Abstract
Change of mitochondrial DNA (mtDNA) copy numbers is considered to be an important hallmark of cancers. However, whether quantitative changes in mtDNA occur during the initiation and progression of colorectal cancer remains to be determined. Quantitative change in mtDNA was observed during the initiation and progression of colorectal cancer and possible correlations of the mtDNA copy number in colorectal cancer with the clinicopathological stage were investigated. Tumor tissues and the corresponding non-cancerous tissues were surgically resected from 24 colon and 20 rectal patients between 2008 and 2009. β-actin expression was quantified in all of the specimens, and the copy numbers were calculated. In colorectal cancer, the quantitative changes of mtDNA exhibited a significant increase. In 24 cases of colon cancer, the average relative mtDNA copy number ratios were 115.15±31.57 in cancer tissues and 54.09±13.22 in the corresponding non-cancerous tissues (p<0.01). Furthermore, in 20 cases of rectal cancer, the ratios were 145.6±43.83 in cancer tissues and 55.58±12.47 in the corresponding non-cancerous tissues (p<0.001). Following correlation with clinicopathological data, change of the mtDNA copy number in colorectal cancer exhibited a significant association with clinicopathological stage, but no association with gender. Moreover, this increase was particularly marked in stages Ⅰ and Ⅱ. Our results indicate that mtDNA copy number plays a significant role during the initiation and progression of colorectal cancer, particularly during early clinicopathological stages.
Introduction
Colorectal cancer is one of the most common malignant tumors in humans. It is also the third most common cause of cancer-related death in the Western world (1). Numerous studies showed that genetic and epigenetic alterations play a significant role during the initiation and progression of colorectal cancer. Human mitochondrial DNA (mtDNA) exists inside the mitochondrion within the cytoplasm. It is the only genetic material outside the cellular nucleus. mtDNA is a closed circular duplex species of 16,569 bases, encoding 13 respiratory chain polypeptides, and 2 ribosomal and 22 transfer RNAs (2).
Under normal conditions, unlike nuclear DNA, mtDNA is present at extremely high levels: the copy number is 103-104 copies per cell (3). However, this number is altered during the aging process or under pathological states. During the aging process, the mtDNA copy number was found to increase in the brain and lung (4,5). Under pathological states, the conditions are complex. Subsequently, alterations in mtDNA copy number have been observed in a variety of human cancers. The copy number is reduced in hepatocellular and renal cell carcinomas and breast cancer (6-8). However, it is increased in head and neck and ovarian cancers and esophageal squamous cell carcinomas (9-11). However, whether and and the manner in which mtDNA copy number changes in colorectal cancer remains to be determined.
In this study, the relative mtDNA copy number of colon and rectal tumor tissues, and their corresponding non-cancerous tissues obtained from 24 colon and 20 rectal cancer patients were tested. Additionally, the correlation between the change of mtDNA copy number and clinicopathological stage was investigated.
Materials and methods
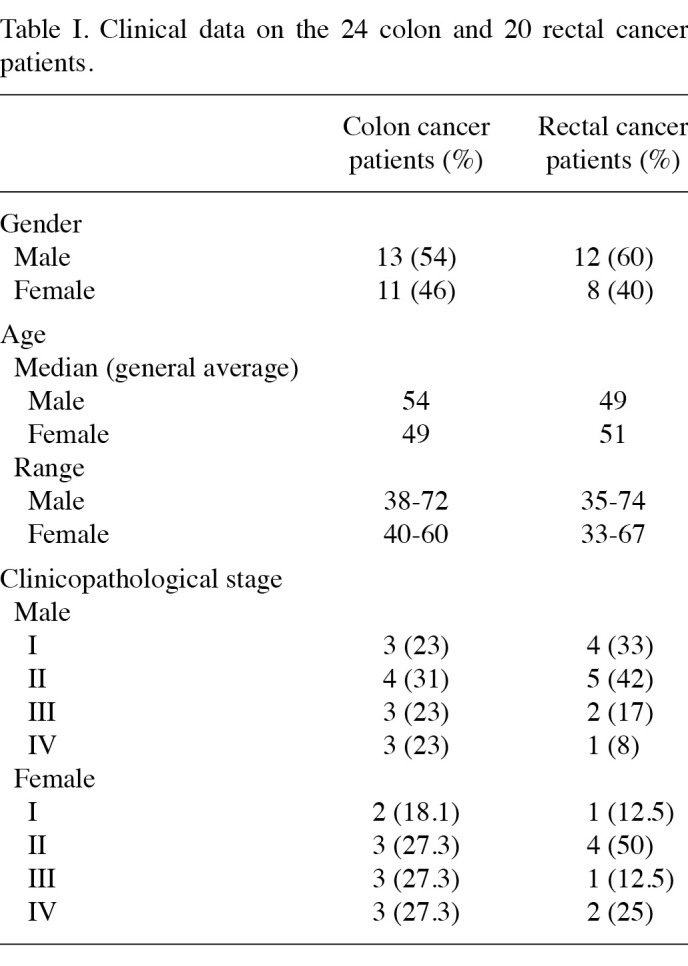
Patients and specimens. The tumor tissues and the corresponding non-cancerous tissues from 24 colon and 20 rectal cancer patients were surgically resected in the West China Hospital, Sichuan University, China, between 2008 and 2009. The 24 colon cancer patients comprised 13 males and 11 females, aged 38-72 years (median 51.9±8.7). The 20 rectal cancer patients included 12 males and 8 females, aged 33-74 years (median 50.4±11.2) (Table Ⅰ). Following resection, the specimens were immediately fresh-frozen and maintained in liquid nitrogen. The specimens of tumor tissues were cut from the edge of the tumors and their corresponding non-cancerous tissues were removed at least 5 cm away from the tumors.
Table 1. Clinical data on the 24 colon and 20 rectal cancer patients.
DNA extraction and real-time quantitative PCR analysis. Corresponding non-cancerous and tumor DNA were prepared as previously described (12). For standardization purposes, the expression of β-actin of each specimen was quantified (Table Ⅱ). The PCR cycling conditions were: activation step at 95˚C for 15 min, followed by 40 cycles of 20 sec at 95˚C, 1 min at 60˚C and 30 sec at 72˚C. To produce the standard curves in each real-time PCR, a UV spectrophotometer was used to measure the concentration of the amplification products of the mtDNA and β-actin, serially diluted as 105, 106, 107, 108 and 109. The real-time quantitative PCR of each concentration was then run. A negative control was included in each reaction.
Table 2. Sequences of primer pairs of mtDNA and internal control gene β-actin.
Statistical analysis. Statistical calculations were performed using the statistical package SPSS version 13.0. The relative mtDNA copy number (mtDNA/β-actin) of each specimen was calculated and the Student's t-test was used to analyse the difference of the mtDNA copy numbers between the tumor and their corresponding non-cancerous tissues and the difference between genders. Quantitative data were expressed as the means ± standard deviation. The presence of a statistically significant difference was denoted by p<0.05.
Results
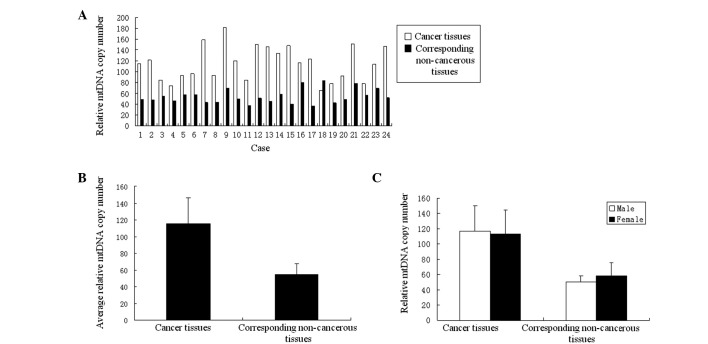
Relative mtDNA copy number of colon cancer. To identify the manner in which the mtDNA copy number alters within the tumor tissue of the colon cancer, the copy numbers of mtDNA of the tumor tissues and the corresponding non-cancerous tissues were tested by real-time quantitative PCR. Following statistical analysis, compared to the non-cancerous tissues, the relative mtDNA copy numbers of the tumor tissues exhibited a marked increase in 23 colon cancer cases (95.8%) (Fig. 1A). The average relative mtDNA copy number ratios were 115.15±31.57 in tumor tissues and 54.09±13.22 in the corresponding non-cancerous tissues. This difference was significant (p<0.01) (Fig. 1B). No significant difference (p>0.05) was noted in the test results of between the tumor tissues or their corresponding non-cancerous tissues (Fig. 1C).
Figure 1. (A) Comparisons of relative mtDNA copy number between colon cancer tissues and corresponding non-cancerous tissues of each case. (B) Comparisons of the average relative mtDNA copy number ratios between the 24 cases of colon cancer tissues and their corresponding non-cancerous tissues. The ratio of tumor tissues was significantly higher than that of the corresponding non-cancerous tissues (p<0.01, Student's t-test). (C) Comparisons of the average relative mtDNA copy number ratios of the 24 cases of colon cancer tissues and their corresponding non-cancerous tissues between different genders. The ratio of tumor tissues showed no significant difference (p=0.778). In corresponding non-cancerous tissues, there was also no significant difference (p=0.116).
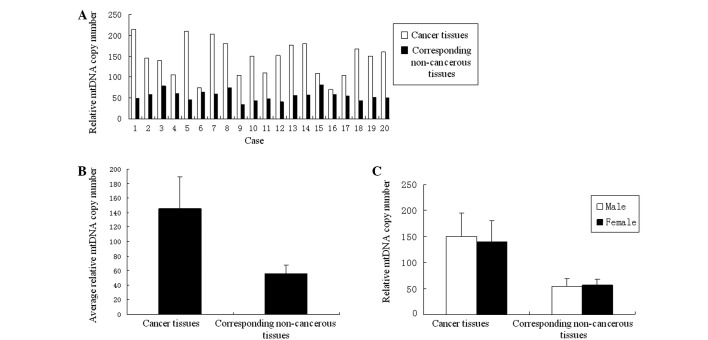
Relative mtDNA copy number of rectal cancer. The test results of rectal cancer revealed a similar situation to that observed in colon cancer. The relative copy number of mtDNA of the rectal tumor tissues and their corresponding non-cancerous tissues were also tested by real-time quantitative PCR. The relative mtDNA copy numbers of the 20 cases of rectal tumor tissues exhibited a marked increase compared to the non-cancerous tissues (Fig. 2A). The average relative mtDNA copy number ratios were 145.6±43.83 in rectal tumor tissues and 55.58±12.47 in the corresponding non-cancerous tissues. This difference was significant (p<0.01) (Fig. 2B). Again, gender revealed no significant difference between the relative mtDNA copy number of the tumor tissues and the corresponding non-cancerous tissues between males and females (Fig. 2C); similar to colon cancer.
Figure 2. (A) Comparisons of relative mtDNA copy number between rectal cancer tissues and corresponding non-cancerous tissues of each case. (B) Comparisons of the average relative mtDNA copy number ratios between the 20 cases of rectal cancer tissues and their corresponding non-cancerous tissues. The ratio of tumor tissues was significantly higher than that of corresponding non-cancerous tissues (p<0.01, Student's t-test). (C) Comparisons of the average relative mtDNA copy number ratios of the 20 cases of rectal cancer tissues and their corresponding non-cancerous tissues between different genders. The ratio of tumor tissues exhibited no significant difference (p=0.647). In corresponding non-cancerous tissues, there was also no significant difference (p=0.798).
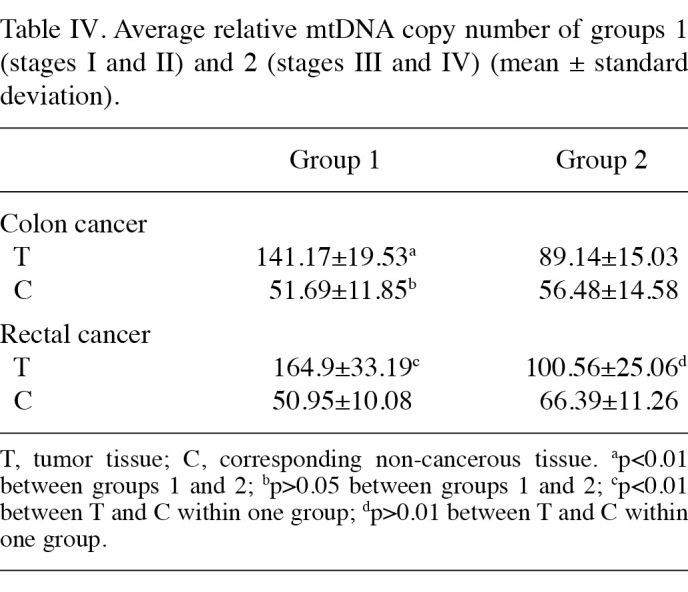
Correlation between the relative mtDNA copy number and clinicopathological stage. The correlation between the clinicopathological stage and relative mtDNA copy number of 24 colon and 20 rectal cancer cases was analysed. The average ratios of each stage of the tumor tissues and their corresponding non-cancerous tissues are shown in Table Ⅲ. From stage Ⅰ to stage Ⅳ in both colon and rectal cancer, the relative mtDNA copy number of tumor tissues was higher than that of the corresponding non-cancerous tissues in each stage, and this difference exhibited a marked significance in stages Ⅰ and Ⅱ. In stages Ⅲ and Ⅳ, this difference was not as evident (Fig. 3). Stages Ⅰ and Ⅱ were defined as group 1, and stages Ⅲ and Ⅳ as group 2. In colon cancer, a significant difference was noted between groups 1 and 2 with regards to the relative mtDNA copy number between tumor tissues (p<0.01). However, no significant difference was found between the corresponding non-cancerous tissues (p=0.386). In rectal cancer group 1, the difference of the relative mtDNA copy number of the tumor tissues between corresponding non-cancerous tissues was markedly significant (p<0.01), whereas in group 2 this difference was markedly reduced (p=0.012) (Table Ⅳ).
Table 3. Relative mtDNA copy number of different clinicopathological stages (mean ± standard deviation).
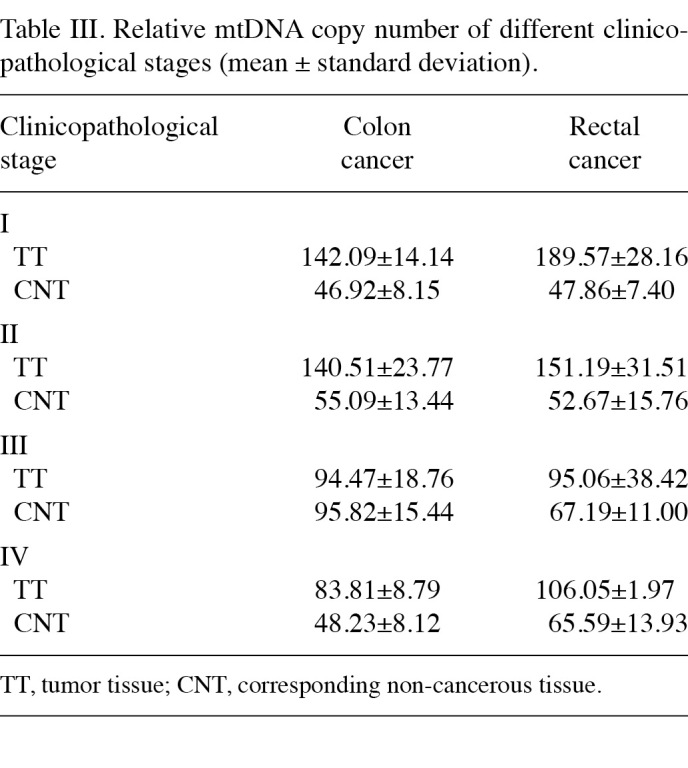
Figure 3. Average relative mtDNA copy number of (A) colon cancer and (B) rectal cancer of each clinicopathological stage. Ⅰ-Ⅳ, clinicopathological stage; T, tumor tissue; C, corresponding non-cancerous tissue.
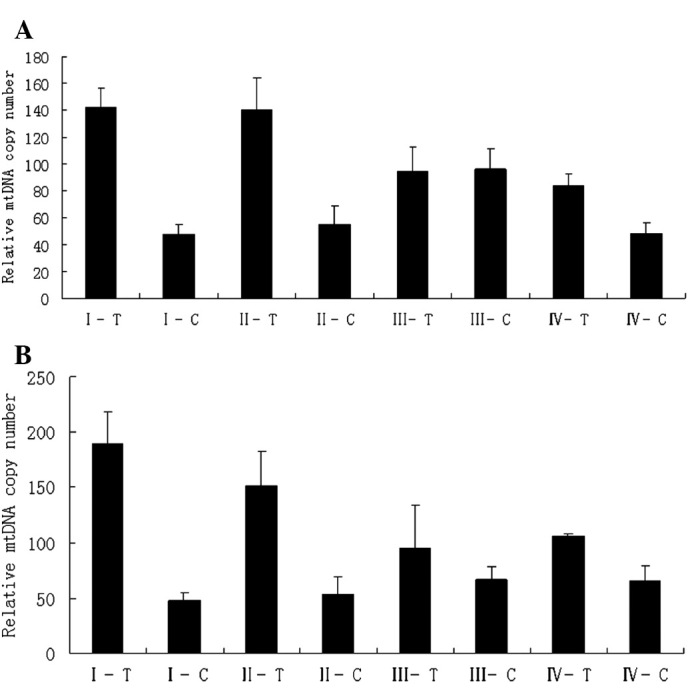
Table 4. Average relative mtDNA copy number of groups 1 (stages Ⅰ and Ⅱ) and 2 (stages Ⅲ and Ⅳ) (mean ± standard deviation).
Discussion
Human cells, with the exception of erythrocytes, contain mitochondria, which produce almost 95% energy through the procedure of oxidative phosphorylation. Unlike nuclear DNA, mtDNA is present at a consistently high level in each cell, usually 103-104 copies (13). Within various cells, tissues and organs, the copy numbers of mtDNA are different, and this difference can also occur in a given type of cell under different conditions or internal or external microenvironments, including hypoxia and steroid hormone stimulation (14-16). Numerous reports have indicated that the copy number of mtDNA changes in various types of cancer (6-11,17). In 1989, Yamamoto et al first reported the increased mitochondrial genomic expression in pre-malignant lesions of the colon in patients with pre-cancerous familial polyposis coli (18). Consequently, Modica-Napolitano et al selectively induced the damage of mtDNA using MKT-007, a derivant of rhodacyanine. Findings by these authors showed that the mtDNA copy number was significantly reduced in the cell line (19).
In our study, real-time quantitative PCR was carried out to measure the relative mtDNA copy number of the tumor tissues and their corresponding non-cancerous tissues. Figs. 1A and B, and 2A and B show the tendencies of the relative mtDNA copy number between tumor tissues and their corresponding non-cancerous tissues in colon and rectal cancer to be almost identical. On the other hand, the average ratios of tumor tissues and their corresponding non-cancerous tissues exhibited statistically significant differences in colon and rectal cancer.
It is known that cancer cells require more energy than normal cells, and it has been reported that the levels of a number of mitochondrial and nuclear gene transcripts were increased in rat hepatoma (20). At present, whether increasing mtDNA copy number is a method by which cancer cells improve the energy level during the progression of colon and rectal cancer remains to be determined. However, this method may be considered to be a significant diagnostic standard. In the study of hepatocellular carcinoma (HCC), it has been reported that different alterations in the mtDNA of male and female HCC may contribute to different clinical manifestations (21). In our study, the ratios of tumor tissues and corresponding non-cancerous tissues were compared individually, between males and females. No significant difference was observed in colon and rectal cancer cases. Thus, it appeared that there was no gender effect on mtDNA copy numbers in colon and rectal cancer (Figs. 1C and 2C).
Numerous studies have revealed that the change of mtDNA copy number is correlated with tumor progression (6,22). In this study, the relative mtDNA copy number was measured by real-time quantitative PCR and the relative mtDNA copy numbers were found to be significantly increased in colon and rectal tumor tissues compared to corresponding non-cancerous ones. The correlation between the relative mtDNA copy number and clinicopathological staging was also analysed. Fig. 3A and B shows that the differences of the relative mtDNA copy numbers between tumor tissues and the corresponding non-cancerous tissues of stages Ⅰ and Ⅱ are more evident than those of stages Ⅲ and Ⅳ in both colon and rectal cancers. To analyse these differences we divided the specimens into two groups for each cancer type. Stages Ⅰ and Ⅱ were defined as group 1, and stages Ⅲ and Ⅳ as group 2. Table Ⅳ shows that the average relative mtDNA copy numbers of tumor tissues in group 1 were higher than those of group 2, but the ratios of the corresponding non-cancerous tissues were almost identical, particularly in colon cancer. Early in 1953, Slaughter et al produced a theory known as ‘Field Cancerization’. These authors believed that corresponding non-cancerous tissues may have alterations at the molecular level without histopathological changes, and that these molecular changes may lead to cancer (23). In our experiment, we observed that in stages Ⅰ and Ⅱ the differences of mtDNA copy number between tumor tissues and corresponding non-cancerous tissues were particularly significant in both colon and rectal cancer. Moreover, the ratios of the two stages in tumor tissues were much higher than those in stages Ⅲ and Ⅳ. These results indicate that the change of mtDNA copy number may be an early event during the initiation and progression of colorectal cancer.
In conclusion, our study shows that the mtDNA copy number increases in colorectal cancer. This increase is particularly marked in clinicopathological stages Ⅰ and Ⅱ. We conjecture that the increased mtDNA copy number is an early molecular event during the initiation and progression of colorectal cancer. This observation supports the hypotheseis of measuring the mtDNA copy number as a method to help diagnose colorectal cancer in early clinicopathological stages.
Acknowledgments
The authors thank the doctors of the Gastrointestinal Surgery Department of West China Hospital for collecting the specimens. We extend thanks to Professor Zhou HongYing for her excellent technical assistance, as well as to Dr Chen Guoqing and Dr Wen Shilei for suggestions in writing the manuscript.
Contributor Information
Shi Feng, Department of Human Anatomy, Academy of Preclinical and Forensic Medicine, West China Medicine College, Sichuan University, Chengdu, Sichuan, P.R. China.
Lili Xiong, Department of Human Anatomy, Academy of Preclinical and Forensic Medicine, West China Medicine College, Sichuan University, Chengdu, Sichuan, P.R. China.
Zhenni Ji, Department of Human Anatomy, Academy of Preclinical and Forensic Medicine, West China Medicine College, Sichuan University, Chengdu, Sichuan, P.R. China.
Wei Cheng, Department of Human Anatomy, Academy of Preclinical and Forensic Medicine, West China Medicine College, Sichuan University, Chengdu, Sichuan, P.R. China.
Huijun Yang, Department of Human Anatomy, Academy of Preclinical and Forensic Medicine, West China Medicine College, Sichuan University, Chengdu, Sichuan, P.R. China.
References
- Jemal A, Siegel R, Xu J and Ward E: Cancer statistics. CA Cancer J Clin 60: 277-300, 2010. [DOI] [PubMed]
- Grievell LA: Mitochondrial DNA. Small, beautiful and essential. Nature 341: 569-571, 1989. [DOI] [PubMed]
- Lightowlers RN, Chinnery PF, Turnbull DM and Howell N: Mammalian mitochondrial genetics: heredity, heteroplasty and disease. Trends Genet 13: 450, 1997. [DOI] [PubMed]
- Barrientos A, Casademont J, Cardellach F, Estivill X, Urbano‑Marquez A and Nunes V: Reduced steady-state levels of mitochondrial RNA and increased mitochondrial DNA amount in human brain with aging. Mol Brain Res 52: 284-289, 1997. [DOI] [PubMed]
- Lee HC, Yin PH, Lu CY, Chi CW and Wei YH: Increase of mitochondria and mitochondrial DNA in response to oxidative stress in human cells. Biochem J 348: 425-432, 2000. [PMC free article] [PubMed]
- Yamada S, Nomoto S, Fujii T, Kaneko T, Takeda S, Inoue S, Kanazumi N and Nakao A: Correlation between copy number of mitochondrial DNA and clinico-pathologic parameters of hepatocellular carcinoma. EJSO 32: 303-307, 2006. [DOI] [PubMed]
- Xing J, Chen M, Wood CG, Lin J, Spitz MR, Ma J, Amos CI, Shields PG, Benowitz NL, Gu J, de Andrade M, Swan GE and Wu X: Mitochondrial DNA content: its genetic heritability and association with renal cell carcinoma. J Natl Cancer Inst 100: 1104-1112, 2008. [DOI] [PMC free article] [PubMed]
- Yu M, Zhou Y, Shi Y, Ning L, Yang Y, Wei X, Zhang N, Hao X and Niu R: Reduced mitochondrial DNA copy number is correlated with tumor progression and prognosis in Chinese breast cancer patients. IUBMB Life 59: 450-457, 2007. [DOI] [PubMed]
- Kim MM, Clinger JD, Masayesva BG, Ha PK, Zahurak ML, Westra WH and Califano JA: Mitochondrial DNA quantity increases with histopathologic grade in premalignant and malignant head and neck lesions. Clin Cancer Res 10: 8512‑8515, 2004. [DOI] [PubMed]
- Lin CS, Chang SC, Wang LS, Chou TY, Hsu WH, Wu YC and Wei YH: The role of mitochondrial DNA alterations in esophageal squamous cell carcinomas. J Thorac Cardiovasc Surg 139: 189-197, 2010. [DOI] [PubMed]
- Wang Y, Liu VW, Xue WC, Cheung AN and Ngan HY: Association of decreased mitochondrial DNA content with ovarian cancer progression. Br J Cancer 95: 1087-1091, 2006. [DOI] [PMC free article] [PubMed]
- Obtułowicz T, Winczura A, Speina E, et al: Aberrant repair of etheno-DNA adducts in leukocytes and colon tissue of colon cancer patients. Free Radical Biology and Medicine 9: 1064‑1071, 2010. [DOI] [PubMed]
- Clay Montier LL, Deng JJ and Bai Y: Number matters: control of mammalian mitochondrial DNA copy number. Genet Genomics 36: 125-131, 2009. [DOI] [PMC free article] [PubMed]
- Weitzel JM, Iwen KA and Seitz HJ: Regulation of mitochondrial biogenesis by thyroid hormone. Exp Physiol 88: 121-128, 2003. [DOI] [PubMed]
- Robin ED and Wong R: Mitochondrial DNA molecules and virtual number of mitochondria per cell in mammalian cells. J Cell Physiol 136: 507-513, 1988. [DOI] [PubMed]
- Hoppeler H, Vogt M, Weibel ER and Fulk M: Response of skeletal muscle mitochondria to hypoxia. Exp Physiol 88: 109-119, 2003. [DOI] [PubMed]
- Tseng LM, Yin PH, Chi CW, Lee LM, Wei YH and Lee HC: Mitochondrial DNA mutations and mitochondrial DNA depletion in breast cancer. Genes Chromosomes Cancer 45: 629-638, 2006. [DOI] [PubMed]
- Yamamoto A, Horai S and Yuasa Y: Increased level of mitochondria gene expression in polyps of familial polyposis coli patients. Biochem Biophys Res Commun 159: 1100-1106, 1989. [DOI] [PubMed]
- Modica-Napolitano JS, Koya K, Weisberg E, Brunelli BT, Li Y and Chen LB: Selective damage to carcinoma mitochondria by the rhodacyanine MKT-077. Cancer Res 56: 544, 1996. [PubMed]
- Luciakova K and Kuzela S: Increased steady-state levels of several mitochondrial and nuclear gene transcripts in rat hepatoma with a low content of mitochondria. Eur J Biochem 205: 1187, 1992. [DOI] [PubMed]
- Yin PH, Lee HC, Chau GY, Wu YT, Li SH, Lui WY, Wei YH, Liu TY and Chi CW: Alteration of the copy number and deletion of mitochondrial DNA in human hepatocellular carcinoma. Br J Cancer 90: 1-7, 2004. [DOI] [PMC free article] [PubMed]
- Park SY, Shin MG, Kim HR, Oh JY, Kim SH, Shin JH, Cho YB, Suh SP and Ryang DW: Alteration of mitochondrial DNA sequence and copy number in nasal polyp tissue. Mitochondrion 9: 318-325, 2009. [DOI] [PubMed]
- Slaughter DP, Southwick HW and Smejkal W: Field cancerization in oral stratified squamous epithelium; clinical implications of multicentric origin. Cancer 6: 963-968, 1953. [DOI] [PubMed]