria-dependent and -independent antiapoptotic pathways. Overexpression of SVV at the transcriptional and translational level has been associated with cancer, a multifactorial disorder in which the occurrence of a -31G to C polymorphism in the promoter region may significantly contribute to the development of this pathology. To verify this hypothesis, the occurrence of a single nucleotide polymorphism (SNP) in cis-acting cell cycle-dependent elements (CDEs) and in cell cycle homology regions (CHRs) of the survivin TATA-less promoter was investigated. A total of 23 oral squamous cell carcinoma (OSCC) cell lines and normal epithelium-derived normal human epidermal keratinocyte (NHEK) cell lines were analyzed by RFLP and direct DNA sequencing of their promoter region. Furthermore, survivin expression at the transcriptional and translational levels was evaluated in these cells by RT-PCR and Western blotting, respectively. The findings indicate that the presence of a G or C allele is not directly correlated to survivin expression, at the mRNA or at the protein level, at least in the OSCC lines analyzed in this study.
Introduction
Survivin was discovered by Altieri's group in 1997 (1). The gene coding for survivin was also identified by the same group during a search for the unknown receptor (effector cell protease receptor, epr-1) of blood coagulation factor Xa (2).
Survivin is an essential protein that acts as a passenger protein involved in cell division, and belongs to the inhibitor of apoptosis proteins (IAP) family (1-3). Its antiapoptotic zinc-binding domain, baculoviral IAP repeat (BIR), was identified for the first time in a baculoviral protein (4). The human locus is indicated as BIR-containing 5 (BIRC5) and lies on the long arm of chromosome 17 in the telomeric region 25 (17q25) (5). This locus is the source of numerous variant transcripts (survivin, survivin-1α, survivin-2α, survivin-2β, 3β and Δ3), where survivin is the most abundantly transcribed and translated variant in a 142 amino acid-long polypeptide, whereas the minor variants are poorly expressed and investigated (6-9). Furthermore, the occurrence of a natural survivin antisense strand, defined as a distinct gene, codifying for a hypothetical receptor of Xa coagulation factor, epr-1, is well documented (2-10). All of the splicing isoforms share a common promoter belonging to the housekeeping family promoter that is known to be TATA‑less and CpG-rich (11).
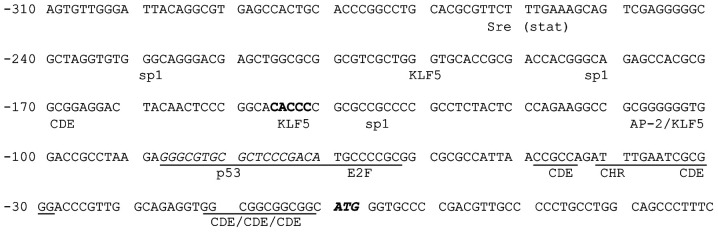
The survivin promoter region was investigated and the minimal promoter region, detected within the proximal -220 nt of the human survivin gene, was found to contain numerous Sp1 sites, three cell cycle-dependent elements (CDEs) and one cell cycle homology region (CHR) implicated in G1 transcriptional repression in a number of S/G2-regulated genes (11,12). Xu et al refined the map of the regulatory consensus sequences by identifying two other CDEs flanking the unique CHR box, indicated as CDE1 and CDE2. In the latter, these authors highlighted the outcome of the G/C single nucleotide polymorphism (SNP) at position -31 (13). The rationale was that this repressor ligand zone binds the putative repressor complex with different affinity depending on whether G or C lies in position -31. Studies by these authors, performed on cancer-derived or normal cell lines, showed that the presence of the C allele was associated with normal cell lines, whereas the G allele was related to cancer. Subsequently, by using a luciferase reporter gene construct transfected in normal breast MCF-10A and cancer MDA-MB221 cell lines, Xu et al demonstrated that the -31G survivin promoter upstream of the reporter gene was more effective in stimulating gene expression at all cell cycle stages, even though the greatest effect was found, as expected, in the G2/M phase (13). These data suggest that the presence of G at position -31 derepresses survivin gene transcription, favoring the cancer phenotype.
Successive studies, performed in cancer-bearing patients, indicate that the negative and positive risk of cancer is associated with the -31C/G polymorphism (14-16). In particular, where the association with SNP was positive, the -31C allele appears to be related to increased cancer risk, which is in contrast to the results by Xu et al.
On the basis of these data and considerations, we analyzed a possible correlation between the -31G/C SNP and survivin expression in various oral squamous cell carcinoma (OSCC) cell lines in comparison to a normal human epidermal keratinocyte (NHEK) epithelial cell line.
Materials and methods
Cell cultures. The cell lines shown in Table I were grown in the recommended media at 37˚C in a humidified atmosphere of 5% CO2. NHEK cells were obtained from Lonza (Lonza, Switzerland), cultured in Clonetics® KGM complete medium (Lonza), and utilized up to the fourth passage. After washing with phosphate-buffered saline (PBS), the cultured cells were harvested with a cell scraper, and centrifuged at 2,000 rpm for 10 min. The cell pellets were stored at -80˚C prior to protein, RNA and genomic DNA isolation. The NHEK cell line was derived from normal epithelium.
Table 1. Survivin promoter polymorphism, mRNA and protein in OSCC cell lines.
Extraction, amplification and sequencing of the survivin promoter region. Genomic DNA was isolated using a DNA microextraction kit (Stratagene, La Jolla, CA, USA) according to the manufacturer's instructions. The PCR was carried out in 40 µl master mixture containing 10% DMSO for extension through the GC-rich region. The thermal cycling process included initial denaturation at 95˚C for 10 min, followed by 35 cycles (denaturation at 94˚C for 45 sec, annealing at 60˚C for 45 sec, and extension at 72˚C for 1 min). Sequencing was performed at 5 µl (Primm, Naples, Italy), and 30 µl were used for restriction fragment length polymorphism (RFLP) as described in the following paragraph.
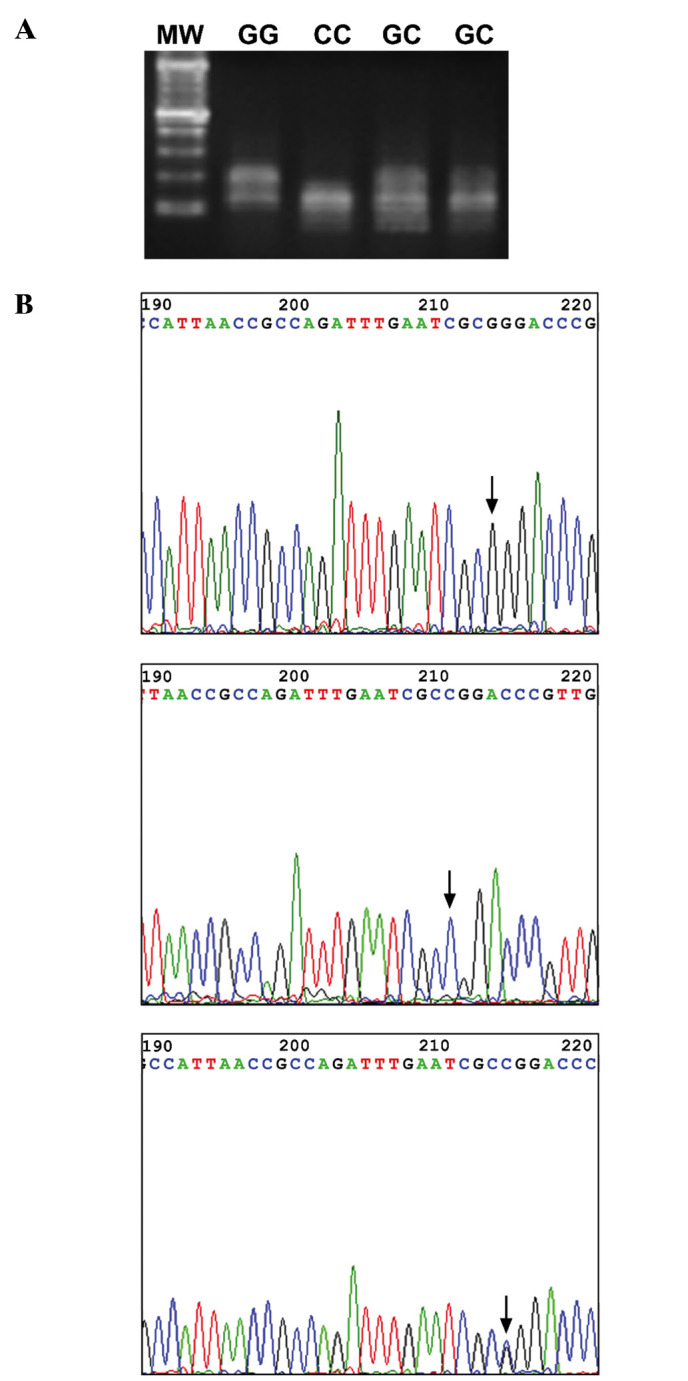
Restriction fragment length polymorphism. The PCR products were subjected to electrophoresis on a 2% agarose gel. The 315 bp amplimers were extracted and then digested with the restriction enzyme MspI (New England Biolabs, MA, USA) at 37˚C for 1 h. Digestion patterns were analyzed by electrophoresis in a 3% ethidium bromide-stained agarose gel. The G allele was cleaved by the restriction enzyme MspI, generating a 190 and a 125 bp-long fragment (upper band, U, and middle band, M), whereas the cleaved C allele generated 3 fragments, two of which were 120 and 125 bp long, comigrating as a middle band (M band) and the third one, 70 bp long, as a lower band (L band). The heterozygote G + C generated a G and C digestion pattern showing a mixture of U, M and L bands (Fig. 1A).
Figure 1. -31G/C polymorphism of PCR- amplified SVV promoter DNA. (A) RFLP profile of the polymorphic pattern obtained by electrophoretic analysis (see Materials and methods). (B) Direct sequencing analysis of the survivin promoter: arrows indicate the -31 position of the G/G, C/C or G/C polymorphism.
Western blot analysis. Cell pellets were suspended in lysis buffer (~5x106/ml) containing 50 mM Tris HCl, 2% sodium dodecyl sulfate (SDS) and 10% glycerol for 15 min in ice-cold water, and heated twice to 95˚C for 3 min. Lysates were microfuged at room temperature for 10 min and supernatants were collected. Protein concentration was measured using Bradford's method (Sigma-Aldrich, St. Louis, MO, USA). Proteins (50 µg/lane) were electrophoresed on 15% SDS-polyacrylamide gels and transferred to nitrocellulose membranes (Bio-Rad Laboratories, Hercules, CA, USA). Immunoreactive proteins were detected by chemiluminescence (SuperSignal West Pico, Pierce Chemical Co., Rockford, IL, USA) according to the manufacturer's instructions using Hyperfilm ECL (Amersham Biosciences, Piscataway, NJ, USA). Semi-quantitative values of proteins were measured by densitometry of Western blotting using β-actin as an internal control.
RT-PCR analysis. Total RNA was isolated from 5x106 cells by TRIzol reagent (Invitrogen, Inc., Carlsbad, CA, USA). Total RNA (300 ng) was used for RT-PCR, using the SuperScript III One-Step RT-PCR kit (Invitrogen). Full-length specific primers were 5'-GACCACCGCATCTCTACATTC-3' (forward) and 5'-TGC TTTTTATGTTCCTCTATGGG-3' (reverse). GAPDH was used as a housekeeping gene: 5'-TTG GTATCGTGGAAGGACTCA-3' (forward) and 5'-TGTCATC ATATTTGGCAGGTTT-3' (reverse). RT-PCR conditions were as follows: reverse transcription at 55˚C for 30 min, terminated by 94˚C for 2 min and followed by 35 cycles of denaturation for 15 sec at 94˚C, annealing for 30 sec at 55˚C and extension for 1 min at 68˚C. The final extension was carried out at 68˚C for 5 min. Semi‑quantitative analysis of mRNA was performed by densitometry of RT-PCR amplification products using GAPDH as an internal control.
Statistical analysis. Statistical analysis was carried out using the two-way ANOVA test, followed by a post-hoc (Newman‑Keuls) test to form multiple comparisons. P<0.05 was considered to be statistically significant: p<0.05, p<0.01, p<0.001.
Results
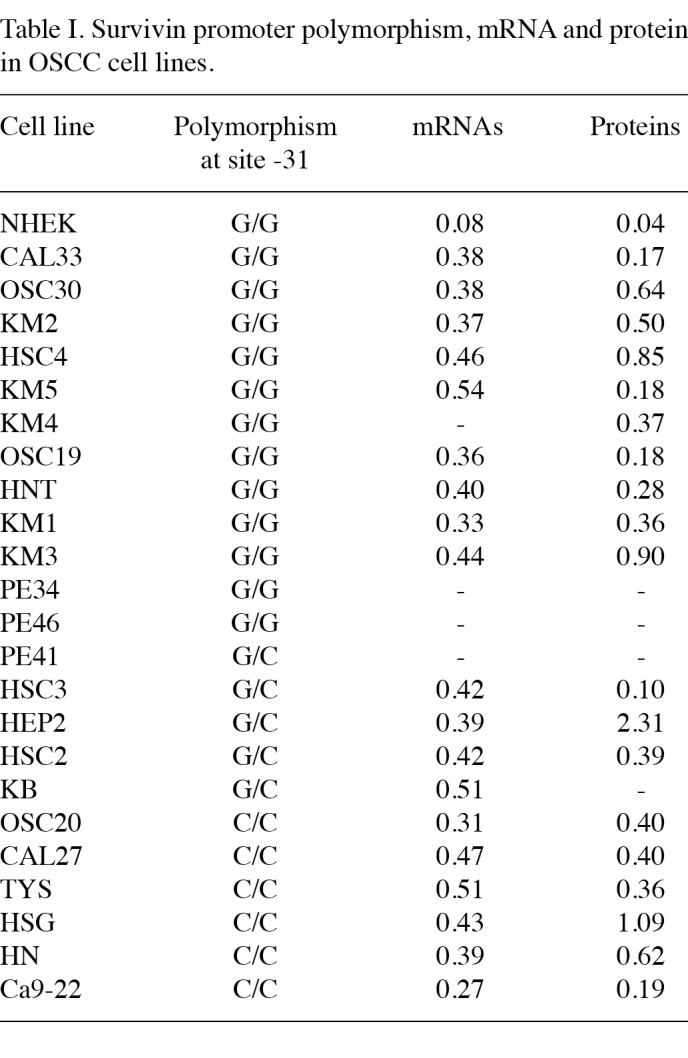
The -31C/G polymorphism in the OSCC survivin promoter is significantly related to cancer phenotype. The characterization of the G/C polymorphism at position -31 of the survivin promoter was evaluated by RFLP and direct DNA sequencing in 22 OSCC cell lines and in one NHEK cell line. The results are shown in Table I, and indicate that the -31G polymorphism in the OSCC survivin promoter is significantly (73% of G alleles vs. 37% of C alleles) correlated to the cancer phenotype, as previously reported by Xu et al (13).
Survivin mRNA expression is not associated with the -31C/G polymorphism and is always higher in OSCC as compared to NHEK cell lines. The mRNA survivin level, as evaluated by densitometry, was similar in the GG and CC genotypes (Table I). The values obtained do not suggest any significant association with the -31G/C polymorphism, although the amount of survivin mRNA was always higher in the OSCC cell lines as compared to the NHEK ones. The results obtained suggest that the survivin promoter polymorphism at position -31 is not associated with survivin expression, at least in the OSCC cell lines analyzed.
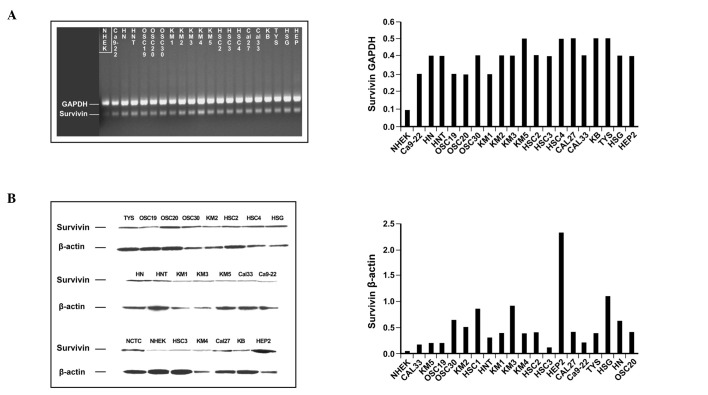
Protein levels of survivin do not correlate with the -31G/C polymorphism and survivin is always overexpressed in OSCC as compared to NHEK cell lines. The protein level of survivin, as analysed by Western blotting and quantified by densitometry, was similar in the GG and CC genotypes in 20 OSCC cell lines (Table I), although the amount of survivin protein was always found to be higher in OSCC cell lines than in NHEK ones. The data are consistent with a direct involvement of protein expression in the cancerous phenotype; however, no correlation was found between expression and G/C polymorphism. Moreover, the protein expression was varied when correlated to the mRNA expression (Fig. 2).
Figure 2. Survivin expression in OSCC cell lines. (A) Survivin mRNA expression as evaluated by RT-PCR. (B) Survivin protein expression as assessed by Western blot analysis. The histograms (left panel) show the semi-quantitative values of mRNA and protein normalized with GAPDH, and β-actin used as reporter genes (right panel), respectively.
Figure 3. Mapping of a putative transcription factor for consensus sequences in the proximal promoter of the survivin gene.
Discussion
In the present study, we examined polymorphisms in the promoter of the survivin gene at position -31, in 22 OSCC and NHEK cell lines. The association between -31G/C SNP and survivin expression was investigated. The -31G allele occurred in 63% (22 OSCC: 12 GG, and 5 GC and 6 CC) of the oral cancer cells analyzed (Fig. 1). These results were in line with the findings of two other authors who demonstrated that the G allele was present in 70% (22 different carcinomatous cell lines: 15 GG, 6 CC and 1 GC) and 75% (12 cell lines: 7 GG, 1 CC and 4 GC) of the cancer cell lines analyzed (13,18). In order to explore whether this polymorphism was associated with survivin expression, the expression was investigated using RT-PCR and Western blotting (Fig. 2). The data obtained do not confirm any direct relationship between the G (or C) allele at position -31 and enhanced mRNA expression (see Table I). In addition, consistent with our previous data on oral cancer patients, the mRNA expression was always significantly higher in OSCC cells than in normal tissues (21). Similar results were obtained by other authors using breast cancer cells and MCF-10A normal cells as controls (18).
The -31G/C polymorphism was also investigated in human epidemiological studies to determine whether there was any possible association between this polymorphism and cancer development. Association with cancer risk has been demonstrated in lung carcinoma (Korea), sporadic colorectal cancer (Greece), urothelial carcinoma (Taiwan) and gastric carcinoma (China). When an association was found, cancer risk was always correlated to the C allele. In contrast, no association was detectable in acute myeloid leukemia (Germany), cervical cancer (Hungary), breast cancer (France) and esophageal SCC (China) (14-19). Concerning sporadic colorectal cancer, urothelial carcinoma and epithelial SCC, a positive correlation between -31C SNP and cancer risk was found when the clinical and pathological grade of the tumor was taken into consideration (15,16,19,20).
In conclusion, we believe that in defined human populations the -31C allele dependence of the survivin expression in certain tumors may be associated with cancer risk, although more studies, particularly population-based studies, are required to elucidate the role of survivin in cancer development.
Acknowledgments
To the memory of Professor Salvatore Metafora.
Contributor Information
Salvatore de Maria, Department of Experimental Medicine, Medical School, Second University of Naples, I-80138 Naples, Italy.
Lorenzo lo Muzio, Department of Surgical Sciences, University of Foggia, Foggia, Italy.
Alessandra Braca, Department of Experimental Medicine, Medical School, Second University of Naples, I-80138 Naples, Italy.
Paolo Rega, Department of Experimental Medicine, Medical School, Second University of Naples, I-80138 Naples, Italy.
Amalia Cassano, Department of Biomedical Sciences and Human Oncology, Medical School, University of Bari, Bari, Italy.
Angela Vinella, Department of Biomedical Sciences and Human Oncology, Medical School, University of Bari, Bari, Italy.
Ruggiero Fumarulo, Department of Biomedical Sciences and Human Oncology, Medical School, University of Bari, Bari, Italy.
Rosario Serpico, Department of Odontostomatological, Orthodontical and Surgical Sciences, Second University of Naples, Naples, Italy.
Ernesto Farina, Department of Odontostomatological, Orthodontical and Surgical Sciences, Second University of Naples, Naples, Italy.
Vittoria Metafora, Institute of Genetics and Biophysics 'Adriano Buzzati‑Traverso', CNR, Naples, Italy.
Giuseppe Pannone, Department of Surgical Sciences, University of Foggia, Foggia, Italy.
Gian Pietro Ravagnan, Department of Environmental Sciences, Ca' Foscari Venezia University, Venice, Italy.
Salvatore Metafora, Institute of Genetics and Biophysics 'Adriano Buzzati‑Traverso', CNR, Naples, Italy.
Corrado Rubini, Department of Pathology Ospedale Le Molinette Ancona, Ancona, Italy.
Maria Carteni, Department of Experimental Medicine, Medical School, Second University of Naples, I-80138 Naples, Italy.
Maria Addolorata Mariggiò, Department of Biomedical Sciences and Human Oncology, Medical School, University of Bari, Bari, Italy.
References
- Ambrosini G, Adida C and Altieri DC: A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med 3: 917-921, 1997. [DOI] [PubMed]
- Altieri DC: Molecular cloning of effector cell protease receptor-1, a novel cell surface receptor for the protease factor Xa. J Biol Chem 269: 3139-3142, 1994. [PubMed]
- Skoufias DA, Mollinari C, Lacroix FB and Margolis RL: Human survivin is a kinetochore-associated passenger protein. J Cell Biol 151: 1575-1582, 2000. [DOI] [PMC free article] [PubMed]
- Crook NE, Clem RJ and Miller LK: An apoptosis-inhibiting baculovirus gene with a zinc finger-like motif. J Virol 67: 2168‑2174, 1993. [DOI] [PMC free article] [PubMed]
- Risk JM, Evans KE, Jones J, et al: Characterization of a 500 kb region on 17q25 and the exclusion of candidate genes as the familial tylosis oesophageal cancer (TOC) locus. Oncogene 21: 6395-6402, 2002. . [DOI] [PubMed]
- Mahotka C, Wenzel M, Springer E, Gabbert HE and Gerharz CD: Survivin-deltaEx3 and survivin-2B: two novel splice variants of the apoptosis inhibitor survivin with different antiapoptotic properties. Cancer Res 59: 6097-6102, 1999. [PubMed]
- Badran A, Yoshida A, Ishikawa K, Goi T, Yamaguchi A, Ueda T and Inuzuka M: Identification of a novel splice variant of the human anti-apoptopsis gene survivin. Biochem Biophys Res Commun 314: 902-907, 2004. [DOI] [PubMed]
- Caldas H, Honsey LE and Altura RA: Survivin 2α: a novel survivin splice variant expressed in human malignancies. Mol Cancer 4: 11, 2005. [DOI] [PMC free article] [PubMed]
- Zheng W, Ma X, Wei D, Wang T, Ma Y and Yang S: Molecular cloning and bioinformatics analysis of a novel spliced variant of survivin from human breast cancer cells. DNA Seq 16: 321-328, 2005. [DOI] [PubMed]
- Zaman GJ and Conway EM: The elusive factor Xa receptor: failure to detect transcripts that correspond to the published sequence of EPR-1. Blood 96: 145-148, 2000. [PubMed]
- Li F and Altieri DC: Transcriptional analysis of human survivin gene expression. Biochem J 344: 305-311, 1999. [DOI] [PMC free article] [PubMed]
- Zwicker J, Lucibello FC, Wolfraim LA, Gross C, Truss M, Engeland K and Müller R: Cell cycle regulation of the cyclin A, cdc25C and cdc2 genes is based on a common mechanism of transcriptional repression. EMBO J 14: 4514-4522, 1995. [DOI] [PMC free article] [PubMed]
- Xu Y, Fang F, Ludewig G, Jones G and Jones D: A mutation found in the promoter region of the human survivin gene is correlated to overexpression of survivin in cancer cells. DNA Cell Biol 23: 527-537, 2004. [DOI] [PubMed]
- Wang YH, Chiou HY, Lin CT, Hsieh HY, Wu CC, Hsu CD and Shen CH: Association between survivin gene promoter -31 C/G polymorphism and urothelial carcinoma risk in Taiwanese population. Urology 73: 670-674, 2009. [DOI] [PubMed]
- Gazouli M, Tzanakis N, Rallis G, et al: Survivin -31G/C promoter polymorphism and sporadic colorectal cancer. Int J Colorectal Dis 24: 145-150, 2009. [DOI] [PubMed]
- Cheng ZJ, Hu LH and Huang SJ: Correlation of -31G/C polymorphisms of survivin promoter to tumorigenesis of gastric carcinoma. Ai Zheng 27: 258-263, 2008. [PubMed]
- Wagner M, Schmelz K, Dörken B and Tamm I: Epigenetic and genetic analysis of the survivin promoter in acute myeloid leukemia. Leuk Res 32: 1054-1060, 2008. [DOI] [PubMed]
- Boidot R, Vegran F, Jacob D, et al: The expression of BIRC5 is correlated with loss of specific chromosomal regions in breast carcinomas. Genes Chromosomes Cancer 47: 299-308, 2008. [DOI] [PubMed]
- Jang JS, Kim KM, Kang KH, et al: Polymorphisms in the survivin gene and the risk of lung cancer. Lung Cancer 60: 31-39, 2008. [DOI] [PubMed]
- Borbély AA, Murvai M, Szarka K, Kónya J, Gergely L, Hernádi Z and Veress G: Survivin promoter polymorphism and cervical carcinogenesis. J Clin Pathol 60: 303-306, 2007. [DOI] [PMC free article] [PubMed]
- De Maria S, Pannone G, Bufo P, et al: Survivin gene-expression and splicing isoforms in oral squamous cell carcinoma. J Cancer Res Clin Oncol 135: 107-116, 2009. [DOI] [PubMed]