Abstract
Topoisomerase I (TOP-I) mutations have been shown to be correlated to irinotecan resistance in vitro. However, the prevalence of TOP-I germline mutations has yet to be systematically elucidated. On the other hand, polymorphisms of UGT1A1 have been shown to be associated with CPT-11 toxicity in clinical situations. The primary aim of this study was to investigate the prevalence of mutations in the TOP-I exons associated with CPT-11 resistance, including untreated cancer tissue. A secondary aim was to confirm the less frequent UGT1A1*28 and more frequent UGT1A1*6 in individuals of Asian descent compared to Caucasians and individuals of African descent. The prevalence of 5 reported TOP-I mutations in exons was investigated in volunteers (n=236) using DNA sequencing of the PCR products. The prevalence of TOP-I mutations in untreated lung cancer tissues (n=16) was also investigated. Additionally, 3 UGT1A1 polymorphisms, UGT1A1*6, *27 and *28, were investigated in volunteers (n=126). There were no mutations of TOP-I in any of the 236 subjects or in the untreated lung tissues. Among 128 subjects, the distribution of homozygous polymorphisms of UGT1A1 was: UGT1A1*28 in 3 (2.4%) and UGT1A1*6 in 4 (3.2%) subjects, and co-occurrence of heterozygous polymorphisms for both UGT1A1*6 and UGT1A1*28 in 4 (3.2%) subjects, and for UGT1A1*27 and UGT1A1*28 in 1 subject (0.8%). The Hardy-Weinberg deviation test showed there was no significant deviation from the equilibrium, and the association analysis indicated no significant linkage between UGT1A1*6 and UGT1A1*28. In conclusion, TOP-I genetic mutations correlated to CPT-11 resistance were not detected in any of the subjects and untreated lung cancer tissues. Less frequent UGT1A1*28 and more frequent UGT1A1*6 were confirmed in East Asian individuals compared to Caucasians and individuals of African descent. Linkage disequilibrium was not detected between UGT1A1*6 and UGT1A1*28.
Introduction
Irinotecan hydrochloride (CPT-11), a water-soluble semi-synthetic derivative of camptothecin, has been shown to exert marked antitumor activity (1). It is an inactive prodrug, and its major metabolite SN-38 is a potent topoisomerase I (TOP-I) inhibitor. SN-38 stabilizes covalent TOP-I-DNA complexes, causing DNA strand breaks. Several point mutations of TOP-I were identified as being associated with resistance to CPT-11 (2-4), and structural models were introduced to explain how the mutations of TOP-I hinder the docking of camptothecin derivatives in the ternary complex of TOP-I-DNA (5-7).
The cultured cells with the TOP-I mutation showed no obvious or only minor defects in cell function and proliferation (2-4). Consequently, such mutations may be innocent or not fatal to the cells. Therefore, it is reasonable to consider that the mutations may even occur in germlines.
CPT-11 exhibits inter-individual variations in terms of both pharmacokinetic and pharmacodynamic behavior (8). CPT-11 is hydrolyzed to yield active SN-38 (9) and detoxified via glucuronidation of SN-38 by uridine diphosphate glucuronosyltransferase (UGTs) to yield its β-glucuronides, SN-38G (10). UGT1A1 is the main isoform of UGTs involved in the formation of SN-38G. Genetic polymorphisms of the UGT1A1 gene were revealed to explain the variability of CPT-11-related toxicity among patients, particularly UGT1A1*28 [(TA)7TAA], the existence of which is known to be predictive of CPT-11-induced neutropenia (11). In addition, the UGT1A1*6 and *27 alleles, two variants in exon 1 of the UGT1A1 gene, are found mainly among individuals of Asian descent, and have also been indicated to affect enzyme function (12,13). The associated phenotype of UGT1A1*28, UGT1A1*6, or UGT1A1*27 is the Gilbert syndrome (14), and a strong association between 5 single nucleotide polymorphisms (SNPs; 4 introns, and 1 exon: UGT1A1*6) within the UGT1A1 gene and serum total bilirubin levels was noted in Japanese subjects (15).
Although the TOP-I mutation-related CPT-11 resistance has been elucidated, the prevalence of TOP-I germline mutation has yet to be investigated. The prevalence of TOP-I germline mutation requires investigation to elucidate the group of TOP-I-resistant patients prior to cancer chemotherapy. On the other hand, polymorphisms of UGT1A1 have been revealed to be associated with CPT-11 toxicity in clinical situations. Additionally, less frequent (~50%) UGT1A1*28 and frequent UGT1A1*6 have been reported in individuals of Asian descent compared to Caucasians and individuals of African descent (16-21). The prevalence of UGT1A1 polymorphisms in Asian individuals requires further confirmation studies to avoid toxicity-related death in cancer patients.
The primary aim of this study was to investigate the prevalence of mutation in the TOP-I exons associated with CPT-11 resistance, including untreated cancer tissue. A secondary aim was to confirm the frequency of polymorphisms in the UGT1A1 genes related to CPT-11 toxicity in a Japanese population.
Materials and methods
Materials. The study protocol was approved by the Ethics Committee of Kitasato University School of Medicine, Japan (B03-07 for healthy volunteers, B03-28 and G03-04 for patients with lung cancer). Blood samples of healthy volunteers (n=236 for TOP-I mutations and n=126 for UGT1A1 polymorphism) as well as lung cancer tissue specimens from untreated patients (n=16) were examined. The lung cancer tissue samples comprising 6 resected tissues and 10 biopsy specimens constituted of 8 cases of small cell carcinoma, 7 of adenocarcinoma and 1 of squamous cell carcinoma. All of the subjects provided written informed consent prior to their being enrolled in this study.
DNA extraction and polymerase chain reaction. DNA was extracted from 200 µl whole blood using the QIAamp® DNA blood mini kit (Qiagen, Valencia, CA, USA). Paraffin‑embedded lung cancer tissues obtained from lung cancer patients were stained with hematoxylin and eosin, and the DNA was extracted from LASER-captured microdissected tumor tissue (AS LMD, Leica, Tokyo, Japan) using the QIAamp® DNA Micro kit (Qiagen).
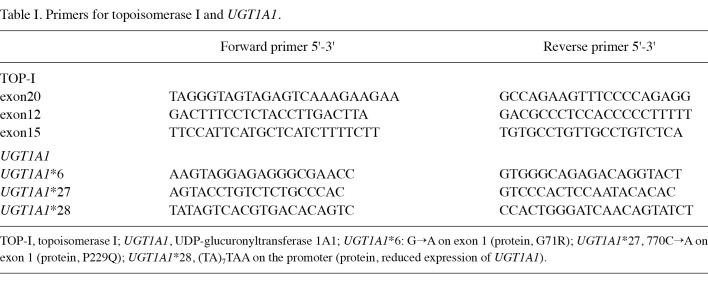
Polymerase chain reaction (PCR) was performed to amplify the targeted regions of TOP-I (or UGT1A1) with a thermal cycler (Takara Thermal Cycler SP, Takara, Shiga, Japan; or PTC-200 DNA Engine, MJ Research, Watertown, MA, USA) using each primer (Table I), HotStarTaq DNA polymerase and the Q-Solution Kit (Qiagen) in accordance with the manufacturer's protocol, with the exception of the case of UGT1A1*28 (see below). The PCR products were identified by gel electrophoresis. After excising the DNA fragment band from the agarose gel, high final concentrations of DNA were extracted using a MinElute® gel extraction kit (Qiagen).
Table 1. Primers for topoisomerase I and UGT1A1.
Genetic analyses of TOP-I. Five TOP-I mutations correlated to CPT-11 resistance were previously reported (2-7), as follows [see Redinbo et al (6) for the numbering of each region]: region 1, exon 20 (541-558); region 2-1, exon 12 (501-530); region 2-2, exon 15 (186-194); region 3, exon 20 (577-579); and region 4, exon 15 (99-101) (Fig. 1A and B). The 5 mutation-hotspot regions of TOP-I were amplified using the relevant primer pairs (Table I). Direct sequencing of the PCR products was performed using ABI PRISM 3100 Genetic Analyzer (Life Technologies Corporation, Carlsbad, CA, USA). Sequencing reactions were performed in both the forward and reverse directions.
Figure 1A. TOP I mutation corrrelated to CPT-11 resistance. NCBI reference sequence: NM_003286.1. (A) Protein sequence; Y, active catalytic tyrosine site; shaded location with underline: mutation site related to TOP I resistance; 1, region 1; 2-1 and 2-1, region 2; 3, region 3; 4, region 4 [see Redinbo et al (6) for the numbering of each region].
Figure 1B. (B) cDNA sequence; *tat, active catalytic tyrosine site. Shaded location with bold character(s) and underline: mutation site correlated to TOP I resistance.
Genetic analyses of UGT1A1. The following variant sequences were investigated: a 2-extra-nucleotide insertion (TA) within the TATA box in the promoter, resulting in the sequence (TA)7TAA (-39 to -53, UGT1A1*28: rs8175347); transition (+295 from the initial site of transcription, G to A) at codon 71 in exon 1 that is associated with a substitution of glycine to arginine (G71R, UGT1A1*6: rs4148323); and a transversion (+770, C to A) at codon 229 in exon 1 that alters proline to glutamine (P229Q, UGT1A1*27: rs35350960). The variant longer sequence of UGT1A1*28 was distinguished from the wild-type sequence using poly-acrylamide gel electrophoresis (PAGE) with a DNA-sequencer (Long-Read Tower™, Visible Genetics, Suwanee, GA, USA) following amplification of the targeted sequence using PCR with Pfu DNA Polymerase (native) (Fermentas Life Sciences, Crt Burlington, Ontario, Canada). The primers (Table II) were designed to amplify a 216-bp segment of UGT1A1*28 variant-type sequence as compared with the 214-bp segment of the wild-type sequence. The separation ability was previously verified using guaranteed wild-type and variant-type genome DNA obtained from Daiichi Pure Chemicals (now Sekisui Medical Co. Ltd., Japan). For analysis of UGT1A1*6 and UGT1A1*27, direct sequencing of the PCR amplification product obtained using specific primers (Table I) was performed using the ABI PRISM 3100 genetic analyzer.
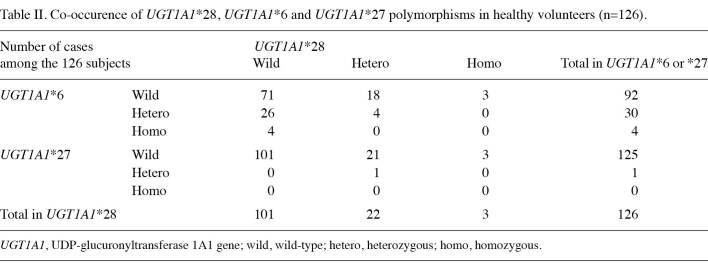
Table 2. Co-occurence of UGT1A1*28, UGT1A1*6 and UGT1A1*27 polymorphisms in healthy volunteers (n=126).
Results
Mutations of TOP-I related to CPT-11 resistance. Since no TOP-I mutations were observed in any of the 126 subjects, an additional 110 healthy volunteers, as well as untreated lung cancer tissue specimens in patients (n=16), were investigated. The results revealed that no genetic mutations correlated to CPT11-resistance in exons 12, 15, 16 or 20 of TOP-I.
UGT1A1 genetic polymorphism associated with CPT-11 toxicity. Homozygous polymorphisms of UGT1A1 were detected in 126 subjects, and were distributed as follows: UGT1A1*28 in 3 (2.4%) subjects and UGT1A1*6 in 4 (3.2%) subjects (Table II), and co-occurrence of heterozygous polymorphisms for both UGT1A1*6 and UGT1A1*28 in 4 subjects (3.2%), and for both UGT1A1*27 and UGT1A1*28 in 1 (0.8%) subject (Table III). The frequency of the UGT1A1*28 variant allele was found to be 19.8%. In general, either homozygous or heterozygous polymorphisms of UGT1A1*6 were detected in 30 (23.8%) subjects, and either homozygous or heterozygous polymorphisms of UGT1A1*6, *27 or *28 were detected in 55 (43.7%) subjects.
The Hardy-Weinberg deviation test showed that UGT1A1*6 (p=0.43), *27 (p=0.96) and *28 (p=0.19) was not significantly deviated from the equilibrium. The linkage analysis revealed that normalized linkage disequilibrium coefficient D' for UGT1A1*6 and UGT1A1*28 was 0.05. However, the linkage disequilibrium was not detected (p= 0.64).
Discussion
This is the first study to exmaine the frequency of TOP-I mutation associated with CPT-11 resistance in healthy subjects and in untreated lung cancer tissue specimens. Although in vitro reports have demonstrated that CPT-11-resistant cancer cell lines exposed to CPT-11 possessed TOP-I mutations with no obvious or only minor defects in cell function, these changes were undetectable in healthy subjects and in untreated lung cancer tissue as mutation. Therefore, in the initial chemotherapy with CPT-11, the resistance-related TOP-I mutation is unlikely to occur.
Tsurutani et al (22) examined 16 samples obtained from 8 CPT-11-treated patients with lung cancer, and detected 2 types of TOP-I mutations in exon 21 in 1 tumor specimen. In a human colon cancer cell line (HCT-15) (23), one exonic mutation was detected in a heterozygous state in exon 19. Since the mutations coded on exon 19 and 21 have not been reported in previous in vitro CPT-11-resistant cancer cell lines, these regions were not examined in our study. However, the results indicated that the development of some acquired CPT-11 mutations was possible in patients in the course of treatment with CPT-11.
According to the SNP database (http://www.ncbi.nlm.nih.gov/SNP/), missense mutations in exons 4 (1 locus), 9 (1 locus), 15 (3 loci), and 21 (1 locus) are currently registered. These SNPs are not correlated to CPT-11 resistance, although a haplotype-tagging SNP in the intervening sequence region has been found to be associated with toxicity (grade 3/4 neutropenia) in patients treated with CPT-11 (17).
Determination of the UGT1A1 genotypes is clinically significant for the prediction of CPT-11-related severe toxicity (11). Individuals who have at least 1 variant (heterozygous) allele for the UGT1A1*28 may exhibit reduced elimination of SN-38 and increased probability of development of dose-limiting neutropenia (11).
The frequency of the UGT1A1*28 variant allele has been reported to be 30-45% in Caucasian, African and Indian populations, which is approximately twice that of the 10-20% reported in East Asian populations (17-22), as confirmed by the frequency of this study (19.8%). On the other hand, UGT1A1*6 has been detected in 16-40% of Asian individuals, as confirmed by the frequency of 23.8% found in this study; UGT1A1*6, however, was extremely rare in the Caucasian and African populations (17). The UGT1A1*27 allele detected only in Asian individuals was reported to be harboured exclusively by a UGT1A1*28 haplotype, as findings of this study show. Either homozygous or heterozygous polymorphisms of UGT1A1*6, *27 or *28 were detected in 55 (43.7%) subjects in this study; the risk of UGT1A1 polymorphism-related toxicity of CPT-11 is considered to be high in East Asian individuals.
The Food and Drug Administration in the United States has approved an amendment of the label for Camptosar (irinotecan hydrochloride), to which a warning to reduce the starting dose of irinotecan for UGT1A1*28 homozygous patients has been added. In East Asian individuals, both UGT1A1*6 and UGT1A1*28 require examination, since there is no linkage disequilibrium between the two polymorphisms (24), as indicated in the present study.
As a clinical relevance, the risk of TOP-I mutation-related resistance to CPT-11 is unlikely and it is not necessary to test for TOP-I mutation prior to chemotherapy with CPT-11. However, the risk of UGT1A1 polymorphism-related toxicity of CPT-11 is markedly higher in East Asian individuals, and UGT1A1 polymorphisms, not only of UGT1A1*28, but also of UGT1A1*6, should be tested prior to treatment with CPT-11 to avoid severe adverse effects.
In conclusion, the main findings in this study were twofold. First, TOP-I genetic mutations related to CPT-11 resistance were not detected in any of the subjects or in untreated lung cancer tissues. Second, compared to Caucasians and individuals of African descent, it was observed that UGT1A1*28 was less frequent and UGT1A1*6 was more frequent in Japanese subjects. Moreover, linkage disequilibrium was not noted between UGT1A1*6 and UGT1A1*28.
Acknowledgments
We thank Yakult Honsha Co., Ltd., and Masumi Tanaka (Department of Respiratory Medicine) for their partial technical support. This study was supported by Grant-in-Aid No. 15590826 and Grant-in-Aid for Development of leading-edge cancer professionals in the South Kanto area, from the Ministry of Education, Culture, Sports, Science and Technology.
Contributor Information
Tomoya Fukui, Department of Respiratory Medicine, School of Medicine, Kitasato University, Kanagawa 252-0373, Japan.
Hisashi Mitsufuji, Department of Respiratory Medicine, School of Medicine, Kitasato University, Kanagawa 252-0373, Japan.
Masaru Kubota, Department of Respiratory Medicine, School of Medicine, Kitasato University, Kanagawa 252-0373, Japan.
Hidenori Inaoka, Department of Medical Engineering and Technology, School of Allied Health Sciences, Kanagawa 252-0373, Japan.
Minoru Hirose, Department of Medical Engineering and Technology, School of Allied Health Sciences, Kanagawa 252-0373, Japan.
Keiichi Iwabuchi, Department of Pathology, School of Medicine, Kitasato University, Kanagawa 252-0373, Japan.
Noriyuki Masuda, Department of Respiratory Medicine, School of Medicine, Kitasato University, Kanagawa 252-0373, Japan.
Hirosuke Kobayashi, Department of Medical Engineering and Technology, School of Allied Health Sciences, Kanagawa 252-0373, Japan.
References
- Pizzolato JF and Saltz LB: The camptothecins. Lancet 361: 2235-2242, 2003. [DOI] [PubMed]
- Arakawa Y, Suzuki H, Saito S and Yamada H: Novel missense mutation of the DNA topoisomerase I gene in SN-38-resistant DLD-1 cells. Mol Cancer Ther 5: 502-508, 2006. [DOI] [PubMed]
- Kubota N, Kanzawa F, Nishio K, Takeda Y, Ohmori T, Fujiwara Y, Terashima Y and Saijo N: Detection of topoisomerase I gene point mutation in CPT-11 resistant lung cancer cell line. Biochem Biophys Res Commun 188: 571-577, 1992. [DOI] [PubMed]
- Chang JY, Liu JF, Juang SH, Liu TW and Chen LT: Novel mutation of topoisomerase I in rendering cells resistant to camptothecin. Cancer Res 62: 3716-3721, 2002. [PubMed]
- Chrencik JE, Staker BL, Burgin AB, Pourquier P, Pommier Y, Stewart L and Redinbo MR: Mechanisms of camptothecin resistance by human topoisomerase I mutations. J Mol Biol 339: 773-784, 2004. [DOI] [PubMed]
- Redinbo MR, Stewart L, Kuhn P, Champoux JJ and Hol WGJ: Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA. Science 279: 1504-1513, 1998. [DOI] [PubMed]
- Laco GS, Collins JR, Luke BT, Kroth H, Sayer JM, Jerina DM and Pommier Y: Human topoisomerase I inhibition: docking camptothecin and derivatives into a structure-based active site model. Biochemistry 41: 1428-1435, 2002. [DOI] [PubMed]
- Mathijssen RH, van Alphen RJ, Verweij J, Loos WJ, Nooter K, Stoter G and Sparreboom A: Clinical pharmacokinetics and metabolism of irinotecan (CPT-11). Clin Cancer Res 7: 2182‑2194, 2001. [PubMed]
- Humerickhouse R, Lohrbach K, Li L, Bosron WF and Dolan ME: Characterization of CPT-11 hydrolysis by human liver carboxylesterase isoforms hCE-1 and hCE-2. Cancer Res 60: 1189-1192, 2000. [PubMed]
- Kawato Y, Aonuma M, Hirota Y, Kuga H and Sato K: Intracellular roles of SN-38, a metabolite of the camptothecin derivative CPT-11, in the antitumor effect of CPT-11. Cancer Res 51: 4187-4191, 1991. [PubMed]
- Ando Y, Saka H, Ando M, Sawa T, Muro K, Ueoka H, Yokoyama A, Saitoh S, Shimokata K and Hasegawa Y: Polymorphisms of UDP-glucuronosyltransferase gene and irinotecan toxicity: a pharmacogenetic analysis. Cancer Res 60: 6921-6926, 2000. [PubMed]
- Ando Y, Fujita K, Sasaki Y and Hasegawa Y: UGT1A1*6 and UGT1A1*27 for individualized irinotecan chemotherapy. Curr Opin Mol Ther 9: 258-262, 2007. [PubMed]
- Araki K, Fujita K, Ando Y, Nagashima F, Yamamoto W, Endo H, Miya T, Kodama K, Narabayashi M and Sasaki Y: Pharmacogenetic impact of polymorphisms in the coding region of the UGT1A1 gene on SN-38 glucuronidation in Japanese patients with cancer. Cancer Sci 97: 1255-1259, 2006. [DOI] [PMC free article] [PubMed]
- Strassburg CP: Pharmacogenetics of Gilbert’s syndrome. Pharmacogenomics. 9: 703-715, 2008. [DOI] [PubMed]
- Saito A, Kawamoto M and Kamatani N: Association study between single-nucleotide polymorphisms in 199 drug-related genes and commonly measured quantitative traits of 752 healthy Japanese subjects. J Hum Gen 54: 317-323, 2009. [DOI] [PubMed]
- Kaniwa N, Kurose K, Jinno H, Tanaka-Kagawa T, Saito Y, Saeki M, Sawada K, Tohkin M and Hasegawa R: Racial variability in haplotype frequencies of UGT1A1 and glucuronidation activity of a novel single nucleotide polymorphism 686C> T (P229L) found in an African-American. Drug Metab Dispos 33: 458-465, 2005. [DOI] [PubMed]
- Hoskins JM, Marcuello E, Altes A, Marsh S, Maxwell T, Van Booven DJ, Paré L, Culverhouse R, McLeod HL and Baiqet M: Irinotecan pharmacogenetics: influence of pharmacodynamic genes. Clin Cancer Res 14: 1788-1796, 2008. [DOI] [PubMed]
- Chowbay B, Zhou S and Lee EJ: An interethnic comparison of polymorphisms of the genes encoding drug-metabolizing enzymes and drug transporters: experience in Singapore. Drug Metab Rev 37: 327-378, 2005. [DOI] [PubMed]
- Mercke Odeberg J, Andrade J, Holmberg K, Hoglund P, Malmqvist U and Odeberg J: UGT1A polymorphisms in a Swedish cohort and a human diversity panel, and the relation to bilirubin plasma levels in males and females. Eur J Clin Pharmacol 62: 829-837, 2006. [DOI] [PubMed]
- Liu JY, Qu K, Sferruzza AD and Bender RA: Distribution of the UGT1A1*28 polymorphism in Caucasian and Asian populations in the US: a genomic analysis of 138 healthy individuals. Anticancer Drugs 18: 693-696, 2007. [DOI] [PubMed]
- Innocenti F, Grimsley C, Das S, Ramirez J, Cheng C, Kuttab‑Boulos H, Ratain MJ and Di Rienzo A: Haplotype structure of the UDP-glucuronosyltransferase 1A1 promoter in different ethnic groups. Pharmacogenetics 12: 725-733, 2002. [DOI] [PubMed]
- Tsurutani J, Nitta T, Hirashima T, Komiya T, Uejima H, Tada H, Syunichi N, Tohda A, Fukuoka M and Nakagawa K: Point mutations in the topoisomerase I gene in patients with non-small cell lung cancer treated with irinotecan. Lung Cancer 35: 299-304, 2002. [DOI] [PubMed]
- Moisan F, Longy M, Robert J and Le Morva V: Identification of gene polymorphisms of human DNA topoisomerase I in the National Cancer Institute panel of human tumour cell lines. Br J Cancer 95: 906-913, 2006. [DOI] [PMC free article] [PubMed]
- Saito Y, Maekawa K, Ozawa S and Sawada J. Genetic polymorphisms and haplotypes of major drug metabolizing enzymes in east Asians and their comparison with other ethnic populations. Curr Pharmacogenomics 5: 49-78, 2007.