Abstract
The analysis of molecular markers in the biological field has been proposed as a useful tool for cancer diagnosis. MicroRNAs (miRNAs) may regulate diverse biological processes and play a significant role in tumorigenesis. The potential use of blood-based miRNAs as a biomarker of cancer and as a target for therapeutics is promising. The purpose of this study was to determine whether aberrant miRNA expression can be used as a molecular marker in peripheral blood for the diagnosis of lung cancer. Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) was performed to analyze the expression levels of mature microRNA‑21 (miR‑21). Blood samples from 20 lung cancer patients and 10 healthy individuals were collected. The data were compared for the diagnosis of lung cancer. The results demonstrated that miR-21 was present in the peripheral blood in a markedly stable form and could be detected by real-time PCR sensitively and specifically. A significant difference was observed between the lung cancer cases and controls regarding miR-21 levels in peripheral blood (1947.26±930.56 pg/ml vs. 943.42±314.12 pg/ml, P=0.005). Furthermore, the over-expression of miR-21 showed a highly discriminative receiver operating characteristic (ROC) curve profile, clearly distinguishing cancer patients from cancer-free subjects with an area under the ROC curve (AUROC) of 0.912±0.045. The detection of miR‑21 expression yielded 78.80% sensitivity and 100.00% specificity in the diagnosis of lung cancer. These findings indicate that in peripheral blood miR‑21 may serve as a potential molecular marker for lung cancer and provide a new approach in the diagnosis of lung cancer.
Introduction
Lung cancer is the leading cause of cancer death in China and worldwide (1). The five-year survival rate of lung cancer patients is 13%. A crucial reason for this is that lung cancer has often already metastasized at the time of detection, rendering the long-term prognosis as poor (2). Therefore, early diagnosis of lung cancer is a realistic approach in reducing mortality associated with lung cancer. Although advanced technology, such as computed tomography (CT), positron emission tomography (PET) and X-ray appear to be promising methods of detection, technology is not capable of precisely realizing the early detection of lung cancer. The development of diagnostic tools is necessary for the early detection of lung cancer (3,4).
MicroRNAs (miRNAs) are a class of small RNAs (~21-24 nt in length) that are capable of post-transcriptionally regulating hundreds of target genes, thereby controlling a wide variety of biological processes, including cell growth, proliferation, differentiation and apoptosis (5). Furthermore, miRNA expression is involved in cancer development and progression as a tumor suppressor or oncogene (6,7). Aberrant miRNA expression as a biomarker provides a new approach to improving the accuracy of the early diagnosis of lung cancer. However, miRNA expression pattern biomarkers for disease management, personalized therapy, diagnosis and prognosis remain in their infancy (8). The expression of mature microRNA‑21 (miR‑21) has been and to play a role in the underlying pathogenesis of lung cancer in human tissue and serum (9,10). However, the feasibility of examining aberrant miR‑21 expression levels in peripheral blood for a non-invasive diagnosis of lung cancer has yet to be investigated.
The objective of this study was to determine whether an altered miR‑21 expression detected in peripheral blood could be a potential biomarker for the diagnosis of lung cancer. Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) was performed on lung cancer patients and healthy controls in the Chinese population.
Materials and methods
Patients. A total of 20 patients diagnosed with lung cancer and 10 controls without cancer were enrolled in the present study. All 20 patients were recruited from the Hunan Provincial Tumor Hospital in Changsha, China. The research study was approved by an institutional review board. Of the 20 tumor cases, 4 were stage I, 4 were stage II, 8 were stage III and 4 were stage IV. Histologically, 8 of the 20 tumors were squamous cell carcinomas (SCC), 6 were adenocarcinomas and 6 were other types of carcinomas.
RNA extraction. RNA was extracted from peripheral blood using the BioTeKe miRNA extraction kit (BioTeKe Corporation, China), according to the manufacturer's instructions. The qualification and quantification of RNA were assessed using a Photodiode Array Bio-spectrophotometer (Hutchinson Technology Inc, Hutchinson, MN, USA) and an Electrophoresis Bioanalyzer (Agilent Technologies, Foster City, CA, USA).
Reverse transcription (RT) with miRNA-specific stem‑loop primer. RNA was applied for RT using an AMV first strand cDNA synthesis kit (Sangon Co., Shanghai, China), according to the manufacturer's instructions. The reaction includes 20 nM stem-loop RT primer, 1X RT buffer, 0.2 mM each of the dNTPs, 1 U/µl RNase inhibitor and 1 U/µl AMV reverse transcriptase in a total volume of 20 µl. The specific stem‑loop reverse transcription primer of mature miR‑21 was 5'-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCAACA-3' (11).
Quantification of mature miRNAs by real-time PCR. Stem‑loop qRT-PCR analysis for the detection of miR-21 levels was performed as previously described (12). The primers for miR‑21 were: forward, 5'-GCCCGCTAGCTTATCAGACTGATG-3' and reverse 5'-GTGCAGGGTCCGAGGT-3'. Real‑time PCR was performed to measure expression levels of target miRNAs using a SYBR premixture kit (BioTeKe Corporation) on a Bio-Rad IQ2 real-time PCR detection system (Bio-Rad, Hercules, CA, USA). The 25 µl PCR reaction included RT product, 2X SYBR real-time premixture (BioTeKe Corporation) and the corresponding primers. The reactions were incubated in a 94-well plate at 95˚C for 7 min, followed by 40 cycles of 95˚C for 15 sec, 60˚C for 30 sec and 72˚C for 30 sec. The threshold cycle (Ct) was defined as the fractional cycle number at which the fluorescence passed the fixed threshold. The expression of target miR-21 was normalized by miRNA mimics (RiboBio, Guangzhou, China) (3). No miRNA was used as a negative control and all tests were performed in triplicate.
Statistical analysis. Statistical analyses were performed using the statistical software SPSS 18.0. The Mann-Whitney U-test and the Kruskal-Wallis test were used to evaluate the difference in miRNA expression between cancer cases and healthy controls. The receiver operating characteristic (ROC) curve was used to analyze the specificity and sensitivity, and the area under the ROC curve (AUROC) was used to represent an overall summary of diagnostic accuracy (8). All tests of statistical significance were two‑sided. P<0.05 was considered to be statistically significant. Values were shown as the mean ± standard deviation (SD).
Results
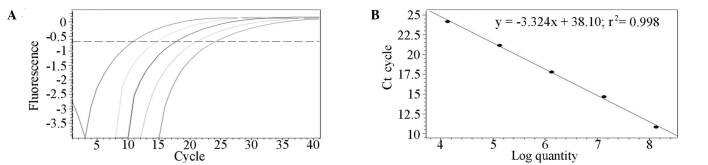
miR-21 expression profile. RNA was extracted from peripheral blood and the mean ratio of absorbance at 260 and 280 nm was 1.89 (range 1.71-2.13±0.16). The quality of RNA samples was sufficient for qRT-PCR under identical conditions. Synthesized miR-21 mimics were diluted by 10 orders of magnitude in DEPC‑treated H2O to generate a standard curve, the measurement range of which was from 1.34 pg/ml to 134 ng/ml.
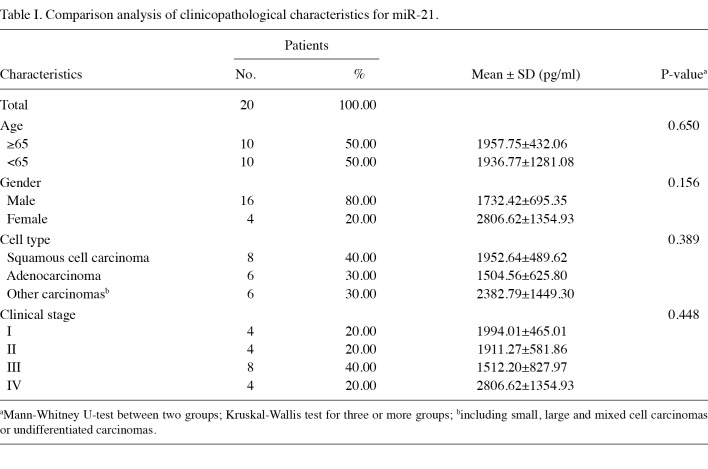
The qRT-PCR assay revealed excellent linearity between the log of target input and the Ct value, demonstrating that the assay had a strong range of at least 4 orders of magnitude (R2=0.9980) (Fig. 1). The concentration of miR-21 in all of the samples can be derived from the Ct in the standard curve. miR-21 expression levels in peripheral blood were significantly higher in cancer patients (1947.26±930.56 pg/ml) than in cancer-free individuals (943.42±314.12 pg/ml) (Fig. 2).
Figure 1. Dynamic range and sensitivity of the miR-21 qRT-PCR assay in peripheral blood. (A) Amplification plot of synthetic miR-21 over four orders of magnitude. (B) Standard curve.
Figure 2. The absolute expression of miR-21 in peripheral blood between normal and cancer specimens (bars 1-20 indicate lung cancer patients, bars 21-30 indicate healthy individuals).
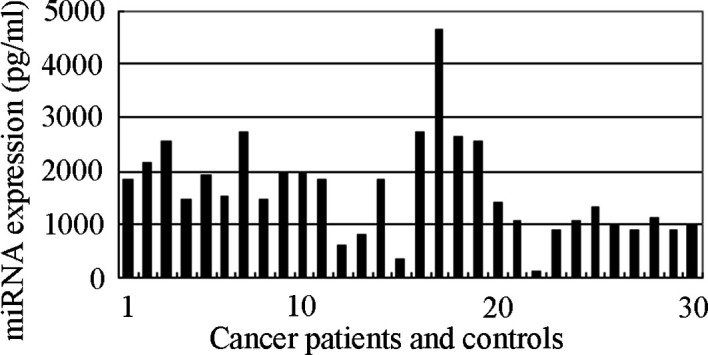
Sensitivity, specificity and dynamic range of miRNA quantification in peripheral blood. To determine the dynamic range, sensitivity and specificity of miRNA quantification in peripheral blood by qRT-PCR assay, miR-21 mimics (RiboBio) were synthesized and diluted by 10 orders of magnitude to generate a standard curve. The ROC curve analysis was carried out on cancer patients and cancer-free controls using SPSS 18.0. The ROC curve is a plot of the diagnostic test's sensitivity, or true positive rate vs. 1-specificity or the false positive rate (13). The AUROC is an overall summary of diagnostic accuracy (13,14). Using this approach, the AUROC identified optimal sensitivity and specificity levels, which can be used to distinguish normal subjects from cancer patients. The ROC curve with AUROC for miR-21 expression in the blood of lung cancer and control cases is shown in Fig. 3. The results revealed a highly discriminative ROC curve profile that distinguished cancer patients from cancer-free subjects with an AUROC of 0.912±0.045. The detection of miR‑21 expression yielded a 78.80% sensitivity and 100.00% specificity in the diagnosis of lung cancer at optimum conditions.
Figure 3. Receiver operating characteristic (ROC) curve analysis of the expression of miR-21 in blood ROC curve with corresponding area under the ROC curve (AUROC) for each gene expression conveys its accuracy in distinguishing cancer-free subjects from cancer patients in terms of sensitivity and specificity.
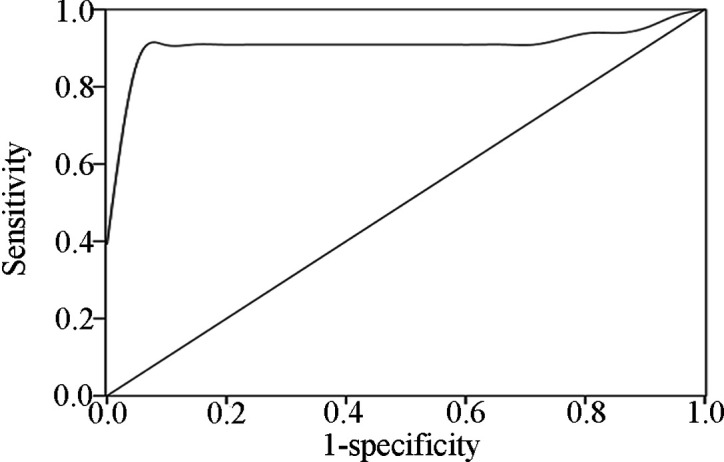
Comparison analysis of altered miR-21 expression levels and their clinicopathological characteristics. miRNA-specific qRT-PCR was used to test miR-21 expression levels in the peripheral blood samples of the 20 lung cancer patients with varying clinical stages and histological types, and the 10 healthy controls. The expression level of miR‑21 differed significantly between lung cancer patients and healthy controls (P=0.005). In the present study, miR-21 was up-regulated by 85% (17/20). No association was found between the expression levels of miR‑21 and clinicopathological characteristics (clinical stage, age, gender and cell type) (Table I).
Table 1. Comparison analysis of clinicopathological characteristics for miR-21.
Discussion
miR-21 is located in chromosome 17q23.1, which is the site of translocation breakpoints or amplifications in all types of cancer (15,16). It may play an oncogenic role in tumorigenesis by specifically regulating target and tumor suppressor genes. Chan et al observed that miR-21 was highly overexpressed in glioblastoma tumors compared with normal tissues (15). Xie et al reported that miR-21 was significantly elevated in the sputum of patients with non-small cell lung cancer (NSCLC) and was therefore a potential non-invasive molecular marker for NSCLC (17). Moreover, Yu et al demonstrated that miR-21 may be used as a non-invasive and cost-effective diagnostic tool for early lung adenocarcinoma (8). Gao et al reported the potential of miR-21 as a novel diagnostic or prognostic biomarker for NSCLC in tissue samples (18). Various scientific studies have reported that the expression of miR-21 plays a significant role in the development of lung cancer (19,20). In the present study, miR-21 expression profiles were detected by qRT-PCR assay in blood with lung cancer. The expression level of miR-21 was 1947.26 pg/ml in lung cancer patients and 943.42 pg/ml in controls. A significant difference in the expression levels of miR-21 was detected between lung cancer patients at diagnosis and controls (P=0.005). Subsequently, the result was compared to previous studies. The assessment of miR-21 expression may serve as a potential diagnostic biomarker for lung cancer. Furthermore, all 4 stage I lung cancer patients were identified by measuring miR‑21 expression in peripheral blood compared with controls, indicating that miR‑21 may serve as a biomarker for the early detection of lung cancer (8,17).
No association was observed between the overexpression of miR-21 in peripheral blood of lung cancer patients and the various histological types and clinical stages, suggesting that the genetic change is not specific to histologic type (P=0.389) and clinical stage (P=0.448). Similar results were obtained for age and gender. These results were consistent with other scientific studies (8,17,19). However, the small sample size used in this study caused the broad confidence intervals observed in the regression model. Accordingly, further study is necessary to evaluate the association between miR-21 expression and clinical features in a large sample size, to determine whether or not such a correlation exists.
Although the detection of miR-21 expression in the peri-pheral blood appears to be more sensitive and specific than in sputum in the detection of lung cancer (8,17), the 78.80% sensitivity rate is not sufficiently efficient for routine clinical application. It may be that a single miRNA is not sufficient to identify a tumor, thus more miRNAs should be used to detect lung carcinoma. Therefore, panels of tumor-specific miRNA biomarkers in peripheral blood should be developed as tools for the highly sensitive and specific diagnosis of lung cancer. Once confirmed, the peripheral blood miRNA panel would improve the detection rate for lung cancer, as it is more difficult to realize using conventional techniques.
In conclusion, although a small sample size was used in this study, the detection of miR-21 expression in peripheral blood distinguished patients with lung cancer from controls with a high specificity in the Chinese population. miR-21 levels in peripheral blood may serve as a potential molecular marker for lung cancer and provide a new approach for the diagnosis of lung cancer.
Acknowledgments
This study was supported by a research grant (no. 60871007) from the National Natural Science Foundation of China and research grants (nos. 10JJ2049 and 10JJ3083) from the Natural Science Foundation of Hunan Province of China.
Contributor Information
Yanzhao Li, Key Laboratory of Green Packaging and Application of Biological Nanotechnology of Hunan Province, Hunan University of Technology, Zhuzhou, Hunan 412007, P.R. China.
Wen Li, Key Laboratory of Green Packaging and Application of Biological Nanotechnology of Hunan Province, Hunan University of Technology, Zhuzhou, Hunan 412007, P.R. China.
Quchang Ouyang, Department of Pathology, Hunan Tumor Hospital, Changsha, Hunan 410113, P.R.China.
Shunqin Hu, Key Laboratory of Green Packaging and Application of Biological Nanotechnology of Hunan Province, Hunan University of Technology, Zhuzhou, Hunan 412007, P.R. China.
Jianxin Tang, Key Laboratory of Green Packaging and Application of Biological Nanotechnology of Hunan Province, Hunan University of Technology, Zhuzhou, Hunan 412007, P.R. China.
References
- Greenlee RT, Hill-Harmon MB, Murray T and Thun M: Cancer statistics. CA Cancer J Clin 51: 15-36, 2001. [DOI] [PubMed]
- Li W, Deng J, Jiang P and Tang J: Association of 5'-CpG island hypermethylation of the FHIT gene with lung cancer in southern-central Chinese population. Cancer Biol Ther 10: 997‑1000, 2010. [DOI] [PubMed]
- Mitchell PS, Parkin RK, Kroh EM, et al: Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA 105: 10513-10518, 2008. [DOI] [PMC free article] [PubMed]
- Gilad S, Meiri E, Yogev Y, et al: Serum microRNAs are promising novel biomarkers. PLoS One 3: e3148, 2008. [DOI] [PMC free article] [PubMed]
- Esquela-Kerscher A and Slack FJ: Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer 6: 259-269, 2006. [DOI] [PubMed]
- Yanaihara N, Caplen N, Bowman E, et al: Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 9: 189-198, 2006. [DOI] [PubMed]
- Ortholan C, Puissegur MP, Ilie M, Barbry P, Mari B and Hofman P: MicroRNAs and lung cancer: new oncogenes and tumor suppressors, new prognostic factors and potential therapeutic targets. Curr Med Chem 16: 1047-1061, 2009. [DOI] [PubMed]
- Yu L, Todd NW, Xing L, et al: Early detection of lung adenocarcinoma in sputum by a panel of microRNA markers. Int J Cancer 127: 2870-2878, 2010. [DOI] [PMC free article] [PubMed]
- Manikandan J, Aarthi JJ, Kumar SD and Pushparaj PN: Oncomirs: the potential role of non-coding microRNAs in understanding cancer. Bioinformation 2: 330-334, 2008. [DOI] [PMC free article] [PubMed]
- Pillai RS: MicroRNA function: Multiple mechanisms for a tiny RNA? RNA 11: 1753-1761, 2005. [DOI] [PMC free article] [PubMed]
- Chen C, Ridzon DA, Broomer AJ, et al: Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 33: e179, 2005. [DOI] [PMC free article] [PubMed]
- Gao W, Shen H, Liu L, Xu J, Xu J and Shu Y: MiR‑21 overexpression in human primary squamous cell lung carcinoma is associated with poor patient prognosis. J Cancer Res Clin Oncol 137: 557-566, 2011. [DOI] [PMC free article] [PubMed]
- Dodd LE and Pepe MS: Partial AUC estimation and regression. Biometrics 59: 614-623, 2003. [DOI] [PubMed]
- Zou KH, O'Malley AJ and Mauri L: Receiver-operating characteristic analysis for evaluating diagnostic tests and predictive models. Circulation 115: 654-657, 2007. [DOI] [PubMed]
- Chan JA, Krichevsky AM and Kosik KS: MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res 65: 6029‑6033, 2005. [DOI] [PubMed]
- Selcuklu SD, Donoghue MT and Spillane C: miR-21 as a key regulator of oncogenic processes. Biochem Soc Trans 37: 918‑925, 2009. [DOI] [PubMed]
- Xie Y, Todd NW, Liu Z, et al: Altered miRNA expression in sputum for diagnosis of non-small cell lung cancer. Lung Cancer 67: 170-176, 2010. [DOI] [PMC free article] [PubMed]
- Gao W, Yu Y, Cao H, et al: Deregulated expression of miR-21, miR-143 and miR-181a in non small cell lung cancer is related to clinicopathologic characteristics or patient prognosis. Biomed Pharmacother 64: 399-408, 2010. [DOI] [PubMed]
- Seike M, Goto A, Okano T, et al: MiR-21 is an EGFR-regulated anti-apoptotic factor in lung cancer in never-smokers. Proc Natl Acad Sci USA 106: 12085-12090, 2009. [DOI] [PMC free article] [PubMed]
- Rosell R, Wei J and Taron M: Circulating microRNA signatures of tumor-derived exosomes for early diagnosis of non-small-cell lung cancer. Clin Lung Cancer 10: 8-9, 2009. [DOI] [PubMed]